Abstract
The transcription factor NF-κB plays a central role in the human immunodeficiency virus type 1 (HIV-1) activation pathway. HIV-1 transcription is also regulated by protein acetylation, since treatment with deacetylase inhibitors such as trichostatin A (TSA) or sodium butyrate (NaBut) markedly induces HIV-1 transcriptional activity of the long terminal repeat (LTR) promoter. Here, we demonstrate that TSA (NaBut) synergized with both ectopically expressed p50/p65 and tumor necrosis factor alpha/SF2 (TNF)-induced NF-κB to activate the LTR. This was confirmed for LTRs from subtypes A through G of the HIV-1 major group, with a positive correlation between the number of κB sites present in the LTRs and the amplitude of the TNF-TSA synergism. Mechanistically, TSA (NaBut) delayed the cytoplasmic recovery of the inhibitory protein IκBα. This coincided with a prolonged intranuclear presence and DNA binding activity of NF-κB. The physiological relevance of the TNF-TSA (NaBut) synergism was shown on HIV-1 replication in both acutely and latently HIV-infected cell lines. Therefore, our results open new therapeutic strategies aimed at decreasing or eliminating the pool of latently HIV-infected reservoirs by forcing viral expression.
The persistence of latently human immunodeficiency virus (HIV)-infected cellular reservoirs, despite prolonged treatment with highly active antiretroviral therapy (HAART), represents the major hurdle to virus eradication. These latently infected cells are a permanent source for virus reactivation and lead to a rebound of the viral load after interruption of HAART (reviewed in reference 42). Therefore, a greater understanding of the molecular mechanisms regulating viral latency and reactivation should lead to rational strategies aimed at purging the latent HIV reservoirs (6). At the cellular level, two major forms of HIV type 1 (HIV-1) latency have been described: preintegration latency and postintegration latency (reviewed in reference 33). Several cell lines selected in vitro have served as models for studying the latter type of latency. In these cell lines, production of viral particles can be induced at the transcriptional level by a variety of agents, including phorbol esters and cytokine tumor necrosis factor SF2 (TNF) (16). Several explanations have been proposed for the low level of transcription observed during postintegration latency, including the following. (i) The first explanation involves the site of integration of the provirus into the host cell genome and the cellular chromatin environment at this site (26, 60). (ii) A second explanation involves the presence of a potentially repressive nucleosome (nuc-1) located immediately downstream of the HIV transcription start site under latency conditions. Nuc-1 is remodeled upon activation of the HIV promoter located in its 5′ long terminal repeat (LTR) in response to Tat, phorbol esters, and deacetylase inhibitors (12, 54, 58). Nuc-1 could pose a unique elongation barrier for the polymerase, and its remodeling could play a significant role in the transcriptional reactivation of the HIV promoter from latency (reviewed in reference 52). (iii) A third explanation involves the absence of the viral trans-activator Tat, which binds to TAR, an RNA hairpin loop formed at the 5′ termini of all nascent HIV-1 transcripts (1). (iv) A fourth explanation involves the presence of mutations in the integrated provirus, including interruption of the Tat-TAR axis (13, 14). (v) A fifth explanation involves the absence of the inducible transcription factor NF-κB. The enhancer in the U3 region of the LTR contains two binding sites for NF-κB, which plays a central role in the activation pathway of the HIV-1 provirus. Various studies have reported that the NF-κB-binding sites as well as the NF-κB proteins are critical for LTR promoter activity and important for optimal HIV-1 replication (reviewed in reference 44). Recently, West et al. have shown that p65 stimulates transcriptional elongation of the HIV-1 promoter (59).
NF-κB is an inducible transcription factor complex that plays a role in the expression of a variety of genes involved in immune and inflammatory responses and cell survival (reviewed in references 18 and 28). In mammalian cells, there are five known members of the NF-κB/Rel family: p65 (RelA), c-Rel, RelB, p50, and p52. The most widely studied and most abundant form of NF-κB is a heterodimer of p50 and p65. In unstimulated cells, NF-κB is sequestered in the cytoplasm in an inactive form through interaction with inhibitory IκB proteins including IκBα, IκBβ, and IκBɛ. Upon activation of NF-κB by various stimuli (including inflammatory cytokines [TNF and interleukin-1], bacterial lipopolysaccharides, viral proteins, and mitogens [phorbol esters and UV light]), IκBs are rapidly phosphorylated by a macromolecular IκB kinase complex (22, 27), ubiquitinated, and degraded by the 26S proteasome. The released NF-κB then translocates to the nucleus, where it can activate transcription from a wide variety of promoters, including that of its own inhibitor, IκBα. The newly synthesized IκBα enters the nucleus, enhances NF-κB removal from DNA, and takes it back to the cytoplasm, thus restoring the inducible cytoplasmic pool of NF-κB. Thus, the de novo expression of IκBα proteins, which display nucleocytoplasmic shuttling properties, participates in a negative feedback system ensuring a transient NF-κB transcriptional response (reviewed in references 28 and 32).
There is now strong evidence that both transcriptional activation and silencing are mediated through the recruitment of enzymes that control protein acetylation. Acetylation of specific lysine residues within nucleosomal histones is closely linked to chromatin disruption and transcriptional activation in many genes. Consistent with their role in altering chromatin structure, many transcriptional coactivators (including GCN5, CBP/p300, p300/CBP-associated factor [P/CAF], and SRC-1) possess intrinsic histone acetyltransferase activity that is critical for their function (reviewed in reference 46). Similarly, corepressor complexes include proteins that have histone deacetylase (HDAC) activity (reviewed in reference 29). Importantly, reversible acetylation is also a critical posttranslational modification of nonhistone proteins, including general and specific transcription factors, coactivators, nonhistone structural chromosomal proteins, and nuclear import factors. Protein acetylation regulates many diverse functions, including DNA binding, protein-protein interaction, protein stability, and cellular localization (reviewed in references 3 and 31). Hence, acetylation may rival phosphorylation as a mechanism for the transduction of cellular regulatory signals.
In the case of HIV-1, we and others have previously reported ample evidence that viral transcription is regulated by protein acetylation. We have shown that treatment of latently HIV-infected cell lines with deacetylase inhibitors [trichostatin A (TSA), trapoxin, and sodium butyrate (NaBut)] induces viral transcription and nuc-1 remodeling (54; reviewed in reference 52). Transcriptional activation of the HIV-1 promoter in response to TSA has also been demonstrated in ex vivo transiently or stably transfected HIV LTR reporter constructs (12, 26, 30) and on in vitro chromatin-reconstituted HIV-1 templates (48, 49). Moreover, acetylation of Tat by p300, by P/CAF, and by human GCN5 is important for its transcriptional activity (7, 30).
To better understand the molecular mechanisms regulating HIV-1 reactivation from latency, we here extended our studies on the TSA inducibility of the viral promoter (LTR) and focused on the functional role of the κB sites in this TSA response. In this report, we demonstrate that deacetylase inhibitors (TSA and NaBut) synergized with both ectopically expressed p50/p65 and TNF-induced NF-κB to activate the HIV-1 promoter. This synergism required intact κB sites and was observed with LTRs from subtypes A through G of the HIV-1 group M (major). TSA and NaBut drugs prolonged TNF-induced NF-κB binding to DNA and the intranuclear presence of p65. Remarkably, a marked delay in the cytoplasmic recovery of IκBα coincided with the prolonged binding activity and presence of NF-κB in the nucleus. Importantly, the physiological relevance of the TNF-TSA (NaBut) synergism was shown both on HIV-1 reactivation in a model cell line for postintegration latency and on HIV-1 replication in the context of a de novo viral infection. Therefore, our results open new therapeutic strategies aimed at forcing viral expression and at contributing, in the presence of an efficient HAART, to a reduction of the pool of latently HIV-infected cellular reservoirs.
MATERIALS AND METHODS
Cell culture
The U1 and SupT1 cell lines were obtained from the AIDS Research and Reference Reagent Program (National Institute of Allergy and Infectious Diseases [NIAID], National Institutes of Health [NIH]). The monocytic cell lines U937 (reference no. 85011440) and HL60 (reference no. 98070106) were obtained from the European Collection of Cell Cultures. All cell lines were grown as reported previously (56).
Plasmid constructs.
The plasmids pLTR(1-789)-luc, pLTR(292-789)-luc, pLTR(1-531)-luc, pLTR(292-531)-luc, and pLTR(345-531)-luc contain the HIV-1LAI 5′ LTR (nucleotides indicated parenthetically) cloned into the reporter vector pGL2-Basic (Promega). To construct pLTR(345-531)mut κB-luc, pLTR(345-531)-luc was used as substrate for mutagenesis of the two κB sites (5′-AACTCACTTTCCGCTGCTCACTTTCCA-3′; the mutations are shown in boldface type, and the κB sites are underlined on the coding strand) by the QuikChange site-directed mutagenesis method (Stratagene).
The pLTR(A, B, C1, D, E, F, G, AG)-luc plasmids were previously described (23).
The plasmids pRSV-p50 and pRSV-p65 were obtained from G. Nabel and N. Perkins through the AIDS Research and Reference Reagent Program (Division of AIDS, NIAID, NIH).
Transient transfection and luciferase assays.
SupT1 cells were transfected using the DEAE-dextran procedure as previously described (56). At 20 h posttransfection, the cells were treated or mock treated with TSA (450 nM when a single dose was used) (Sigma Chemical Co.), NaBut (5 mM) (Sigma Chemical Co.), TNF (10 ng/ml; catalog no. 210-TA; R&D Systems) or a combination of these drugs. At 42 h posttransfection, cells were lysed and assayed for luciferase activity (Promega). Luciferase activities derived from the HIV-1 LTRs were normalized with respect to protein concentration using the detergent-compatible protein assay (Bio-Rad).
EMSAs.
