Abstract
The isolation of a capsid intertypic poliovirus recombinant from a child with vaccine-associated paralytic poliomyelitis is described. Virus 31043 had a Sabin-derived type 3-type 2-type 1 recombinant genome with a 5′-end crossover point within the capsid coding region. The result was a poliovirus chimera containing the entire coding sequence for antigenic site 3a derived from the Sabin type 2 strain. The recombinant virus showed altered antigenic properties but did not acquire type 2 antigenic characteristics. The significance of the presence in nature of such poliovirus chimeras and the consequences for the current efforts to detect potentially dangerous vaccine-derived poliovirus strains are discussed in the context of the global polio eradication initiative.
Poliovirus, the agent responsible for paralytic poliomyelitis, is a human enterovirus member of the family Picornaviridae, a group of nonenveloped positive-strand RNA viruses. The coding region of the genome is preceded by a long 5′ noncoding region (NCR) of approximately 740 nucleotides that contains important determinants of virulence. The 3′ end of the genome consists of a short NCR of about 70 nucleotides that is also implicated in RNA replication (33). The coding region is translated as a single polyprotein and is then processed to generate the viral capsid (VP1 to VP4) and the nonstructural proteins. The antigenic determinants for poliovirus-neutralizing antibodies are located on surface-exposed loops in the capsid proteins VP1, VP2, and VP3. Four main antigenic sites have been identified by the use of murine monoclonal antibodies (MAbs) (30).
Genomic RNA recombination is a common event during poliovirus evolution. The oral poliovaccine (OPV) consists of live attenuated strains of the three polio serotypes, which replicate in the guts of immunocompetent vaccinees for periods that range between several weeks and 3 months. A high proportion of type 2 and type 3 polio isolates excreted by healthy vaccinees and patients with vaccine-associated paralytic poliomyelitis (VAPP), have been found to be intertypic recombinants (10, 16, 32). Sabin type 1 (Sabin 1) intertypic recombinants are rare but do occur, and indeed, type 1 sequences encoding nonstructural proteins are frequently found in type 2 and type 3 recombinant strains (10). Natural poliovirus recombinants containing different combinations of vaccine and wild polio and/or nonpolio enterovirus sequences have also been described (1, 14, 16, 20, 25, 41). Of great significance are the recent reports of vaccine-derived poliovirus strains that have been associated with outbreaks of poliomyelitis in Egypt, the island of Hispaniola (Dominican Republic and Haiti), and the Philippines. In all three cases the epidemic strains have been identified as recombinant polioviruses containing sequences of unidentified enteroviral origin spanning most of the nonstructural genomic region (1, 7, 20).
Recombination junctions have almost invariably been found in genomic regions coding for nonstructural proteins. There are, however, at least two known exceptions in the literature. A type 2 vaccine-wild recombinant poliovirus isolated from a child with VAPP in Romania had a genome with a central capsid core of vaccine origin flanked by two units of nonvaccine origin from an unidentified enterovirus (15). The crossover point in the 5′ end involved a short sequence in the capsid region. More recently, evidence has been found that type 1 poliovirus strains isolated from poliomyelitis patients in China from 1991 to 1993 contained a 367-nucleotide block of sequence derived from the Sabin 1 OPV strain spanning the 3′ end of VP1 and the 5′ half of the protease 2A inserted in a wild type 1 poliovirus genomic background (25). Neither of those two recombination events resulted in amino acid changes in the capsid proteins with respect to their parental strains. Intertypic poliovirus recombinants with crossovers in the capsid region seem, therefore, to be very rare in nature, possibly because it is important to maintain the integrity of the capsid shell.
Intertypic capsid recombinants involving genomic sequence exchanges corresponding to the four polio antigenic sites among the three Sabin strains have been readily generated by DNA recombination techniques (6, 27, 34, 35, 37, 38). Furthermore, a number of foreign amino acid sequences have been introduced into the VP1 BC loop of the Mahoney strain, resulting in viable laboratory viruses (11). Many of these viruses have poor growth characteristics in cell culture, and early reports suggested that certain in vitro intertypic recombinant chimeras were nonviable when recombination junctions were located in the capsid coding region (21, 44). Similarly, a laboratory intratypic recombinant with a recombination junction in the capsid coding region was also shown to be unstable compared to its parental strains (22).
Here, we report, to the best of our knowledge, the first poliovirus capsid chimera isolated in nature in which the recombination event has introduced novel amino acids. The virus was isolated in Belarus from a child with vaccine-associated paralysis. In recent years, the vaccination rate for children under 1 year of age in Belarus was >98%. The last indigenous wild poliovirus was isolated in 1964, and the last imported wild virus was reported in 1986 (World Health Organization, presented at the thirteenth meeting of the Regional Commission for the Certification of Poliomyelitis Eradication, Copenhagen, Denmark, 13 to 15 March 2002).
The results are discussed in the context of the global polio eradication effort with an emphasis on the risks posed by vaccine-derived strains and the implications for the design of efficient vaccination and surveillance strategies to interrupt polio immunization after eradication.
MATERIALS AND METHODS
Case history.
