Abstract
Equine arteritis virus (EAV) is an enveloped, positive-stranded RNA virus belonging to the family Arteriviridae of the order Nidovirales. Four envelope proteins have hitherto been identified in EAV particles: the predominant membrane proteins M and GL, the unglycosylated small envelope protein E, and the nonabundant membrane glycoprotein GS. In this study, we established that the products of EAV open reading frame 3 (ORF3) and ORF4 (designated GP3 and GP4, respectively) are also minor structural glycoproteins. The proteins were first characterized by various analyses after in vitro translation of RNA transcripts in a rabbit reticulocyte lysate in the presence and absence of microsomal membranes. We subsequently expressed ORF3 and -4 in baby hamster kidney cells by using the vaccinia virus expression system and, finally, analyzed the GP3 and GP4 proteins synthesized in EAV-infected cells. The results showed that GP4 is a class I integral membrane protein of 28 kDa with three functional N-glycosylation sites and with little, if any, of its carboxy terminus exposed. Both after independent expression and in EAV-infected cells, the protein localizes in the endoplasmic reticulum (ER), as demonstrated biochemically by analysis of its oligosaccharide side chains and as visualized directly by immunofluorescence studies. GP3, on the other hand, is a heavily glycosylated protein whose hydrophobic amino terminus is not cleaved off. It is an integral membrane protein anchored by either or both of its hydrophobic terminal domains and with no parts detectably exposed cytoplasmically. Also, GP3 localizes in the ER when expressed independently and in the context of an EAV infection. Only a small fraction of the GP3 and GP4 proteins synthesized in infected cells ends up in virions. Most, but not all, of the oligosaccharides of these virion glycoproteins are biochemically mature. Our results bring the number of EAV envelope proteins to six.
Equine arteritis virus (EAV), the etiological agent of equine viral arteritis (9, 11, 45), has been assigned to the family Arteriviridae. The Arteriviridae constitute the single genus Arterivirus. Other members of this genus are lactate dehydrogenase-elevating virus (LDV), porcine reproductive and respiratory syndrome virus (PRRSV), and simian hemorrhagic fever virus (SHFV). Although their physicochemical properties, genome sizes, and virion architectures suggest otherwise, on the basis of similarities in genomic organization and replication strategy, the Arteriviridae were grouped together with the Coronaviridae in the order Nidovirales (2, 3, 6, 41).
The EAV genome consists of a single, positive-stranded RNA molecule of 12.7 kb that is 5′ capped and 3′ polyadenylated (3). The 5′ three-quarters of the genome contains two open reading frames (ORFs), ORF1a and -1b, that encode the proteins involved in viral RNA replication and transcription (3). Downstream of these ORFs, the genome contains a set of seven smaller ORFs (ORF2a, -2b, and -3 through -7) that are expressed from a 3′-coterminal nested set of subgenomic mRNAs (4, 46). Most of these ORFs code for the known structural proteins of the virion (5, 42).
EAV virions have a diameter of 40 to 60 nm and possess a putatively icosahedral core surrounded by a lipid-containing envelope with tiny surface projections (22, 30). The core particle is composed of the viral genome and the phosphorylated nucleocapsid protein (N), which is encoded by ORF7 (24, 54). Four viral proteins have been identified in the viral envelope: a 16-kDa nonglycosylated membrane protein (M), a relatively large envelope glycoprotein (GL) of 30 to 42 kDa, a small envelope glycoprotein (GS) of 25 kDa (5), and the recently discovered 8-kDa unglycosylated envelope protein (E) (42). The M and GL proteins are the major structural polypeptides and are present in virus particles as disulfide-linked heterodimers (7). The GS and E proteins occur in virions in minor and intermediate amounts, respectively (8, 42). The proteins M, GL, GS, and E are encoded by ORF6, -5, -2b, and -2a, respectively.
Nothing is known about the significance or function of the EAV ORF3 and -4 products, except that both are essential in the viral life cycle. When the expression of these ORFs was separately blocked by mutagenesis with a full-length cDNA clone, infectious virus was no longer produced (36). The products of ORF3 and -4 have not been demonstrated in EAV-infected cells or in virions. Recently, an in vitro translation study has shown that ORF3 encodes an extensively glycosylated, membrane-associated protein of 36 to 42 kDa, antibodies to which occur in infected horses (21).
Also, for other arteriviruses, the role of the ORF3 and -4 products is far from clear. In the Lelystad strain of PRRSV, it was found that their products (designated GP4 and GP3, respectively) are minor glycoproteins of the virus (47). In contrast, the ORF3-encoded protein of the Quebec strain of PRRSV (IAF-Klop) has been shown to code for a soluble nonstructural protein (20, 31). In vitro translation experiments showed that the ORF3-encoded protein of LDV is also a soluble protein (14). The ORF3 and -4 homologues of SHFV have not been experimentally investigated.
In this study, we performed a detailed characterization the EAV ORF3 and -4 products. We raised specific antibodies to these proteins and analyzed their synthesis and intracellular transport, both in EAV-infected cells and when they are expressed independently. In addition, we studied the membrane topology of the ORF4-encoded protein. Finally, we determined whether the ORF3 and -4 proteins are structural components of the virion.
MATERIALS AND METHODS
Cells and viruses.
Two baby hamster kidney cell lines were used, BHK-21 C13 (American Type Culture Collection) and BSR T7/5 (1). These cells were grown and maintained in Glasgow minimal essential medium (GMEM; Invitrogen-Life Technologies) supplemented with 10% heat-inactivated fetal calf serum (FCS), 100 IU of penicillin per ml, and 100 μg of streptomycin per ml (GMEM-10% FCS), supplemented in the case of BSR T7/5 cells with 1 mg of G-418 (Geneticin; Invitrogen-Life Technologies) per ml. Virus stocks of the Utrecht variant of the Bucyrus strain of EAV (EAV Utr) were grown in BHK-21 C13 cells.
The recombinant vaccinia viruses vTF7.3 and MVA-T7 expressing bacteriophage T7 RNA polymerase were propagated in rabbit kidney (RK-13) cells and chicken embryonic fibroblasts, respectively, as described previously (16, 44).
Plasmid construction.
Recombinant DNA techniques were performed essentially as described by Sambrook et al. (40). Unless indicated otherwise, bacterial transformations were carried out with Escherichia coli strain PC2495 (Phabagen). EAV ORF3 was cloned into pBluescript KS(−) (Stratagene) by ligating the blunt-ended 0.8-kb PvuII-HinfI fragment from EAV cDNA clone PB535 (5) into the SmaI site of the vector. Subsequently, the 0.8-kb BamHI-EcoRI fragment of the latter plasmid was inserted into BamHI- and EcoRI-digested pBluescript SK(−) (Stratagene), yielding pAVI13. The orientation and nucleotide sequence of the insert were verified by restriction enzyme digestions and sequencing of alkali-denatured plasmid DNA with a T7 DNA polymerase sequencing kit (Amersham Pharmacia Biotech) and [α-35S]dATP (>1,000 Ci/mmol; Amersham Pharmacia Biotech). The construction of EAV ORF4-expressing vector pMRI14 has been described elsewhere (51).
To generate ORF4-specific antigen for immunization, we constructed a plasmid encoding a His-tagged version of the putative GP4 ectodomain (FP24). For this purpose, the relevant region of EAV cDNA clone PB535 (5) was amplified by PCR with primers 679 (5′-dCGGGATCCTGGTGCACTTTCTACCATG-3′; corresponding to nucleotides 10757 to 10776 of EAV Utr) and 680 (5′-dCGGGATCCACCAAGCGGTAAAGCC-3′; corresponding to nucleotides 11095 to 11080 of EAV Utr) (3, 18). Both primers contain a 5′ extension introducing a BamHI restriction enzyme recognition site (underlined). The PCR product was digested with BamHI and ligated into BamHI-digested pBluescript SK(+). The nucleotide sequence of the PCR product was confirmed by sequencing as described above. After sequence verification, the ORF4-specific PCR fragment was excised from the pBluescript SK(+) backbone with BamHI and cloned into the BamHI site of prokaryotic expression vector pQE9 (Qiagen), yielding pQE9-ORF4. The latter construct was transfected, together with plasmid pREP4 (Qiagen), into E. coli strain M15 (Qiagen).
Bacterial fusion proteins.
A large batch of purified FP24 was prepared by affinity chromatography essentially as described by Nugent et al. (37). Following elution of FP24 from the affinity column, the antigen solution was transferred to a 20-ml Spectra/Por CE dialysis membrane (Spectrum) with a molecular weight cutoff of 5,000 and dialyzed overnight at 4°C against phosphate-buffered saline containing 0.05% (first dialysis) and 0.01% (second dialysis) sodium dodecyl sulfate (SDS). The concentration of FP24 in the sample obtained after dialysis was both estimated from the gel and determined with the aid of the Micro BCA Protein Assay Reagent (Perbio).
Antibodies.
To generate an antiserum directed against the ectodomain of the ORF4 product (αGP4E), two 3-month-old New Zealand White rabbits were injected subcutaneously with 2 ml of an antigen emulsion containing 1 ml of Freund's complete adjuvant and 125 μg of FP24 in 1 ml of PBS-0.01% SDS. At 4, 8, 12, 16, and 20 weeks after the primary immunization, the animals were boosted with approximately 500 μg of the antigen in incomplete Freund's adjuvant. Two weeks after the third, fourth, and fifth booster immunizations, blood plasma was collected from both rabbits and stored at −20°C until further use.