Nuclear extracts were prepared, and electrophoretic mobility shift assays (EMSAs) with the HIV-1 NF-κB probe were performed as previously described (54). For supershift assays, monoclonal antibody against p52 (catalog no. 05-361; Upstate Biotechnology) and polyclonal antibodies against p50 (catalog no. 06-886; Upstate Biotechnology), p65 (catalog no. sc-109X; Santa Cruz Biotechnology, Inc.), RelB (catalog no. sc-226X; Santa Cruz Biotechnology, Inc.), and c-rel (catalog no. sc-6955X; Santa Cruz Biotechnology, Inc.) were added to the binding reaction at a final concentration of 2 μg/reaction mixture. As loading controls, the same nuclear extracts were tested for binding of Oct-1 to an Oct-1 consensus probe (5′-TGTCGAATGCAAATCACTAGAA-3′).
RNase protection assays.
Total RNA samples were prepared from SupT1 cells or U1 cells using the commercial RNAqueous phenol-free total RNA isolation kit (Ambion) from 5 × 106 cells treated or mock treated with TSA or NaBut or/and TNF during different periods of time. The HIV-1-specific antisense riboprobe was obtained as previously described (54). An IκBα-specific 32P-labeled antisense riboprobe was synthesized in vitro by transcription of AflIII-restricted pcDNA3-IκBα with SP6 polymerase according to the protocol provided with the riboprobe in vitro transcription systems (Promega). As control, a glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific antisense probe was synthesized by the same method and used on the same RNA samples.
The RNase protection assays were performed with the RPA II kit (Ambion) according to the manufacturer's recommendations. Briefly, hybridization reaction mixtures (20 μl) containing 12 μg of total cellular RNA and 200,000 cpm of probe in hybridization buffer were heated to 95°C for 4 min to denature the RNA and then incubated at 42°C for 16 h. The reaction mixtures were diluted by the addition of 200 μl of digestion buffer, and the single-stranded sequences were digested with RNase T1 and RNase A for 1 h at 37°C. Following addition of 300 μl of inactivation buffer and ethanol precipitation, the protected RNA fragments were analyzed by electrophoresis through 6% urea polyacrylamide gels.
Western blot analysis.
Nuclear and cytoplasmic extracts were prepared as previously described (in references 37 and 47, respectively). Western blots were realized (47) with antibodies against the following proteins (each at a 1:1,000 dilution): IκBα (catalog no. 06-494; Upstate Biotechnology), IκBβ (catalog no. sc-945; Santa Cruz Biotechnology, Inc.), IκBɛ (catalog no. sc-7156; Santa Cruz Biotechnology, Inc.), and p65 (catalog no. sc-109X; Santa Cruz Biotechnology, Inc.).
HIV-1 infections and CA-p24 measurements.
HIV-1 infectious DNA was prepared from the pHIV (57). Infectious viral stocks and infections were performed as previously described (57). One day after infection, the cells were treated or mock treated with TSA or/and TNF. Every 2 days, aliquots of 200 μl were removed from the infected cultures and replaced by complete RPMI 1640 medium. The aliquots were assayed for CA-p24 antigen concentration by an enzyme-linked immunosorbent assay (Innogenetics) in order to monitor the kinetics of viral replication.
Immunofluorescence confocal microscopy.
SupT1 cells were treated or not with TSA (450 nM) and/or TNF (50 ng/ml). After a 30-min or 2-h treatment, cells were centrifuged (with cytospin 3; Shandon) and fixed for 5 min with Immunohistofix (Bio-Rad) at room temperature followed by 6 min with 100% methanol at −20°C. After two washes with phosphate-buffered saline (PBS), the samples were saturated with PBS containing 0.5% gelatin and 0.25% bovine serum albumin for 1 h and stained for 1 h with a 1/100 dilution of an anti-human p65 rabbit polyclonal immunoglobulin G (IgG) (C-20; catalog no. sc-372; Santa Cruz Biotechnology, Inc.). The samples were then washed three times with PBS containing 0.2% gelatin and incubated for 1 h with a 1/200 dilution of the secondary antibody: Alexa-488-coupled goat anti-rabbit IgG (Molecular Probes). The samples were then washed three times in PBS with 0.2% gelatin and mounted for analysis on a Zeiss LSM510 laser scanning confocal microscope.
RESULTS
TSA inducibility of different deleted HIV-1 LTRs.
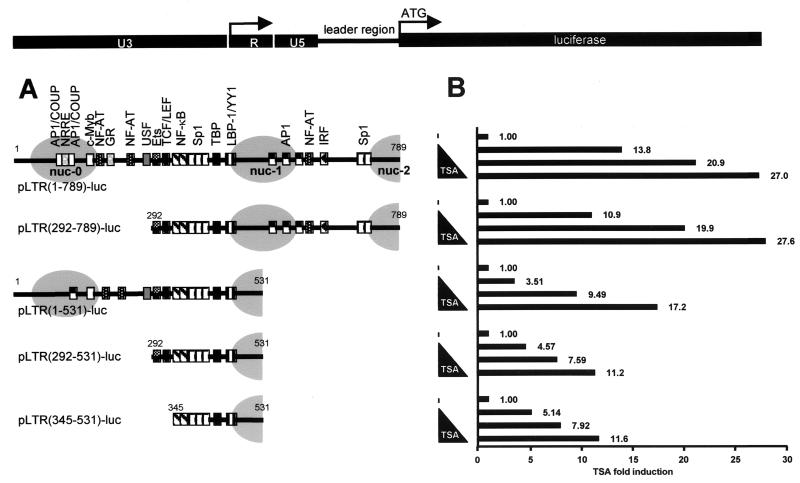
In order to delineate the LTR sequences responsible for activation of the HIV-1 promoter activity in response to TSA, we generated a series of pLTR-luciferase reporter constructs containing various 5′ and/or 3′ deletions within the viral promoter region (the prototype LAI strain of HIV-1 subtype B). The five resulting plasmids were designated pLTR(1-789)-luc (containing the complete 5′ LTR plus the leader region up to the beginning of the GAG open reading frame), pLTR(292-789)-luc, pLTR(1-531)-luc, pLTR(292-531)-luc, and pLTR(345-531)-luc, respectively (Fig. 1A). These plasmids were transiently transfected into the human CD4+ T-lymphoid cell line SupT1. Transfected cells were mock treated or treated with increasing concentrations of TSA (0, 250, 500, and 1,000 nM) and assayed for luciferase activity. Results presented in Fig. 1B show the TSA inductions for each construct to eliminate the variations due to the differences in basal activity observed with the various deleted LTRs. All LTR constructs were activated by TSA in a dose-dependent manner. pLTR(1-789)-luc and pLTR(292-789)-luc were induced 13.8- to 27.0-fold and 10.9- to 27.6-fold, respectively. This induction by TSA is likely to be explained by histone hyperacetylation (48, 49, 54, 55) and/or by acetylation and deacetylation phenomena involved in the regulation of transcription factors binding to the LTR.
FIG. 1.
TSA responsiveness of different HIV-1 LTRs with deletions. (A) Organization of the 5′ LTR region of the prototype HIV-1 virus LAI (subtype B). The LTR is composed of the U3, R, and U5 regions. The complete LTR and the leader region (nt 1 to 789) (nt +1 is the start of U3 in the 5′ LTR) control the luciferase reporter gene. The transcription factor binding sites present in the different LTRs with deletions are detailed for each construct and are aligned with nucleosome positioning (grey ovals). The following transcription factor binding sites are present in the full-length LTR construct (reviewed in reference 38): two AP1/COUP sites, the nuclear receptor-responsive element (NRRE), one c-Myb site, three NF-AT sites, one GR site, one USF site, one Ets-1 site, one TCF/LEF site, two κB sites, five Sp1 sites, the TATA box, one LBP-1/YY1 site, three AP-1 sites, and one IRF site. (B) SupT1 cells were transiently transfected with 500 ng of each reporter construct containing the different HIV-1 LTRs with deletions, and cells were treated with increasing concentrations of TSA. Luciferase activities were measured in cell lysates. To compare the TSA responsiveness of the different LTRs, basal LTR activity of each construct was arbitrarily set at a value of 1. Results are presented as histograms indicating the TSA inductions of the constructs with respect to their respective basal activity. Values represent the means of duplicate samples. An experiment representative of three independent transfections is shown. Variation for a given plasmid between different experiments was <15% in most cases.
The constructs pLTR(1-531)-luc, pLTR(292-531)-luc, and pLTR(345-531)-luc presented TSA activations from 3.51- to 17.2-fold, from 4.57- to 11.2-fold, and from 5.14- to 11.6-fold, respectively (Fig. 1B), indicating a decrease in TSA inducibility associated with the deletion of the 3′ region encompassing nucleotides (nt) 532 to 789. A possible explanation for this decrease in TSA inducibility would be the absence of the nucleosome nuc-1 in the LTR constructs with 3′ deletions when assembled into chromatin, since an important part of the DNA sequence wrapped around nuc-1 was lacking in these constructs (Fig. 1A).
Importantly, a significant TSA inducibility was still observed with the smallest LTR (nt 345 to 531), containing the two κB sites, the three Sp1 sites, the TATA box and the ligand binding protein 1 (LBP-1)/YY1 site (Fig. 1A and B). This could be explained by the recruitment at the level of these sites of different factors implicated in acetylation and deacetylation events: TATA-binding protein (TBP)-associated factor II250 (34), transcription factor IIEβ (TFIIEβ) and TFIIF (21), Sp1 (10, 50), YY1 (9), and p65 (17, 40).
Synergistic activation of HIV-1 promoter activity by NF-κB and TSA.