The patient was a boy born on 22 November 1998 in Belarus. The child suffered from postnatal asphyxia due to intrauterine hypoxia of the fetus and was followed up by a neuropathologist because of the risk of his developing perinatal encephalopathy. At the age of 2 months, he had acute rhinitis; from 26 March 1999 to 2 April 1999, he was in the hospital with the diagnosis of acute bronchitis. He was immunized with OPV on 30 April 1999. On 2 May 1999, he had fever and cough. From 4 May 1999 to 17 May 1999, the child was treated in the hospital for tracheobronchitis. He was discharged from the hospital in a healthy state. On 19 May 1999, he showed sudden fever and a slight cough. Two days later, his mother noticed that the child could not lean on his right leg. Neurological examination confirmed that active movements were limited in the right leg, which was bent at the knee joint. In the standing position, diffuse atrophy of the muscles of the calf was observed. Reflex in the right knee was scarcely seen, but abdominal and Achilles reflexes were maintained. Serological investigation on 15 February 2000 confirmed normal levels of immunoglobulins (7.8 g of immunoglobulin G [IgG]/liter, 0.58 g of IgA/liter, and 1.9 g of IgM/liter). On 28 May 1999, 7 days after the onset of paralysis, the titers of poliovirus-neutralizing antibodies were 1:512, 1:192, and 1:24 for types 1, 2, and 3, respectively. Fecal specimens for virological examination were obtained on 27 May and 1 June 1999, 6 and 11 days after the onset of paralysis, respectively.
Virus isolation.
The fecal specimens were processed at the National Polio Laboratory in Belarus according to standard protocols for virus isolation and characterization (47). Virological investigation identified polioviruses of serotypes 1, 2, and 3 in stool samples from 27 May 1999 and 28 May 1999. Viruses of individual serotypes were purified by neutralization with different combinations of polyclonal sera against each of the three poliovirus serotypes. Working virus preparations were collected by growth of the viruses in HEp-2C cells at 35°C in minimal essential medium without fetal calf serum. Poliovirus vaccine strains Sabin 1, Sabin 2, and Sabin 3 and the wild type 3 strain Leon/37 were used as references in our experiments. Virus stocks were stored at −70°C.
ITD test.
Intratypic differentiation (ITD) assays are used to investigate the wild or vaccine origin of poliovirus field isolates. A MAb-based neutralization assay was used. This method is currently used at the National Institute for Biological Standards and Control in its activities as a World Health Organization (WHO) Global Specialized Laboratory of the Polio Laboratory Network to monitor poliovirus field isolates sent from Ireland and the United Kingdom National Laboratories as part of the polio-free certification process in both countries. The method was validated in a WHO collaborative study (46). Neutralization with a mixture of Sabin-specific MAbs of the three poliovirus serotypes (National Institute for Biological Standards and Control numbers 955, 1233, and 889 for poliovirus serotype 1 [PV1], PV2, and PV3, respectively) was carried out as an additional typing reaction when isolates were serotyped in the standard neutralizing antibody test (47). A 1:100 dilution of the trivalent MAb ascites mixture was used in the assay. If an isolate is neutralized by the MAb mixture, it is identified as Sabin-like. If an isolate is not neutralized, it is either a Sabin-derived strain with antigenic drift at the epitope recognized by the MAb or a wild-type virus. To distinguish between these two possibilities, genomic ITD tests are required. When necessary, the nucleotide sequence of the entire genomic region coding for the VP1 capsid protein plus a short fragment of the protease 2A (nucleotides ∼2,450 to 3,500) of the relevant poliovirus isolate was determined.
RT, PCR, and nucleotide sequencing of poliovirus genomes.
Poliovirus RNA was purified from HEp-2c cell culture supernatants and used for reverse transcription (RT) and PCRs using standard procedures. PCR restriction fragment length polymorphism (PCR-RFLP) assays were performed similarly to the original method (3). RT-PCR products (1/10 of the total volume) were incubated with the corresponding restriction enzyme under the conditions recommended by the manufacturer. The digestion products were analyzed by agarose gel electrophoresis.
Sequencing of the purified viral RT-PCR DNA products was carried out using the ABI Prism 310 Genetic Analyzer as specified by the manufacturer. Primers were designed by the “primer-walking” strategy. Sequence data were stored as standard chromatogram format (∗.scf) files and analyzed using the Wisconsin Package version 10.0-UNIX (Genetics Computer Group) and AlignIRV11 (LI-COR) software.
Antigenic characterization.
The antigenic properties of virus 31043 were studied by a microneutralization assay using poliovirus Sabin-specific MAbs corresponding to antigenic sites 1 to 4 as described before (36). One hundred 50% cell culture infective doses (CCID50) of the challenge virus were used in the test. Polyclonal sera specific for each polio serotype were also included in the neutralization assay. Monoclonal and polyclonal sera were used at 1:50 and 1:200 dilutions.
Temperature sensitivity.
Temperature sensitivity was assayed by comparison of plaque formation on HEp-2c cells at 35.0, 39.5, and 40.0°C as described before (31).
Neurovirulence in transgenic mice.
Tg21-Bx transgenic mice expressing the human poliovirus receptor were used for these experiments. Tg21-Bx mice are the product of crossing TgPVR21 mice (23) with BALB/c mice followed by repeated backcrossing of the offspring with BALB/c mice, interbreeding, and selection by PCR screening of tail DNA. The mice are homozygous for PVR and class II IA-β genes (H2d). The mice were inoculated intramuscularly (in the left hind limb) with 50 μl of 10-fold viral dilutions, and the daily clinical score was monitored for 14 days. The probit method was used to calculate the 50% paralytic dose (PD50).
Titration of human sera for poliovirus-neutralizing antibodies.
The procedure for titration of human sera for poliovirus-neutralizing antibodies was as recommended by WHO (47). The serum antibody titer was considered to be the highest dilution of serum that protected 50% of the cultures against 100 CCID50 of challenge virus. Antibody titers were expressed as reciprocals of that dilution. The challenge virus dose preparation ±100 CCID50 (50 to 200) was confirmed using the Karber formula. The statistical comparison of the titers was carried out using the Student paired t test on the log2 titers and confirmed with the Wilcoxon signed-rank test.
RESULTS
ITD.
