Abstract
Functional domains of the strikingly conserved envelope (Env) glycoproteins of bovine leukemia virus (BLV) and its close relative, human T-cell leukemia virus type 1 (HTLV-1), are still being defined. We have used BLV Env protein variants to gain insights into the structure and function of this important determinant of viral infectivity. Each of 23 different single amino acid variants found in cDNA clones of env transcripts present after short-term culture of peripheral blood mononuclear cells from BLV-infected sheep was expressed in COS-1 cells and tested for the ability to mediate cell fusion and to be cleaved to surface (SU) and transmembrane (TM) protein subunits. Of 11 Env variants that failed to induce syncytia or did so poorly, 7 contained changes in amino acids identical or chemically conserved in the HTLV-1 Env protein. These seven included the four variants that showed aberrant proteolytic cleavage and poor cell surface expression, underscoring their importance for Env structure. Ten of 12 variants that retained wild-type syncytium-inducing ability clustered in the N-terminal half of BLV SU, which forms the putative receptor-binding domain (RBD). Several variants in the RBD showed evidence of subtle misfolding, as judged by reduced binding to monoclonal antibodies recognizing conformational epitopes F, G, and H formed by the N terminus of SU. We modeled the BLV RBD by aligning putative structural elements with known elements of the ecotropic Friend murine leukemia virus RBD monomer. All the variant RBD residues but one are exposed on the surface of this BLV model. These variants as well as function-altering, antibody-reactive residues defined by other investigators group on one face of the molecular model. They are strikingly absent from the opposite face, implying that it is likely to face inward in Env complexes. This surface might interact with the C-terminal domain of SU or with an adjacent monomer in the Env oligomer. This location suggests an orientation for the monomer of ecotropic Friend murine leukemia virus RBD.
Envelope (Env) proteins confer infectivity on retroviral particles. Oligomers of these proteins bind to receptors on the surfaces of host cells and mediate entry of the viral genome into the cell. The Env protein complex is composed of surface glycoprotein (SU) subunits, which are anchored to virions by their association with transmembrane (TM) protein subunits. SU molecules recognize and bind to cellular receptors, thereby initiating a complex series of conformational changes that lead to fusion of viral and cellular membranes by TM oligomers (reviewed in reference 30). Upon successful synthesis of the DNA provirus and its integration into host cell DNA, expression of viral genes ensues. When the newly synthesized polyprotein Env precursor is cleaved in the Golgi apparatus of the virus-producing host cell, SU and TM subunits remain associated in a metastable state (18). Oligomers of mature Env proteins are transported to the cell surface membrane, where they can be incorporated into budding viral particles.
What is known about the structure and function of the Env protein of the deltaretrovirus bovine leukemia virus (BLV) is derived from delineation of epitopes recognized by monoclonal antibodies (2, 6, 8) and from identification of antipeptide antibodies that block syncytium formation or neutralize the infectivity of pseudotype virus (10, 49). Two models of BLV SU have previously been developed, the first based on protein-folding patterns (39) and the second based on hydrophobic cluster analysis and comparison with the known structures of influenza virus hemagglutinin-1 protein and the HLA-A2 protein of the human major histocompatibility complex (9).
Evidence that the N-terminal half of mature gp51-SU plays an important role in virus infectivity and syncytium formation (6, 48) suggests that it most probably contains the receptor-binding domain (RBD), analogous to the RBD of gammaretroviruses. This region (Fig. 1) forms the epitopes F, G, and H (5), which are designated conformational because their recognition by specific monoclonal antibodies depends on disulfide bonding (48) and glycosylation (7). Antibodies from naturally infected cattle also recognize only the glycosylated form of SU, suggesting a specificity for conformation-dependent epitopes in vivo (47). The differential binding of monoclonal antibodies specific for the F, G, and H epitopes to Env proteins encoded by a number of BLV isolates from different geographical origins led to the identification of amino acids potentially affecting SU conformation (39, 48). Antibodies raised against peptides located near these amino acids neutralize infectivity and inhibit syncytium formation (10, 49).
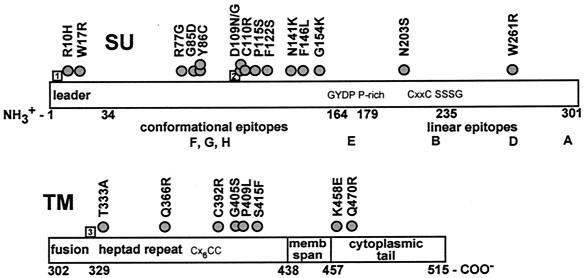
FIG. 1.
Distribution of amino acid substitutions encoded in env cDNA clones. The BLV Env protein is represented approximately to scale, with amino acids numbered according to their positions in the Env precursor protein. In SU, aa 1 to 33 form the signal peptide. The strong turn GYDP (aa 164 to 167) separates the N-terminal conformational epitopes, F, G, and H, from the C-terminal linear epitopes, A, B, D, and E. A conserved P-rich region begins at aa 179. A second conserved strong turn, SSSG, is located at aa 235 to 238. In TM, the extracellular domain includes an N-terminal fusion peptide followed by a 4-3 heptad repeat motif. The predicted membrane-spanning region is formed by aa 437 to 457, and the cytoplasmic tail encompasses aa 458 to 515. Circles, sites of amino acid substitutions encoded in individual cDNAs. Numbered squares represent the sites of strain-specific substitutions encoded in all env clones derived after sheep passage of the Bat2cl6 strain of BLV; relative to the FLK North American prototype BLV, the Bat2cl6 substitutions are as follows: 1, K4E; 2, Y108H; 3, A326S.
Putative strong turn GYDP, which is conserved in all oncogenic retroviruses (22), precedes a sequence resembling the conserved proline-rich (P-rich) region in SU of murine retroviruses. The putative P-rich region of BLV SU is followed by the C terminus containing the epitopes A, B, D, and most of E (5, 8). These epitopes are classified as linear because they were mapped with peptides (2, 8). This segment of SU is thought to interact with TM; one component is likely to be the formation of a labile disulfide bond (33, 43, 59) between a conserved CXXC motif (22) in SU and a conserved CX6CC motif (53) in TM. Just after the CXXC motif in SU is the sequence SSSG, which has been suggested to be a second strong turn in SU (22).
The extracellular domain of BLV gp30-TM includes an N-terminal hydrophobic fusion peptide (63) as well as a 4-3 heptad amino acid repeat (23, 34) characteristic of coiled coils that are important for oligomerization of Env proteins of Rous sarcoma virus and human immunodeficiency virus type 1 (3, 19). A stretch of 20 uncharged residues (amino acids [aa] 438 to 457) in BLV TM is believed to span host cell membranes (53). Finally, the cytoplasmic domain includes a PXXPX4-5P protein interaction motif (11, 52) upstream of and interspersed with two YXXL immunoreceptor tyrosine activation motifs (1).
While investigating differences in syncytium-inducing ability of peripheral blood mononuclear cells (PBMCs) from two groups of BLV-infected sheep (32), we generated 17 env cDNA clones from viral transcripts present in cultured PBMCs (31). Individual clones encoded between zero and four amino acid substitutions. To learn how these amino acid alterations affect Env protein synthesis and function, we constructed env gene chimeras encoding single mutations in a wild-type (WT) env backbone. We then determined the effect of individual amino acid substitutions on the ability of variant Env proteins to induce syncytia and to be proteolytically processed. Variants displaying reduced or no syncytium induction were further assessed for their ability to be expressed on the cell surface. Variants in the N-terminal putative RBD of SU were tested for their ability to attain a conformation displaying the F, G, and H epitopes. To understand how the substitutions in the N terminus of BLV SU might affect its configuration, we developed a model of the putative RBD of BLV based on the crystal structure (20) of the monomeric RBD of ecotropic Friend murine leukemia virus (F-MuLV). All but one of the variant residues were exposed on the same face of the BLV model as residues identified by others as being involved in conformational epitopes. The opposite face showed a conspicuous absence of variant residues, suggesting that this face is not accessible to antibodies. We propose that the latter side of the BLV RBD faces inward in native Env complexes. This surface might interact with the C-terminal domain of SU or with an adjacent monomer in the Env oligomer. This location suggests an orientation for the structure solved for the monomeric RBD of ecotropic F-MuLV.