The HIV-1 NF-κB binding sites confer a high rate of transcription to the viral promoter in activated T cells and monocytes/macrophages (reviewed in reference 44). In order to investigate the functional role of NF-κB in the inducibility of the LTR by TSA, SupT1 cells were transiently cotransfected with the pLTR(345-531)-luc construct and with increasing amounts (from 6 ng each to 1,600 ng each) of expression vectors for p50 and p65 (pRSV-p50/pRSV-p65). Cells were treated with TSA or mock treated and assayed for luciferase activity (Table 1). As expected, in the absence of TSA, p50/p65 trans-activated the HIV-1 promoter in a dose-dependent manner up to 7.66-fold (Table 1, p50/p65-fold activation, rows 2 to 13). Treatment of cells with TSA alone resulted in a 51.8-fold activation of transcription (Table 1, row 1). Remarkably, when cells were both cotransfected with increasing amounts of expression vectors for p50/p65 and treated with TSA, a strong synergism was observed between the two activators, resulting in transactivations ranging from 95.0- to 2,655-fold (Table 1, p50/p65 plus TSA activation, rows 2 to 13). Transcriptional activators synergize when their combination produces a transcriptional rate that is greater than the sum of the effects produced by the individual activators. Transfection of 1,200 ng each of pRSV-p50/pRSV-p65 led to a 7.38-fold stimulation of transcription in the absence of TSA, whereas in presence of TSA, it led to a 2,655-fold stimulation (Table 1, row 11). This amount of transcription is 45-fold greater (45-fold synergism) than the sum of the effects produced by each activator separately (51.8-fold + 7.38-fold) (Table 1, rows 1 and 11). This synergism between p50/p65 and TSA persisted even at saturating amounts of p50/p65 proteins (see data for 1,400 and 1,600 ng each of cotransfected p50/p65 plasmid DNAs), indicating that the observed effect was not the consequence of increased p50/p65 expression due to activation of the RSV promoter by TSA.
TABLE 1.
Synergistic activation of HIV-1 promoter by NF-κB (p50/p65) and TSAa
| Row no. | Plasmid | Amt of p50/p65b (ng) | Result of luciferase assay (RLU)
|
Activation (fold) with:
|
Synergism (fold) | ||
|---|---|---|---|---|---|---|---|
| −TSA | +TSA | p50/p65 | p50/p65 + TSA | ||||
| 1 | pLTR(345-531)-luc | 2.58 | 133 | 1.00 | 51.8 | ||
| 2 | 6 | 2.99 | 245 | 1.16 | 95.0 | 1.8 | |
| 3 | 25 | 2.43 | 530 | 1.94 | 205 | 3.9 | |
| 4 | 50 | 2.92 | 811 | 1.13 | 314 | 6.0 | |
| 5 | 100 | 8.01 | 2,961 | 3.11 | 1,148 | 21 | |
| 6 | 200 | 6.15 | 2,723 | 2.39 | 1,055 | 20 | |
| 7 | 400 | 11.8 | 2,631 | 4.59 | 1,020 | 18 | |
| 8 | 600 | 11.5 | 4,102 | 4.45 | 1,590 | 28 | |
| 9 | 800 | 10.9 | 6,129 | 4.24 | 2,376 | 43 | |
| 10 | 1,000 | 14.1 | 5,809 | 5.46 | 2,252 | 39 | |
| 11 | 1,200 | 19.0 | 6,851 | 7.38 | 2,655 | 45 | |
| 12 | 1,400 | 19.7 | 5,394 | 7.66 | 2,091 | 35 | |
| 13 | 1,600 | 18.3 | 3,189 | 7.10 | 1,236 | 21 | |
| 14 | pLTR(345-531)mut κB-luc | 1.55 | 101 | 1.00 | 65.0 | ||
| 15 | 6 | 2.02 | 115 | 1.31 | 74.2 | 1.1 | |
| 16 | 25 | 1.87 | 90.0 | 1.21 | 58.1 | 0.9 | |
| 17 | 50 | 2.05 | 130 | 1.32 | 84.1 | 1.3 | |
| 18 | 100 | 3.30 | 161 | 2.13 | 104 | 1.6 | |
| 19 | 200 | 1.80 | 82.5 | 1.16 | 53.2 | 0.8 | |
| 20 | 400 | 2.10 | 98.8 | 1.36 | 63.7 | 1.0 | |
| 21 | 600 | 1.88 | 70.2 | 1.21 | 45.3 | 0.7 | |
| 22 | 800 | 1.55 | 71.6 | 1.00 | 46.2 | 0.7 | |
| 23 | 1,000 | 1.79 | 95.7 | 1.16 | 61.8 | 0.9 | |
| 24 | 1,200 | 1.51 | 97.8 | 0.98 | 61.2 | 0.9 | |
| 25 | 1,400 | 1.76 | 129 | 1.14 | 83.2 | 1.3 | |
| 26 | 1,600 | 0.97 | 66.0 | 0.63 | 42.6 | 0.7 | |
SupT1 cells were transiently cotransfected with 500 ng of either pLTR(345-531)-luc (rows 1 to 13) or pLTR(345-531)mut κB-luc (rows 14 to 26) and increasing amounts of expression vectors for p50 and p65 (from 6 to 1,600 ng of each vector). Cells were mock treated (−TSA) or treated with TSA (+TSA). Luciferase assay results are represented as relative light units (RLU). For rows 1 to 13, p50/p65 activations were obtained by dividing the RLUs in the −TSA column for pLTR(345-531)-luc by the RLU result obtained for this construct in the absence of both TSA and p50/p65. The p50/p65 + TSA activations were obtained by dividing the RLUs in the +TSA column for pLTR(345-531)-luc by the RLU result obtained for this construct in the absence of both TSA and p50/p65. Similarly, for rows 14 to 26, p50/p65 activations were obtained by dividing the RLUs in the −TSA column for pLTR(345-531)mut κB-luc by the RLU result obtained for this construct in the absence of both TSA and p50/p65. The p50/p65 + TSA activations were obtained by dividing the RLUs in the +TSA column for pLTR(345-531)mut κB-luc by the RLU result obtained for this construct in the absence of both TSA and p50/p65. The synergism was determined as previously described (19) using the following formula: (fold activation by TSA + p50/p65)/(fold activation by TSA alone + fold activation by p50/p65 alone). An experiment representative of five repeated transfections is shown.
Same amount for each.
Synergistic activation by ectopically expressed p50/p65 and TSA required intact NF-κB binding sites in the HIV-1 proximal enhancer, since point mutations in these sites eliminated this effect [Table 1, pLTR(345-531)mut κB-luc, rows 14 to 26]. This implies that the synergistic effect was mediated by interactions at the κB sites and not at the otherwise intact LTR(345-531) DNA sequences.
In conclusion, these results demonstrate that TSA synergistically enhanced NF-κB-dependent transcriptional activation of the HIV-1 promoter, suggesting that the NF-κB signaling pathway can be functionally regulated by posttranslational acetylation in vivo.
Synergistic activation of HIV-1 promoter activity by cytokine TNF and deacetylase inhibitors.
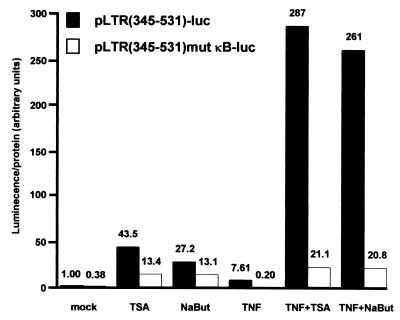
Proinflammatory cytokine TNF stimulates the HIV-1 LTR through activation of NF-κB in both human CD4+ T cells and monocytes/macrophages. To determine whether the synergism between NF-κB and deacetylase inhibitor TSA could be observed in the context of a physiological activation of the NF-κB pathway, we examined the effect of deacetylase inhibitors, TSA and NaBut, on the TNF-induced HIV-1 promoter activity. To this end, SupT1 cells were transiently transfected with the reporter construct pLTR(345-531)-luc or pLTR(345-531)mut κB-luc and subsequently mock treated or treated with TNF alone, TSA alone, NaBut alone, TNF-TSA, or TNF-NaBut. Treatment of SupT1 cells with TSA, NaBut, or TNF alone resulted in a 43.5-, 27.2-, or 7.61-fold increase, respectively, of luciferase gene expression driven by the wild-type LTR (Fig. 2). Remarkably, when cells were treated with a combination of TNF-TSA or TNF-NaBut, we observed a 287- or 261-fold increase of luciferase expression above the control level obtained in absence of any treatment, demonstrating an important synergism between TNF and deacetylase inhibitors. Mutation in the κB sites [pLTR(345-531)mut κB-luc] eliminated both the activation of the LTR by TNF and its synergistic activation by TSA (NaBut) and TNF (Fig. 2). Moreover, results obtained by transient transfection of the promonocytic cell line HL60 confirmed those observed with the CD4+-T-lymphoid-cell line SupT1 (data not shown). Thus, the synergism between TNF and TSA (NaBut) was thus observed in two cell lines representative of the two major cellular targets for HIV-1 infection.
FIG. 2.
Synergistic activation of HIV-1 promoter by cytokine TNF and inhibitors of deacetylases, TSA, and NaBut. SupT1 cells were transiently transfected with 500 ng of the indicated constructs. Cells were mock treated or treated with TSA, NaBut, TNF, TNF-TSA, or TNF-NaBut. The mock-treated value of the wild-type LTR reporter construct was arbitrarily set at a value of 1. Values represent the means of duplicate samples. A representative experiment of three independent transfections is shown. Variation for a given plasmid between different experiments was <15% in most cases.
In conclusion, our functional results demonstrate that the deacetylase inhibitors TSA and NaBut functionally synergized with TNF to activate the HIV-1 LTR. The synergism we observed between TSA (NaBut) and TNF was strictly dependent on the presence of intact κB sites in the HIV-1 proximal enhancer.
Synergistic activation by TSA and TNF of LTR activity from the HIV-1 subtypes A through G of group M.