Poliovirus strain 31043 was part of a collection of poliovirus isolates from VAPP patients in Belarus analyzed to investigate their genetic and antigenic drift from the Sabin vaccine strain. The virus was obtained from a 6-month-old boy 27 days after OPV immunization and 6 days after the onset of paralysis. This strain was characterized as a type 3 non-Sabin-like poliovirus, since it was not neutralized by Sabin-specific MAbs in the ITD neutralization assay described above but was neutralized by polyclonal serum specific for type 3 and not with polyclonal sera against type 1 and type 2 polioviruses (data not shown). Virus 31043 was therefore selected for further molecular and phenotypic analyses.
Nucleotide sequence analysis.
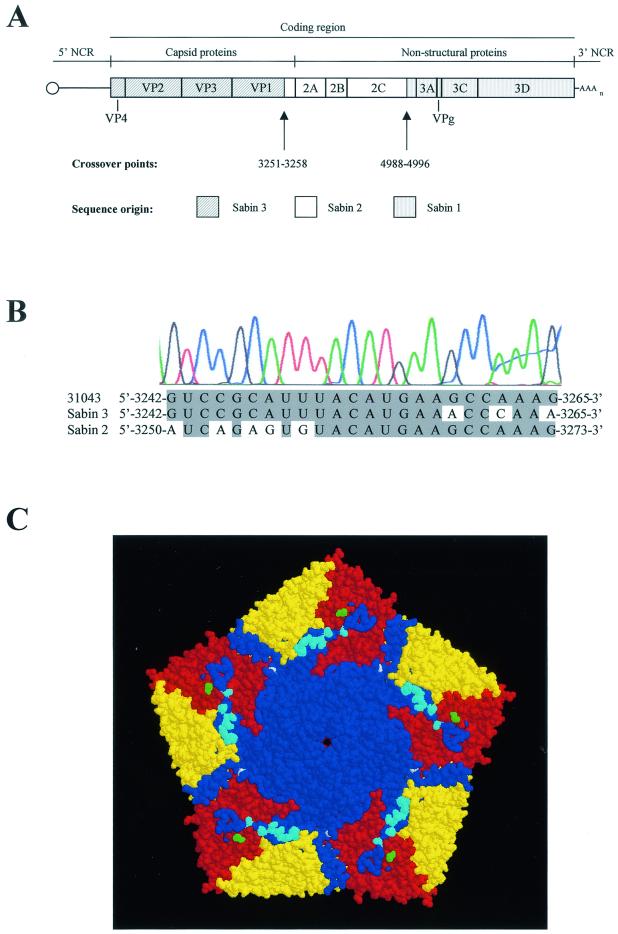
The genomic sequence of strain 31043 was initially determined between nucleotides 2477 and 3450, which includes the entire coding region for VP1 capsid protein and part of that for protease 2A. The sequence analysis revealed an uncommon genomic intertypic (type 3-type 2) recombinant structure with a crossover junction within the capsid coding region (see Fig. 2A and B). This recombination event resulted in the insertion of a 120-nucleotide sequence in the 3′ end of the VP1 coding region from the Sabin 2 strain in a Sabin 3 genomic background, effectively resulting in six amino acid changes at positions VP1-279, VP1-286, VP1-287, VP1-288, VP1-290, and VP1-293. All six amino acids were located on the surface of the virion and comprised the entire antigenic site 3a and residues implicated in receptor binding (see Fig. 2C) (4, 18, 30). Further nucleotide sequence analysis revealed a Sabin-derived type 3-type 2-type 1 tripartite genomic structure with crossover points located at nucleotides 3251 to 3258 and 4988 to 4996 (Fig. 2A). The genome of virus 31043 (sequenced from nucleotide 48 to the end) contained 19 nucleotide mutations with respect to the corresponding Sabin sequences for each region, summarized in Table 1. Mutations at nucleotide 472 in domain V of the 5′ NCR and at 6194 (6203 in Sabin 1) in the codon for amino acid 3D-73 were direct reversions to sequences present in the Sabin 3 and Sabin 1 wild parental viruses, the Leon/37 and Mahoney strains, respectively. Reversions at these positions are commonly observed in isolates from healthy vaccinees and VAPP patients and have been associated with the attenuation phenotype of both vaccine strains (32). An additional mutation was found in domain V of the 5′ NCR at nucleotide 510 that resulted in the weakening of the G-C predicted base pair between nucleotides 488 and 510 (43). Mutations were also found at capsid amino acid VP1-232 at the protomer interface; at VP2-30, which is located next to the seven-stranded β sheet that forms part of the interface between the pentameric subunits; and at residue VP3-59, which is part of antigenic site 3b (12, 30).
FIG. 2.
Structure of virus 31043. (A) Genomic structure and localization of crossover points (Sabin 3 numbering). (B) Nucleotide sequence alignment of 31043, Sabin 2, and Sabin 3 genomes at the capsid crossover junction. An image of the actual sequence chromatogram of virus 31043 is shown. Common nucleotides are shaded. (C) Space-filled diagram of the three-dimensional structure of Sabin 3 virus viewed from the outside of the virion (12). A pentameric unit is represented. The virus particle consists of 60 protomers. Each protomer contains a single copy of VP1 (blue), VP2 (yellow), VP3 (red), and VP4 (internal) arranged in icosahedral symmetry. Sabin 2-derived amino acid sequences are represented in cyan, and single amino acid mutations with respect to the Sabin 3 strain are shown in white (VP1-232) or green (VP3-59). The image was generated using RasWin Molecular Graphics version 2.4 software (42).
TABLE 1.