An antiserum specific for the ORF3 protein (αGP3) was obtained by using the synthetic peptide SP03 (NH2-Ser-Phe-Val-Asp-Glu-Asp-Leu-Arg-Leu-His-Ile-Arg-Pro-Thr-Leu-Ile-Cys-COOH). This peptide corresponds to amino acids 129 through 145 of the ORF3 coding sequence and was produced by 9-fluorenylmethoxy carbonyl solid-phase peptide synthesis (43). The peptide was coupled to keyhole limpet hemocyanin (Calbiochem) via the carboxy-terminal Cys residue by using m-maleimidobenzoyl-N-succinimide ester as a cross-linker (40). Approximately 200 μg of the antigen in Freund's complete adjuvant was subcutaneously injected into a 3-month-old New Zealand White rabbit. The animal was boosted at monthly intervals with 500 μg of the conjugate in incomplete Freund's adjuvant and bled after the fourth booster.
Transfection/infection experiments.
Subconfluent monolayers (10 cm2) of BHK-21 C13 or BSR T7/5 cells were washed with GMEM and infected with vTF7.3 or MVA-T7 in GMEM for 50 min at 37°C at a multiplicity of infection (MOI) of ≥10. The cells were then washed with GMEM and overlaid with 200 μl of plasmid-liposome mixture. For this purpose, 200 μl of GMEM at room temperature (RT) was mixed with 10 μl of Lipofectin reagent (Invitrogen-Life Technologies) and incubated for 5 min at RT. Next, 5 μg of plasmid DNA was added to the mixture, which was then incubated for 15 to 20 min at RT and subsequently added to the cells. After a 10-min incubation at RT, 800 μl of GMEM was added to the transfection medium and the cells were incubated further at 37°C. At 3 h postinfection (p.i.), 1 ml of prewarmed GMEM-10% FCS was added to the cells and the incubation at 37°C was continued.
EAV infection.
Subconfluent monolayers of BHK-21 C13 cells were washed once with phosphate-buffered saline (PBS) containing 50 μg of DEAE-dextran per ml. Subsequently, the cells were infected with EAV at an MOI of ≥10 in GMEM containing 2% FCS and 50 μg of DEAE-dextran per ml. After incubation for 1 h at 37°C, the inoculum was replaced with GMEM-10% FCS at 39°C and the cells were kept at that temperature until the start of the radiolabeling procedures.
Metabolic radiolabeling of intracellular proteins.
At the indicated time points, the culture fluid was removed and the cells were washed with prewarmed starvation medium (Dulbecco's modified Eagle's medium without l-cysteine and l-methionine [Invitrogen-Life Technologies] supplemented with 5% dialyzed FCS, 10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid [HEPES; pH 7.4], and 0.2 mM l-methionine) and subsequently incubated in 800 μl of fresh starvation medium. Following an incubation period of 30 min, 80 μCi of [35S]Cys (ICN) was added and the cells were labeled for the indicated length of time at 39°C. After the labeling, the cells were placed on ice and washed with ice-cold PBS containing 50 mM CaCl2 and 50 mM MgCl2 and in some cases with a 20 mM concentration of the sulfhydryl modifying agent N-ethylmaleimide (NEM; Sigma-Aldrich), as indicated in the figure legends. Next, the cells were lysed in ice-cold lysis buffer (20 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% Nonidet P-40 [NP-40], 0.5% sodium deoxycholate, and 0.1% SDS containing 1 μg each of aprotinin, leupeptin, and pepstatin A per ml) with or without 20 mM NEM. The lysate was cleared by centrifugation in a Microfuge for 15 min at 4°C and 14,000 rpm. The pellet was discarded, and the supernatant was supplemented with EDTA to a final concentration of 5 mM. Alternatively, the labeling was followed by a rapid wash with prewarmed chase medium (GMEM-10% FCS containing 1 mM l-methionine, 2 mM l-cysteine hydrochloride monohydrate, 10 mM HEPES [pH 7.4], and, wherever indicated, 0.5 mM cycloheximide). Subsequently, the cells were incubated in chase medium for different times and the samples were further processed as described above.
Preparation of radiolabeled virions.
Subconfluent monolayers of BHK-21 C13 cells were infected with EAV at a high MOI as described above. At 6 h p.i., the medium was removed and the cells were washed with prewarmed starvation medium and subsequently incubated in 800 μl of fresh starvation medium. Following an incubation period of 30 min at 39°C, 80 μCi of [35S]Cys was added and the cells were further incubated for 4.5 h at 39°C. At 11 h p.i., the medium was harvested and cleared by centrifugation in a Microfuge for 10 min at RT and 4,000 rpm. The labeled virus was pelleted through a cushion of 20% (wt/wt) sucrose in TNE (20 mM Tris-HCl [pH 7.6], 100 mM NaCl, 1 mM EDTA) by centrifugation for 2 h in an SW 50.1 rotor (Beckman) at 28,000 rpm and 4°C. The pellet was then dissolved in 1 ml of ice-cold lysis buffer containing 20 mM NEM.
In vitro transcription and translation.
Five micrograms of plasmid pMRI14 or pAVI13 was digested with EcoRV. The linearized plasmid DNA was purified by phenol-chloroform extraction and ethanol precipitation and dissolved in 5 μl of water. In vitro transcription reactions were carried out by using T7 mMESSAGE mMACHINE (Ambion) in accordance with the manufacturer's instructions in 20-μl volumes. After a 1.5-h incubation period at 37°C, the template DNA was degraded by treatment of the transcription reaction mixture with 1 μl of DNase I (Ambion) for 15 min at 37°C. Next, 7.5 μl of ammonium acetate stop solution (Ambion) was added. The RNA was subsequently purified by phenol-chloroform extraction, precipitated with isopropanol, and dissolved in water. Translations of the mRNAs were done for 1.25 h at 30°C in the Promega rabbit reticulocyte lysate system in the presence or absence of canine pancreatic microsomal membranes (Promega) by using [35S]Cys or, for translation of the Saccharomyces cerevisiae α-factor RNA (Promega), Redivue Pro-mix l-[35S] in vitro cell labeling mix ([35S]Met plus [35S]Cys; >1,000 Ci/mmol; Amersham Pharmacia Biotech). When indicated, the samples were subjected to immunoprecipitation (IP).
Proteinase K digestion and membrane association assay.
For protease protection assays, 40-μl aliquots of the in vitro translation sample were mixed on ice with 80 μl of 50 mM Tris-HCl (pH 7.6)-25 mM CaCl2 (TC buffer) and split into three equal portions. The first aliquot was adjusted to a final volume of 66.6 μl with water, the second portion was supplemented with 13.3 μl of water and 13.3 μl of proteinase K (2 mg/ml; Roche), and the third aliquot received 13.3 μl of water and 13.3 μl of 10% Triton X-100 (TX-100). The samples were then incubated for 60 min at 4°C. The protease was then inactivated by addition of 6.3 μl of 12.5 mg of phenylmethylsulfonyl fluoride per ml and 10 mg each of leupeptin, pepstatin, and aprotinin per ml and incubation for 5 min at 0°C. Subsequently, the samples were subjected to IP with the indicated antisera. For the membrane association assays, 18-μl aliquots of the in vitro translation sample were split into three equal portions. The first aliquot was supplemented with 200 μl of TC buffer; the second portion was supplemented with 200 μl of 100 mM sodium carbonate (pH 11.5), and 200 μl of 100 mM sodium carbonate (pH 11.5)-2% TX-100 was added to the third aliquot. The samples were then incubated for 60 to 120 min at 4°C. After centrifugation for 60 min at 4°C and 75,000 rpm in a 100.2 rotor (Beckman), the supernatants were subjected to IP with the indicated antisera. The pellets were resuspended in sample buffer (LSB) containing 5% β-mercaptoethanol (5).
IP and gel electrophoresis.
Crude protein samples were diluted in IP buffer (20 mM Tris-HCl [pH 7.6], 150 mM NaCl, 5 mM EDTA [pH 8.0], 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS containing 1 μg each of aprotinin, leupeptin, and pepstatin per ml) to a final volume of 1 ml. The samples were supplemented with 3 μl of rabbit serum and incubated overnight at 4°C. When indicated, dithiothreitol (DTT) was added to a final concentration of 5 mM. On the next day, 20 μl of Pansorbin (Calbiochem) was added to each sample. After incubation for ≥1 h at 4°C, the immune complexes were collected by centrifugation and washed three times in wash buffer I (20 mM Tris-HCl [pH 7.6], 150 mM NaCl, 5 mM EDTA, 0.1% NP-40) and once in wash buffer II (20 mM Tris-HCl [pH 7.6], 0.1% NP-40). Next, the immune complexes were resuspended in 20 μl of LSB containing 50 mM DTT or 5% β-mercaptoethanol and incubated for 5 min at 96°C. After centrifugation in a Microfuge for 15 min at RT and 14,000 rpm, the supernatants were analyzed by SDS-polyacrylamide (PAA) gel electrophoresis (PAGE). Following SDS-PAGE, the gels were fixed in 10% acetic acid-50% methanol-0.005% Coomassie brilliant blue for 30 min and incubated for another 30 min in 1 M sodium salicylate. Finally, the gels were dried on Whatman 3MM paper and exposed to Kodak X-ray films at −80°C.