HIV-1 isolates have been classified into three genetic groups: the major group (M), the outlier group (O), and the non-M, non-O group (N). All groups are thought to have arisen from independent zoonotic transmissions. The group M isolates, which are responsible for more than 99% of all infections, have diversified during their worldwide spread. These isolates have been grouped according to their genomic sequences and can be divided into 10 distinct subtypes termed A through J. Although there is some variation in the exact position and in the sequence of κB sites, two similar sites are present in the LTRs of most described HIV-1 isolates, including isolates from subtypes A and B (used in the above experiments and most extensively studied in laboratories) and subtypes D, F, G, and AG (the last of which is a recombinant between subtypes A and G). On the other hand, subtype C viruses generally contain three κB sites, whereas subtype E viruses contain one functional κB site (23).
To examine the impact on the TNF-TSA synergism of these differences among the various HIV-1 group M subtypes, we performed transient transfections of SupT1 cells with reporter luciferase constructs containing LTRs from subtypes A, B, C1, D, E, F, G, and AG. Transfected LTRs were assayed for their responsiveness to TSA, to TNF and to both agents in combination. Results presented in Fig. 3 show the induction for each subtype (obtained by dividing the induced luciferase activities of subtype X by the basal activity of this same subtype X) in order to eliminate subtype-specific differences in basal activity of the LTRs.
FIG. 3.
Synergistic activation by TSA and TNF of the LTRs from the HIV-1 group M subtypes A through G. SupT1 cells were transiently transfected with 500 ng of an LTR luciferase construct containing the fragment from nt −147 to +63 of the subtype LTRs (A through G and AG as indicated). Results are presented as histograms indicating inductions by TSA, TNF, or TNF-TSA with respect to the basal activity of each subtype LTR construct, which was assigned a value of 1. Values represent the means of duplicate samples. An experiment representative of three independent transfections is shown. Variation for a given plasmid between different experiments was <15% in most cases.
LTR activity of each subtype tested was induced by TNF alone from 2.55- to 6.63-fold and by TSA alone from 26.2- to 63.6-fold depending on the subtype (Fig. 3). Importantly, TNF-TSA together synergized to activate all the subtype LTRs. Subtype E, containing one κB site, was induced 169-fold by TNF-TSA, corresponding to a 3.7-fold synergism. The subtypes A, B, D, F, G, and AG, each containing two κB sites, presented inductions from 216- to 596-fold, corresponding to synergisms from 5.1- to 11-fold. Subtype C1, containing three κB sites, was activated 802-fold by TNF-TSA, corresponding to a 11.8-fold synergism, a synergism three times higher than that observed for subtype E (Fig. 3).
From these experiments, we conclude that the synergistic transcriptional activation of the LTR by TNF-TSA is a common feature of HIV-1 subtypes A through G. In addition, we measured a positive correlation between the number of κB sites present in the respective LTRs and the amplitude of the synergism between TNF and TSA.
Synergistic activation by TNF and TSA of HIV-1 replication following infection of U937 cells.
To assess the biological relevance of the TNF-TSA synergism, we tested the effects of these drugs on viral replication in the context of an infection by HIV-1. To this end, we infected U937 monocytic cells with an HIV-1 NL4-3 stock. One day after infection, cells were mock treated or treated with either TSA, TNF, or both activators. HIV-1 replication was monitored by measuring the concentration of CA-p24 gag antigen in the culture supernatants over a 15-day period (Fig. 4). Our results indicated that, in absence of any treatment, infection resulted in progressive virus production. Following treatment with TSA alone or TNF alone, HIV-1 NL4-3 replicated more efficiently with levels of virus production higher than the control level. Importantly, TNF-TSA together synergized to enhance virus production at each time point. At day 15, TSA alone, TNF alone, and TNF-TSA increased CA-p24 levels by two-, three-, and eightfold, respectively, above the control level obtained in the absence of any treatment.
FIG. 4.
Synergistic effect of TSA and TNF on HIV-1 replication in monocytic cells. U937 cells were infected with an HIV-1 NL4-3 infectious stock. One day after infection, cells were mock treated or treated with TSA, TNF, or TNF-TSA. Virus replication was monitored at different intervals (every 2 days) by measuring the CA-p24 concentration in the culture supernatants. For each time point, CA-p24 was quantified from independent triplicate infections and the means of the triplicate samples are presented. An experiment representative of four independent infections performed in triplicate is shown.
Our data indicate that TNF and TSA synergistically increased the replicative capacity of the HIV-1 NL4-3 virus in U937 cells. These results were confirmed in three independent infection experiments performed in triplicate and were consistent with the results of the LTR-luciferase assays.
Thus, while the transcriptional activation of the HIV-1 promoter in response to TSA had been previously demonstrated in ex vivo transiently or stably transfected HIV LTR reporter constructs (26, 30), in latently HIV-infected cell lines (54), and on in vitro chromatin-reconstituted HIV-1 templates (48, 49), the results presented here constitute the first demonstration of the activating effect of a deacetylase inhibitor in the context of a natural HIV-1 infection. Importantly, we demonstrated a synergistic effect of TNF and TSA on the level of HIV-1 replication from a monocytic cell line, a cell lineage substantially contributing to simian immunodeficiency virus latent reservoirs in monkeys (20).
Synergistic activation of HIV-1 transcription and replication by TNF and inhibitors of deacetylases in latently infected cells.
Different culture systems have served as in vitro models for postintegration latency, and the study of these cells has provided important insight into the mechanism of transcriptional reactivation and pathogenesis of HIV. The U1 monocytic cell line (cloned from a population of chronically HIV-1-infected U937 cells) is one of the most-studied models of postintegration latency. The inducing effect of TNF on endogenous HIV-1 replication in U1 cells has been correlated with the activation of NF-κB binding to the viral enhancer and the stimulation of newly transcribed HIV-1 RNAs (43).
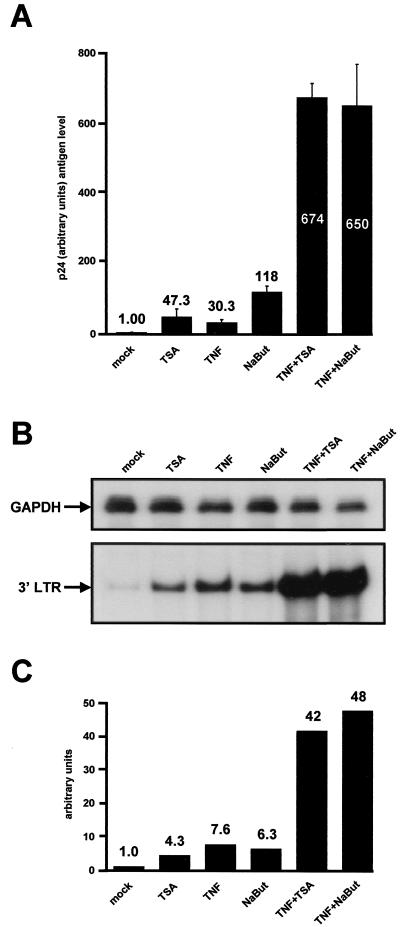
To study the effect of deacetylase inhibitors on HIV reactivation, we treated the latently infected cell line U1 for 24 h with TSA, TNF, NaBut, TNF-TSA, or TNF-NaBut. Treatment with TSA alone, TNF alone, or NaBut alone resulted in increases in p24 antigen release of 47.3-, 30.3-, and 118-fold, respectively, confirming the previous observations from our laboratory (54) (Fig. 5A ). Induction by TNF-TSA and TNF-NaBut caused a 674- and 650-fold activation of virus production, respectively (Fig. 5A). This synergistic activation by TSA (NaBut) and TNF of virus production in U1 cells took place at the transcriptional level. Indeed, RNase protection analysis with an antisense riboprobe corresponding to the HIV LTR showed that treatment with TSA or NaBut resulted in a 4.3- or 6.3-fold increase of HIV-1 transcription, to a degree similar to that observed following TNF treatment (Fig. 5B and C). After treatment with TNF-TSA and TNF-NaBut, we observed a 42- and 48-fold induction of the steady-state HIV mRNA level, respectively, above the mRNA level measured in the absence of any treatment. Our data thus demonstrated a synergistic activation by TNF and deacetylase inhibitors of HIV-1 transcription in latently infected U1 cells. As an internal control, RNase protection analysis of the same RNA samples using an antisense riboprobe corresponding to the housekeeping gene for GAPDH showed no change in the level of mRNA following treatment with any drug alone or in combination (Fig. 5B).
FIG. 5.
Synergistic activation of HIV-1 transcription and virus production by deacetylase inhibitors and TNF in the latently infected cell line U1. (A) U1 cells were mock treated or treated with TSA, TNF, NaBut, TNF-TSA, or TNF-NaBut. At 24 h posttreatment, viral production was estimated by measuring CA-p24 antigen concentration in supernatants. The mock-treated value was arbitrarily set at a value of 1. Each point is the average of triplicate cultures performed in the same experiment. The error bars show the standard errors of the mean. A representative experiment out of four independent experiments is shown. (B) RNase protection analysis after a 6 h-treatment of U1 cells with the same activators as in panel A. To detect HIV-1 RNA, total RNA samples were incubated with an antisense riboprobe corresponding to the HIV-1 LTRs. The figure shows the 3′ LTR protected band (bottom panel). As a control, the same RNA samples were incubated with a specific probe corresponding to the GAPDH gene (top panel). (C) Relative levels of HIV mRNA shown in panel B (bottom) were quantified by radioimaging analysis using an Instant Imager (Packard). The HIV mRNA level in untreated U1 cells was assigned a value of 1.
These results demonstrate that the combination of TNF with a deacetylase inhibitor has a synergistic effect on reactivation of HIV-1 transcription and replication in the latently infected U1 cell line.
Deacetylase inhibitors TSA and NaBut prolong TNF-induced NF-κB binding to DNA.