Nucleotide and amino acid changes between virus 31043 and Sabin strainsa
| Origin | Region | Nucleotide
|
Amino acid
|
||||
|---|---|---|---|---|---|---|---|
| Position | Sabin | 31043 | Position | Sabin | 31043 | ||
| Sabin 3 | 5′ NCR | 472 | U | C | |||
| 510 | C | U | |||||
| 610 | U | C | |||||
| 625 | U | C | |||||
| VP2 | 1037 | A | G | 30 | Asn | Asp | |
| VP3 | 1938 | G | A | 59 | Ser | Asn | |
| 2281 | U | C | |||||
| VP1 | 2479 | U | C | ||||
| 3170 | A | G | 232 | Thr | Ala | ||
| 3175 | U | C | |||||
| 3238 | C | U | |||||
| Sabin 2 | 2A | 3670 (3678) | U | C | |||
| 3676 (3684) | U | C | |||||
| 3A | 5119 (5127) | A | G | ||||
| 2C | 4120 (4128) | U | C | ||||
| 4342 (4350) | A | G | |||||
| Sabin 1 | 3D | 6194 (6203) | C | U | 73 | His | Tyr |
| 6211 (6220) | A | G | |||||
| 6725 (6734) | G | A | 249 | Glu | Lys | ||
The nucleotide position in the original Sabin strain is indicated in parentheses when different from 31043.
Additional molecular analyses.
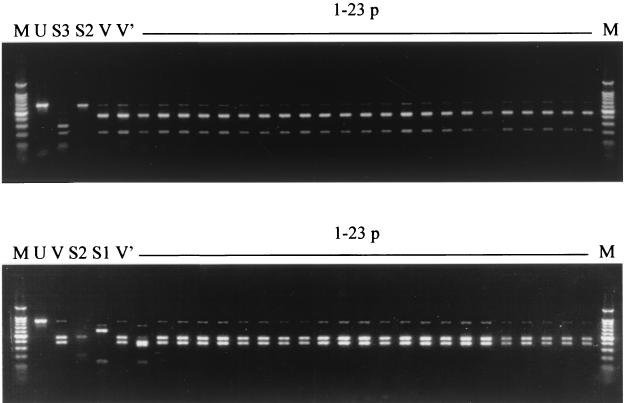
To exclude the possibility that strain 31043 was a mixture of Sabin 1-, 2-, and 3-derived viruses, a plaque purification assay was carried out; 23 virus plaques were purified and analyzed by PCR-RFLP. Two sets of PCR fragments that included the sequences of either of the two crossover junctions were used. As shown in Fig. 1, all 23 plaques gave restriction band patterns identical to those of the original isolate and different from those of the Sabin reference strains with the exception of plaque 1, which gave a different restriction pattern with AluI. On further analysis, a single synonymous mutation (C to U) was identified at position 4555 (Sabin 3 numbering) in plaque 1 with respect to strain 31043, which introduced an additional AluI recognition site. The restriction band patterns shown by strains 31043 and 31044 and plaques 1 to 23 were compatible with what was expected from the nucleotide sequence of strain 31043 described above. Both RT-PCR products of five randomly selected plaques were sequenced, and a type 3-type 2-type 1 recombinant structure identical to that of strain 31043 was confirmed. Strain 31044, a second type 3 poliovirus obtained from a stool sample from the same patient 1 day after that of strain 31043, was also analyzed. The restriction band patterns (Fig. 1, lane V′) were identical to those of the previous strain, 31043. Partial nucleotide sequencing through the crossover junctions revealed the same genomic structure of strain 31043 shown in Fig. 2. Taken together, these results strongly indicate that viral recombination took place in the patient. The possibility that this recombinant virus was generated during its passage in cell culture is very unlikely but cannot be completely ruled out.
FIG. 1.
Ethidium bromide-stained 2% agarose gels showing the results of PCR-RFLP analysis of poliovirus strains 31043 and 31044 and 23 plaque isolates of strain 31043 compared with Sabin 1, 2, and 3 reference strains. The upper gel shows PCR products spanning from nucleotide 2843 to 3513 (Sabin 3 numbering) digested with DdeI. The bottom gel shows PCR products spanning from nucleotide 4427 to 5268 (Sabin 3 numbering) digested with AluI. Lanes: U, example of an undigested PCR product; S1, Sabin 1 reference strain; S2, Sabin 2 reference strain; S3, Sabin 3 reference strain; V, strain 31043; V′, strain 31044; 1-23 p, plaque isolates of strain 31043; M, 100-bp DNA ladder (New England Biolabs).
Antigenic properties.
The antigenic properties of virus 31043 were determined by studying its reactivity with a panel of monoclonal and polyclonal sera of known specificities. The results are shown in Table 2. Virus 31043 failed to react with all nine Sabin 3 MAbs against antigenic site 3, which was consistent with the observed amino acid changes at the predefined antigenic site (30). In addition, isolate 31043 also lost reactivity with two of the five Sabin 3 MAbs specific for antigenic site 2 that were tested. Sabin 3 MAbs against sites 1 and 4 neutralized 31043 virus infectivity. Virus 31043 reacted with polyclonal serum raised against type 3 poliovirus and not with type 1 or 2 polyclonal sera even when high concentrations of sera (dilution, 1:50) were employed. Virus 31043 did not react with Sabin 2-specific MAbs (data not shown).
TABLE 2.
Reactivity of virus 31043 with polio monoclonal and polyclonal sera
| Virus | Sabin 3-specific MAbs (no. of neutralizing antibodies/total no. of antibodies)
|
Polyclonal seraa
|
|||||
|---|---|---|---|---|---|---|---|
| Site 1 | Site 2 | Site 3 | Site 4 | PV1 | PV2 | PV3 | |
| Sabin 3 | 9/9 | 5/5 | 9/9 | 1/1 | − | − | + |
| 31043 | 9/9 | 3/5 | 0/9 | 1/1 | − | − | + |
| Sabin 2 | 0/9 | 0/5 | 0/9 | 0/1 | − | + | − |
+, neutralization; −, no neutralization.