Endoglycosidase treatment.
Two different methods of endoglycosidase H (endo H) and N-glycosidase F (PNGase F) digestion were used. For the experiment described in Fig. 1, washed immunoprecipitates were either resuspended in 300 μl of endo H buffer (50 mM sodium acetate [pH 5.5], 10 mM EDTA, 10% bovine serum albumin, 1 mg each of aprotinin, leupeptin, and pepstatin A per ml) or PNGase F buffer (50 mM sodium phosphate [pH 7.0], 10 mM EDTA, 10% bovine serum albumin, 1 mg each of aprotinin, leupeptin, and pepstatin A per ml). The samples were then incubated with or without endo H (New England Biolabs) or PNGase F (New England Biolabs) under continuous mixing by rotation for ≥16 h at 37°C. Subsequently, the samples were centrifuged for 2 min at 20,000 × g and RT. The pellets were resuspended in 20 μl of LSB containing 50 mM DTT and further processed as described above. For the other experiments, the endo H and PNGase F digestions were performed in accordance with the instructions of the manufacturer except that the reactions were done overnight at 37°C. In all experiments, 0.05 U of endo H or PNGase F was used per digestion.
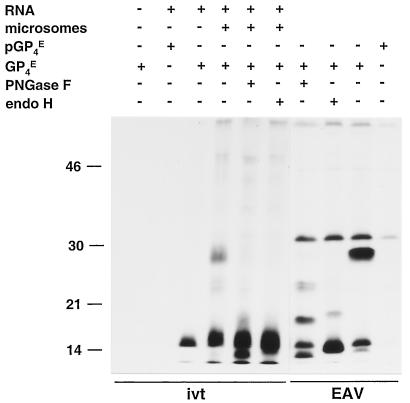
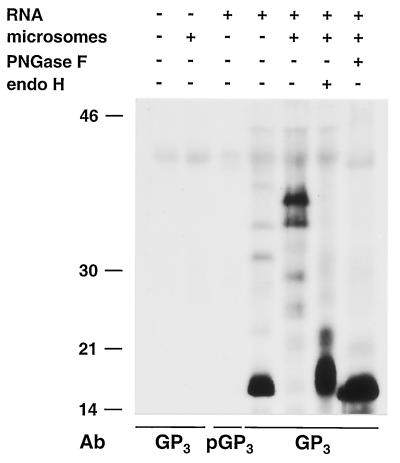
FIG. 1.
Identification of the ORF4-encoded protein. ORF4 RNA transcripts were translated in vitro (ivt) in the absence or presence of canine pancreatic microsomes, and the expression products were immunoprecipitated with the GP4-specific antipeptide serum (GP4E) or its preimmune serum (pGP4E). The precipitated proteins were treated or mock treated with PNGase F or endo H. For comparison, EAV-infected BHK-21 C13 cells were pulse-labeled for 15 min with [35S]cysteine at 8.25 h p.i. and chased for 30 min. Cells were lysed, and IPs were performed with the GP4-specific antipeptide serum (EAV). The precipitated proteins were also treated or mock treated with PNGase F or endo H. All samples were dissolved in LSB containing 5% β-mercaptoethanol and analyzed in SDS-15% PAA gels. The values on the left are the molecular sizes, in kilodaltons, of marker proteins analyzed in the same gel.
Indirect immunofluorescence assay.
BHK-21 C13 cells grown on glass coverslips were infected with MVA-T7 at a low MOI and transfected with pMRI14 or pAVI13 as described above. At 7 h p.i., the cells were rinsed twice with PBS and fixed with ice-cold methanol at −20°C for 10 min. The fixed cells were washed twice with PBS and incubated for 30 min with αGP4E or αGP3 diluted 1/100 in PBS-5% FCS. Following three 5-min washes with PBS-5% FCS, the cells were stained with Cy2-conjugated donkey anti-rabbit immunoglobulin G (heavy and light chains) antibodies (Jackson) diluted 1/100 in PBS-5% FCS. After three 5-min washes with PBS-5% FCS, the cells were stained with the Alexa 594-conjugated endoplasmic reticulum (ER) marker concanavalin A (Molecular Probes) diluted 1/400 in PBS-5% FCS. After three 5-min washes with PBS, the samples were mounted on glass slides in FluorSave (Calbiochem). The samples were examined with a confocal microscope (Leica TCS SP2).
RESULTS
Identification of the EAV ORF4 product.
To identify the ORF4 product, RNA was transcribed in vitro from plasmid pMRI14 and translated in a reticulocyte lysate system in the absence and presence of canine pancreatic microsomal membranes. The radiolabeled translation products were immunoprecipitated with the antiserum αGP4E, which is directed against the putative ectodomain of GP4, and their electrophoretic mobilities were compared with those of the ORF4 proteins immunoprecipitated with the same antiserum from lysates of EAV-infected cells (Fig. 1). After in vitro translation in the absence of microsomes, the GP4-specific antiserum precipitated a protein with an apparent molecular mass of about 15 kDa. This molecular mass is slightly smaller than that predicted for the unprocessed ORF4 translation product (17 kDa) (5). In the presence of microsomal membranes, the size of the ORF4 product increased to about 28 kDa. A large amount of the unprocessed 15-kDa form of the protein was also observed. Since protein species of 28 and 15 kDa that comigrated with the in vitro translation products were also immunoprecipitated with αGP4E from lysates of EAV-infected cells, we conclude that the 15- and 28-kDa proteins represent the primary and processed products of EAV ORF4, respectively.
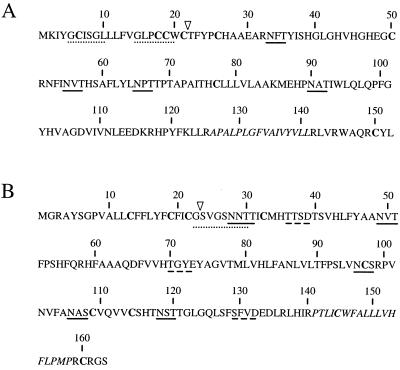
The amino acid sequence of the polypeptide encoded by ORF4 contains a predicted amino-terminal signal sequence and four potential N-glycosylation sites (Fig. 2A), the third of which contains a Pro residue at its +1 position and is therefore presumably not used. To find out whether the increase in molecular mass observed in the presence of microsomes resulted from N glycosylation and whether the signal sequence was cleaved off, the in vitro translation products of ORF4 were treated with PNGase F or endo H.
FIG. 2.
Deduced amino acid sequences of the polypeptides specified by EAV ORF4 (A) and ORF3 (B). The Cys residues are in boldface, potential N-glycosylation sites are underlined, potential N-myristoylation sites are indicated by dotted lines, and potential casein kinase II phosphorylation sites are represented by broken lines. The triangles indicate the most likely signal sequence cleavage sites, as determined by the method of von Heijne (50). The putative membrane-spanning segments are in italics.
PNGase F treatment of the ORF4 products reduced the molecular mass of the 28-kDa species to approximately 13 kDa but did not affect the unprocessed ORF4-encoded protein of 15 kDa. A similar pattern was observed when the immunoprecipitate from EAV-infected cells was treated with PNGase F. However, in addition to the 13- and 15-kDa protein species, partially deglycosylated products of 19 and 24 kDa were observed. The reduction in size of GP4 from 28 kDa to approximately 13 kDa and the presence of two partly deglycosylated forms after digestion with PNGase F indicate that the ORF4-encoded polypeptide, indeed, contains three functional N-glycosylation sites. After endo H digestion, the protein migrated slightly slower than after PNGase F digestion. This difference is explained by the different cleavage specificities of these enzymes. While PNGase F removes the N-linked sugar chains completely, endo H leaves the first monosaccharide—the N-acetylglucosamine that tethers each side chain to the Asn residue—on the polypeptide.
The PNGase F- and endo H-treated GP4 species from EAV-infected cells and from the in vitro translation in the presence of microsomal membranes were smaller than the protein synthesized in the absence of microsomes, which indicated that the amino-terminal signal sequence is cleaved off.
Membrane association and topology of the GP4 protein.
Its amino-terminal signal sequence, its N-glycosylation pattern, and its predicted carboxy-terminal hydrophobic membrane anchor (Fig. 2A) (5) strongly suggest that the GP4 protein is a typical class I (NexoCcyt) integral membrane protein. To address whether the GP4 protein is, indeed, membrane associated, the polypeptide was synthesized again in vitro in the presence of microsomal membranes. The reaction mixture was then split into three equal portions. One portion was diluted and incubated in sodium carbonate buffer at pH 11.5. This treatment disrupts the microsomal vesicles and consequently causes the release of the superficially membrane associated, but not the membrane-anchored, proteins (17). The second portion was diluted and incubated in the same buffer supplemented with TX-100 to dissolve the membranes and membrane proteins. The third portion was mixed with TC buffer, under which condition the microsomal vesicles should remain intact. After the incubations, the membranes were pelleted by ultracentrifugation. The membrane pellets were analyzed directly by SDS-PAGE (Fig. 3; pellet), while the supernatants were subjected to IP (Fig. 3; supernatant). The yeast α-factor was in vitro synthesized in parallel and used as a soluble protein control (26). Because of the lack of an antiserum for this protein, the membrane pellets and supernatants were both analyzed directly.
