Abstract
The viral infectivity factor (Vif) of human immunodeficiency virus type 1 (HIV-1) neutralizes an unidentified antiviral pathway that occurs only in nonpermissive (NP) cells. Using a yeast two-hybrid screen of a human lymphocyte cDNA library, we identified several potential Vif partners. One, the nuclear body protein Sp140, was found specifically in all NP cells (n = 12 cell lines tested; P ≤ 0.001), and HIV-1 infection induced its partial dispersal from nuclear bodies into cytosolic colocalization with Vif. Our results implicate Sp140 in a response to HIV-1 that may be related to or coordinated with the pathway that inactivates HIV-1 lacking vif.
The Vif protein encoded by human immunodeficiency virus type 1 (HIV-1) is a 23,000-Mr phosphoprotein that has been detected in the cytosol and nucleus in association with membranes, intermediate filaments, the viral Gag polyprotein, and the viral genomic RNA (8, 10, 17, 23, 25, 27, 43, 55). It enhances the infectivity but not the quantity of HIV-1 virions released from nonpermissive (NP) cells, including T lymphocytes and macrophages and some leukemic T-cell lines, by 20- to 100-fold, but it is unnecessary in permissive (P) cells (7, 13, 22, 39, 44, 53). Although the HIV-1 lacking vif that is made in P cells efficiently infects NP cells, the NP cells release noninfectious HIV-1 virions that appear to have a normal protein and RNA composition but that are irreversibly altered in a manner that inhibits reverse transcription in target cells (9, 13, 17, 24, 36, 44, 53).
Two lines of evidence suggest that for Vif to function, the protein must interact with an unknown cellular factor that occurs in NP cells. Recently, we and others found that the NP phenotype is dominant in P × NP heterokaryons (31, 42). This implies that NP cells contain a pathway for inactivating HIV-1, that this pathway is absent or incomplete in P cells, and that Vif neutralizes this pathway. Additionally, it was reported that Vif proteins of primate lentiviruses function only in the lymphocytes of species closely related to the viral hosts (41). This finding implies that for Vif to function, rather than associating with a viral factor, the protein must bind to a cellular factor that changes during evolution (41). Consequently, we hypothesized that P cells lack this Vif-binding target factor and/or another component or components of the pathway that inactivates HIV-1 lacking vif.
We used the L40 yeast system (26) with full-length HXB2 strain Vif as bait in a two-hybrid screen of a human leukocyte Matchmaker cDNA library (Clontech, Palo Alto, Calif.). This system provides high-level expression of LexA-B domain (BD)-bait and Ga14-A domain (AD)-prey proteins, thereby enhancing weak bait-prey associations. Although several prey were of potential interest, we focused principally on Sp140 because it is lymphocyte and macrophage specific and is induced by gamma interferon (3, 4, 15). Additionally, Sp140 is closely related to the widely expressed protein Sp100, which has been implicated in defenses against other viruses and associates with the promyelocytic leukemia protein (PML) in nuclear-matrix-associated nuclear bodies (NBs), also called PML oncogenic domains or nuclear domains 10 (6, 11, 12, 16, 18, 20, 34, 48, 49, 54, 57). Ku70, which is a component of HIV-1 preintegration complexes (14, 29), was also selected in our screen. These Vif-prey interactions were specific, as indicated by the activation of multiple reporter genes that contain the LexA promoter and by negative results obtained with an unrelated bait derived from the Sin Nombre hantavirus nucleocapsid and with unrelated prey.