MATERIALS AND METHODS
Derivation of env cDNA clones.
env cDNAs were prepared from transcripts synthesized in cultured PBMCs from three BLV-infected sheep (31). Freshly purified PBMCs were cultured for 14 h in medium supplemented with 10% fetal bovine serum (FBS) and 10 μg of lipopolysaccharide per ml, and then RNA was purified with guanidinium thiocyanate. First-strand env cDNAs were synthesized from env mRNAs with gene-specific primers and RNase H-negative reverse transcriptase from Moloney MuLV (Superscript preamplification system kit; Invitrogen-Life Technologies). First-strand cDNAs were subjected to nested amplification with Taq polymerase. First-round primers were 4448 forward (5′-GGAGAAACACCCAAGGGCTCTGAT; where the nucleotide number corresponds to the first 5′ virus-complementary base of the primer and nucleotide [nt] 1 begins the viral R region) and 6288 reverse (5′-GAGGCTCAGGGTAGGGCTGTGTTC); second-round primers were 4501 forward (5′-GGGGTACCAAACAATCGTCGGTGGCTAGGA) and 6252 reverse (5′-GGAATTCGGCAGCAAGAAGAGGCTTGTG), in which the underlined bases were added for cloning with KpnI and EcoRI, respectively. For each animal's cDNA, PCR products from two first-round reactions were amplified in five second-round reactions and then were combined to obtain a total of 1.2 to 1.5 μg of DNA. Purified env DNAs were cloned into the pcDNA3 expression vector (Invitrogen) for transient expression from the immediate-early promoter of cytomegalovirus. Clones were sequenced manually (Silver Sequence DNA sequencing system; Promega) and with an automated DNA sequencer (Prism; Applied Biosystems, Inc.).
Construction of chimeric env clones.
Fragments encoding single amino acid substitutions were obtained by digestion once or serially with the following restriction enzymes: HindIII (cleavage in vector multiple cloning site 11 nt upstream of env) and BamHI (cleavage at env nt 5021) for Y86C (where aa 1 is the initiator Met of the Env precursor protein), D109N, P115S, F121S, N141K, G154K, N203S, and G405S; HindIII and BamHI and then BstXI (cleavage at env nt 4927) for R10H and C110R; SmaI (cleavage at env nt 5507) for W261R and C392R; BamHI and then BsmI (cleavage at env nt 5799) for T333A and S415F; BsmI alone for Q366R, P409L, K458E, and Q470R; and BsmI and then BstXI for R77G and D109G. These digested fragments were isolated by gel electrophoresis and then ligated in place of the corresponding fragments of pcDNA3-ENV (BLV 468-22) containing the WT env sequence derived after animal passage from the Bat2cl6 strain (25) of BLV.
Transfection of COS-1 cells.
COS-1 cells obtained from the American Type Culture Collection were maintained in high-glucose Dulbecco's modified Eagle medium (DMEM) supplemented with 10% heat-inactivated FBS (DMEM-10F). To express env or control constructs for assessment by immunocytochemistry, cell fusion, immunoblotting, or immunoprecipitation, COS-1 cells were transfected with the appropriate env plasmid by using Lipofectamine and PLUS reagent (Invitrogen-Life Technologies) in serum-free DMEM as recommended by the manufacturer. Transfected cells were then incubated at 37°C for 20 to 24 h before being tested for Env protein expression and syncytium formation or for 48 h before being lysed for immunoblotting or immunoprecipitation.
Syncytium assay.
F81 cells used as fusion partners in syncytium assays are a subline of CC81, a feline kidney cell line transformed with murine sarcoma virus (21). They were maintained in minimal essential medium with Earle's salts supplemented with 5% FBS. At 24 h posttransfection, 2 × 104 COS-1 cells were trypsinized and resuspended in DMEM-10F and then were mixed with trypsinized F81 cells (105) and seeded into 12-well plates. After being incubated at 37°C for 24 h, cells were fixed with 100% methanol for 10 min and then were stained with Giemsa. Syncytia containing >5 nuclei were counted under ×100 magnification. Nuclei in individual syncytia were counted under ×200 or ×400 magnification, and the numbers were expressed as percentages of nuclei in WT syncytia from the same experiment. WT syncytia contained an average of 214 nuclei per syncytium. COS-1 cells transfected with the expression vector pcDNA3 encoding chloramphenicol acetyltransferase (CAT; Invitrogen-Life Technologies) served as negative controls.
Immunocytochemical assays.
To enumerate cells expressing SU, one set of COS-1 cells was plated at 24 h after transfection on eight-well glass chamber slides. After 16 h at 37°C, cells were fixed for 20 min with 2% paraformaldehyde in buffer (60 mM PIPES [piperazine-N,N′-bis{2-ethanesulfonic acid}], 25 mM HEPES, 10 mM EDTA, 2 mM MgCl2 [pH 6.9]) and then were permeabilized for 10 min with acetone. Nonspecific antibody binding was blocked by incubation with 5% FBS in BLOTTO (5% [wt/vol] nonfat dry milk suspended in 25 mM Tris-HCl [pH 8.0], 150 mM NaCl, and 0.075% [vol/vol] Tween 20) prior to incubation with mouse monoclonal antibodies specific for the G or D epitope of SU (BLV1 and BLV2 from Veterinary Medical Research and Development, Inc., Pullman, Wash.) diluted 1:400. For comparative staining, mouse monoclonal antibodies specific for the F, G, or H epitope of SU (a kind gift from D. Portetelle, Faculté Universitaire des Sciences Agronomiques, Gembloux, Belgium) were diluted 1:100. Primary antibodies were detected by incubation with a 1:400 dilution of biotinylated sheep anti-mouse Ig and then with biotinylated alkaline phosphatase linked to avidin (Vector Labs). Positive cells were visualized with HistoMark red substrate (Kirkegaard & Perry Laboratories); cells were counterstained with 0.5% methyl green and counted under ×400 magnification.
To enumerate cells expressing Env protein on their surfaces, a second set of cells that had been plated and incubated in parallel was fixed with paraformaldehyde. To prevent permeabilization and detection of cytoplasmic proteins, the cells were not treated with acetone and blocking was with BLOTTO lacking Tween 20. Cells were stained with a G epitope-specific monoclonal antibody. Negative controls included staining only with a secondary antibody as well as staining nontransfected cells with both primary and secondary antibodies. To enumerate CAT expression in the negative-control cells transfected with pcDNA3-CAT, one sample of the fixed cells was incubated with a rabbit anti-CAT polyclonal antibody (1:800; 5 Prime-3 Prime, Inc.) and then with biotinylated goat anti-rabbit immunoglobulin (Ig; 1:1,500). Staining was visualized as described above.
Immunoblots.
At 48 h posttransfection, COS-1 cells were disrupted with 1 ml of lysis buffer (25 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% [wt/vol] Nonidet P-40, 1% [wt/vol] sodium deoxycholate, 2 mM EDTA, 10 mM N-ethylmaleimide, 1 mM phenylmethylsulfonyl fluoride, 1 mg of ovalbumin per ml, 0.5 μg of leupeptin per ml, 2 μg of aprotinin per ml, 10 μg of pepstatin A per ml). Lysates were incubated on ice for 20 min, and then insoluble material was removed by centrifugation at 16,000 × g for 30 min at 4°C. To enrich glycoproteins, cleared lysates (1 ml) were gently rocked for 4 h at 4°C with 60 μl of 50% (wt/vol) Sepharose 4B conjugated to lentil lectin (Sigma) and washed once with lysis buffer and then glycosylated proteins were eluted with an equal volume of double-strength gel sample buffer (125 mM Tris-HCl [pH 6.8], 4% [wt/vol] sodium dodecyl sulfate [SDS], 20% [vol/vol] glycerol, 0.01% [wt/vol] bromophenol blue, 100 mM dithiothreitol) and boiled for 4 min. The supernatant was electrophoresed on an SDS-12% polyacrylamide gel. Proteins were transferred onto polyvinylidene difluoride membranes (Bio-Rad). Blots were probed with a hyperimmune rabbit serum raised against the 16-aa C-terminal peptide of TM (12) (a kind gift from G. Cantor, Washington State University, Pullman) diluted 1:8,000 in 25 mM Tris-HCl [pH 8.0]-150 mM NaCl-0.1% (vol/vol) Tween 20. The primary antibody was detected by incubation first with a 1:10,000 dilution of biotinylated goat anti-rabbit Ig in the same buffer and then with biotinylated alkaline phosphatase linked to avidin (Vector Labs) and finally with Lumi-Phos Plus substrate (Invitrogen-Life Technologies) for 1 min prior to exposure to X-ray film for 10 to 30 min.
Nucleotide sequence accession number.
The env gene sequence of cDNA clone 468-22, which has the consensus sequence of the Bat2cl6 strain of BLV after passage through sheep, was assigned GenBank accession no. AY078387.
RESULTS
Derivation of clones expressing chimeric BLV Env proteins.
We previously used BLV derived from a clone (Bat2cl6) of infected bat lung cells (25) to experimentally infect a group of sheep (36, 50). Peripheral blood cells from two of those animals were injected into a second group of sheep (32). To determine the env sequence of the sheep-passaged BLV, we prepared PBMC DNA from three sheep of the first group and four sheep of the second group, amplified the env gene, and sequenced the resulting PCR products (31). We also prepared cDNA clones from transcripts extracted from cultured PBMCs and sequenced 17 clones that had been derived from three animals in the first group. The env sequences of two cDNA clones (468-22 and 468-4) were identical to the proviral consensus sequence, which we designated the WT. The predicted Env protein contains three strain-specific amino acid substitutions, K4E, Y108H, and A326S (Fig. 1), relative to Env encoded by BLV from fetal lamb kidney (FLK) cells (GenBank accession no. M35242) (39). The FLK strain represents the North American BLV prototype.