Temperature sensitivity and neurovirulence.
The results for the temperature sensitivity and neurovirulence of isolate 31043 are shown in Table 3. Virus 31043 lost the temperature-sensitive phenotype of Sabin at 39.5°C but showed some degree of temperature sensitivity for growth in HEp-2c cells at 40°C with respect to the Sabin 3 wild parental virus, the Leon/37 strain. Similarly, virus 31043 partially regained the levels of neurovirulence characteristic of the Leon/37 strain as determined by PD50 measurements in Tg21-Bx mice expressing the human poliovirus receptor.
TABLE 3.
Temperature sensitivity and neurovirulence of virus 31043
| Virus | Log titer reduction
|
Neurovirulence in TgPVR21 mice
|
|||
|---|---|---|---|---|---|
| 35-39.5°C | 35-40.0°C | Dose (log CCID50/ml) | No. of clinically affected mice/total no. | PD50a | |
| Sabin 3 | 3.4 | >5.0 | 7.7 | 0/12 | >7.7 |
| 31043 | 0.5 | 2.0 | 8.0 | 9/10 | 7.3 (7.1-7.6) |
| 7.5 | 6/10 | ||||
| 6.5 | 3/10 | ||||
| 5.5 | 0/10 | ||||
| Leon | 0.2 | 0.5 | 6.5 | 6/6 | 5.7 (5.3-6.4) |
| 5.5 | 2/6 | ||||
| 4.5 | 1/6 | ||||
| 3.5 | 0/6 | ||||
Estimates of the PD50 with associated 95% confidence intervals were calculated by the probit method.
Neutralization assays with human sera.
In order to assess the potential risks of virus 31043 spreading in the population, the titers of neutralizing antibodies against strain 31043 in the sera of 154 individuals from Belarus were determined and compared to that against the Sabin 3 vaccine strain for each individual serum. The sample included individuals of both sexes, from three different geographical regions, and with ages ranging between 0 and 65 years old. The vaccination history was known for 74 individuals. All of them, except for one, had been given three or more OPV immunizations. No significant differences in polio antibody levels were observed between these individuals and those with no vaccination records (data not shown). Figure 3A shows the relationship between the neutralization titers against poliovirus strains Sabin 3 and 31043 in sera from the 154 individuals. The neutralization titers against the recombinant virus 31043 were significantly lower than those obtained against the Sabin 3 vaccine strain when compared for each individual separately (P < 0.001). As shown in Fig. 2B, the proportion of sera with antibody responses of <1:8 against virus 31043 was 26.0%, whereas only 5.8% of the sera showed neutralization activities of <1:8 against Sabin 3 virus. The geometric mean neutralization titers against Sabin 3 and virus 31043 expressed as the reciprocals of log2 dilution values were 4.17 and 3.40, respectively.
FIG. 3.
In vitro neutralization titers against the recombinant virus 31043 and Sabin 3 in sera from 154 individuals from Belarus. (A) The data are represented as the ratio between the neutralization titer against virus 31043 and that against the Sabin 3 strain for each individual serum. (B) Neutralization titers against each virus classified by the number of individual sera in each group. The geometric mean neutralization titer against each virus and the corresponding standard error of the mean are shown.
DISCUSSION
In this paper, we describe intertypic recombination in the capsid coding region of poliovirus as a novel mechanism to generate vaccine-derived variants with altered antigenic properties previously unknown in natural poliovirus isolates. Virus 31043 was identified as a Sabin-derived type 3-type 2-type 1 poliovirus recombinant with crossover junctions at nucleotides 3251 to 3258 (within the VP1 coding region) and 4988 to 4996 (within the coding region for nonstructural protein 2C).
The recombinant virus lost the attenuated and temperature-sensitive phenotypes characteristic of Sabin 3 to a significant extent. This was most likely due to direct reversion of the change at nucleotide 472 in domain V of the 5′ NCR present in the Sabin 3 strain, which affects protein translation, and to secondary site mutations at capsid residues VP2-30 and/or VP1-232, located at pentameric and protomeric interfaces in the virion, respectively. Either of these two mutations alone or both in combination may have reversed the effect of the mutation at VP3-91 in Sabin 3, which is known to have an effect on capsid assembly and which, together with the mutation at 472, is the main determinant of the attenuated and temperature-sensitive phenotypes of Sabin 3 (32). Similar secondary site mutations in capsid residues have been described in isolates from healthy vaccinees and VAPP patients (26). The recombinant virus also had a direct reversion at nucleotide 6194 (6203 in Sabin 1) in the codon for amino acid 3D-73 that has also been associated with the temperature sensitivity and attenuation phenotypes of the Sabin 1 strain (5, 8, 29, 45).
As expected, virus 31043 failed to react with MAbs specific for Sabin 3 antigenic site 3. The recombinant strain also failed to react with some site 2 antibodies, and as no mutations were found in sequences responsible for this antigenic site, it is possible that insertion of the type 2 sequence of antigenic site 3a in the capsid of virus 31043 could have had some effect on the conformation of antigenic site 2 of this virus. Single amino acid mutations in antigenic site 3 have been shown to have a partial effect on the conformation of antigenic site 2 of type 3 and type 1 Sabin mutant strains (39, 40). Virus 31043 did not express type 2 antigenic properties, as its infectivity was not neutralized by monoclonal and polyclonal sera specific for the Sabin type 2 strain. This is somewhat in agreement with the properties that have been described for Sabin intertypic chimeras generated by DNA recombination technology. The experiments have shown that whereas antigenic site 1, and to some extent antigenic site 2, could be expressed in infectious poliovirus chimeras of a different serotype, antigenic site 3 was not recreated in its native conformation (6, 27, 34, 35, 37, 38). Poliovirus antigenic variation is thought to be restricted by structural constraints and the involvement of neutralization sites in receptor binding (4, 9, 17, 18), which has been proposed as a possible explanation of why poliovirus exists in only three serotypes (17).