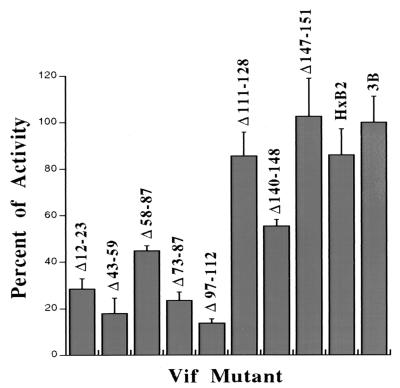
We analyzed small deletion mutations distributed throughout Vif that were previously shown to reduce its activity by greater than 93% (45). Mutations in the amino-terminal region (resulting in mutants Δ12-23, Δ43-59, and Δ73-87) and especially in the central region (resulting in mutant Δ97-112) strongly inhibited the protein's two-hybrid association with Sp140, whereas mutations further toward the carboxyl terminus (resulting in mutants Δ111-128, Δ140-148, and Δ147-158) had negligible effects (Fig. 1). The Vif domain involved in the interaction with Sp140 does not include the conserved SLYQXLA sequence that has been implicated in RNA binding (17). Despite considerable differences in sequence, Vif proteins encoded by the IIIB and HXB2 strains of HIV-1 were similarly active in associating with Sp140. The Ga14-AD-Sp140 prey cDNAs that we isolated encoded the common carboxyl-terminal region between amino acids 527 and 836, which begins with the sequence DGQVV and includes a SAND domain, a PHD-type zinc finger, and a bromodomain (15). Preliminary evidence suggested that the SAND and PHD domains interact and that one or both of these regions associate with Vif (data not shown).
FIG. 1.
Effects of inactivating small deletion mutations in Vif on its interaction with Sp140 in the yeast two-hybrid system. LexA-BD-Vif plasmids encoding the wild-type or mutant HIV-1 strain IIIB Vif proteins or the wild-type HXB2 strain Vif were cotransfected with the Ga14-AD-Sp140 plasmid into L40 yeast cells, and colonies were selected for growth in medium lacking tryptophan, uridine, leucine, and lysine. Three individual colonies in each plate were then tested for their rates of growth in liquid culture medium lacking histidine as well as the above-mentioned omissions and supplemented with 3 mM 3-amino-1,2,4-triazole (Sigma). Growth rates were measured in the triplicate samples by adsorption at 600 nm at 24-h intervals. The results are plotted as the averages of values from two independent assays relative to the growth rate of strain IIIB, considered to be standard, and with the error bars showing the ranges of the two assays. The two-hybrid interaction enables cell replication in the absence of histidine.
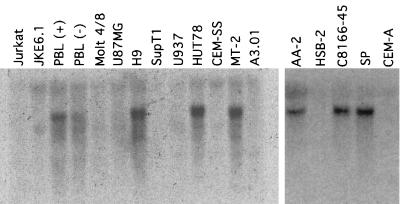
Northern blot analyses of RNAs from P and NP leukemic cell lines and from stimulated and unstimulated peripheral blood lymphocytes (PBLs) were done as previously described (50). As shown in Fig. 2, there was a precise correspondence between the NP phenotype (7, 21, 22, 38, 39, 44, 46, 53) and the presence of an approximately 3.0-kb component of Sp140 mRNA. Equal amounts of RNA were loaded in the lanes, and identical results were obtained with independent preparations of the RNAs. Thus, in an initial study (left panel), we found this Sp140 mRNA in PBLs, especially after the PBLs were stimulated with interleukin-2 plus phytohemagglutinin-P, and in H9, HUT78, and MT-2 cells but not in Jurkat, JKE6.1, Molt4/8, U87MB, SupT1, U937, CEM-SS, or A3.01 cells. Sp140 mRNA is also expressed when HL60 and NB4 myeloid cells are induced to differentiate, and it appears to be absent in nonhematopoietic cells (3, 4, 15). Subsequently, we confirmed the correspondence by testing additional leukemic cell lines for their NP or P phenotype and for the presence of Sp140 mRNA (Fig. 1, right panel). Although C8166 cells initially appeared to be an exception since they were reported to be semipermissive or P (13, 47), the C8166-45 clone that we received from a repository contains Sp140 and is NP by our assay system (30). If we consider only the independently derived cell lines as described in the legend to Fig. 2, these results demonstrate a correlation between the presence of Sp140 mRNA and the NP phenotype that is extremely unlikely to have occurred by chance (n = 12 cell lines; P ≤ 0.001).
FIG. 2.