To learn how individual amino acid alterations found in the cDNA clones (Fig. 1) affect Env protein synthesis and function, we constructed env gene chimeras encoding single mutations in the WT Bat2cl6 sequence. All the chimeric clones reported here supported production of the Env protein in transfected COS-1 cells, as revealed by immunocytochemical staining of fixed and permeabilized cells with one or more monoclonal antibodies specific for the SU protein.
Individual amino acid substitutions affected the efficiency of syncytium induction and the size of syncytia.
As a measure of function, each variant Env protein containing a single altered amino acid was tested for its ability to induce membrane fusion. Transfected COS-1 cells were cocultured with F81 indicator cells, which form multinucleated syncytia after fusing with cells displaying the BLV Env protein on the surface (26, 29). To account for differences in numbers of cells expressing particular Env variants, the efficiency of syncytium induction (Fig. 2) is expressed as the ratio of the number of syncytia induced to the number of fixed and permeabilized COS cells that immunostained for SU in parallel cultures. Syncytium size was assessed by counting nuclei as described in Materials and Methods.
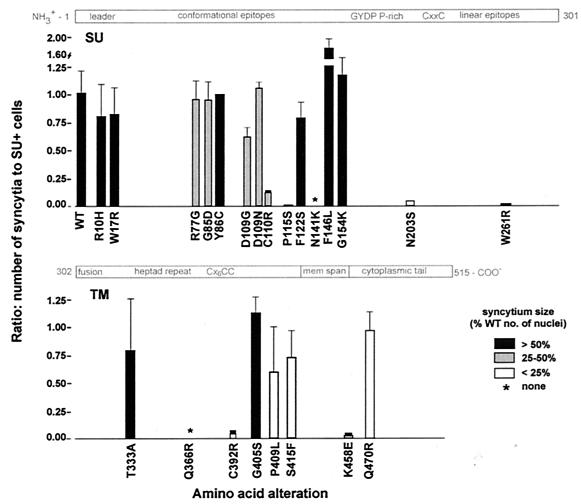
FIG. 2.
Induction of syncytia by variant Env proteins. COS-1 cells were trypsinized 24 h after transfection with env plasmids. A portion of each population was then cocultured with F81 cells for 24 h to assess the ability of the expressed Env protein to induce syncytium formation. Another portion was replated for 24 h, after which COS-1 cells were fixed, permeabilized, and immunostained to reveal the presence of the SU protein. The plot shows the averages from two or three experiments of the ratios of the number of syncytia obtained from 2 × 104 transfected cells to the corresponding number of cells containing the SU protein. The average sizes of individual syncytia are indicated by fill color as percentages of the number of nuclei present in syncytia induced in parallel by WT Env. Amino acid substitutions are depicted along the length of the SU and TM proteins, drawn approximately to scale. ∗, no syncytia ever observed.
Most of the amino acid substitutions within the first 164 residues (aa 1 to the GYPD strong turn) of the Env precursor protein had little or no effect on syncytium induction, even when the changes were nonconservative (Fig. 2). Variants containing substitutions R10H or W17R within the signal sequence (aa 1 to 33) and Y86C, F122S, or G154K induced large syncytia at levels comparable to that for the WT Env protein. Large syncytia were also induced by variant F146L but with an efficiency greater than that for the WT Env protein. Variant Envs containing R77G or G85D substitutions induced WT ratios of syncytia, but these syncytia contained reduced numbers of nuclei. Amino acid 109 was the site of two substitutions: both D109N and D109G induced syncytia of reduced size, but the D109G change appears to be more severe since it reduced the number of syncytia somewhat.
In contrast to these mild effects, three N-terminal region variants and two C-terminal segment variants of SU showed substantially impaired syncytium induction. Variant C110R induced small syncytia among only 13% of SU-positive cells, P115S induced only a few very small syncytia, and N141K induced no syncytia at all. Syncytium induction was also profoundly affected by the N203S and W261R substitutions located in the C-terminal half of SU, which includes the linear epitopes A, B, D, and E (2, 8). Variant protein N203S induced syncytia at less than 5% of the level of the WT Env protein, and the few syncytia that formed were very small. Notably, this substitution removes a potential glycosylation site. The W261R substitution, located within a sequence containing the D epitope (8), induced essentially no syncytia.
Most of the substitutions in the extracellular domain of TM also impaired cell-cell fusion. Envs carrying one of three substitutions, T333A, I336T, which was not characterized further (data not shown), and G405S, induced syncytia similarly to WT Env. In contrast, the Env protein with a Q366R substitution well within the 4-3 heptad repeats important for Env oligomerization did not induce any syncytia in any of six independent experiments, and the C392R variant exhibited drastically impaired syncytium formation (Fig. 2). While the P409L and S415F proteins induced 60 to 74% as many syncytia as WT Env, the variant-induced syncytia were very small.
The cytoplasmic domain of TM harbored two amino acid substitutions. The substitution of negatively charged glutamate for K458 greatly reduced the efficiency of syncytium formation, and the few resulting syncytia were small. The substitution Q470R altered a residue in the cytoplasmic domain of TM midway between the membrane and the C-terminal YXXL motifs; the variant Q470R protein induced WT numbers of syncytia that were very small.
Cell surface expression and proteolytic processing of variant Env proteins.
Since amino acid substitutions that prevent localization of the BLV Env protein on the cell surface or that interfere with proteolytic processing of the Env precursor protein would reduce syncytium formation, we determined whether variant Env proteins that were severely compromised for syncytium formation were present on the cell surface. To quantify cells expressing SU, transfected cells were fixed and permeabilized and then were stained with a monoclonal antibody that recognizes the G epitope of SU (Fig. 3A). To enumerate cells expressing Env on the cell surface, parallel cultures of transfected cells were fixed without permeabilization and were immunostained (Fig. 3B).
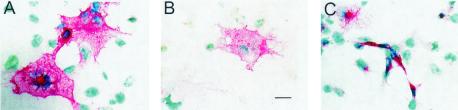
FIG. 3.
Staining of Env protein by anti-G monoclonal antibody. Whole-cell staining of WT Env protein (A) and variant N141K protein (C) is shown for transfected COS-1 cells that were fixed with paraformaldehyde and permeabilized with acetone before staining. Surface staining typical of the WT Env protein (B) is shown for transfected cells that were fixed only in paraformaldehyde before staining. The red precipitate results from alkaline phosphatase activity; nuclei are counterstained with methyl green. Cells were photographed at ×100 magnification, and images from 35-mm slide film were scanned. Bar, 20 μm.
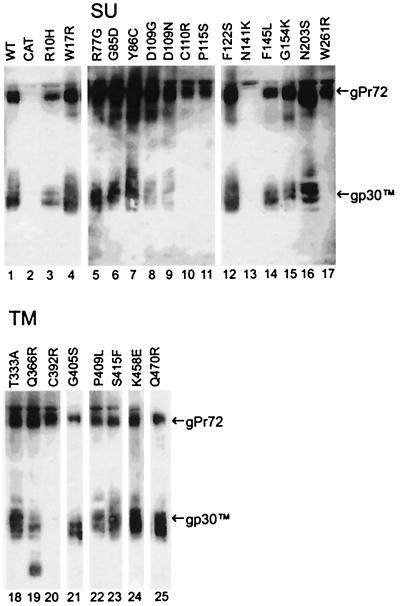
Next, we determined whether the Env precursor protein (gPr72) was being proteolytically processed to SU and TM proteins in cells expressing variants. Cleavage of the WT Env precursor protein was confirmed on immunoblots (Fig. 4) probed with an antibody specific for the C terminus of TM that detects both the Env precursor and TM. The Env variants fell into one of three classes. (i) Loss of syncytium induction is attributable to defects in protein expression. Variants C110R, P115S, W261R, C392R, and N141K fell into this class. Variant Envs C110R, P115S, W261R, and C392R were not detected at the cell surface by immunostaining (Table 1) although proteins were being synthesized within transfected cells, since permeabilized cells were immunostained. These variants induced very few syncytia. Although the G monoclonal antibody stained only a few permeabilized cells expressing variants C110R and P115S, many cells stained well with the anti-D monoclonal antibody (Table 2), demonstrating that ample Env protein was present. Unfortunately, the anti-D antibody could not be used to assess cell surface expression because it did not bind detectably to the Env protein of any type on the surfaces of nonpermeabilized cells. Only gPr72 was detected in lysates containing C110R, P115S, W261, and C392 variant proteins (Fig. 4, lanes 10, 11, 17, and 20), revealing that little if any cleavage was taking place. Since none of these four amino acid substitutions directly alters the protease cleavage site, each appears to induce misfolding that prevents transport of the variant protein to the Golgi apparatus and/or access of the protease to the cleavage site. Notably, the substitution C392R alters the third cysteine of the CX6CC motif, thought to function in the association of SU with TM. Even though the N141K protein was displayed on the cell surface (Table 1), fewer than 3% of the COS-1 cells expressed N141K in each of several transfections. The band representing its precursor was extremely light on immunoblots, and the processed TM protein could not be detected (Fig. 4, lane 13). The N141K protein may compromise the survival of expressing cells, or it may be turned over quickly. N141K-expressing cells that stained with SU-specific antibodies tended to be small, and many were elongated and thin (Fig. 3C). This was in striking contrast to most COS-1 cells expressing syncytium-inducing Env variants. Such cells were typically very spread out on the substrate (Fig. 3A) and often included several nuclei, indicating that COS-1 cells expressing Env could fuse to other COS-1 cells.