Significant differences were found in the neutralization activities of sera from 154 individuals in Belarus in that the recombinant virus 31043 was neutralized less efficiently than Sabin 3 vaccine virus, confirming the differences between the two strains found in some neutralizing epitopes. The biological consequences of those differences in terms of the potential of such a strain to spread and cause disease in the population are not easy to assess (13). It is generally accepted that routine and mass vaccinations with OPV under optimal conditions can interrupt the circulation of any poliovirus strain in human populations. This assumption is the result of many years of intensive vaccination campaigns and surveillance for poliovirus transmission and has been the basis for the highly successful program of poliomyelitis eradication (2). However, the possibility that antigenic poliovirus variants such as strain 31043 could establish widespread infections in suboptimally vaccinated populations cannot be excluded. Low levels of protective antibodies against poliovirus in the population as a consequence of inadequate vaccination and the atypical antigenic properties of epidemic strains were identified as key factors in the establishment of outbreaks of type 3 poliomyelitis in Poland (1968) (28) and Finland (1984) (19). The outbreak in Poland involved poliovirus strains derived from the USOL-D-bac vaccine virus, which was used in a small trial. In both cases, the spread of the disease was interrupted by mass vaccination campaigns with OPV and improved routine vaccine coverage (19, 24). Following the case described here, two more VAPP cases were identified in Belarus, where polio 3 virus was isolated, but neither of them was temporally or geographically associated with this case. Both type 3 isolates were classified as Sabin-like by ITD antibody neutralization assays and molecular characterization (E. Samoilovich, unpublished results).
Intriguingly, it has only recently become clear, after 40 years of intense vaccination campaigns with Sabin OPV, that evolved vaccine-derived strains can, in some circumstances, be responsible for poliomyelitis outbreaks. Sabin-derived type 2 strains circulated in Egypt from 1982 to 1993 and were responsible for at least 32 poliomyelitis cases (7). More recently, in 2000 and 2001, two unrelated outbreaks of type 1 poliomyelitis on the island of Hispaniola (Dominican Republic and Haiti) (20) and in the Philippines (1) have been linked to Sabin 1-derived strains. All three Sabin-derived outbreaks occurred in populations where vaccination levels had dropped significantly. As in Poland and Finland, the transmission of the disease was interrupted by improving the vaccination coverage (1, 7, 20).
Isolates from the outbreaks in Egypt, Hispaniola, and the Philippines have been identified as recombinant polioviruses containing sequences of unidentified enteroviral origin spanning most of the nonstructural genomic region (1, 20). The viruses also contained vaccine-derived capsid sequences with 2 to 3% nucleotide sequence divergence from Sabin, which was evidence of an estimated 2 to 3 years of sustained circulation in humans (1, 20). It is believed that at some point in their evolution these vaccine-derived polioviruses reverted to virulence and transmissibility characteristics typical of wild poliovirus strains (20).
The possible implications of poliovirus capsid recombination events for the generation of potentially dangerous vaccine-derived strains are not clear at present. In view of the findings presented here, the possibility of poliovirus chimeras containing vaccine-derived, wild, and even nonpoliovirus capsid sequences that could result in more dramatic antigenic modifications, although less likely, cannot be completely ruled out.
The results presented here confirm the importance of using two different ITD methods as recommended by WHO, one based on viral nucleotide sequence differences and another based on viral antigenic properties (46, 47).
REFERENCES
- 1.Anonymous. 2001. Acute flaccid paralysis associated with circulating vaccine-derived poliovirus—Philippines, 2001. Morb. Mortal. Wkly. Rep. 50:874-875. [PubMed] [Google Scholar]
- 2.Aylward, R. B., H. F. Hull, S. L. Cochi, R. W. Sutter, J. M. Olive, and B. Melgaard. 2000. Disease eradication as a public health strategy: a case study of poliomyelitis eradication. Bull. W. H. O. 78:285-297. [PMC free article] [PubMed] [Google Scholar]
- 3.Balanant, J. G. S., A. Candrea, F. Delpeyroux, and R. Crainic. 1991. The natural genomic variability of poliovirus analyzed by a restriction fragment length polymorphism assay. Virology 184:645-654. [DOI] [PubMed] [Google Scholar]
- 4.Belnap, D. M., B. M. McDermott, Jr., D. J. Filman, N. Cheng, B. L. Trus, H. J. Zuccola, V. R. Racaniello, J. M. Hogle, and A. C. Steven. 2000. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl. Acad. Sci. USA 97:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouchard, M. J., D. H. Lam, and V. R. Racaniello. 1995. Determinants of attenuation and temperature sensitivity in the type 1 poliovirus Sabin vaccine. J. Virol. 69:4972-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burke, K. L., G. Dunn, M. Ferguson, P. D. Minor, and J. W. Almond. 1988. Antigen chimaeras of poliovirus as potential new vaccines. Nature 332:81-82. [DOI] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2001. Circulation of a type 2 vaccine-derived poliovirus—Egypt, 1982-1993. JAMA 285:1148-1149. [PubMed] [Google Scholar]
- 8.Christodoulou, C., I. Pelletier, and F. Colbere-Garapin. 1989. Genetic stability of poliovirus insertion mutants with a foreign oligopeptide on the capsid surface. Res. Virol. 140:501-509. [DOI] [PubMed] [Google Scholar]