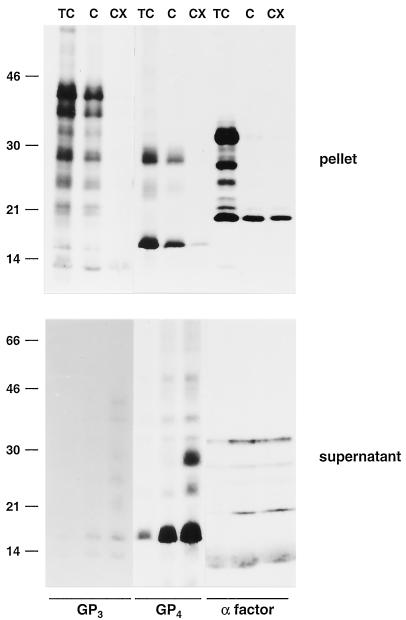
FIG. 3.
Membrane association of the GP4 and GP3 proteins. ORF4 and -3 RNA transcripts were translated in a rabbit reticulocyte lysate in the presence of canine pancreatic microsomes. Equal portions of the in vitro translation mixtures were incubated for 60 to 120 min at 4°C in three different buffers: 50 mM Tris-HCl (pH 7.6)-25 mM CaCl2 (TC), 100 mM sodium carbonate (pH 11.5) (C), and 100 mM sodium carbonate (pH 11.5)-2% TX-100 (CX). After subsequent high-speed centrifugation of the samples, the pellets were dissolved directly in LSB containing 5% β-mercaptoethanol and analyzed in SDS-15% PAA gels while the GP3 and GP4 proteins released into the supernatants were immunoprecipitated with the αGP4E and αGP3 antisera, respectively. The yeast α-factor was used as a soluble-protein control. Since a specific antibody for this protein was not available, the supernatants were not subjected to IP but dissolved directly in LSB containing 5% β-mercaptoethanol. The values on the left are the molecular sizes, in kilodaltons, of marker proteins analyzed in the same gels.
The pellets obtained after incubation in the TC buffer should contain the vesicular membrane proteins, as well as the lumenal proteins. As is clear from Fig. 3, both the soluble α-factor species and the ORF4 products were present almost exclusively in the pellet fraction. After the sodium carbonate extraction, the glycosylated forms of the α-factor were observed mainly in the supernatant while its unprocessed 18-kDa form remained largely associated with the membrane pellet. In contrast, the glycosylated GP4 protein was still found in the pellet, indicating its membrane association. Most of the unprocessed GP4 protein had been released into the supernatant, demonstrating that this form was loosely associated with the lipid bilayer. When the microsomal vesicles were incubated in the presence of TX-100, which solubilizes the membranes, virtually of all the GP4 molecules were found in the supernatant.
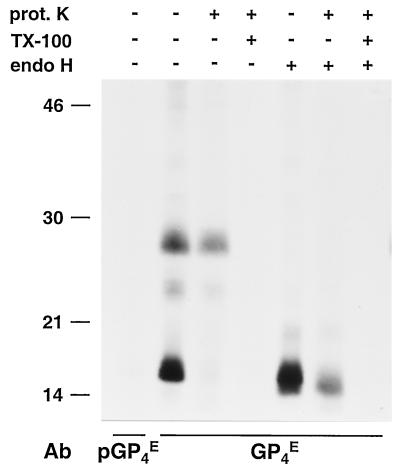
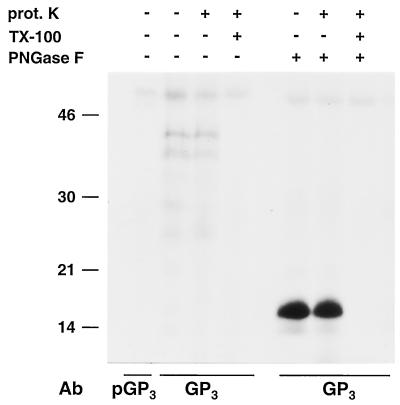
To investigate the membrane topology of the ORF4-encoded protein, we treated the ORF4 proteins synthesized in the presence of microsomal membranes with proteinase K (Fig. 4). This had no effect on the electrophoretic mobility of the glycosylated GP4 protein, whereas the unprocessed 15-kDa ORF4 product was completely degraded. This implies that the unprocessed GP4 protein is present outside of the microsomes. Proteinase K treatment in the presence of TX-100 resulted in complete degradation of all of the GP4 species, indicating that none of the ORF4 products is intrinsically resistant to protease treatment. Since the predicted GP4 endodomain is very short (only about 10 residues; Fig. 2A), its degradation would only have a minor effect on the protein's molecular mass that might hence remain unnoticed in a comparison of the electrophoretic mobilities of the proteinase K- and mock-treated glycosylated GP4 protein. We therefore deglycosylated the GP4 protein with endo H after treatment or mock treatment with proteinase K. This still did not reveal a difference in migration between the mock- and proteinase K-treated GP4 molecules, indicating that little, if any, of the carboxy terminus of the mature ORF4-encoded protein is accessible to the protease and thus protrudes from the microsomal membranes. Altogether, these data indicate that GP4 is a type I membrane glycoprotein without an exposed C terminus.
FIG. 4.
Membrane topology of the GP4 protein. ORF4 RNA transcripts were translated in vitro in the presence of canine pancreatic microsomes. Subsequently, the samples were treated or mock treated with proteinase K (prot. K) in the absence of a detergent or after disruption of the microsomal membranes with TX-100. After a 1-h incubation at 4°C, the proteinase K was inactivated by addition of protease inhibitors. Next, the samples were split into three equal portions. One aliquot was subjected to IP with a GP4-specific antipeptide serum (GP4E), the second aliquot was mixed with the corresponding preimmune serum (pGP4E), and the third aliquot was subjected to IP with αGP4E and treated with endo H. The samples were dissolved in LSB containing 5% β-mercaptoethanol and analyzed in SDS-15% PAA gels. The values on the left are the molecular sizes, in kilodaltons, of marker proteins analyzed in the same gel. Ab, antibody.
Immunoprecipitation of the ORF3 product.
The predicted ORF3 product has a molecular mass of 18 kDa, contains six potential N-glycosylation sites (Fig. 2B) (5), and is predicted to possess a cleavable amino-terminal signal sequence. To investigate the specificity of the αGP3 antiserum that we raised against amino acids 129 though 145 of the predicted ORF3-encoded protein, RNA was transcribed in vitro from plasmid pAVI13 and translated in a rabbit reticulocyte lysate system. The radiolabeled translation products were subsequently incubated with αGP3, and IPs were carried out.
After translation in the absence of microsomal membranes, the GP3-specific antiserum precipitated a protein with an apparent molecular mass of about 16 kDa (Fig. 5), which is also slightly less than that predicted for the primary translation product. In the presence of microsomal membranes, a number of GP3 species were produced, with proteins migrating at about 37 and 42 kDa being most prevalent. After PNGase F treatment, a major product was obtained with about the same mobility as the protein made in the absence of microsomes membranes. These data indicate that the GP3 protein contains multiple functional N-glycosylation sites and that its signal sequence is not cleaved. Like GP4, after endo H treatment, the GP3 protein migrated slightly more slowly in SDS-PAGE than after PNGase F treatment. These results are consistent with those of Hedges et al. (21), who found a primary in vitro translation product of approximately 17 kDa that, in the presence of microsomal membranes, was extensively N glycosylated, migrating as a smear of 36 to 42 kDa. They also concluded that no signal sequence was cleaved off.
FIG. 5.
Identification of the ORF3 product. ORF3 RNA transcripts were translated in vitro in the absence or presence of canine pancreatic microsomes, and the expression products were immunoprecipitated with a GP3-specific antipeptide serum (GP3) or its preimmune serum (pGP3). Subsequently, the precipitates were treated or mock treated with PNGase F or endo H. The samples were dissolved in LSB containing 5% β-mercaptoethanol and analyzed in SDS-15% PAA gels. The values on the left are the molecular sizes, in kilodaltons, of marker proteins analyzed in the same gel. Ab, antibody.
Membrane association and topology of the GP3 protein.
In parallel with that of the GP4 protein, we also investigated the membrane association of the GP3 protein (Fig. 3). When the ORF3 translation products synthesized in vitro in the presence of microsomes were incubated in TC or sodium carbonate buffer, the glycosylated GP3 protein was pelleted together with the membranes. In contrast, after solubilization of the microsomal membranes in the sodium carbonate buffer supplemented with TX-100, the different glycoforms of the GP3 protein were found in the supernatant. Note that the apparent loss of GP3 signal in the TX-100 supernatant relative to that in the TC pellet is due to the less efficient retrieval of the protein by IP with the anti-GP3 serum. These results indicate that the GP3 protein is membrane anchored, which is also in accordance with the conclusions of Hedges et al. (21).