FIG. 4.
Proteolytic processing of Env precursor proteins. Immunoblots of glycoproteins from lysates of COS-1 cells transfected with WT or variant env constructs or with CAT were prepared by using a polyclonal antiserum specific for the C terminus of TM. The WT envelope protein (lane 1) was derived from 5 × 104 transfected cells; glycoproteins from 105 CAT-transfected cells (lane 2) served as the negative control. The remaining lanes contained glycoproteins from 1 × 105 or 2 × 104 (lanes 3, 13, and 16) transfected cells. Alkaline phosphatase-labeled sites of antibody binding were visualized with a luminescent reaction product. Shown are scans prepared from 10- (lanes 21 and 25) or 30-min exposures of blots to X-ray film. gPr72-Env and gp30-TM are identified on the right.
TABLE 1.
Cell surface expression of SU by cells producing BLV envelope proteins having low-to-moderate syncytium-inducing capacity
| Envelope variant | Syncytium inductiona | SU surface expressionb |
|---|---|---|
| WT | 100 | 100 (51) |
| C110R | 2 | 12 (6) |
| P115S | 0 | 2 (1) |
| N141K | 0 | 104 (52) |
| W261R | 0 | 10 (5) |
| C392R | 6 | 10 (5) |
| N203S | 6 | 86 (43) |
| Q366R | 0 | 58 (29) |
| K458E | 10 | 48 (24) |
| P409L | 22 | 80 (40) |
| S415F | 55 | 60 (30) |
| Q470R | 91 | 124 (62) |
Values are the percentages of cells (enumerated as SU+ in parallel immunostaining) incorporated into syncytia normalized to the percent observed for WT Env.
Nonpermeabilized cells and parallel cultures of permeabilized cells stained with anti-G or anti-D (C110R and P115S) monoclonal antibodies were enumerated by light microscopy. Values are percentages normalized to cells transfected with WT BLV env; values in parentheses are the data before normalization.
TABLE 2.
Reactivity of SU-specific monoclonal antibodies with variant Env proteins
| Env variant | Degree of staininga with epitope-specific antibodies
|
|||
|---|---|---|---|---|
| Conformational
|
Linear (D) | |||
| F | G | H | ||
| WT | +++ | +++ | +++ | ++ |
| Y86C | +++ | +++ | +++ | ++ |
| F122S | +++ | +++ | +++ | + |
| G154K | +/− | +++ | +++ | ++ |
| D109N | + | +++ | ++ | + |
| F146L | + | ++ | + | + |
| D109G | +/− | ++ | + | + |
| N141K | +/− | ++ | + | ++ |
| R77G | − | + | + | ++ |
| G85D | − | + | + | ++ |
| C110R | − | + | + | +++ |
| P115S | − | +/− | + | ++ |
Percentages of fixed and permeabilized cells that stained for SU: −, none; +/−, <0.5%; +, 0.5 to 5%; ++, 5.1 to 10%; +++, >10%.
(ii) Loss of syncytium induction is not attributable to a defect in protein expression or proteolytic cleavage. The N203S, Q366R, and K458E variants showed greatly reduced syncytium induction even though they were expressed well on the cell surface (Table 1). The N203S substitution did not appreciably reduce surface expression of the variant or affect proteolytic cleavage (Fig. 4, lane 16), even though it removed a potential N-linked glycosylation site in SU. Env variant Q366R was present on the cell surface (Table 1), and its precursor protein was proteolytically processed at the correct site, since a TM band at 30 kDa was present on immunoblots (Fig. 4, lane 19). However, an additional lower-molecular-weight protein containing the C terminus of TM was also present at a level equivalent to that for the 30-kDa protein. The substitution of arginine at residue 366 creates a second RXRR cleavage site of the type recognized by the cellular kexin-like protease responsible for processing the BLV Env precursor protein (64); the presence of the small anti-TM reactive species suggests that this cleavage site is accessible in an appreciable number of Q366R Env proteins. Since this site is located in the 4-3 heptad repeat domain of TM, it is not surprising that its cleavage might interfere with Env protein fusion. Unexpectedly, this secondary cleavage appeared to have little effect on the stability of SU association with its TM anchor, since the intensity of SU staining on the cell surface was comparable to that for the WT Env protein (data not shown). Perhaps the association was mediated by the CX6CC motif present in the remaining membrane-spanning segment of the Q366R TM protein. K458 is the first positively charged residue following 20 uncharged amino acids that form the putative membrane-spanning domain; all retroviral TM proteins contain such a positively charged amino acid, which is thought to act as a signal to stop transfer of newly synthesized peptide chains through the membrane of the endoplasmic reticulum. The K458E variant precursor was proteolytically cleaved (Fig. 4, lane 24). While a negatively charged glutamate in this position might fail to stop transfer, a second positively charged residue, R463, is located just downstream and could function as a signal in the absence of K458.
(iii) Reduction in the size of syncytia is not attributable to protein expression. Variant S415F, P409L, and Q470R proteins were on the surface at 60, 80, and 124%, respectively, of WT levels (Table 1). Precursors of these variant proteins were proteolytically processed into SU and TM (Fig. 4, lanes 22, 23, and 25). Thus, some other aspect of these proteins interferes with syncytium induction.
Conformation of Env variants with changes in N-terminal amino acids.
The F, G, and H conformational epitopes of BLV SU are important for Env function, since monoclonal antibodies specific for these epitopes block syncytium induction and neutralize viral infectivity (6, 48). To determine how individual N-terminal amino acid substitutions affect antibody recognition of these epitopes, we immunostained Env-expressing COS-1 cells that had been fixed and permeabilized. A similar percentage of cells transfected with WT env were stained by each of the conformation-dependent monoclonal antibodies (Table 2). Thus, the strain-specific substitutions K4E, Y108H, and A326S in the Env protein of Bat2cl6-BLV do not alter recognition of the F, G, and H epitopes of SU. A monoclonal antibody specific for the linear D epitope stained slightly fewer cells containing the WT Env protein than did the conformation-specific antibodies.
In general, recognition by all the conformation-specific antibodies correlated with syncytium-inducing ability of an Env variant. Variants that induced few syncytia and that were not proteolytically processed were recognized poorly by the conformation-dependent monoclonal antibodies, although they stained very well with the D-specific antibody. One exception was the F146L variant, which showed reduced staining, particularly with anti-F and anti-H antibodies. Two other N-terminal variants inducing syncytia at close to WT efficiency, Y86C and F122S, had staining profiles like those of WT Env, indicating that the F, G, and H conformational epitopes were preserved. Syncytium-inducing variant G154K was also similar to WT except that it displayed a strikingly reduced staining by the F-specific monoclonal antibody. D109N and D109G variants, which showed reduced syncytium induction, displayed reduced recognition by both anti-F and anti-H antibodies. The R77G and G85D variants, which induced small syncytia, and the C110R and P115S variants, which almost lacked syncytium-inducing ability, were poorly recognized by the conformation-dependent monoclonal antibodies, suggesting that these four substitutions profoundly altered SU conformation. Surprisingly, the syncytium-negative variant N141K was better recognized by the conformation-dependent antibodies than were C110R and P115S. Altogether, these results demonstrate that some syncytium-inducing variant proteins can nonetheless display globally altered conformation-dependent epitopes.
DISCUSSION
Comparison of BLV variants with HTLV mutants.
The env genes of BLV and its close relative, human T-cell leukemia virus type 1 (HTLV-1), are each highly conserved. Nucleotide variation is 6% among BLV env proviral sequences from different parts of the world (39) and is similarly low for HTLV-1 env (24, 35, 38, 58). The amino acid sequences of BLV and HTLV-1 Env proteins are 36% identical (53) and can be aligned by motifs common to retroviral Env proteins (Fig. 5). Information about the structure or function of these proteins may thus be reciprocally informative. Most amino acid substitutions introduced by mutagenesis into the HTLV-1 Env protein resulted in diminished function, as assessed by reductions in syncytium induction, infectivity, or susceptibility to proteolytic processing (14, 15, 46, 54). Substitutions within the middle of gp46-SU of HTLV-1 interfere with precursor cleavage, while syncytium formation is affected by clusters of amino acids, mainly aa 76 to 101 and aa 181 to 286 (14, 15). Most amino acid changes made within the external domain of TM of HTLV-1 also affect either precursor cleavage or syncytium formation. However, three changes within the 30 aa just external to the membrane-spanning region of TM enhance syncytium formation but interfere with viral transmission, indicating their importance for postfusion events (54). Truncation of the cytoplasmic domain of TM reduces total Env protein expression in cells but enhances syncytium induction over that for WT Env (45). Thus, both the internal and external domains of TM as well as SU of HTLV-1 are important for fusion of viral and cellular membranes.
FIG. 5.