Detection of Sp140 mRNA by Northern blot analyses of RNAs from diverse human leukemic cell lines and from PBLs. RNAs were extracted from cultured cells and analyzed as previously described (50) by using a fragment of Sp140 cDNA corresponding to nucleotides 1350 to 2198 of the full-length cDNA (4) as the hybridization probe. Equal loadings of the lanes were verified by observing the quenching of autofluorescence caused by rRNAs, and sample integrity was shown by probing for the S2 ribosomal protein mRNA (results not shown). The cell lines were from the American Type Culture Collection (Manassas, Va.) or were generously provided by the AIDS Research and Reference Reagent Program at the National Institutes of Health. The P and NP phenotypes of the cell lines in the right panel were tested as previously described (30). The following pairs of cell lines had a common origin: Jurkat and JKE6.1, H9 and HUT78, and CEM-SS and A3.01. Moreover, C8166 cells were previously reported to be P or semipermissive (13, 47), implying a difference in our C8166-45 sample perhaps caused by divergence or contamination. If we consider only the other cells that were independently derived, there was a precise correspondence of the NP phenotype with the presence of Sp140 mRNA (n = 12 cell lines; P ≤ 0.001).
Although it could be postulated that Sp140 expression and the NP phenotype are not functionally coupled but are only coincidentally restricted to a subset of leukemic cells that are arrested at a specific stage of differentiation, this idea is difficult to reconcile with the fact that these NP cell lines are heteroploid and derive from different hematopoietic lineages. Sp140 occurs in T and B lymphocytes and in NB4 and HL60 myeloid cells (3, 15). Nevertheless, it is possible that the NP phenotype develops in the late stages of differentiation in several hematopoietic lineages and that Sp140 is specifically expressed at that same stage. In any case, Sp140 occurs in all natural cellular targets of HIV-1 in close association with the antiviral NP phenotype.
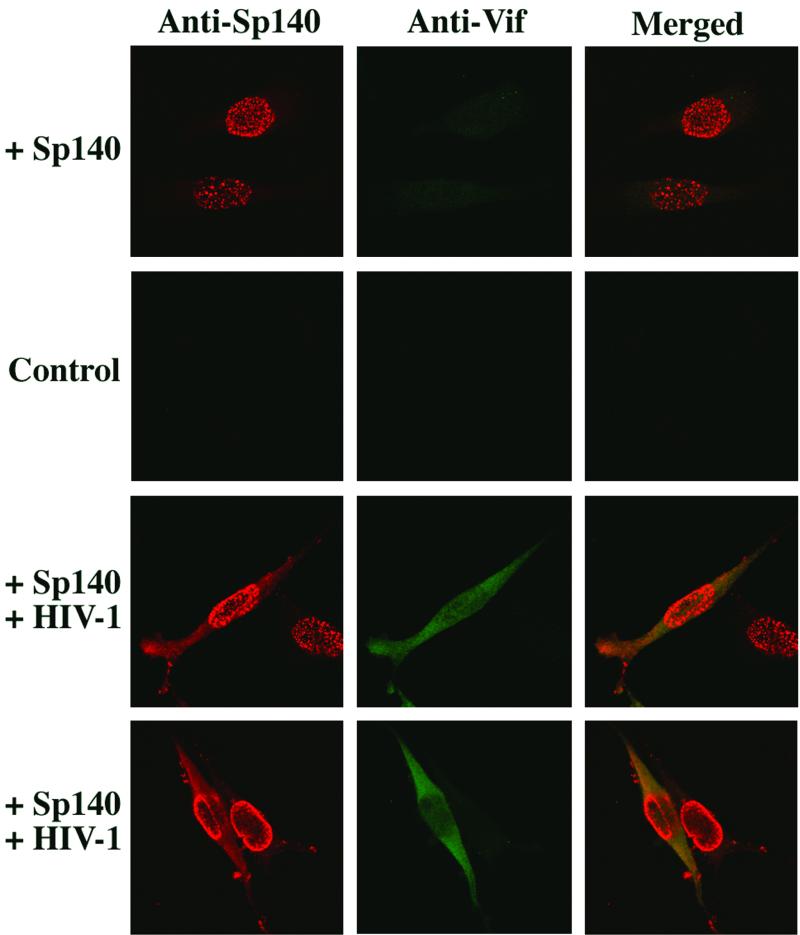
Previous studies have shown that Sp140 occurs almost exclusively in NBs (3, 4, 15) but that Vif is mainly cytosolic (23, 25, 27, 43). We found, however, that HIV-1 induces significant relocalization of Sp140 between 16 and 20 h postinfection. Because the endogenous Sp140 in NP cell lines occurs in small amounts that were difficult to detect, and because recombinant Sp140 damages P cell lines and therefore has not been stably expressed (see below), we transiently transduced HeLa-CD4 cells with a previously described adenoviral-Sp140 (Ad-Sp140) vector (3-5). As expected, Sp140 localized in clusters that were randomly situated within the nuclei (Fig. 3, top panels). However, by 24 h following exposure to HIV-1, at a time when Vif became evident in approximately 3 to 5% of the cells, several changes occurred in the Sp140 distribution. First, in the cells that expressed Vif, a portion of Sp140 was dispersed throughout the cytosol (Fig. 3). Indeed, in these cells, Vif appeared to colocalize with this dispersed form of Sp140. In addition, in these and other HIV-1-infected cells, the Sp140-containing nuclear clusters often appeared to have partially or completely migrated toward the nuclear peripheries, where they eventually formed a layer underlying the nuclear membranes (Fig. 3, lower panels). Although Vif does not induce these changes in the nuclear clusters, our results suggest that it may enhance the cytosolic retention of Sp140.