The topology of the GP3 protein was investigated in the same manner as that of the GP4 protein (Fig. 6). Proteinase K treatment of the glycosylated GP3 species had no effect on their electrophoretic mobility, whereas in the presence of TX-100, the ORF3 translation products were completely degraded by the protease. To further confirm this result, the immunoprecipitated proteins were deglycosylated with PNGase F after proteinase K digestion. Again, the mock- and protease-treated GP3 species exactly comigrated, which implies that little or nothing of the GP3 protein is exposed on the outside of the microsomal membranes.
FIG. 6.
Membrane topology of the GP3 protein. ORF3 RNA transcripts were translated in vitro in the presence of canine pancreatic microsomes. Subsequently, the translation products were treated or mock treated with proteinase K (prot. K) in the absence of detergent or after disruption of the microsomal membranes with TX-100. After a 1-h incubation at 4°C, the proteinase K was inactivated by protease inhibitors. Next, the samples were immunoprecipitated with a GP3-specific antipeptide serum (GP3) or its preimmune serum (pGP3). The resulting immune complexes were each split into two equal portions that were treated or mock treated with PNGase F. The samples were dissolved in LSB containing 5% β-mercaptoethanol and analyzed in SDS-15% PAA gels. The values on the left are the molecular sizes, in kilodaltons, of marker proteins analyzed in the same gel. Ab, antibody.
Intracellular fate of the individually expressed GP4 and GP3 proteins.
To study the intracellular processing of GP4, BHK-21 C13 cells were infected with the recombinant vaccinia virus vTF7.3 (16) and transfected with ORF4 expression plasmid pMRI14. Cells were labeled for 15 min with [35S]cysteine and then chased for different times. Next, the cells were lysed and the GP4 proteins were immunoprecipitated with αGP4E and analyzed by SDS-PAGE under reducing conditions. The glycosylation status and intracellular transport of the proteins were monitored biochemically by assaying the acquisition of resistance to endo H cleavage of their N-linked oligosaccharide chains. N-glycosylated proteins are initially sensitive to this enzyme but become resistant upon passage through the medial Golgi compartment (28).
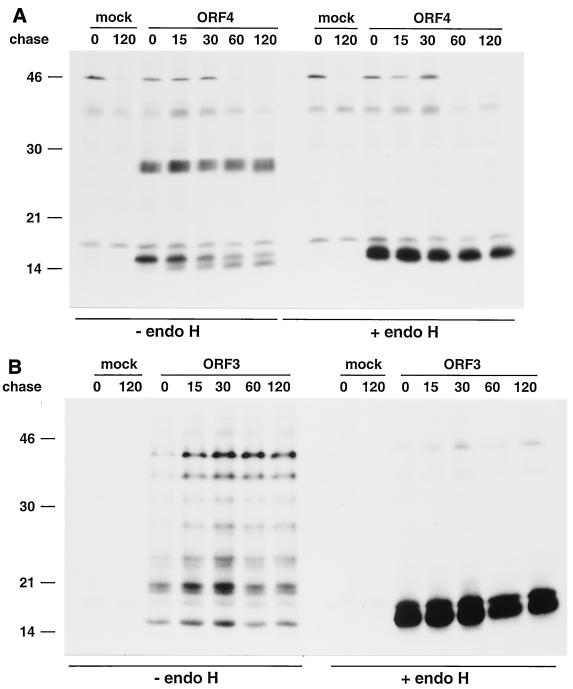
As shown in Fig. 7A, after pulse-labeling, the N-glycosylated 28-kDa GP4 protein, the unprocessed 15-kDa primary translation product of ORF4, and even some unglycosylated 13-kDa GP4 protein with no signal sequence were detected. The fuzziness of the N-glycosylated GP4 band indicates that the protein is heterogeneously glycosylated, probably because of its continuously undergoing modifications while in the ER. The analysis of the chase samples revealed that the glycoprotein is fairly stable. Most of the GP4 protein labeled during the pulse was still present after the 2-h chase period, and the protein did not undergo detectable mobility changes. In contrast, the amount of unprocessed GP4 protein slowly decreased while the amount of unglycosylated GP4 protein that had lost the signal sequence slightly increased. Since the removal of the signal sequence halves the cysteine content of the polypeptide, the decrease in the amount of the unprocessed 15-kDa GP4 protein seems to be fully accounted for by its conversion to the 13-kDa GP4 species. The glycosylated GP4 protein synthesized during the pulse remained fully endo H sensitive throughout the chase. Apparently, the independently expressed glycoprotein does not leave the ER.
FIG. 7.
Kinetics of endo H resistance acquisition of individually expressed GP4 and GP3 proteins. BHK-21 C13 and BSR T7/5 cells were infected with vTF7.3 and transfected or mock transfected with ORF4-specific plasmid pMRI14 (A) or ORF3-encoding plasmid pAVI13 (B). The cells were pulse-labeled for 15 min with [35S]cysteine at 4.5 h p.i. and chased in the presence of 0.5 mM cycloheximide for the times indicated (in minutes). The GP4 and GP3 proteins were then immunoprecipitated from cell lysates with the αGP4E and αGP3 antisera, respectively, in the presence of 5 mM DTT. The immunoprecipitates were treated with endo H (+) or mock treated (−). The samples were finally dissolved in LSB containing 50 mM DTT and analyzed in SDS-15% PAA gels. The values on the left are the molecular sizes, in kilodaltons, of marker proteins analyzed in the same gels.
A similar experimental setup was used to study the intracellular fate of the GP3 protein. However, since the GP3 protein was poorly expressed in BHK-21 C13 cells, we used BSR T7/5 cells, which constitutively express the bacteriophage T7 RNA polymerase, in combination with ORF3 expression plasmid pAVI13. A series of protein species ranging in apparent molecular mass from 16 to 42 kDa was specifically immunoprecipitated by αGP3 serum after the 15-min pulse (Fig. 7B). These differently sized GP3 molecules represent distinct glycoforms of the GP3 protein, as confirmed by the endo H sensitivity of all except the lowest-molecular-weight species. The analysis of the chase samples demonstrated that GP3 is stable, that the glycosylation pattern does not change over time, and that the glycoproteins remain endo H sensitive at all chase times.
Subcellular localization of the individually expressed GP4 and GP3 proteins.
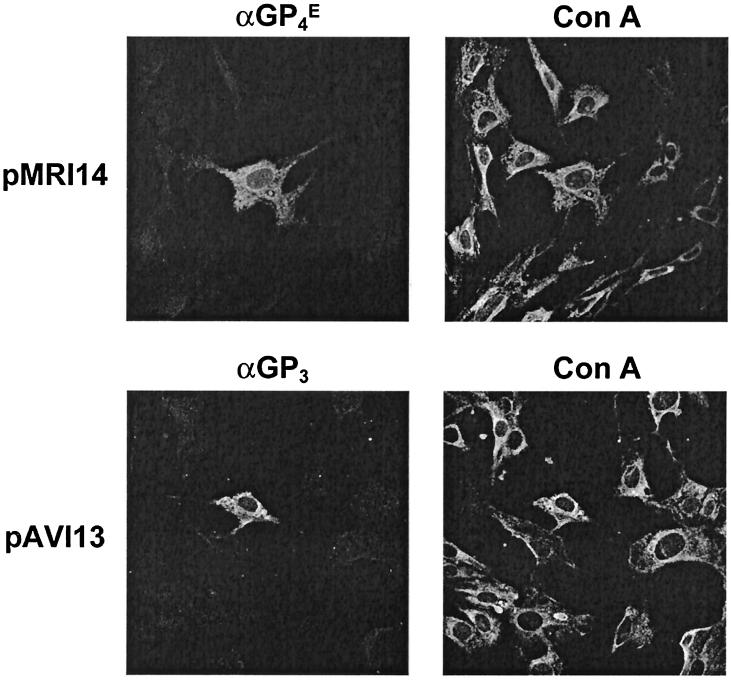
The previous biochemical observations suggested that the individually expressed GP4 and GP3 proteins were not transported through the Golgi complex. To study their subcellular localization more directly, we expressed ORF3 and -4 in BHK-21 C13 cells from plasmids pAVI13 and pMRI14, respectively, with the aid of recombinant vaccinia virus MVA-T7 (44). At 7 h p.i., the cells were fixed with methanol and stained with GP4- or GP3-specific antibodies and with the ER marker concanavalin A (Fig. 8). Immunofluorescence studies with the αGP4E and αGP3 sera showed a reticular staining pattern that overlapped the intracellular distribution of the ER marker concanavalin A. These results demonstrate that the individually expressed GP4 and GP3 proteins localize in the ER.
FIG. 8.
Intracellular localization of expressed GP3 and GP4 proteins by immunofluorescence assay. BHK-21 C13 cells were infected with MVA-T7 and transfected with pMRI14 or pAVI13. At 7 h p.i., the cells were fixed with methanol and labeled with the ER marker concanavalin A (Con A) and with antibodies specific for the GP4 (αGP4E) or GP3 (αGP3) protein as described in Materials and Methods.