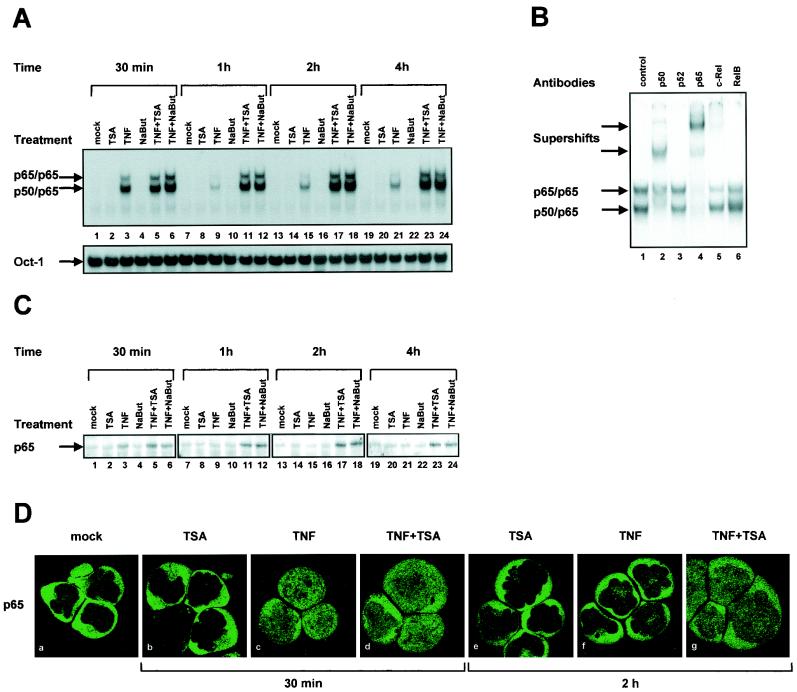
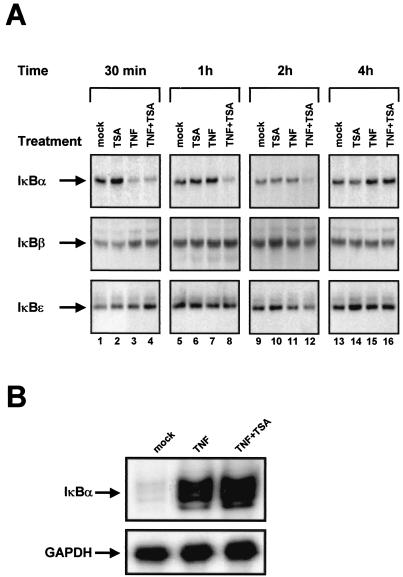
To examine the effect of deacetylase inhibitors on NF-κB binding to DNA, EMSAs were performed by using as a probe an oligonucleotide corresponding to the two κB sites from the prototype virus LAI of subtype B (54). This probe was incubated with nuclear extracts prepared from SupT1 cells either mock treated or treated with TSA, TNF, NaBut, TNF-TSA, or TNF-NaBut for different periods of time (30 min, 1 h, 2 h, and 4 h) (Fig. 6A). As expected, a rapid appearance of NF-κB binding activity was observed in response to TNF (Fig. 6A, lane 3). We showed by supershift assays using antibodies directed against individual members of the NF-κB family that these two retarded complexes corresponded to p50/p65 heterodimers and to p65/p65 homodimers, respectively (Fig. 6B). NF-κB appeared after a 30-min treatment and faded away after a 1-h treatment (Fig. 6A, lane 9). Treatment of cells with TSA alone or NaBut alone caused no induction of NF-κB binding activity even after a 4-h treatment (Fig. 6A, lanes 2, 8, 14, and 20 or lanes 4, 10, 16, and 22, respectively). A 30-min treatment with TNF-TSA or TNF-NaBut caused an induction of NF-κB binding activity identical to that obtained with TNF alone (Fig. 6A, lanes 5 or 6). Remarkably, when we compared induction by TNF alone with the inductions by TNF-TSA or TNF-NaBut at later times (1, 2, and 4 h), we observed that NF-κB binding activity was prolonged up to 4 h in the presence of TNF-TSA or TNF-NaBut (Fig. 6A, compare lane 9 with lanes 11 and 12, lane 15 with lanes 17 and 18, and lane 21 with lanes 23 and 24). This finding indicated a sustained NF-κB binding to DNA after TNF-TSA or TNF-NaBut versus TNF treatment. TSA (NaBut) did not alter the binding of the constitutively expressed Oct-1 transcription factor in either the presence or absence of TNF (Fig. 6A, lower panel).
FIG. 6.
Sustained NF-κB binding to DNA after TNF-TSA (NaBut) versus TNF treatment. (A) EMSA analysis of NF-κB binding activity. Nuclear extracts were prepared from SupT1 cells mock treated or treated with TSA, TNF, NaBut, TNF-TSA, or TNF-NaBut for different periods of time. An oligonucleotide corresponding to the LTR κB sites was used as probe. As a control for equal loading, the lower panel shows comparability of the various nuclear extracts assessed by EMSA with an Oct-1 consensus probe. (B) Supershift assays. The HIV-1 κB site oligonucleotide probe was incubated with 5 μg of nuclear extracts from SupT1 cells treated for 2 h with TNF-TSA. Next, antibodies directed against different members of the NF-κB family (lanes 2 to 6) or purified rabbit IgG as a negative control (lane 1) were added to the binding reaction. (C) Western blot analysis of levels of the p65 protein in SupT1 cells mock treated or treated with TNF and/or TSA (NaBut). The same nuclear extracts used in panel A were fractionated by electrophoresis, and the Western blots were probed with an anti-p65-specific antibody. (D) Immunofluorescence analysis of the p65 protein in SupT1 cells mock treated or treated with TNF and/or TSA. Subcellular localization of endogenous p65 was assessed by indirect immunofluorescence with a rabbit polyclonal anti-p65 IgG antibody and a goat anti-rabbit IgG antibody coupled with Alexa-488 (green color). SupT1 cells were left unstimulated or were stimulated with TNF and/or TSA for the indicated periods of time.
Taken together, these in vitro binding studies demonstrate that deacetylase inhibitors TSA and NaBut prolonged TNF-induced NF-κB binding to DNA but did not themselves stimulate NF-κB binding.
The presence of p65 is sustained in the nuclei of TNF-stimulated SupT1 cells in response to TSA or NaBut.
The same nuclear extracts used in EMSAs were also examined by Western blotting with an anti-p65 antibody in order to monitor the presence of p65 as a function of time in the nucleus after treatment with TSA, TNF, NaBut, TNF-TSA, or TNF-NaBut. Immunoblotting revealed sustained nuclear p65 presence after TNF-TSA (NaBut) versus TNF treatment (Fig. 6C).
To study the effect of deacetylase inhibitors on the subcellular distribution of p65, we monitored by confocal microscopy the localization of the endogenous p65 protein during stimulation with TSA, TNF, or both in combination (Fig. 6D). In unstimulated SupT1 cells, p65 was localized predominantly in the cytoplasmic compartment (Fig. 6D, panel a). Treatment with TSA for 30 min or 2 h did not alter this subcellular distribution (Fig. 6D, panel b or e, respectively). Treatment with TNF led after 30 min to the migration of p65 to the nucleus (Fig. 6D, panel c). This was transient, since, following 2 h of treatment with TNF, we observed the return of the nuclear p65 to the cytoplasm (Fig. 6D, panel f). A 30-min treatment with TNF-TSA caused a translocation of p65 into the nucleus identical to that observed with TNF alone (Fig. 6D, compare panels c and d). In contrast, comparison between both treatments at the 2 h-time point indicated that TSA treatment prolonged the TNF-induced nuclear translocation of p65 (Fig. 6D, compare panels f and g).
These results indicate that the prolonged NF-κB binding to DNA we observed after TNF-TSA (NaBut) versus TNF treatment (Fig. 6A) coincided with a prolonged intranuclear presence of p65.
Delay in the cytoplasmic recovery of IκBα in response to TNF-TSA versus TNF treatment.
The nuclear expression and action of NF-κB require signal-coupled phosphorylation and degradation of the IκB inhibitory proteins, which normally bind and sequester NF-κB in the cytoplasm. The activation of de novo IκBα gene expression by NF-κB likely plays a key role in the termination of nuclear NF-κB action, thereby ensuring a transient NF-κB transcriptional response. Therefore, we reasoned that the prolonged nuclear binding activity and presence of NF-κB we observed in response to TNF-TSA versus TNF treatment could result from a delay in cytoplasmic IκBα recovery.
To test this hypothesis, we monitored the presence of the IκBs as a function of time in the cytoplasm after treatment with TSA, TNF, or TNF-TSA. Cytoplasmic extracts were prepared from SupT1 cells treated with these activators for different periods of time (30 min, 1 h, 2 h, and 4 h) and analyzed for IκBα, IκBβ, and IκBɛ expression by Western blotting. TSA alone did not induce IκBα degradation (Fig. 7A, lanes 2, 6, 10, and 14). As expected, TNF induced a rapid degradation of the IκBα protein (Fig. 7A, lane 3) followed by its recovery, which was completed 1 h after stimulation (Fig. 7A, lane 7). After TNF-TSA treatment, rapid IκBα degradation was also observed (Fig. 7A, lane 4), but in contrast to what we saw with TNF alone, its recovery was delayed up to more than 2 h (Fig. 7A, compare lane 7 with lane 8 and lane 11 with lane 12). No change in IκBβ and IκBɛ cytoplasmic concentration was observed after any treatment. Similar results were obtained when examining the combined effect of TNF and NaBut (data not shown). Moreover, we confirmed the delay in cytoplasmic IκBα recovery in response to TNF-TSA versus TNF treatment using cytoplasmic extracts prepared from U1 cells (data not shown).
FIG. 7.