FIG. 3.
Confocal immunofluorescence microscopy of HeLa-CD4 cells expressing Sp140 in the presence or absence of Vif. The HeLa-CD4 cells (clone HTC.15) were transduced with an Ad-Sp140 expression vector (3-5) and were then infected where indicated with wild-type NL4-3 strain HIV-1 (30) for 24 h prior to fixation and staining with a rat antiserum to Sp140 (3-5). The cells were then treated with Alexa 594-conjugated anti-rat immunoglobulin (Molecular Probes, Eugene, Oreg.) or with rabbit antiserum to Vif followed by Alexa-488 conjugated secondary antibody from the same source. Confocal microscopy was done as previously described (28).
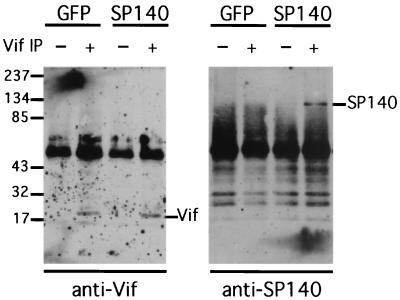
We used the same cell culture system to determine whether Vif-Sp140 complexes could be immunoprecipitated from cytosolic extracts. In this case, HeLa-CD4 cells that had been transduced with either Ad-Sp140 or a negative control Ad-green fluorescent protein (GFP) vector (3-5) were infected for 24 h with wild-type HIV-1 prior to lysis with a buffer containing 1.0% Triton X-100 at 0°C. After the removal of nuclei and debris by centrifugation, the extracts were incubated with preimmune rabbit serum and protein A-Sepharose beads (Sigma, St. Louis, Mo.) prior to recentrifugation. Aliquots of the supernatants were incubated either with more preimmune serum or with antiserum to Vif prior to the addition of more protein A-Sepharose beads and centrifugation. Proteins extracted from these beads were used for Western immunoblotting as described previously (33). Consistent with our immunofluorescence microscopy results, small amounts of Vif were immunoprecipitated by our Vif antiserum but not by preimmune serum, and the Vif immunoprecipitates specifically contained a small quantity of Sp140 (Fig. 4). Although the quantities of these proteins were small compared to the background levels, this was expected because only 3 to 5% of the cells expressed a detectable amount of Vif and because only approximately 10% of the Sp140 in these few cells was dispersed into the cytosol (Fig. 3). Control experiments using cells lacking HIV-1 established that these extraction procedures solubilized approximately 5 to 10% of the total cellular Sp140 and that our Vif antiserum did not directly immunoprecipitate Sp140 (results not shown). Based principally on the congruence of these results with our immunofluorescence microscopy data, we propose that Vif can at least weakly interact with a form of Sp140 that disperses into the cytosol following HIV-1 infections.
FIG. 4.