Alignment of BLV and HTLV-1 Env precursors by amino acid sequences and the shared motifs indicated above the sequences. Residues predicted to span the membrane are in italics. Dark gray, residues identical in both proteins; light gray, conserved residues (D↔E, K↔R, S↔T, Y↔F, and I↔L↔V). Numbering begins with the first amino acid of each Env precursor protein. The BLV sequence is that of our WT BLV (GenBank accession no. AY078387), so H108 represents the strain-specific Y108H substitution. BLV residues altered in variants are underlined. The HTLV sequence shown is the consensus from accession no. J02029 (60), D13784 (38), M37747 (27), and M67514 (42).
Many of the variant BLV Env proteins that failed to induce syncytia or that did so poorly had sustained substitution of an amino acid conserved at analogous positions (Fig. 5) in the HTLV-1 Env protein: C110BLV/108HTLV, P115BLV/113HTLV, Q366BLV/377HTLV, C392BLV/401HTLV, and P409BLV/418HTLV. In addition, W261BLV and K458BLV are conserved as aromatic (F277HTLV) and positively charged (R467HTLV) amino acids. This conservation provides a basis for comparison of the phenotypes of the BLV variants reported here with those of mutant HTLV-1 envelope proteins created in the corresponding residues by site-directed mutagenesis (14, 15, 46, 54).
Two classes of BLV Env variants showed altered syncytium formation. The first class reduced the size of syncytia and, in some cases, the total number of cells fused, although the gPr72 Env precursor was processed normally. This class consists of variants R77G, G85D, D109G/N, P409L, S415F, and Q470R. The R77G, G85D, and D109G/N variants have substitutions in residues within the putative RBD of SU, suggesting that syncytium induction is altered because Env binding is affected.
The remaining variants in this class had substitutions in the TM protein. P409L and S415F induced fewer syncytia than WT Env, and the syncytia were very small. These variants were expressed on the cell surface. Thus, residues more membrane proximal than the fusion domain and 4-3 heptad repeat of BLV TM influence membrane fusion. Interestingly, syncytium induction is also lost when the leucine residue in the HTLV-1 Env protein (L419HTLV), which corresponds to the nearby L410BLV, is mutated to arginine (54). The W418QWPWNWDLGLTAWV sequence immediately downstream from P409 and S415 in BLV TM is strikingly similar to a highly conserved tryptophan-rich motif in the corresponding region of the human immunodeficiency virus type 1 TM protein, which is essential for membrane fusion (56, 61). Since proline residues are often found in turns, the P409L change might act by disrupting the folding of the nearby tryptophan-rich domain. Perhaps the aromatic phenylalanine of S415F also disturbs the structure of this domain in some way.
The other class of variants showed marked reductions in the total number of syncytia as well as in the size of an individual syncytium. These variants were of two distinctive types, distinguished by the extent of processing of the Env precursor. The first type, for which gPr72 was processed into SU and TM at levels comparable to that for the WT BLV Env protein, included variants N203S and K458E. The asparagine at position 203 is a putative N-linked glycosylation site located close to the CXXC motif, thought to be responsible for covalent bonding between SU and TM. Mutation of the corresponding residue, N222HTLV, in HTLV-1 SU reduced syncytium formation by fivefold (44). However, in contrast to the BLV N203S mutation, the N222Q mutation impairs the processing of the HTLV-1 Env precursor, perhaps because N222HTLV is closer to the CXXC motif. The substitution of negatively charged glutamate for BLV Env residue K458 changed the charge of an amino acid at the C terminus of the membrane-spanning domain of TM, suggesting that the membrane topology of TM is affected in this variant. The alteration was very detrimental to syncytium induction, and surface expression of the K458E Env variant was one-half that of WT Env. A mutant with the arginine at the C terminus of the membrane-spanning domain of HIV TM changed to glycine gave a similar reduction in syncytium formation (41).
The non-syncytium-inducing Q366R protein was processed into correctly sized SU and TM, but some TM was cleaved aberrantly. Q366 lies within a highly conserved Q366NRRGLD sequence in the middle of the putative heptad repeat of TM. Substitution of arginine alters the sequence of this oligomerization domain and introduces a protease cleavage site that is utilized in about one-half the molecules. Uncoupling the fusion domain from the rest of TM provides an obvious block to fusion. However, the remaining TM molecules that were not cleaved were also nonfunctional. In HTLV-1 Env, substitution of leucine for glutamine in the analogous Q377NRRGLD sequence did not introduce a cleavage site but did give a syncytium-negative envelope protein (54), suggesting that disruption of the coiled-coil structure of the heptad repeat (34) might be as detrimental as its cleavage. Notably, the HTLV-1 mutant was noninfectious in a cell-to-cell transmission assay (54).
The second type of BLV Env variant exhibiting markedly reduced cell-cell fusion showed severely impaired processing of gPr72, suggesting that induction of syncytia was reduced because the bulk of the fusion peptides on these variants were not free. These variants consisted of C110R, C392R, P115S, N141K, and W261R. Cysteine residues are important for viral membrane protein folding (16). Altering any of the conserved cysteines of the CX6CC motif in the MuLV TM protein prevents proteolytic processing (62) as did the C392R substitution in BLV TM (this work). Immune serum from BLV-infected sheep precipitated the C110 cleavage mutant (data not shown), suggesting that the structural changes in this variant are less profound than those in variant P115S, which was not recognized by immune sheep serum (data not shown). Interestingly, the addition of a cysteine residue in variant Y86C did not interfere with correct proteolytic processing; syncytia were formed at WT levels, suggesting that this variant residue does not form an inappropriate disulfide bond. The model we propose below positions this amino acid on the surface of SU, distant from other cysteines.
In terms of retaining function, the N-terminal half of BLV SU, which forms the RBD, tolerated amino acid substitution better than did the C terminus of SU and the ectodomain of TM. Six of 13 changes in the N-terminal half of SU retained WT syncytium-inducing ability, even though the chemical natures of 5 of those were not conserved. However, a number of the variants whose syncytium induction was not appreciably diminished showed reduced binding by monoclonal antibodies specific for the conformational epitopes F, G, and H, suggesting that protein structure was subtly altered.
A model for the BLV RBD structure.
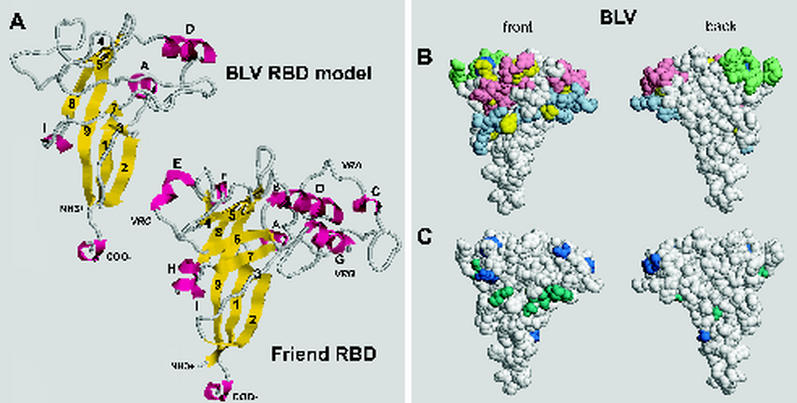
To gain some idea of where the variant amino acids might be located in the BLV RBD, we modeled this domain using the crystal structure for the RBD of SU from ecotropic F-MuLV (20). WT BLV sequences were aligned with those of F-MuLV, and then the alignment was submitted to Swiss-Model (28) for modeling. The following guidelines were used to generate the initial alignment: sequences within WT BLV SU that were judged likely to form α helices and β strands were aligned with known structural elements of the F-MuLV RBD; the start of the putative P-rich region in each SU molecule was aligned and set as the C terminus; and, wherever possible, conserved cysteines and glycosylation sites were aligned. The initial alignment failed to generate a model because the program encountered too many gaps that could not be resolved and were thus judged misalignments and because too many amino acid side chains clashed. Misaligned residues were reiteratively moved one position amino or carboxy terminal with respect to the F-MuLV sequence, and the adjusted alignments were resubmitted until one was successfully modeled. The final alignment is shown in Fig. 6. Further adjustments of this alignment by as much as three positions gave small changes in the positions of side chains but did not alter the local structure predicted for adjacent residues and did not influence distant structure predictions. In the final step, the side chain rotamers giving the maximum number of hydrogen bonds between neighboring residues were chosen, resulting in the model shown in Fig. 7A. For this discussion, amino acids are numbered in the model according to their positions in the BLV Env precursor. W1 in mature BLV SU is W34 in the Env precursor.
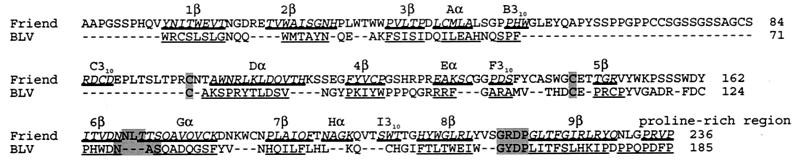
FIG. 6.