(A) Delay in cytoplasmic IκBα recovery in response to TNF-TSA versus TNF treatment. Cytoplasmic extracts were prepared from SupT1 cells untreated or treated with TNF and/or TSA for various times, and Western blot analyses of levels of the IκBα, IκBβ and IκBɛ were performed with specific antibodies against these proteins. (B) TSA did not prevent the TNF-dependent transcriptional activation of IκBα. RNase protection analysis after a 2 h-treatment of SupT1 cells with TNF or TNF-TSA. To detect IκBα RNA, total RNA samples were incubated with an antisense riboprobe corresponding to the IκBα gene (top panel). The figure shows the 162-nt IκBα protected band. As a control, the same RNA samples were incubated with a specific probe corresponding to the GAPDH gene (bottom panel).
We analyzed the steady-state level of IκBα mRNA by RNase protection assay using an IκBα-specific antisense riboprobe and RNA samples prepared from SupT1 cells treated or not for 2 h with either TNF or TNF-TSA (Fig. 7B). We observed similar strong activations of IκBα transcription following TNF or TNF-TSA treatment (Fig. 7B), thereby indicating that TSA did not prevent the TNF-dependent transcriptional activation of IκBα. Control RNase protection analysis of the same RNA samples using a GAPDH riboprobe showed no difference in the mRNA levels (Fig. 7B).
Our results thus demonstrate a marked delay in the cytoplasmic recovery of the NF-κB inhibitor, IκBα, after TNF-TSA versus TNF treatment. This delay was not due to a defect in the steady-state level of IκBα mRNA and coincided with the sustained NF-κB binding activity and the sustained intranuclear presence of p65 that we observed after TNF-TSA versus TNF treatment by EMSAs, immunoblotting, and confocal microscopy. This delay could explain the strong transcriptional synergism we observed between NF-κB and TSA on the HIV-1 promoter.
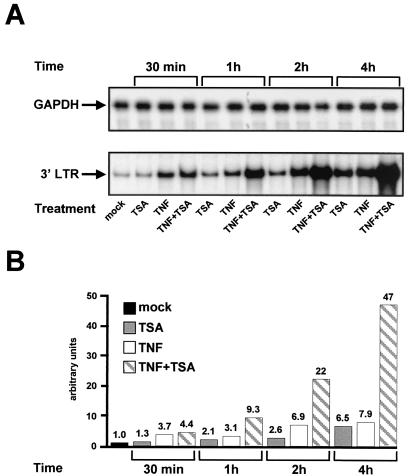
The TNF-TSA synergistic activation of HIV-1 transcription coincides with the prolonged nuclear level of NF-κB.
To determine whether a temporal correlation existed between the synergistic transcriptional activation of the HIV-1 promoter and the prolonged nuclear level of NF-κB, we examined by RNase protection the time course of HIV transcription in response to TNF and/or TSA. Total RNA samples were prepared from latently infected U1 cells treated or not with TSA, TNF, or TNF-TSA for different periods of time (30 min, 1 h, 2 h, and 4 h) (Fig. 8). The steady-state level of HIV-1 mRNA was evaluated using an HIV-1 promoter-specific probe. Treatment with TSA alone caused 1.3-, 2.1-, 2.6-, and 6.5-fold inductions of HIV-1 transcription after 30-min, 1-h, 2-h, and 4-h treatments, respectively (Fig. 8A [bottom panel] and B). Treatment with TNF alone caused 3.7-, 3.1-, 6.9-, and 7.9-fold HIV-1 mRNA inductions after 30 min-, 1 h-, 2 h- and 4 h-treatments, respectively (Fig. 8A [bottom panel] and B). Treatment with TNF-TSA caused 4.4-, 9.3-, 22-, and 47-fold inductions of LTR transcriptional activity after 30-min, 1-h, 2-h, and 4-h treatments, respectively (Fig. 8A [bottom panel] and B). GAPDH mRNA levels were unaffected by treatment with any drug alone or in combination (Fig. 8A). Therefore, the TNF-TSA synergistic effect at the mRNA level was detectable 1 h after addition of drugs to the cells and gradually increased over time. Our results thus demonstrate that the TNF-TSA synergistic activation of HIV-1 transcription was temporally parallel to the prolonged presence and binding of NF-κB in the nucleus (observed in Fig. 6).
FIG. 8.
Time course of HIV-1 transcriptional activation in response to TNF and/or TSA. (A) RNase protection analysis after 30-min, 1-h, 2-h, and 4-h treatments of U1 cells with TSA, TNF, or TNF-TSA. To detect HIV-1 RNA, total RNA samples were incubated with an antisense riboprobe corresponding to the HIV-1 LTRs. The figure shows the 3′ LTR protected band (bottom panel). As a control, the same RNA samples were incubated with a specific probe corresponding to the GAPDH gene (top panel). (B) Relative levels of HIV mRNA shown in panel A (bottom) were quantified by radioimaging analysis using an Instant Imager (Packard). The HIV mRNA level in untreated U1 cells was assigned a value of 1.
DISCUSSION
In summary, our results demonstrate a synergistic activation of the HIV-1 transcriptional promoter activity by NF-κB and inhibitors of deacetylases in transient transfection reporter assays and in acutely and latently infected cell lines. We provide a molecular explanation for this synergism by demonstrating a marked delay in the cytoplasmic recovery of IκBα in response to TNF-TSA (NaBut) versus TNF treatment and a temporal correlation between this delayed recovery and a prolonged binding activity and presence of NF-κB in the nucleus.
In this study, we have shown by deletion analysis that the TSA inducibility of the HIV-1 promoter required diverse sequences scattered throughout the LTR. The TSA response of the HIV LTR is likely to be explained by histone hyperacetylation. Indeed, our laboratory has previously demonstrated by chromatin mapping experiments that a nucleosome (nuc-1) positioned immediately downstream of the transcription start site is remodeled upon activation of the HIV promoter in response to HDAC inhibitors (54). Nuc-1 is likely to be the unique nucleosome target of action of the deacetylases since it is the only nucleosome whose structure or conformation is affected when deacetylases are inhibited (54). Although we did not investigate in the present study the chromatin organization of the transiently transfected LTR templates, we speculated that our LTR templates with deletions exhibited nucleosome positioning similar to that observed in vivo. Indeed, it is known that transfected DNA rapidly assembles into minichromosomes with histones attached (45). Moreover, the DEAE-dextran transfection technique we used here allows the typical 160-bp DNA ladder characteristic of the physiological nucleosomal DNA (24). Finally, in vitro chromatin-reconstituted HIV-1 templates corroborate the native nucleosomal organization found in latently infected cells (48, 49).
Importantly, because of the numerous nonhistone protein substrates for acetylation, the TSA response of the HIV-1 promoter could be explained in large part by acetylation and deacetylation events involved in the regulation of nuclear factors binding to the LTR. On one hand, several of these factors, including AP-1, ligand-bound nuclear hormone receptors, c-Myb, glucocorticoid receptor (GR), NF-AT, E-box binding proteins, Ets-1, TCF/LEF, NF-κB, Sp1, interferon regulatory factor (IRF), and the HIV trans-activator Tat have been shown to interact with acetyltransferases. On the other hand, several transcription factors that bind to the LTR, including unliganded nuclear hormone receptors, GR, E-box binding proteins, YY1, Sp1, TCF/LEF have been shown to interact with deacetylases. These factors therefore represent good candidates for the specific targeting of acetyltransferases and deacetylases to the HIV promoter, thereby regulating the acetylation level of histones (in particular nuc-1 histones) and/or of transcription factor substrates binding to the LTR (such as c-Myb, Sp1, IRF, TFIIEβ and TFIIF, and Tat) (43a). The addition and removal of acetyl groups on these histone and nonhistone proteins could be crucial in controlling transcription initiation and elongation.
Thus, the HIV promoter appears to contain numerous cis-regulatory DNA elements involved in the inducibility of the LTR by TSA. As such, the HIV promoter is representative of a small subset (<2%) of cellular genes that have their expression upregulated by deacetylase inhibitors (55) and constitutes a unique regulatory model system to study the complex relationship between acetylation processes and transcriptional activity.
Importantly, a significant TSA inducibility was still observed with a reduced LTR (nt 345 to 531), containing the two κB sites, the three Sp1 sites, the TATA box, and the LBP-1/YY1 site (Fig. 1A and B). This could be explained by the recruitment at the level of these sites of different factors presenting linkages with deacetylation and acetylation processes. (i) At the Sp1 sites, Sp1 is acetylated in vitro by p300 and interacts with p300, which acts as a coactivator for Sp1-mediated transcriptional activation (50). Sp1 has also been shown to interact directly with HDAC-1 (10). (ii) At the TATA box, the general transcription factors TFIIEβ and TFIIF are acetylated in vitro by P/CAF and p300 (21). The TFIID subunit TBP-associated factor II250 is a histone acetyltransferase (34). (iii) At the LBP-1 site, LBP functions as a docking molecule for YY1, which in turn acts by recruiting HDAC-1. This ternary complex represses the HIV-1 promoter, probably via the HDAC activity since this repression is blocked by TSA (9). (iv) At the κB sites, NF-κB-dependent gene expression requires the function of transcriptional coactivator proteins, including CBP/p300, P/CAF, and SRC-1, which possess acetyltransferase activity (17, 35, 40). Moreover, there is some evidence to suggest that deacetylase inhibitors may function to positively regulate NF-κB transcriptional activity (53).
In the present study, we focused on the potential functional role of these κB sites in the TSA inducibility of the HIV-1 LTR and demonstrated a strong transcriptional synergism between deacetylase inhibitors and NF-κB.