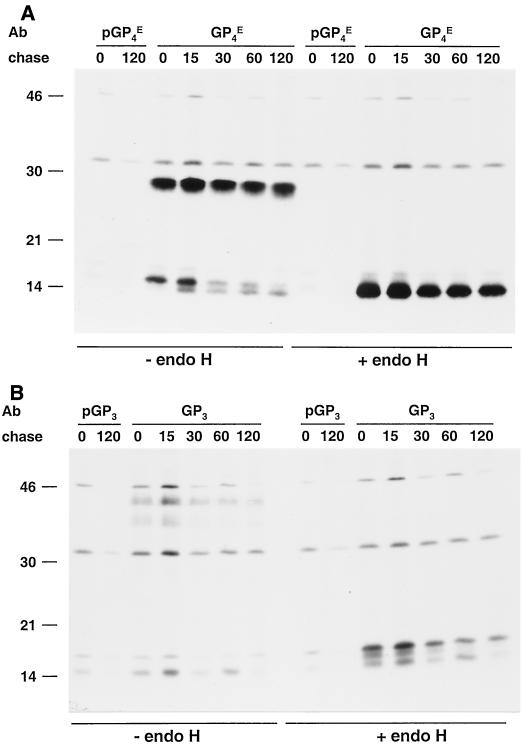
Fate of the GP4 and GP3 proteins in EAV-infected cells.
Next, we analyzed the appearance, maturation, and stability of the GP4 and GP3 proteins in the presence of the other EAV proteins. For this purpose, EAV-infected BHK-21 C13 cells were labeled for 15 min and chased for various times. As when expressed independently, the GP4 protein appeared in EAV-infected BHK-21 cells in its fully glycosylated form and in two unglycosylated forms (Fig. 9). This pattern remained unchanged during the chase period, during which time also no endo H-resistant forms became detectable intracellularly. The overall intensity of labeled GP4 species appeared to decrease slightly, as was seen most clearly after deglycosylation by endo H. This observation most likely reflects the secretion of GP4 protein with virions (see below).
FIG. 9.
Intracellular processing of the GP4 and GP3 proteins in EAV-infected cells. EAV-infected BHK-21 C13 cells were pulse-labeled for 15 min with [35S]cysteine at 8.25 h p.i. and chased in the presence of 0.5 mM cycloheximide for the times indicated (in minutes). The GP4 (A) and GP3 (B) molecules were then immunoprecipitated from cell lysates with the αGP4E and αGP3 antisera, respectively, or with the corresponding preimmune sera in the presence of 5 mM DTT. The immunoprecipitates were treated with endo H (+) or mock treated (−). The samples were finally dissolved in LSB containing 50 mM DTT and analyzed in SDS-15% PAA gels. The values on the left are the molecular sizes, in kilodaltons, of marker proteins analyzed in the same gels. Ab, antibody.
Of the characteristic set of GP3 species observed after independent expression, only the largest two glycoproteins (apparent molecular masses of 37 and 42 kDa) were clearly detected in lysates of EAV-infected cells. During the chase, the intensities of the GP3-specific bands decreased, which might be due to either proteolytic degradation or secretion from the cells, independently or with viral particles. The results of the endo H treatments showed that no detectable intracellular accumulation of the GP3 glycoprotein with mature N-linked glycans took place during the chases.
Detection of the GP4 and GP3 proteins in EAV particles.
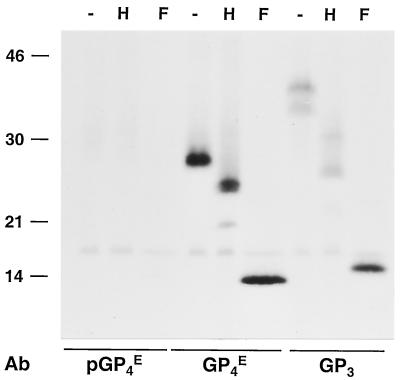
To determine whether GP4 and GP3 are structural proteins, BHK-21 cells were infected with EAV and labeled with [35S]cysteine. The EAV particles released into the supernatant were concentrated by sedimentation through a sucrose cushion, dissolved in lysis buffer, and subjected to IP with antiserum directed against GP4 or GP3 or, as a negative control, with the GP4 preimmune serum. In order to determine their glycosylation status, we mock treated the samples or treated them with endo H or PNGase F and analyzed them by SDS-PAGE. As shown in Fig. 10, both GP4 and GP3 are present in virions.
FIG. 10.
Identification of the GP4 and GP3 proteins in virions. EAV-infected BHK-21 C13 cells were labeled with [35S]cysteine from 6.5 to 11 h p.i. After removal of cell debris by low-speed centrifugation, the labeled virus present in the cell culture medium was pelleted through a cushion of 20% (wt/wt) sucrose. The pellet was then dissolved in lysis buffer containing 20 mM NEM and subjected to IP with αGP4E or αGP3 antiserum or, as a negative control, αGP4E preimmune serum (pGP4E) in the presence of 5 mM DTT. The immunoprecipitates were mock treated (−) or treated with endo H (H) or PNGase F (F). The samples were finally dissolved in LSB containing 50 mM DTT and analyzed in SDS-15% PAA gels. The values on the left are the molecular sizes, in kilodaltons, of marker proteins analyzed in the same gel. Ab, antibody.
The IP with the GP4-specific antiserum demonstrated that of the three ORF4 species present in EAV-infected cells, only the glycosylated 28-kDa protein is incorporated into virus particles. After endo H treatment, its apparent molecular mass decreased by about 3 kDa, to 25 kDa, while a small fraction of the GP4 molecules was converted to a 21-kDa protein. As expected, the N-linked glycans were entirely removed by incubation with PNGase F, which cleaves off N-linked sugars irrespective of their maturation state. These results indicate that the virions contain GP4 molecules with mature and immature N-linked oligosaccharide side chains.
The antipeptide serum directed against GP3 typically immunoprecipitated both the 37- and the 42-kDa N-glycosylated GP3 forms observed in EAV-infected cells. None of the other GP3 species was detected. After endo H treatment, the 37- and 42-kDa GP3 species were converted to molecules of 27 and 32 kDa, respectively, which shows that part of the N-linked glycans were immature. PNGase F digestion reduced the 37- and 42-kDa species to 16 kDa, the molecular mass of the deglycosylated polypeptide. Thus, the GP3 protein incorporated into EAV particles also carries a mixture of mature and immature N-linked oligosaccharides.
DISCUSSION
The analysis of the structure and composition of EAV has a long history. Initially, a complex pattern of eight (25) or nine (24) structural proteins was observed and the virus was classified as a Togavirus on the basis of morphology, size, protein composition, and genome type (38, 52). Subsequent reinvestigation of viral preparations by several groups with available antibodies gradually reduced the putative number of structural components (46, 54), eventually reaching a minimum of five proteins (5, 42). Meanwhile, the nature of these proteins, genomic nucleotide sequence information, and increasing insights into the viral replication strategy also had its consequences for the taxonomic position of the virus, EAV being assigned to the family Arteriviridae within the order Nidovirales (2). In the present study, we established that glycoproteins GP3 and GP4, encoded by ORF3 and -4, respectively, are also virion components. This brings the number of structural proteins back to seven, six of which are associated with the viral membrane. Hence, all of the ORFs downstream of the polymerase gene appear to specify structural proteins.
Until now, three envelope proteins have been identified for LDV (VP-3 M, VP-3P, and M) and SHFV (p42, p54, and M) (13, 19) while six were reported for PRRSV (GP2, 2b, GP3, GP4, GL, and M) (32-34, 47, 53). The composition of PRRSV particles is therefore consistent with that of EAV particles, although the presence of the PRRSV GP3 protein in the viral envelope is still controversial (see below).
Judging by the large number of different, small-sized membrane proteins, arterivirus envelopes have a unique composition among RNA viruses. The GL and M proteins are the most abundant proteins in the EAV envelope, while the E and GS proteins occur in intermediate and minor amounts, respectively (5, 42). Our data do not allow firm conclusions about the relative presence of the GP3 and GP4 proteins. However, all available information indicates that they are minor virion components. The initial inability to detect these proteins in unlabeled purified virus preparations supports this conclusion, as does reinspection of published analyses of radiolabeled virions. The 28-kDa GP4 protein, for instance, can be observed as a band in gel just above the GS protein in an electropherogram of [35S]cysteine-labeled EAV particles that we previously published (42). Quantitative analysis of this electropherogram revealed that the GP4 protein occurs in virions in similar molar amounts as the minor GS protein (data not shown). For the 38- and 42-kDa glycosylated GP3 species, such an interpretation is not possible as these proteins are generally obscured in SDS-PAGE by the prominent heterogeneously glycosylated GL protein. Consistently, also in the Lelystad strain of PRRSV, the GP3 and GP4 proteins are considered minor structural proteins (47).
Our studies demonstrate that the GP4 protein is a type I membrane glycoprotein and that three of its four predicted N-glycosylation motifs are used to produce a fully glycosylated protein with a molecular mass of 28 kDa. However, N glycosylation of the GP4 protein is a rather inefficient process. After in vitro translation, independent expression with the vTF7.3 expression system, or synthesis in EAV-infected cells, a significant portion of the protein does not acquire N-linked glycans. In virus particles, however, only the fully glycosylated form of GP4 is observed. Our immunolocalization experiments, together with the biochemical data on the acquisition of endo H resistance, revealed that the independently expressed GP4 protein is unable to leave the ER. Also, in EAV-infected cells, most of the GP4 protein is retained in the ER, as judged by the invariably immature state of its N-linked sugars. This is similar to what we observed earlier for the GS protein (7). Since the GP4 protein of virions contains mature, as well as partially mature, sugars, we conclude that only a small fraction of the ORF4 products synthesized in EAV-infected cells ends up in virus particles. The presence of both endo H-resistant and endo H-sensitive oligosaccharides on viral glycoproteins is not unusual (49). Most likely, not all of the N-linked glycans are accessible for further processing during passage through the Golgi apparatus because of the conformation of the protein or as a result of steric hindrance.