Coimmunoprecipitation of a Vif-Sp140 complex from cellular extracts. HeLa-CD4 cells were transduced with Ad-Sp140 or with a negative control Ad-GFP vector (3-5) and were then infected with wild-type HIV-1 (NL 4-3 strain) for 24 h as described in the legend for Fig. 2. The cells were then lysed with 1% Triton X-100 in 150 mM NaCl and 10 mM Tris-HCl in the presence of protease inhibitors (Sigma) at 0°C. After being centrifuged to remove nuclei and debris, the extracts were incubated with preimmune rabbit serum (1:200) and with protein A-Sepharose beads (30 μl of a 50% slurry/ml of lysate) for 1 h prior to centrifugation at 4,000 rpm for 1 min. Aliquots of the supernatants were then incubated either with more preimmune rabbit serum or with Vif antiserum prior to the addition of more protein A-Sepharose beads and recentrifugation. The pelleted beads were washed, and the extracted proteins were analyzed by electrophoresis and Western immunoblotting by using either antiserum to Vif (left panel) or antiserum to Sp140 (right panel) in comparison with protein standards as described previously (30). IP, immunoprecipitation.
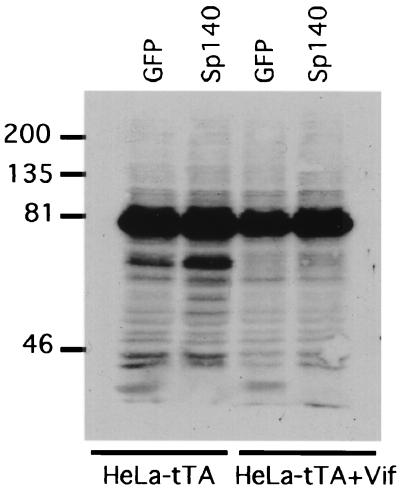
Diverse viruses synthesize proteins that localize in NBs or induce dispersal of NB proteins (6, 11, 16, 18, 20, 35, 49, 54), consistent with evidence that PML and associated proteins, including Sp100 and Mx1, participate in antiviral defenses that the viruses must counteract (12, 19, 52). Moreover, many NB proteins, including PML and Sp100, are conjugated with the small ubiquitin-related modifier protein SUMO-1 (19, 34, 35, 48). Sumoyl transferase occurs in NBs and contributes to NB integrity (19, 20, 34, 35, 48, 56), and several virus-encoded proteins cause desumoylation and/or degradation of Sp100 and PML (1, 11, 20, 34, 37). As shown by the SUMO-1-specific Western immunoblot in Fig. 5, expression of Sp140 in HeLa cells caused several changes in the pattern of sumoylation and the inductive effects were all counteracted by coexpression of Vif. The major ∼80,000-Mr sumoylated protein in mammalian cells is RanGAP1 (2, 32), which is absent from NBs and was unaffected by Sp140 or Vif.
FIG. 5.
Effects of Sp140 and Vif on protein sumoylation. HeLa-tTA cells or HeLa-tTA Vif cells that express a previously described tetracycline-repressible Vif vector (30) were transduced for 24 h with equal titers of either Ad-Sp140 or Ad-GFP, which encodes GFP in the absence of tetracycline. Cell extracts were used for Western immunoblotting with a SUMO-1-specific monoclonal antibody (αGMP-1) (Zymed, Inc., South San Francisco, Calif.). Sp140 reproducibly induced elevated expression of at least one protein (Mr, ∼65,000) that reacted with the monoclonal antibody and of several minor components, and in some cases it also reduced expression of a smaller protein (Mr, ∼30,000). The inductive effects of Sp140 were eliminated by expression of Vif.
It has not been possible to stably express Sp140 in P cell lines. Indeed, we were unable to stably transfect HeLa-tTA cells with a tetracycline-repressible Sp140 expression vector (30) when the cells were maintained throughout selection in a high concentration of tetracycline, implying that even trace levels of Sp140 are damaging to cells. This made it very difficult to determine whether its expression in P cells would convert them to NP cells. Efficient transient expression of Sp140 in a large proportion of cells was achieved in HeLa-CD4 cells by using our Ad-Sp140 vector and in HEK293T cells by transfection with a pcDNA3.1-Sp140 vector but was not achieved by these same vectors in leukemic T-cell lines. Under these conditions, we did not detect conversion of HeLa-CD4 or HEK293T cells to NP cells (results not shown). However, HeLa cells are unrelated to T lymphocytes and may lack multiple components of the NP cell-specific antiviral pathway.