Alignment of putative structural elements within BLV SU with known structural elements forming the RBD of SU from F-MuLV. Amino acids involved in the β strands, α helices, and helix-like elements of the F-MuLV RBD are italicized, underlined, and named as designated by Fass et al. (20); the P-rich region is similarly identified. BLV sequences judged likely to form corresponding elements are underlined. Other aligned residues, including two cysteines, a putative N-linked glycosylation site, and the conserved GxDP strong turn, are shaded in gray. The first amino acid of mature F-MuLV SU is A1; the first amino acid of mature BLV SU is W34.
FIG. 7.
(A) Comparison of the proposed structure of the BLV RBD with a known structure (20) of the F-MuLV RBD monomer. Structures are depicted by using RasMol, version 2.7.1.1 (4, 57). Yellow, β strands; magenta, helices. Of the β strands predicted to be present in the BLV RBD, 1β, 2β, 4β, 8β, and 9β were modeled as such; 3β, 5β, and 7β were modeled as β-like structures, and 6β was not modeled. Of the predicted α helices, Aα, Dα, and Iα were modeled, whereas Eα and Gα were not. The BLV RBD has no helix analogous to Cα of F-MuLV. Coordinates for the BLV model and an annotated version of Fig. 7 are available from the corresponding author upon request. (B) Space-filling models of the BLV RBD illustrating the locations of the variant residues described in this paper (yellow) and the strain-specific Y108H of Bat2cl6 BLV (dark blue), as well as locations of peptides whose polyclonal antibodies neutralize infectivity or block fusion (10, 49). Pink, aa 72 to 81 and 111 to 125; light green, aa 97 to 106; light blue, aa 131 to 150. (C) Locations of strain-specific variant amino acids (A48T, A73P, K74R, S82F, Y108H, and R111H; dark blue) and amino acids varying in individual BLV isolates (S39F, S56F, S58A, D60N, Q95K, V140A, N141D, I144T, and H148Q; dark green) among complete BLV Env RBD sequences in the GenBank database. BLV Env sequences from K02120 (55) and M35242 (39) are the same as the those for the Bat2cl6 strain except for Y108. Accession numbers of BLV sequences encoding the depicted variants are K02251 (53); M35238, M35239, and M35240 (39); D00647 (13); S83530 (40); AF111171 (51); AF257515 (17); and AF067081.
The BLV model predicts a smaller and more compact molecule than the F-MuLV RBD (Fig. 7A). The core of the predicted BLV structure is composed of antiparallel β and β-like strands arranged in an Ig-like fold. The portion of the structure shown at the top of the molecule has segments analogous to the variable domains VRA, VRB, and VRC, which contribute to the differences in receptor-binding specificity of the MuLVs. However, the VRA-like segment in the BLV RBD is minimal, containing only an α helix corresponding to Dα on the F-MuLV RBD followed by three amino acids.
A monoclonal antibody specific for conformational epitope G does not recognize Env proteins of the natural BLV isolates LB285, LB59, and T15-2, all of which contain strain-specific substitutions at A73, K74, and S82 (A40, K41, and S49 in mature SU) (39, 48). Callebaut et al. (9) previously concluded that polyclonal antibodies to a peptide encompassing aa 72 to 81 (aa 39 to 48 in mature SU) and aa 111 to 125 (aa 78 to 92) influence epitope G. Amino acids 72 to 81 are located in or just adjacent to Dα in the VRA-like region in the model (Fig. 7B), suggesting that Dα and the immediately adjacent residues are key to the antigenic shape of the G epitope. Our variants R77G and G85D, located in the middle of Dα and just C-terminal to it, have subtly altered function: they initiate as many syncytia as WT Env, but the extent of fusion is reduced. Both variants are poorly recognized by a G-specific monoclonal antibody. In addition, poor recognition of these two variants by the F- and H-specific monoclonal antibodies suggests that the conformation of this region is also important for the structure of the other two conformational epitopes.
Epitope H-specific monoclonal antibodies poorly recognize the natural variant LB285 Env containing S58A (S25 in mature SU) and do not recognize natural variant LB59 Env, which contains the substitutions S56F and I144T (S23 and I111 in mature SU) (39, 48). Our variant F146L (F113 in mature SU) was also poorly recognized by this monoclonal antibody. A region containing aa 131 to 150 (aa 98 to 117 of mature SU) is thought to affect epitope H (9) because both H-specific monoclonal antibodies and peptide-specific polyclonal antibodies efficiently inhibit syncytium induction (10). In the model of the BLV RBD, S56, S58, I144, and F146 are closely juxtaposed, suggesting that the H epitope is bipartite.
Replacement of Q95 (Q62 in mature SU) by lysine in Env of the natural BLV variant VdM destroys recognition by the F-specific monoclonal antibody (39, 48). Polyclonal antibodies elicited by aa 97 to 106 (aa 64 to 73 of mature SU) are reported to compete with the F-specific antibody for binding to SU in an enzyme-linked immunosorbent assay (10). Our variant G154K (G121 in mature SU) differentially lost recognition by an F-specific monoclonal antibody. G154 is located in helix I310 of the model and is the only amino acid of those affected by our variants to lack surface exposure in the RBD. Since G154 is below and not immediately adjacent to the F epitope area in the model, the substituted lysine appears to exert its effect at a distance. Other variants may also influence the conformational epitopes by acting at a distance.
Comparison with the previous model of BLV developed by HCA.
Our model of the BLV RBD has some similarities to and some major differences from the one developed by Callebaut et al. (9) using hydrophobic cluster analysis (HCA) prior to solution of the F-MuLV RBD structure. The earlier BLV model was based partly on the structure of the influenza virus hemagglutinin type 1 (HA-1) protein and partly on the α3 domain of HLA-A2 (major histocompatibility complex class I). That model included an oligomeric structure based on the influenza virus HA trimer. Based on the site where HA binds to sialic acid, it also predicted a bipartite receptor-binding site containing residues around aa 144 and 210 (aa 111 and 177 in mature SU). In the new model presented here, residue 144 is exposed on the lower front face, well away from the surface corresponding to the F-MuLV receptor-binding site containing D84 and W142 (37, 65), whereas BLV residue 210 is not predicted to be part of the RBD, which ends at aa 185. In addition, the previous model positioned the two parts of epitope H at opposite ends of SU, locations that were difficult to reconcile with the data on monoclonal antibody recognition. The new model proposes a structure for this epitope that is more consistent with the antibody data in that the two segments influencing H-specific antibody recognition fold next to each other, particularly S56 and I144 (S23 and I111 in mature SU), whose side chains appear to pack against each other.
Since HCA does not generate specific alignments and does not predict structural elements, our alignment is not directly comparable to the previous analysis (9). However, since HCA can identify patterns of hydrophobic residues, we examined BLV and F-MuLV RBD sequences for conservation of hydrophobic clusters that could represent conserved segments to use as a basis for an alignment. Four clusters of hydrophobic residues appeared to be conserved. These clusters corresponded to the established β strands 1, 2, 8, and 9 of F-MuLV RBD and to the sequences predicted by our model to be the corresponding β strands of BLV, suggesting that, at least in these regions, our model and HCA agree.
Orientation for RBD in the Env oligomer.
A striking aspect of the new model became apparent when we mapped the positions of variant amino acids on it. All of the residues were visible from a single angle of view, which we refer to as the front face in Fig. 7B. None was located on the opposite or back face of the BLV RBD, suggesting that the back face was conserved. Since our variants were isolated from reverse-transcribed cDNAs that were amplified by Taq polymerase after one round of provirus expression in PBMCs, we cannot be certain that all variants reflect changes occurring in the virus in its host cells (31). We therefore searched GenBank for complete BLV RBD sequences from naturally occurring strains of BLV, identified 15 variant amino acids among 12 isolates (listed in the legend to Fig. 7), and mapped their locations on the model. Six strain-specific substitutions (shown in blue in Fig. 7C) have become fixed in more than one isolate: three are on the surface of the front face of the RBD model, while one (A73P) is exposed on the very top of the back face and another (A48T) is exposed off to one side of the back face. The new strain-specific Y108H substitution that we report here is exposed on the front face. In addition, of nine amino acids that vary only in a single BLV sequence (shown in green in Fig. 7C), just one (S39) is visible on the lower back and its side chain is buried. The lack of naturally occurring variant residues on the back face of the molecular model suggests that the sequence and structure of this face, especially those of β strands 8 and 9, are well conserved.
Since the crystallized RBD fragment of F-MuLV was a monomer (20), its structure gave few clues as to the orientation of RBD in Env trimers, particularly about the location of the surface(s) that faces toward the carboxy-terminal domain of SU and the native trimer interfaces. Since antibodies specific for the F, G, and H conformational epitopes neutralize the infectivity of BLV-vesicular stomatitis virus pseudotypes and block fusion initiated by the BLV Env protein (6), their epitopes are likely to be exposed to solvent in the native Env structure. This predicts that the outward-facing surfaces of the RBD can also be identified by locating these epitopes. We asked where the residues affecting the F, G, and H conformational epitopes are located on the model. Most residues are visible from the front although some can be seen at the top of the back view (Fig. 7B). Moreover, the single N-linked glycosylation site that is conserved between the RBDs of BLV and F-MuLV (Fig. 6) is located on the front face of the BLV model.