Mechanistically, we showed here by gel retardation assays that TSA (or NaBut) prolonged TNF-induced NF-κB DNA-binding activity, whereas TSA (or NaBut) alone caused no induction of NF-κB. These in vitro binding studies coincided with a sustained nuclear p65 presence as revealed by immunoblotting and confocal immunofluorescence microscopy. Importantly, Western blot analysis also revealed a marked delay in the cytoplasmic reappearance of the inhibitory protein IκBα after TNF-TSA versus TNF treatment. This delay correlated temporally with the sustained binding activity and presence of NF-κB in the nucleus and was not due to a defect in the IκBα mRNA production. These data therefore provide a molecular mechanism involving IκBα for the functional synergism we observed between TNF and inhibitors of deacetylases. IκBα plays a pivotal role in the NF-κB signaling pathway. Indeed, the primary level of regulation of NF-κB activity is through its retention in the cytoplasm through interactions with IκBα. Moreover, the resynthesis of de novo IκBα participates in a negative feedback system ensuring a transient NF-κB transcriptional response (reviewed in reference 28). We are currently further investigating the role of TSA in the delayed cytoplasmic reappearance of IκBα reported here. To this end, we are testing whether some proteins involved in the NF-κB/IκB signaling have their expression and/or action modulated by TSA.
The molecular mechanisms mediating the TNF-TSA synergism are likely to be highly complex and to implicate phenomena other than the delayed cytoplasmic recovery of IκBα. On one hand, the direct acetylation of Rel family members could also intervene in the mechanism of synergistic activation by TNF and TSA. In this regard, we have shown in a separate study that the p65 and p50 NF-κB subunits are weakly subject to reversible acetylation (E. Adam, V. Quivy, and C. Van Lint, unpublished results). In agreement, during the preparation of the present work, W. Greene and colleagues reported that p65 is acetylated and that this posttranslational modification governs IκBα binding to p65 and the nuclear export of the NF-κB complex (4). On the other hand, deacetylase corepressor proteins might be involved in the TNF-TSA synergism. Consistent with this, we have recently shown by in vitro interaction assays that HDAC-1, -3, and -4 can interact directly with the p65 and p50 subunits of NF-κB (Y. de Launoit and C. Van Lint, unpublished results). During the preparation of this manuscript, two other groups have separately reported the interaction of p65 either with HDAC-1 (2) or with HDAC-3 (4). These HDACs could repress expression of NF-κB-regulated genes by maintaining histones and/or other proteins in a deacetylated state. TSA or NaBut, which inhibit the HDAC activity, would increase NF-κB-dependent transcription by alleviating the chromatin- and/or factor-mediated block to transcriptional activation.
The application of HAART has resulted in a major reduction of virus loads in individuals tolerating the regimen and complying with its requirements, a stabilization of the clinical course, and a significant decline in mortality. Nonetheless, the persistence of HIV reservoirs has posed a sobering challenge to the long-term control or eradication of HIV in infected individuals receiving HAART (reviewed in reference 42). These latently infected cells are a permanent source for reactivation and lead to a rebound of viral load levels after interruption of HAART (15, 61). Activators of HIV expression combined with HAART could lead to the elimination of the latently infected cells and to the eradication of the infection. Indeed, it is likely that the latently infected cells die upon reactivation of virus (39) and that HAART prevents spread of released virus to adjacent cells (5). In this report, we demonstrate a synergistic effect of TNF and TSA (NaBut) for HIV-1 reactivation in the U1 cell line, a postintegration latency cell culture model. It is important to note that an array of cytokines, including the proinflammatory cytokines TNF and interleukin-1 (inducers of NF-κB), are already copiously expressed in the microenvironment of the lymphoid tissues, which harbor latent viral reservoirs (36). Therefore, our results suggest that the use of deacetylases inhibitors in the treatment of HIV infection may represent a valuable approach for purging the latently infected reservoirs in HAART-treated individuals. These deacetylase inhibitors would synergize with the TNF already present at increased level in the serum of the HIV-infected individuals. The possible clinical use of deacetylase inhibitors raises several issues. First, these drugs do not act in a cell-specific manner. Second, this class of agents is safely administered for other diseases, including beta chain hemoglobinopathies (such as beta-thalassemia and sickle cell anemia) (8, 11) and epilepsy and bipolar disorders (25, 41, 51). Third, an increasing number of non-B HIV-1 subtype infections are currently diagnosed. Here, we have shown that, in addition to the prototypical subtype B LTR, the LTRs from subtypes A through G of the HIV-1 group M were also activated synergistically by TSA and TNF, and the amplitude of the synergism correlated with the number of κB sites in the respective LTRs, which varies from one (subtype E) to three (subtype C). Based on our results, we propose the administration of deacetylase inhibitor(s) together with continuous HAART as a new potential therapeutic perspective to decrease in a subtype-nonspecific manner the pool of latent HIV reservoirs.
In conclusion, the results described in this study provide new insights into HIV-1 transcriptional regulation and more generally into the molecular mechanisms of NF-κB-mediated transactivation.
Acknowledgments
V.Q. and E.A. contributed equally to this work.
We thank Françoise Bex for her help in the confocal microscopy analysis and Fabrice Moore for his help in the transfection assays. The following reagents were obtained through the AIDS Research and Reference Reagent Program, NIAID, NIH: pRSV-p50 and pRSV-p65 from G. Nabel and N. Perkins, the U1 cell line from T. Folks, and the SupT1 cell line from J. Hoxie. We are grateful to L. Vanhamme and C. Calomme for critical reading of the manuscript.
This work was supported by grants to C.V.L. from the Fonds National de la Recherche Scientifique (FNRS, Belgium), the Télévie-Program, the Université Libre de Bruxelles (ULB), the Internationale Brachet Stiftung, the CGRI-INSERM cooperation, the Région Wallonne-Commission Européenne FEDER, the Agence Nationale de Recherches sur le SIDA (ANRS, France), and the Theyskens-Mineur Foundation. V.Q. is an Aspirant of the FNRS. A.C. is a Chercheur Qualifié of the FNRS. J.P. and V.B. are Directeurs de Recherches of the FNRS. C.V.L. is a Maıcirc;tre de Recherches of the FNRS. E.A. and D.D. are supported by postdoctoral fellowships from the ULB (ARC program 98/03-224) and the Région Wallonne (grant 991/4202), respectively. R.C. is supported by a fellowship from the Agence Nationale de Recherches sur le SIDA (France).
REFERENCES
- 1.Adams, M., L. Sharmeen, J. Kimpton, J. M. Romeo, J. V. Garcia, B. M. Peterlin, M. Groudine, and M. Emerman. 1994. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc. Natl. Acad. Sci. USA 91:3862-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashburner, B. P., S. D. Westerheide, and A. S. Baldwin, Jr. 2001. The p65 (RelA) subunit of NF-κB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol. Cell. Biol. 21:7065-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, H., M. Tini, and R. M. Evans. 2001. HATs on and beyond chromatin. Curr. Opin. Cell Biol. 13:218-224. [DOI] [PubMed] [Google Scholar]
- 4.Chen, L., W. Fischle, E. Verdin, and W. C. Greene. 2001. Duration of nuclear NF-κB action regulated by reversible acetylation. Science 293:1653-1657. [DOI] [PubMed] [Google Scholar]
- 5.Chun, T. W., D. Engel, S. B. Mizell, L. A. Ehler, and A. S. Fauci. 1998. Induction of HIV-1 replication in latently infected CD4+ T cells using a combination of cytokines. J. Exp. Med. 188:83-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen, J. 1998. Exploring how to get at and eradicate hidden HIV. Science 279:1854-1855. [DOI] [PubMed] [Google Scholar]
- 7.Col, E., C. Caron, D. Seigneurin-Berny, J. Gracia, A. Favier, and S. Khochbin. 2001. The histone acetyltransferase, hGCN5, interacts with and acetylates the HIV transactivator, Tat. J. Biol. Chem. 276:28179-28184. [DOI] [PubMed] [Google Scholar]
- 8.Collins, A. F., H. A. Pearson, P. Giardina, K. T. McDonagh, S. W. Brusilow, and G. J. Dover. 1995. Oral sodium phenylbutyrate therapy in homozygous beta thalassemia: a clinical trial. Blood 85:43-49. [PubMed] [Google Scholar]
- 9.Coull, J. J., F. Romerio, J. M. Sun, J. L. Volker, K. M. Galvin, J. R. Davie, Y. Shi, U. Hansen, and D. M. Margolis. 2000. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J. Virol. 74:6790-6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doetzlhofer, A., H. Rotheneder, G. Lagger, M. Koranda, V. Kurtev, G. Brosch, E. Wintersberger, and C. Seiser. 1999. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol. Cell. Biol. 19:5504-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dover, G. J., S. Brusilow, and S. Charache. 1994. Induction of fetal hemoglobin production in subjects with sickle cell anemia by oral sodium phenylbutyrate. Blood 84:339-343. [PubMed] [Google Scholar]
- 12.el Kharroubi, A., G. Piras, R. Zensen, and M. A. Martin. 1998. Transcriptional activation of the integrated chromatin-associated human immunodeficiency virus type 1 promoter. Mol. Cell. Biol. 18:2535-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emiliani, S., W. Fischle, M. Ott, C. Van Lint, C. A. Amella, and E. Verdin. 1998. Mutations in the tat gene are responsible for human immunodeficiency virus type 1 postintegration latency in the U1 cell line. J. Virol. 72:1666-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emiliani, S., C. Van Lint, W. Fischle, P. Paras, Jr., M. Ott, J. Brady, and E. Verdin. 1996. A point mutation in the HIV-1 Tat responsive element is associated with postintegration latency. Proc. Natl. Acad. Sci. USA 93:6377-6381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295-1300. [DOI] [PubMed] [Google Scholar]