- 9.Colston, E. M., and V. R. Racaniello. 1995. Poliovirus variants selected on mutant receptor-expressing cells identify capsid residues that expand receptor recognition. J. Virol. 69:4823-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuervo, N. S., S. Guillot, N. Romanenkova, M. Combiescu, A. Aubert-Combiescu, M. Seghier, V. Caro, R. Crainic, and F. Delpeyroux. 2001. Genomic features of intertypic recombinant Sabin poliovirus strains excreted by primary vaccinees. J. Virol. 75:5740-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans, D. J., and J. W. Almond. 1991. Design, construction, and characterization of poliovirus antigen chimeras. Methods Enzymol. 203:386-400. [DOI] [PubMed] [Google Scholar]
- 12.Filman, D. J., R. Syed, M. Chow, A. J. Macadam, P. D. Minor, and J. M. Hogle. 1989. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 8:1567-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fine, P. E., and I. A. Carneiro. 1999. Transmissibility and persistence of oral polio vaccine viruses: implications for the global poliomyelitis eradication initiative. Am. J. Epidemiol. 150:1001-1021. [DOI] [PubMed] [Google Scholar]
- 14.Friedrich, F., E. E. Da Silva, and H. G. Schatzmayr. 1996. Type 2 poliovirus recombinants isolated from vaccine-associated cases and from healthy contacts in Brazil. Acta Virol. 40:27-33. [PubMed] [Google Scholar]
- 15.Georgescu, M. M., F. Delpeyroux, and R. Crainic. 1995. Tripartite genome organization of a natural type 2 vaccine/nonvaccine recombinant poliovirus. J. Gen. Virol. 76:2343-2348. [DOI] [PubMed] [Google Scholar]
- 16.Guillot, S., V. Caro, N. Cuervo, E. Korotkova, M. Combiescu, A. Persu, A. Aubert-Combiescu, F. Delpeyroux, and R. Crainic. 2000. Natural genetic exchanges between vaccine and wild poliovirus strains in humans. J. Virol. 74:8434-8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harber, J., G. Bernhardt, H. H. Lu, J. Y. Sgro, and E. Wimmer. 1995. Canyon rim residues, including antigenic determinants, modulate serotype-specific binding of polioviruses to mutants of the poliovirus receptor. Virology 214:559-570. [DOI] [PubMed] [Google Scholar]
- 18.He, Y., V. D. Bowman, S. Mueller, C. M. Bator, J. Bella, X. Peng, T. S. Baker, E. Wimmer, R. J. Kuhn, and M. G. Rossmann. 2000. Interaction of the poliovirus receptor with poliovirus. Proc. Natl. Acad. Sci. USA 97:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hovi, T., K. Cantell, A. Huovilainen, E. Kinnunen, T. Kuronen, K. Lapinleimu, T. Poyry, M. Roivainen, N. Salama, M. Stenvik, et al. 1986. Outbreak of paralytic poliomyelitis in Finland: widespread circulation of antigenically altered poliovirus type 3 in a vaccinated population. Lancet i:1427-1432. [DOI] [PubMed]
- 20.Kew, O., V. Morris-Glasgow, M. Landaverde, C. Burns, J. Shaw, Z. Garib, J. Andre, E. Blackman, C. J. Freeman, J. Jorba, R. Sutter, G. Tambini, L. Venczel, C. Pedreira, F. Laender, H. Shimizu, T. Yoneyama, T. Miyamura, H. van Der Avoort, M. S. Oberste, D. Kilpatrick, S. Cochi, M. Pallansch, and C. de Quadros. 2002. Outbreak of poliomyelitis in Hispaniola associated with circulating type 1 vaccine-derived poliovirus. Science 296:356-359. [DOI] [PubMed] [Google Scholar]
- 21.Kohara, M., S. Abe, T. Komatsu, K. Tago, M. Arita, and A. Nomoto. 1988. A recombinant virus between the Sabin 1 and Sabin 3 vaccine strains of poliovirus as a possible candidate for a new type 3 poliovirus live vaccine strain. J. Virol. 62:2828-2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kohara, M., T. Omata, A. Kameda, B. L. Semler, H. Itoh, E. Wimmer, and A. Nomoto. 1985. In vitro phenotypic markers of a poliovirus recombinant constructed from infectious cDNA clones of the neurovirulent Mahoney strain and the attenuated Sabin 1 strain. J. Virol. 53:786-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koike, S., C. Taya, J. Aoki, Y. Matsuda, I. Ise, H. Takeda, T. Matsuzaki, H. Amanuma, H. Yonekawa, and A. Nomoto. 1994. Characterization of three different transgenic mouse lines that carry human poliovirus receptor gene—influence of the transgene expression on pathogenesis. Arch. Virol. 139:351-363. [DOI] [PubMed] [Google Scholar]
- 24.Kostrzewski, J., A. Kulesza, and A. Abgarowicz. 1970. The epidemic of type 3 poliomyelitis in Poland in 1968. Epidemiol. Rev. 24:89-103. [PubMed] [Google Scholar]
- 25.Liu, H. M., D. P. Zheng, L. B. Zhang, M. S. Oberste, M. A. Pallansch, and O. M. Kew. 2000. Molecular evolution of a type 1 wild-vaccine poliovirus recombinant during widespread circulation in China. J. Virol. 74:11153-11161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Macadam, A. J., C. Arnold, J. Howlett, A. John, S. Marsden, F. Taffs, P. Reeve, N. Hamada, K. Wareham, J. Almond, et al. 1989. Reversion of the attenuated and temperature-sensitive phenotypes of the Sabin type 3 strain of poliovirus in vaccinees. Virology 172:408-414. [DOI] [PubMed] [Google Scholar]
- 27.Martin, A., C. Wychowski, T. Couderc, R. Crainic, J. Hogle, and M. Girard. 1988. Engineering a poliovirus type 2 antigenic site on a type 1 capsid results in a chimaeric virus which is neurovirulent for mice. EMBO J. 7:2839-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin, J., G. L. Ferguson, D. J. Wood, and P. D. Minor. 2000. The vaccine origin of the 1968 epidemic of type 3 poliomyelitis in Poland. Virology 278:42-49. [DOI] [PubMed] [Google Scholar]
- 29.McGoldrick, A., A. J. Macadam, G. Dunn, A. Rowe, J. Burlison, P. D. Minor, J. Meredith, D. J. Evans, and J. W. Almond. 1995. Role of mutations G-480 and C-6203 in the attenuation phenotype of Sabin type 1 poliovirus. J. Virol. 69:7601-7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minor, P. D. 1990. Antigenic structure of picornaviruses. Curr. Top. Microbiol. Immunol. 161:121-154. [DOI] [PubMed] [Google Scholar]
- 31.Minor, P. D. 1980. Comparative biochemical studies of type 3 poliovirus. J. Virol. 34:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Minor, P. D. 1992. The molecular biology of polio vaccines. J. Gen. Virol. 73:3065-3077. [DOI] [PubMed] [Google Scholar]
- 33.Minor, P. D. 1997. Poliovirus, p. 555-577. In N. Nathason (ed.), Viral pathogenesis. Lippincott-Raven Publishers, Philadelphia, Pa.