The EAV GP4 homologues of other arteriviruses also possess hydrophobic termini and at least four putative N-glycosylation sites, but their topologies have not been experimentally determined. In the Lelystad strain of PRRSV, GP4 was shown to be a structural, highly glycosylated protein of 31 to 35 kDa (47). A study of LDV showed that the in vitro translation product of ORF4 is an N-glycosylated and membrane-associated protein of about 31 kDa (14).
Interestingly, monoclonal antibodies specific for the GP4 protein of the Lelystad strain of PRRSV were found to neutralize viral infectivity (29, 47). These antibodies recognized different epitopes in a variable region of the GP4 protein located between amino acids 39 and 79 (35). In our study, the polyclonal serum αGP4E, directed against a His-tagged protein comprising amino acids 20 to 127 of the EAV ORF4 product, appeared not to neutralize the virus (data not shown).
The results of our in vitro translation studies of the GP3 protein confirmed and extended the conclusions of Hedges et al. (21). We observed that the GP3 protein is heavily glycosylated and membrane associated and that its signal sequence is not cleaved. After in vitro translation in the presence of microsomal membranes, the protein was fully proteinase K resistant. The topology of the GP3 protein is, however, not obvious. Assuming that the amino-terminal hydrophobic domain remains associated with the lipid membrane, the GP3 protein may be either a class II protein—anchored only by its signal sequence—or a class IV protein, being anchored at both of its termini. As indicated in Fig. 2B, the GP3 polypeptide has two potential N-myristoylation sites and three casein kinase II phosphorylation motifs. However, since the amino terminus of ORF3 is not cleaved off by the signal peptidase, it is highly unlikely that GP3 becomes myristoylated at Gly-23 or Gly-26. Furthermore, the three putative casein kinase II motifs are located in the lumenal domain of GP3, which makes phosphorylation of the protein improbable. Further experimentation is required to find out whether GP3 contains any posttranslational modifications other than N-linked glycans. After independent expression in BSR T7/5 cells, the GP3 protein appeared as a set of differently glycosylated species ranging in apparent molecular mass from 16 to 42 kDa. Obviously, most, if not all, of the theoretical N-glycosylation sites are occupied in the 42-kDa GP3 species. Both in EAV-infected cells and in virions, only the two most extensively glycosylated GP3 species of 37 and 42 kDa were observed. A fraction of the oligosaccharide side chains of the GP3 protein incorporated into viral particles remained immature.
The GP3 proteins of arteriviruses are all highly glycosylated and antigenic in their respective hosts (14, 20, 21, 27). For PRRSV, data regarding its presence in virus particles are conflicting. The GP3 protein of the Lelystad strain of PRRSV has been identified as a structural protein (47). In contrast, for the PRRSV IAF-Klop strain, the GP3 protein was characterized as nonstructural. Upon individual expression, as well as in the context of a PRRSV infection, a small fraction of the IAF-Klop GP3 protein was shown to be secreted into the extracellular medium but the protein could not be identified in virions (20, 31). For LDV, in vitro transcription and translation studies revealed that the ORF3-encoded protein was soluble or weakly associated with membranes through an uncleaved signal peptide. Furthermore, it was suggested that the protein might be secreted in the context of an LDV infection (14). Because of its structural nature, we could not establish whether the EAV GP3 protein also occurs in a “free,” secreted form in the extracellular medium; any GP3 protein detected in the medium after removal of viral particles by centrifugation might simply have originated from disintegrated particles or from broken cells.
No specific ER retention motifs have been detected in the amino acid sequence of the EAV GP3 and GP4 proteins. Most likely, the proteins are arrested in the ER by the quality control system, which retains misfolded, incompletely folded, and unassembled proteins in the ER (12, 23). EAV assembly takes place in the ER region of the cell by budding of the viral nucleocapsids into the ER lumen, from which the virus particles are transported through the Golgi complex to be released by exocytosis (30). Most likely, the GP3 and GP4 proteins are only transported through the secretory pathway when correctly assembled into oligomeric complexes and incorporated as such into virions.
Nothing is known about the function of the GP3 and GP4 proteins in the viral life cycle, except that both are essential. When the expression of ORF3 or -4 was blocked by mutagenesis with a full-length cDNA clone, infectious virus was no longer produced (36). Whether any virus particles are formed under these conditions is unknown. If not, the proteins might have a function in virus assembly, as is the case for the coronavirus E protein, which is also incorporated into virions in relatively small amounts. (15, 39, 48). Alternatively, the proteins may play a role in virus entry, e.g., in receptor binding or in membrane fusion. Indeed, fusion of EAV particles has not been attributed to any viral protein. Furthermore, while the entry functions are generally assumed to reside in the GL/M complex, this has by no means been proven. In fact, replacement of the ectodomain of the GL protein with that of another arterivirus did not alter the cell tropism of EAV, indicating that this protein is not involved in receptor binding (10). The receptors for the arteriviruses have not been identified. Also, no conspicuous fusion motifs have been observed in any of the viral membrane proteins. Thus, a role for the minor membrane proteins in viral entry is conceivable and is supported by the neutralizing capabilities of antibodies directed against the PRRSV GP4 protein.
Acknowledgments
We are grateful to Gert-Jan Godeke for technical assistance. We thank Bernard Moss and Gerd Sutter for providing the recombinant vaccinia viruses vTF7.3 and MVA-T7, respectively, and Karl-Klaus Conzelmann for making the BSR T7/5 cell line available.
REFERENCES
- 1.Buchholz, U. J., S. Finke, and K. K. Conzelmann. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cavanagh, D. 1997. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch. Virol. 142:629-633. [PubMed] [Google Scholar]
- 3.den Boon, J. A., E. J. Snijder, E. D. Chirnside, A. A. F. de Vries, M. C. Horzinek, and W. J. M. Spaan. 1991. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J. Virol. 65:2910-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Vries, A. A. F., E. D. Chirnside, P. J. Bredenbeek, L. A. Gravestein, M. C. Horzinek, and W. J. M. Spaan. 1990. All subgenomic mRNAs of equine arteritis virus contain a common leader sequence. Nucleic Acids Res. 18:3241-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Vries, A. A. F., E. D. Chirnside, M. C. Horzinek, and P. J. M. Rottier. 1992. Structural proteins of equine arteritis virus. J. Virol. 66:6294-6303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Vries, A. A. F., M. C. Horzinek, P. J. M. Rottier, and R. J. de Groot. 1997. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin. Virol. 8:33-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Vries, A. A. F., S. M. Post, M. J. B. Raamsman, M. C. Horzinek, and P. J. M. Rottier. 1995. The two major envelope proteins of equine arteritis virus associate into disulfide-linked heterodimers. J. Virol. 69:4668-4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Vries, A. A. F., M. J. B. Raamsman, H. A. van Dijk, M. C. Horzinek, and P. J. M. Rottier. 1995. The small envelope glycoprotein (GS) of equine arteritis virus folds into three distinct monomers and a disulfide-linked dimer. J. Virol. 69:3441-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Vries, A. A. F., P. J. M. Rottier, A. L. Glaser, and M. C. Horzinek. 1996. Equine viral arteritis, p. 171-200. In M. J. Studdert (ed.), Virus infections of equines. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 10.Dobbe, J. C., Y. van der Meer, W. J. M. Spaan, and E. J. Snijder. 2001. Construction of chimeric arteriviruses reveals that the ectodomain of the major glycoprotein is not the main determinant of equine arteritis virus tropism in cell culture. Virology 288:283-294. [DOI] [PubMed] [Google Scholar]
- 11.Doll, E. R., J. T. Bryans, W. H. McCollum, and M. E. W. Crowe. 1957. Isolation of a filterable agent causing arteritis of horses and abortion by mares. Its differentiation from the equine abortion (influenza) virus. Cornell Vet. 47:3-41. [PubMed] [Google Scholar]
- 12.Doms, R. W., R. A. Lamb, J. K. Rose, and A. Helenius. 1993. Folding and assembly of viral membrane proteins. Virology 193:545-562. [DOI] [PubMed] [Google Scholar]
- 13.Faaberg, K. S., and P. G. W. Plagemann. 1995. The envelope proteins of lactate dehydrogenase-elevating virus and their membrane topography. Virology 212:512-525. [DOI] [PubMed] [Google Scholar]
- 14.Faaberg, K. S., and P. G. Plagemann. 1997. ORF 3 of lactate dehydrogenase-elevating virus encodes a soluble, nonstructural, highly glycosylated, and antigenic protein. Virology 227:245-251. [DOI] [PubMed] [Google Scholar]
- 15.Fischer, F., C. F. Stegen, P. S. Masters, and W. A. Samsonoff. 1998. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 72:7885-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fujiki, Y., A. L. Hubbard, S. Fowler, and P. B. Lazarow. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93:97-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glaser, A. L., A. A. F. de Vries, M. J. B. Raamsman, M. C. Horzinek, and P. J. M. Rottier. 1999. An infectious cDNA clone of equine arteritis virus: a tool for future fundamental studies and vaccine development, p. 166-176. In U. Wernery, J. F. Wade, J. A. Mumford, and O.-R. Kaaden (ed.), Proceedings of the 8th International Conference on Equine Infectious Diseases, Dubai 1998. R & W Publications, Ltd., Newmarket, England.