Based on reasoning that the inward-facing surfaces of RBD are likely to be highly conserved and will not be solvent exposed, we propose that the lower back face of the RBD model, consisting of residues from highly conserved β strands 8 and 9 and parts of 1 and 2, faces inward in the native Env structure. This surface might interact with the C-terminal domain of SU or with an adjacent monomer in the Env oligomer. From this supposition, it can be inferred that the corresponding region of the F-MuLV RBD also faces the interior of the Env molecule.
Acknowledgments
We thank Daniel Portetelle for his generous gift of SU monoclonal antibodies and Glen Cantor for the kind gift of antipeptide antibodies specific for BLV TM. We thank Debbie Grossman for expert technical assistance and Victor Dauman for assistance with the figures.
This work was supported by Public Health Service grants CA-46374 from the National Cancer Institute (K.R.) and AI-33410 (L.M.A.) and by the UC Davis Cancer Center (K.R.). E.R.J. was a predoctoral trainee supported by Public Health Service grant GM-07377 from the National Institute of Medicine. Instruments located at the NSF-funded Plant Genetics Facility (UC Davis) were used to acquire DNA sequences and images of autoradiograms.
REFERENCES
- 1.Alber, G., K.-W. Kim, P. Weiser, C. Riesterer, R. Carsetti, and M. Reth. 1993. Molecular mimicry of the antigen receptor signalling motif by transmembrane proteins of the Epstein-Barr virus and the bovine leukaemia virus. Curr. Biol. 3:333-339. [DOI] [PubMed] [Google Scholar]
- 2.Ban, J., S. Czene, C. Altaner, I. Callebaut, V. Krchnak, M. Merza, A. Burny, R. Kettmann, and D. Portetelle. 1992. Mapping of sequential epitopes recognized by monoclonal antibodies on the bovine leukaemia virus external glycoproteins expressed in Escherichia coli by means of antipeptide antibodies. J. Gen. Virol. 73:2457-2461. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein, H. B., S. P. Tucker, S. R. Kar, S. A. McPherson, J. W. Dubay, J. Lebowitz, R. W. Compans, and E. Hunter. 1995. Oligomerization of the hydrophobic heptad repeat of gp41. J. Virol. 69:2745-2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein, H. J. 2000. Recent changes to RasMol, recombining the variants. Trends Biochem. Sci. 25:453-455. [DOI] [PubMed] [Google Scholar]
- 5.Bruck, C., S. Mathot, D. Portetelle, C. Berte, J. D. Franssen, P. Herion, and A. Burny. 1982. Monoclonal antibodies define eight independent antigenic regions on the bovine leukemia virus (BLV) envelope glycoprotein gp51. Virology 122:342-352. [DOI] [PubMed] [Google Scholar]
- 6.Bruck, C., D. Portetelle, A. Burny, and J. Zavada. 1982. Topographical analysis by monoclonal antibodies of BLV gp51 epitopes involved in viral functions. Virology 122:353-362. [DOI] [PubMed] [Google Scholar]
- 7.Bruck, C., N. Rensonnet, D. Portetelle, Y. Cleuter, M. Mammerickx, A. Burny, R. Mamoun, B. Guillemain, M. van der Maaten, and J. Ghysdael. 1984. Biologically active epitopes of bovine leukemia virus glycoprotein gp51: their dependence on protein glycosylation and genetic variability. Virology 136:20-31. [DOI] [PubMed] [Google Scholar]
- 8.Callebaut, I., A. Burny, V. Krchnak, H. Gras-Masse, B. Wathelet, and D. Portetelle. 1991. Use of synthetic peptides to map sequential epitopes recognized by monoclonal antibodies on the bovine leukemia virus external glycoprotein. Virology 185:48-55. [DOI] [PubMed] [Google Scholar]
- 9.Callebaut, I., D. Portetelle, A. Burny, and J. P. Mornon. 1994. Identification of functional sites on bovine leukemia virus envelope glycoproteins using structural and immunological data. Eur. J. Biochem. 222:405-414. [DOI] [PubMed] [Google Scholar]
- 10.Callebaut, I., V. Voneche, A. Mager, O. Fumiere, V. Krchnak, M. Merza, J. Zavada, M. Mammerickx, A. Burny, and D. Portetelle. 1993. Mapping of B-neutralizing and T-helper epitopes on the bovine leukemia virus external glycoprotein gp51. J. Virol. 67:5321-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantor, G. H. 1996. A potential proline-rich motif upstream of the immunoreceptor tyrosine-based activation motif in bovine leukemia virus gp30, Epstein-Barr virus LMP2A, herpesvirus papio LMP2A, and African horsesickness virus VP7. Virology 220:265-266. [DOI] [PubMed] [Google Scholar]
- 12.Cantor, G. H., S. M. Pritchard, O. Orlik, G. A. Splitter, W. C. Davis, and R. Reeves. 1999. Bovine leukemia virus transmembrane protein gp30 physically associates with the down-regulatory phosphatase SHP-1. Cell. Immunol. 193:117-124. [DOI] [PubMed] [Google Scholar]
- 13.Coulston, J., H. Naif, R. Brandon, S. Kumar, S. Khan, R. C. W. Daniel, and M. F. Lavin. 1990. Molecular cloning and sequencing of an Australian isolate of proviral bovine leukemia virus DNA: comparison with other isolates. J. Gen. Virol. 71:1737-1746. [DOI] [PubMed] [Google Scholar]
- 14.Delamarre, L., C. Pique, D. Pham, T. Tursz, and M. C. Dokhelar. 1994. Identification of functional regions in the human T-cell leukemia virus type I SU glycoprotein. J. Virol. 68:3544-3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delamarre, L., A. R. Rosenberg, C. Pique, D. Pham, and M. C. Dokhelar. 1997. A novel human T-leukemia virus type I cell-to-cell transmission assay permits definition of SU glycoprotein amino acids important for infectivity. J. Virol. 71:259-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doms, R. W., R. A. Lamb, J. K. Rose, and A. Helenius. 1993. Folding and assembly of viral membrane proteins. Virology 193:545-562. [DOI] [PubMed] [Google Scholar]
- 17.Dube, S., G. Dolcini, L. Abbott, S. Mehta, D. Dube, S. Gutierrez, C. Ceriani, E. Esteban, J. Ferrer, and B. Poiesz. 2000. The complete genomic sequence of a BLV strain from a Holstein cow from Argentina. Virology 277:379-386. [DOI] [PubMed] [Google Scholar]
- 18.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 19.Einfeld, D., and E. Hunter. 1994. Expression of the TM protein of Rous sarcoma virus in the absence of SU shows that this domain is capable of oligomerization and intracellular transport. J. Virol. 68:2513-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fass, D., R. A. Davey, C. A. Hamson, P. S. Kim, J. M. Cunningham, and J. M. Berger. 1997. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 Angstrom resolution. Science 277:1662-1666. [DOI] [PubMed] [Google Scholar]
- 21.Fischinger, P. J., C. S. Blevins, and S. Nomura. 1974. Simple, quantitative assay for both xenotropic murine leukemia and ecotropic feline leukemia viruses. J. Virol. 14:177-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallaher, W. R., J. M. Ball, R. F. Garry, A. M. Martin-Amedee, and R. C. Montelaro. 1995. A general model for the surface glycoproteins of HIV and other retroviruses. AIDS Res. Hum. Retroviruses 11:191-202. [DOI] [PubMed] [Google Scholar]
- 23.Gatot, J. S., I. Callebaut, J. P. Mornon, D. Portetelle, A. Burny, P. Kerkhofs, R. Kettmann, and L. Willems. 1998. Conservative mutations in the immunosuppressive region of the bovine leukemia virus transmembrane protein affect fusion but not infectivity in vivo. J. Biol. Chem. 273:12870-12880. [DOI] [PubMed] [Google Scholar]
- 24.Gessain, A., R. C. Gallo, and G. Franchini. 1992. Low degree of human T-cell leukemia/lymphoma virus type I genetic drift in vivo as a means of monitoring viral transmission and movement of ancient human populations. J. Virol. 66:2288-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graves, D. C., and J. F. Ferrer. 1976. In vitro transmission and propagation of the bovine leukemia virus in monolayer cell cultures. Cancer Res. 36:4152-4157. [PubMed] [Google Scholar]
- 26.Graves, D. C., and L. V. Jones. 1981. Early syncytium formation by bovine leukemia virus. J. Virol. 38:1055-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray, G. S., M. White, T. Bartman, and D. Mann. 1990. Envelope gene sequence of HTLV-1 isolate MT-2 and its comparison with other HTLV-1 isolates. Virology 177:391-395. [DOI] [PubMed] [Google Scholar]
- 28.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 29.Guillemain, B., R. Mamoun, D. Levy, T. Astier, K. Irgens, and A. A. Parodi. 1978. Early polykaryocytosis inhibition: a simple quantitative in vitro assay for the detection of bovine leukemia virus infection in cattle. Eur. J. Cancer 14:811-827. [DOI] [PubMed] [Google Scholar]
- 30.Hunter, E. 1997. Viral entry and receptors, p. 71-119. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 31.Johnston, E. R. 1999. Ph.D. thesis. University of California, Davis.