- 16.Folks, T. M., K. A. Clouse, J. Justement, A. Rabson, E. Duh, J. H. Kehrl, and A. S. Fauci. 1989. Tumor necrosis factor alpha induces expression of human immunodeficiency virus in a chronically infected T-cell clone. Proc. Natl. Acad. Sci. USA 86:2365-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerritsen, M. E., A. J. Williams, A. S. Neish, S. Moore, Y. Shi, and T. Collins. 1997. CREB-binding protein/p300 are transcriptional coactivators of p65. Proc. Natl. Acad. Sci. USA 94:2927-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghosh, S., M. J. May, and E. B. Kopp. 1998. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16:225-260. [DOI] [PubMed] [Google Scholar]
- 19.Herschlag, D., and F. B. Johnson. 1993. Synergism in transcriptional activation: a kinetic view. Genes Dev. 7:173-179. [DOI] [PubMed] [Google Scholar]
- 20.Igarashi, T., C. R. Brown, Y. Endo, A. Buckler-White, R. Plishka, N. Bischofberger, V. Hirsch, and M. A. Martin. 2001. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): implications for HIV-1 infections of humans. Proc. Natl. Acad. Sci. USA 98:658-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imhof, A., X. J. Yang, V. V. Ogryzko, Y. Nakatani, A. P. Wolffe, and H. Ge. 1997. Acetylation of general transcription factors by histone acetyltransferases. Curr. Biol. 7:689-692. [DOI] [PubMed] [Google Scholar]
- 22.Israel, A. 2000. The IKK complex: an integrator of all signals that activate NF-κB? Trends Cell Biol. 10:129-133. [DOI] [PubMed] [Google Scholar]
- 23.Jeeninga, R. E., M. Hoogenkamp, M. Armand-Ugon, M. de Baar, K. Verhoef, and B. Berkhout. 2000. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J. Virol. 74:3740-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeong, S., and A. Stein. 1994. Micrococcal nuclease digestion of nuclei reveals extended nucleosome ladders having anomalous DNA lengths for chromatin assembled on non-replicating plasmids in transfected cells. Nucleic Acids Res. 22:370-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johannessen, C. U. 2000. Mechanisms of action of valproate: a commentary. Neurochem. Int. 37:103-110. [DOI] [PubMed] [Google Scholar]
- 26.Jordan, A., P. Defechereux, and E. Verdin. 2001. The site of HIV-1 integration in the human genome determines basal transcriptional activity and response to Tat transactivation. EMBO J. 20:1726-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karin, M. 1999. The beginning of the end: IκB kinase (IKK) and NF-κB activation. J. Biol. Chem. 274:27339-27342. [DOI] [PubMed] [Google Scholar]
- 28.Karin, M., and Y. Ben Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 29.Khochbin, S., and H. Y. Kao. 2001. Histone deacetylase complexes: functional entities or molecular reservoirs. FEBS Lett. 494:141-144. [DOI] [PubMed] [Google Scholar]
- 30.Kiernan, R. E., C. Vanhulle, L. Schiltz, E. Adam, H. Xiao, F. Maudoux, C. Calomme, A. Burny, Y. Nakatani, K. T. Jeang, M. Benkirane, and C. Van Lint. 1999. HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J. 18:6106-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, N., and M. Karin. 2000. Signaling pathways leading to nuclear factor-kappa B activation. Methods Enzymol. 319:273-279. [DOI] [PubMed] [Google Scholar]
- 33.McCune, J. M. 1995. Viral latency in HIV disease. Cell 82:183-188. [DOI] [PubMed] [Google Scholar]
- 34.Mizzen, C. A., X. J. Yang, T. Kokubo, J. E. Brownell, A. J. Bannister, T. Owen-Hughes, J. Workman, L. Wang, S. L. Berger, T. Kouzarides, Y. Nakatani, and C. D. Allis. 1996. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell 87:1261-1270. [DOI] [PubMed] [Google Scholar]
- 35.Na, S. Y., S. K. Lee, S. J. Han, H. S. Choi, S. Y. Im, and J. W. Lee. 1998. Steroid receptor coactivator-1 interacts with the p50 subunit and coactivates nuclear factor κB-mediated transactivations. J. Biol. Chem. 273:10831-10834. [DOI] [PubMed] [Google Scholar]
- 36.Navikas, V., J. Link, C. Persson, T. Olsson, B. Hojeberg, A. Ljungdahl, H. Link, and B. Wahren. 1995. Increased mRNA expression of IL-6, IL-10, TNF-alpha, and perforin in blood mononuclear cells in human HIV infection. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 9:484-489. [PubMed] [Google Scholar]
- 37.Osborn, L., S. Kunkel, and G. J. Nabel. 1989. Tumor necrosis factor alpha and interleukin 1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor kappa B. Proc. Natl. Acad. Sci. USA 86:2336-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereira, L. A., K. Bentley, A. Peeters, M. J. Churchill, and N. J. Deacon. 2000. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 28:663-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perelson, A. S., P. Essunger, Y. Cao, M. Vesanen, A. Hurley, K. Saksela, M. Markowitz, and D. D. Ho. 1997. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387:188-191. [DOI] [PubMed] [Google Scholar]
- 40.Perkins, N. D., L. K. Felzien, J. C. Betts, K. Leung, D. H. Beach, and G. J. Nabel. 1997. Regulation of NF-κB by cyclin-dependent kinases associated with the p300 coactivator. Science 275:523-527. [DOI] [PubMed] [Google Scholar]
- 41.Phiel, C. J., F. Zhang, E. Y. Huang, M. G. Guenther, M. A. Lazar, and P. S. Klein. 2001. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J. Biol. Chem. 276:36734-36741. [DOI] [PubMed] [Google Scholar]
- 42.Pierson, T., J. McArthur, and R. F. Siliciano. 2000. Reservoirs for HIV-1: mechanisms for viral persistence in the presence of antiviral immune responses and antiretroviral therapy. Annu. Rev. Immunol. 18:665-708. [DOI] [PubMed] [Google Scholar]
- 43.Poli, G., A. Kinter, J. S. Justement, J. H. Kehrl, P. Bressler, S. Stanley, and A. S. Fauci. 1990. Tumor necrosis factor alpha functions in an autocrine manner in the induction of human immunodeficiency virus expression. Proc. Natl. Acad. Sci. USA 87:782-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.Quivy, V., and C. Van Lint. 2002. Diversity of acetylation targets and roles in transcriptional regulation: the HIV-1 promoter as a model system. Biochem. Pharmacol. 64:925-934. [DOI] [PubMed] [Google Scholar]
- 44.Rabson, A. B., and H. C. Lin. 2000. NF-kappa B and HIV: linking viral and immune activation. Adv. Pharmacol. 48:161-207. [DOI] [PubMed] [Google Scholar]
- 45.Reeves, R., C. M. Gorman, and B. Howard. 1985. Minichromosome assembly of non-integrated plasmid DNA transfected into mammalian cells. Nucleic Acids Res. 13:3599-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 47.Schoonbroodt, S., V. Ferreira, M. Best-Belpomme, J. R. Boelaert, S. Legrand-Poels, M. Korner, and J. Piette. 2000. Crucial role of the amino-terminal tyrosine residue 42 and the carboxyl-terminal PEST domain of I kappa B alpha in NF-kappa B activation by an oxidative stress. J. Immunol. 164:4292-4300. [DOI] [PubMed] [Google Scholar]
- 48.Sheridan, P. L., T. P. Mayall, E. Verdin, and K. A. Jones. 1997. Histone acetyltransferases regulate HIV-1 enhancer activity in vitro. Genes Dev. 11:3327-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Steger, D. J., A. Eberharter, S. John, P. A. Grant, and J. L. Workman. 1998. Purified histone acetyltransferase complexes stimulate HIV-1 transcription from preassembled nucleosomal arrays. Proc. Natl. Acad. Sci. USA 95:12924-12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki, T., A. Kimura, R. Nagai, and M. Horikoshi. 2000. Regulation of interaction of the acetyltransferase region of p300 and the DNA-binding domain of Sp1 on and through DNA binding. Genes Cells 5:29-41. [DOI] [PubMed] [Google Scholar]
- 51.Tunnicliff, G. 1999. Actions of sodium valproate on the central nervous system. J. Physiol. Pharmacol. 50:347-365. [PubMed] [Google Scholar]
- 52.Van Lint, C. 2000. Role of chromatin in HIV-1 transcriptional regulation. Adv. Pharmacol. 48:121-160. [DOI] [PubMed] [Google Scholar]
- 53.Vanden Berghe, W., K. De Bosscher, E. Boone, S. Plaisance, and G. Haegeman. 2098. 1999. The nuclear factor-κB engages CBP/p300 and histone acetyltransferase activity for transcriptional activation of the interleukin-6 gene promoter. J. Biol. Chem. 274:32091-32093. [DOI] [PubMed] [Google Scholar]
- 54.Van Lint, C., S. Emiliani, M. Ott, and E. Verdin. 1996. Transcriptional activation and chromatin remodeling of the HIV-1 promoter in response to histone acetylation. EMBO J. 15:1112-1120. [PMC free article] [PubMed] [Google Scholar]
- 55.Van Lint, C., S. Emiliani, and E. Verdin. 1996. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 5:245-253. [PMC free article] [PubMed] [Google Scholar]
- 56.Van Lint, C., J. Ghysdael, P. Paras, Jr., A. Burny, and E. Verdin. 1994. A transcriptional regulatory element is associated with a nuclease-hypersensitive site in the pol gene of human immunodeficiency virus type 1. J. Virol. 68:2632-2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Lint, C., C. A. Amella, S. Emiliani, M. John, T. Jie, and E. Verdin. 1997. Transcription factor binding sites downstream of the human immunodeficiency virus type 1 transcription start site are important for virus infectivity. J. Virol. 71:6113-6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Verdin, E., P. Paras, Jr., and C. Van Lint. 1993. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J. 12:3249-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.West, M. J., A. D. Lowe, and J. Karn. 2001. Activation of human immunodeficiency virus transcription in T cells revisited: NF-κB p65 stimulates transcriptional elongation. J. Virol. 75:8524-8537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Winslow, B. J., R. J. Pomerantz, O. Bagasra, and D. Trono. 1993. HIV-1 latency due to the site of proviral integration. Virology 196:849-854. [DOI] [PubMed] [Google Scholar]
- 61.Wong, J. K., M. Hezareh, H. F. Gunthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]