- 19.Godeny, E. K., L. Zeng, S. L. Smith, and M. A. Brinton. 1995. Molecular characterization of the 3′ terminus of the simian hemorrhagic fever virus genome. J. Virol. 69:2679-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonin, P., H. Mardassi, C. A. Gagnon, B. Massie, and S. Dea. 1998. A nonstructural and antigenic glycoprotein is encoded by ORF3 of the IAF-Klop strain of porcine reproductive and respiratory syndrome virus. Arch. Virol. 143:1927-1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hedges, J. F., U. B. R. Balasuriya, and N. J. MacLachlan. 1999. The open reading frame 3 of equine arteritis virus encodes an immunogenic glycosylated, integral membrane protein. Virology 264:92-98. [DOI] [PubMed] [Google Scholar]
- 22.Horzinek, M. C., J. Maess, and R. Laufs. 1971. Studies on the substructure of togaviruses. II. Analysis of equine arteritis, rubella, bovine viral diarrhea, and hog cholera viruses. Arch. Gesamte Virusforsch. 33:306-318. [PubMed] [Google Scholar]
- 23.Hurtley, S. M., and A. Helenius. 1989. Protein oligomerization in the endoplasmic reticulum. Annu. Rev. Cell Biol. 5:277-307. [DOI] [PubMed] [Google Scholar]
- 24.Hyllseth, B. 1973. Structural proteins of equine arteritis virus. Arch. Gesamte Virusforsch. 40:177-188. [DOI] [PubMed] [Google Scholar]
- 25.Iwashita, O., and R. Harasawa. 1987. Structural polypeptides of equine arteritis virus. Nippon Juigaku Zasshi 49:923-925. [DOI] [PubMed] [Google Scholar]
- 26.Julius, D., R. Schekman, and J. Thorner. 1984. Glycosylation and processing of prepro-alpha-factor through the yeast secretory pathway. Cell 36:309-318. [DOI] [PubMed] [Google Scholar]
- 27.Katz, J. B., A. L. Shafer, K. A. Eernisse, J. G. Landgraf, and E. A. Nelson. 1995. Antigenic differences between European and American isolates of porcine reproductive and respiratory syndrome virus (PRRSV) are encoded by the carboxy-terminal portion of viral open reading frame 3. Vet. Microbiol. 44:65-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kornfeld, R., and S. Kornfeld. 1985. Assembly of asparagine-linked oligosaccharides. Annu. Rev. Biochem. 54:631-664. [DOI] [PubMed] [Google Scholar]
- 29.Le Gall, A., O. Legeay, H. Bourhy, C. Arnauld, E. Albina, and A. Jestin. 1998. Molecular variation in the nucleoprotein gene (ORF7) of the porcine reproductive and respiratory syndrome virus (PRRSV). Virus Res. 54:9-21. [DOI] [PubMed] [Google Scholar]
- 30.Magnusson, P., B. Hyllseth, and H. Marusyk. 1970. Morphological studies on equine arteritis virus. Arch. Gesamte Virusforsch. 30:105-112. [DOI] [PubMed] [Google Scholar]
- 31.Mardassi, H., P. Gonin, C. A. Gagnon, B. Massie, and S. Dea. 1998. A subset of porcine reproductive and respiratory syndrome virus GP3 glycoprotein is released into the culture medium of cells as a non-virion-associated and membrane-free (soluble) form. J. Virol. 72:6298-6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mardassi, H., B. Massie, and S. Dea. 1996. Intracellular synthesis, processing, and transport of proteins encoded by ORFs 5 to 7 of porcine reproductive and respiratory syndrome virus. Virology 221:98-112. [DOI] [PubMed] [Google Scholar]
- 33.Meulenberg, J. J. M., and A. Petersen-den Besten. 1996. Identification and characterization of a sixth structural protein of Lelystad virus: the glycoprotein GP2 encoded by ORF2 is incorporated in virus particles. Virology 225:44-51. [DOI] [PubMed] [Google Scholar]
- 34.Meulenberg, J. J. M., A. Petersen-den Besten, E. P. de Kluyver, R. J. Moormann, W. M. M. Schaaper, and G. Wensvoort. 1995. Characterization of proteins encoded by ORFs 2. to 7 of Lelystad virus. Virology 206:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meulenberg, J. J. M., A. P. van Nieuwstadt, A. van Essen-Zandbergen, and J. P. M. Langeveld. 1997. Posttranslational processing and identification of a neutralization domain of the GP4 protein encoded by ORF4 of Lelystad virus. J. Virol. 71:6061-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molenkamp, R., H. van Tol, B. C. D. Rozier, Y. van der Meer, W. J. M. Spaan, and E. J. Snijder. 2000. The arterivirus replicase is the only viral protein required for genome replication and subgenomic mRNA transcription. J. Gen. Virol. 81:2491-2496. [DOI] [PubMed] [Google Scholar]
- 37.Nugent, J., R. Sinclair, A. A. F. de Vries, R. Y. Eberhardt, J. Castillo-Olivares, N. Davis Poynter, P. J. M. Rottier, and J. A. Mumford. 2000. Development and evaluation of ELISA procedures to detect antibodies against the major envelope protein [G(L)] of equine arteritis virus. J. Virol. Methods 90:167-183. [DOI] [PubMed] [Google Scholar]
- 38.Porterfield, J. S., J. Casals, M. P. Chumakov, S. Y. Gaidamovich, C. Hannoun, I. H. Holmes, M. C. Horzinek, M. Mussgay, N. Oker-Blom, P. K. Russell, and D. W. Trent. 1978. Togaviridae. Intervirology 9:129-148. [DOI] [PubMed] [Google Scholar]
- 39.Raamsman, M. J. B., J. Krijnse Locker, A. de Hooge, A. A. F. de Vries, G. Griffiths, H. Vennema, and P. J. M. Rottier. 2000. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 74:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Snijder, E. J., and J. J. M. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961-979. [DOI] [PubMed] [Google Scholar]
- 42.Snijder, E. J., H. van Tol, K. W. Pedersen, M. J. B. Raamsman, and A. A. F. de Vries. 1999. Identification of a novel structural protein of arteriviruses. J. Virol. 73:6335-6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stewart, J. M., and J. D. Young. 1984. Solid phase peptide synthesis. Pierce Chemical Co., Rockford, Ill.
- 44.Sutter, G., M. Ohlmann, and V. Erfle. 1995. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 371:9-12. [DOI] [PubMed] [Google Scholar]
- 45.Timoney, P. J., and W. H. McCollum. 1993. Equine viral arteritis. Vet. Clin. N. Am. Equine Pract. 9:295-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Berlo, M. F., P. J. M. Rottier, W. J. M. Spaan, and M. C. Horzinek. 1986. Equine arteritis virus-induced polypeptide synthesis. J. Gen. Virol. 67:1543-1549. [DOI] [PubMed] [Google Scholar]
- 47.van Nieuwstadt, A. P., J. J. M. Meulenberg, A. van Essen-Zanbergen, A. Petersen-den Besten, R. J. Bende, R. J. M. Moormann, and G. Wensvoort. 1996. Proteins encoded by open reading frames 3 and 4 of the genome of Lelystad virus (Arteriviridae) are structural proteins of the virion. J. Virol. 70:4767-4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vennema, H., G. J. Godeke, J. W. A. Rossen, W. F. Voorhout, M. C. Horzinek, D. J. E. Opstelten, and P. J. M. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vennema, H., L. Heijnen, A. Zijderveld, M. C. Horzinek, and W. J. M. Spaan. 1990. Intracellular transport of recombinant coronavirus spike proteins: implications for virus assembly. J. Virol. 64:339-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Heijne, G. 1986. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 14:4683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiland, E., S. Bolz, F. Weiland, W. Herbst, M. J. B. Raamsman, P. J. M. Rottier, and A. A. F. de Vries. 2000. Monoclonal antibodies directed against conserved epitopes on the nucleocapsid protein and the major envelope glycoprotein of equine arteritis virus. J. Clin. Microbiol. 38:2065-2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Westaway, E. G., M. A. Brinton, S. Y. Gaidamovich, M. C. Horzinek, A. Igarashi, L. Kääriäinen, D. K. Lvov, J. S. Porterfield, P. K. Russell, and D. W. Trent. 1985. Togaviridae. Intervirology 24:125-139. [DOI] [PubMed] [Google Scholar]
- 53.Wu, W.-H., Y. Fang, R. Farwell, M. Steffen-Bien, R. R. R. Rowland, J. Christopher-Hennings, and E. A. Nelson. 2001. A 10-kDa structural protein of porcine reproductive and respiratory syndrome virus encoded by ORF2b. Virology 287:183-191. [DOI] [PubMed] [Google Scholar]
- 54.Zeegers, J. J. W., B. A. M. van der Zeijst, and M. C. Horzinek. 1976. The structural proteins of equine arteritis virus. Virology 73:200-205. [DOI] [PubMed] [Google Scholar]