- 32.Johnston, E. R., M. A. Powers, L. C. Kidd, and K. Radke. 1996. Peripheral blood mononuclear cells from sheep infected with a variant of bovine leukemia virus synthesize envelope glycoproteins but fail to induce syncytia in culture. J. Virol. 70:6296-6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnston, E. R., and K. Radke. 2000. The SU and TM envelope proteins of bovine leukemia virus are linked by disulfide bonds, both in cells and in virions. J. Virol. 74:2930-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kobe, B., R. J. Center, B. E. Kemp, and P. Poumbourios. 1999. Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc. Natl. Acad. Sci. USA 96:4319-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koralnik, I. J., E. Boeri, W. C. Saxinger, A. L. Monico, J. Fullen, A. Gessain, H. G. Guo, R. C. Gallo, P. Markham, V. Kalyanaraman, V. Hirsch, J. Allan, K. Murthy, P. Alford, P. Slattery, S. J. O'Brien, and G. Franchini. 1994. Phylogenetic associations of human and simian T-cell leukemia/lymphotropic virus type I strains: evidence for interspecies transmission. J. Virol. 68:2693-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lagarias, D. M., and K. Radke. 1990. Transient increases of blood mononuclear cells that could express bovine leukemia virus early after experimental infection of sheep. Microb. Pathog. 9:147-158. [DOI] [PubMed] [Google Scholar]
- 37.MacKrell, A. J., N. W. Soong, C. M. Curtis, and W. F. Anderson. 1996. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J. Virol. 70:1768-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Malik, K. T., J. Even, and A. Karpas. 1988. Molecular cloning and complete nucleotide sequence of an adult T cell leukaemia virus/human T cell leukaemia virus type 1 (ATLV/HTLV-I) isolate of Caribbean origin: relationship to other members of the ATLV/HTLV-I subgroup. J. Gen. Virol. 69:1695-1710. [DOI] [PubMed] [Google Scholar]
- 39.Mamoun, R. Z., M. Morisson, N. Rebeyrotte, B. Busetta, D. Couez, R. Kettmann, M. Hospital, and B. Guillemain. 1990. Sequence variability of bovine leukemia virus env gene and its relevance to the structure and antigenicity of the glycoproteins. J. Virol. 64:4180-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molteni, E., A. Agresti, R. Meneveri, A. Marozzi, M. Malcovati, L. Bonizzi, G. Poli, and E. Ginelli. 1996. Molecular characterization of a variant of proviral bovine leukemia virus (BLV). Zentbl. Vetmed. Reihe B 43:201-211. [DOI] [PubMed] [Google Scholar]
- 41.Owens, R. J., C. Burke, and J. K. Rose. 1994. Mutations in the membrane-spanning domain of the human immunodeficiency virus envelope glycoprotein that affect fusion activity. J. Virol. 68:570-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paine, E., J. Garcia, T. C. Philpott, G. Shaw, and L. Ratner. 1991. Limited sequence variation in human T-lymphotropic virus type 1 isolates from North American and African patients. Virology 182:111-123. [DOI] [PubMed] [Google Scholar]
- 43.Pinter, A., R. Kopelman, Z. Li, S. C. Kayman, and D. A. Sanders. 1997. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J. Virol. 71:8073-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pique, C., D. Pham, T. Tursz, and M. C. Dokhelar. 1992. Human T-cell leukemia virus type I envelope protein maturation process: requirements for syncytium formation. J. Virol. 66:906-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pique, C., D. Pham, T. Tursz, and M. C. Dokhelar. 1993. The cytoplasmic domain of the human T-cell leukemia virus type I envelope can modulate envelope functions in a cell type-dependent manner. J. Virol. 67:557-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pique, C., T. Tursz, and M. C. Dokhelar. 1990. Mutations introduced along the HTLV-I envelope gene result in a non-functional protein: a basis for envelope conservation? EMBO J. 9:4243-4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Portetelle, D., C. Bruck, M. Mammerickx, and A. Burny. 1980. In animals infected by bovine leukemia virus (BLV) antibodies to envelope glycoprotein gp51 are directed against the carbohydrate moiety. Virology 105:223-233. [DOI] [PubMed] [Google Scholar]
- 48.Portetelle, D., D. Couez, C. Bruck, R. Kettmann, M. Mammerickx, M. van der Maaten, R. Brasseur, and A. Burny. 1989. Antigenic variants of bovine leukemia virus (BLV) are defined by amino acid substitutions in the NH2 part of the envelope glycoprotein gp51. Virology 169:27-33. [DOI] [PubMed] [Google Scholar]
- 49.Portetelle, D., C. Dandoy, A. Burny, J. Zavada, H. Siakkou, H. Gras-Masse, H. Drobecq, and A. Tartar. 1989. Synthetic peptides approach to identification of epitopes on bovine leukemia virus envelope glycoprotein gp51. Virology 169:34-41. [DOI] [PubMed] [Google Scholar]
- 50.Radke, K., T. J. Sigala, and D. Grossman. 1992. Transcription of bovine leukemia virus in peripheral blood cells obtained during early infection in vivo. Microb. Pathog. 12:319-331. [DOI] [PubMed] [Google Scholar]
- 51.Reichert, M. 2002.. Importance of a single point mutation of the bovine leukemia virus (BLV) envelope glycoprotein in the serological detection of BLV infection. Bull. Vet. Inst. Pulawy 46:17-26. [Google Scholar]
- 52.Reichert, M., A. Winnicka, L. Willems, R. Kettmann, and G. H. Cantor. 2001. Role of the proline-rich motif of bovine leukemia virus transmembrane protein gp30 in viral load and pathogenicity in sheep. J. Virol. 75:8082-8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rice, N. R., R. M. Stephens, D. Couez, J. Deschamps, R. Kettmann, A. Burny, and R. V. Gilden. 1984. The nucleotide sequence of the env gene and post-env region of bovine leukemia virus. Virology 138:82-93. [DOI] [PubMed] [Google Scholar]
- 54.Rosenberg, A. R., L. Delamarre, C. Pique, D. Pham, and M. C. Dokhelar. 1997. The ectodomain of the human T-cell leukemia virus type I TM glycoprotein is involved in postfusion events. J. Virol. 71:7180-7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sagata, N., T. Yasunaga, J. Tsuzuku-Kawamura, K. Ohishi, Y. Ogawa, and Y. Ikawa. 1985. Complete nucleotide sequence of the genome of bovine leukemia virus: its evolutionary relationship to other retroviruses. Proc. Natl. Acad. Sci. USA 82:677-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salzwedel, K., J. T. West, and E. Hunter. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73:2469-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sayle, R. A., and E. J. Milner-White. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 20:374.. [DOI] [PubMed] [Google Scholar]
- 58.Schulz, T. F., M. L. Calabro, J. G. Hoad, C. V. Carrington, E. Matutes, D. Catovsky, and R. A. Weiss. 1991. HTLV-1 envelope sequences from Brazil, the Caribbean, and Romania: clustering of sequences according to geographic origin and variability in an antibody epitope. Virology 184:483-491. [DOI] [PubMed] [Google Scholar]
- 59.Schulz, T. F., B. A. Jameson, L. Lopalco, A. G. Siccardi, R. A. Weiss, and J. P. Moore. 1992. Conserved structural features in the interaction between retroviral surface and transmembrane glycoproteins? AIDS Res. Hum. Retroviruses 8:1571-1580. [DOI] [PubMed] [Google Scholar]
- 60.Seiki, M., S. Hattori, Y. Hirayama, and M. Yoshida. 1983. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc. Natl. Acad. Sci. USA 80:3618-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Suarez, T., W. R. Gallaher, A. Agirre, F. M. Goni, and J. L. Nieva. 2000. Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: putative role during viral fusion. J. Virol. 74:8038-8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas, A., and M. J. Roth. 1995. Analysis of cysteine mutations on the transmembrane protein of Moloney murine leukemia virus. Virology 211:285-289. [DOI] [PubMed] [Google Scholar]
- 63.Voneche, V., D. Portetelle, R. Kettmann, L. Willems, K. Limbach, E. Paoletti, J. M. Ruysschaert, A. Burny, and R. Brasseur. 1992. Fusogenic segments of bovine leukemia virus and simian immunodeficiency virus are interchangeable and mediate fusion by means of oblique insertion in the lipid bilayer of their target cells. Proc. Natl. Acad. Sci. USA 89:3810-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zarkik, S., E. Decroly, R. Wattiez, N. G. Seidah, A. Burny, and J. M. Ruysschaert. 1997. Comparative processing of bovine leukemia virus envelope glycoprotein gp72 by subtilisin/kexin-like mammalian convertases. FEBS Lett. 406:205-210. [DOI] [PubMed] [Google Scholar]
- 65.Zavorotinskaya, T., and L. M. Albritton. 1999. A hydrophobic patch in ecotropic murine leukemia virus envelope protein is the putative binding site for a critical tyrosine residue on the cellular receptor. J. Virol. 73:10164-10172. [DOI] [PMC free article] [PubMed] [Google Scholar]