- 34.Minor, P. D., M. Ferguson, K. Katrak, D. Wood, A. John, J. Howlett, G. Dunn, K. Burke, and J. W. Almond. 1990. Antigenic structure of chimeras of type 1 and type 3 poliovirus involving antigenic site 1. J. Gen. Virol. 71:2543-2551. [DOI] [PubMed] [Google Scholar]
- 35.Minor, P. D., M. Ferguson, K. Katrak, D. Wood, A. John, J. Howlett, G. Dunn, K. Burke, and J. W. Almond. 1991. Antigenic structure of chimeras of type 1 and type 3 polioviruses involving antigenic sites 2, 3 and 4. J. Gen. Virol. 72:2475-2481. [DOI] [PubMed] [Google Scholar]
- 36.Minor, P. D., A. John, M. Ferguson, and J. P. Icenogle. 1986. Antigenic and molecular evolution of the vaccine strain of type 3 poliovirus during the period of excretion by a primary vaccinee. J. Gen. Virol. 67:693-706. [DOI] [PubMed] [Google Scholar]
- 37.Murdin, A. D., and E. Wimmer. 1989. Construction of a poliovirus type 1/type 2 antigenic hybrid by manipulation of neutralization antigenic site II. J. Virol. 63:5251-5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murray, M. G., R. J. Kuhn, M. Arita, N. Kawamura, A. Nomoto, and E. Wimmer. 1988. Poliovirus type 1/type 3 antigenic hybrid virus constructed in vitro elicits type 1 and type 3 neutralizing antibodies in rabbits and monkeys. Proc. Natl. Acad. Sci. USA 85:3203-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Page, G. S., A. G. Mosser, J. M. Hogle, D. J. Filman, R. R. Rueckert, and M. Chow. 1988. Three-dimensional structure of poliovirus serotype 1 neutralizing determinants. J. Virol. 62:1781-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reimerink, J. H., H. G. van der Avoort, A. M. van Loon, and M. P. Koopmans. 1999. Genetic basis for immunological aberrations in poliovirus Sabin serotype 3 strains imported in The Netherlands. J. Clin. Microbiol. 37:2393-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rico-Hesse, R., M. A. Pallansch, B. K. Nottay, and O. M. Kew. 1987. Geographic distribution of wild poliovirus type 1 genotypes. Virology 160:311-322. [DOI] [PubMed] [Google Scholar]
- 42.Sayle, R. A., and E. J. Milner-White. 1995. RASMOL: biomolecular graphics for all. Trends Biochem. Sci. 2 0:374.. [DOI] [PubMed] [Google Scholar]
- 43.Skinner, M. A., V. R. Racaniello, G. Dunn, J. Cooper, P. D. Minor, and J. W. Almond. 1989. New model for the secondary structure of the 5′ non-coding RNA of poliovirus is supported by biochemical and genetic data that also show that RNA secondary structure is important in neurovirulence. J. Mol. Biol. 207:379-392. [DOI] [PubMed] [Google Scholar]
- 44.Stanway, G., P. J. Hughes, G. D. Westrop, D. M. Evans, G. Dunn, P. D. Minor, G. C. Schild, and J. W. Almond. 1986. Construction of poliovirus intertypic recombinants by use of cDNA. J. Virol. 57:1187-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tardy-Panit, M., B. Blondel, A. Martin, F. Tekaia, F. Horaud, and F. Delpeyroux. 1993. A mutation in the RNA polymerase of poliovirus type 1 contributes to attenuation in mice. J. Virol. 67:4630-4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Avoort, H. G., B. P. Hull, T. Hovi, M. A. Pallansch, O. M. Kew, R. Crainic, D. J. Wood, M. N. Mulders, and A. M. van Loon. 1995. Comparative study of five methods for intratypic differentiation of polioviruses. J. Clin. Microbiol. 33:2562-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.World Health Organization. 1997. Manual for the virological investigation of poliomyelitis. W. H. O./EP/GEN/97.01. World Health Organization, Geneva, Switzerland.