These results implicate Sp140 in an innate response to HIV-1 that occurs specifically in the natural cellular targets. These cells are termed NP because they contain a potent antiviral pathway that is neutralized by Vif (31, 42). Our results are compatible with evidence that NB proteins may be involved in defenses against diverse viruses (6, 11, 16, 18, 20, 34, 49, 54) and confirm the hypothesis that NP cells contain systems for response to HIV-1 that are absent in P cells (31, 42). However, we also emphasize that our results do not establish a direct role for Sp140 in the pathway that inactivates HIV-1 lacking vif. The interaction of Sp140 with Vif appears to be weak, and it is uncertain whether it is a true Vif target. Conceivably, Vif might bind to a motif that occurs in several proteins, and like Nef or Vpu (51), it might influence multiple cellular pathways.
During final revision of this paper, Sheehy et al. (40) reported that the protein CEM-15 occurs only in NP cells and confers the NP phenotype on P cell lines, including 293T cells. In the model suggested by their results, Vif binds to HIV-1 genomic RNA and shields it in some manner from the antiviral activity of CEM-15. Their model implies that Vif function may not involve direct interaction with CEM-15, and it would be difficult to reconcile with earlier evidence from the same group that Vif proteins of primate lentiviruses function only in the NP cells of species closely related to the viral host (41). Resolution of these issues will require additional investigations.
Acknowledgments
This research was supported by NIH grant AI49729 and by grant 02754-RGT from the American Foundation for AIDS Research.
A rabbit antiserum to Vif (courtesy of D. Gabuzda) was generously provided by the AIDS Research and Reference Reagent Program of the NIH, and plasmids encoding Vif mutants were kindly donated by Michael Malim. We are grateful to Stanley Hollenberg for advice concerning the L40 yeast system, to John Scarborough for assistance with the Northern blotting, and to Kristine Rose for help with the testing of leukemic cell lines.
REFERENCES
- 1.Adamson, A. L., and S. Kenney. 2001. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J. Virol. 75:2388-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernier-Villamor, V., D. A. Sampson, M. J. Matunis, and C. D. Lima. 2002. Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1. Cell 108:345-356. [DOI] [PubMed] [Google Scholar]
- 3.Bloch, D. B., J.-D. Chiche, D. Orth, S. M. de la Monte, A. Rosenzweig, and K. D. Bloch. 1999. Structural and functional heterogeneity of nuclear bodies. Mol. Cell. Biol. 19:4423-4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bloch, D. B., S. M. de la Monte, P. Guigaouri, A. Filippov, and K. D. Bloch. 1996. Identification and characterization of a leukocyte-specific component of the nuclear body. J. Biol. Chem. 271:29198-29204. [DOI] [PubMed] [Google Scholar]
- 5.Bloch, D. B., A. Nakajima, T. Gulick, J.-D. Chiche, D. Orth, S. M. de la Monte, and K. D. Bloch. 2000. Sp110 localizes to the PML-Sp100 nuclear body and may function as a nuclear hormone receptor transcriptional coactivator. Mol. Cell. Biol. 20:6138-6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borden, K. L. B., E. J. Campbell Dwyer, and M. S. Salvato. 1998. An arenavirus RING (zinc-binding) protein binds the oncoprotein promyelocyte leukemia protein (PML) and relocates PML nuclear bodies to the cytoplasm. J. Virol. 72:758-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borman, A. M., C. Quillent, P. Charneau, C. Dauguet, and F. Clavel. 1995. Human immunodeficiency virus type 1 Vif− mutant particles from restrictive cells: role of Vif in correct particle assembly and infectivity. J. Virol. 69:2058-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouyac, M., M. Courcoul, G. Bertoia, Y. Baudat, D. Gabuzda, D. Blanc, N. Chazal, P. Boulanger, J. Sire, R. Vigne, and B. Spire. 1997. Human immunodeficiency virus type 1 Vif protein binds to the Pr55Gag precursor. J. Virol. 71:9358-9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bouyac, M., F. Rey, M. Nascimbeni, M. Courcoul, J. Sire, D. Blanc, F. Clavel, R. Vigne, and B. Spire. 1997. Phenotypically Vif− human immunodeficiency virus type 1 is produced by chronically infected restrictive cells. J. Virol. 71:2473-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterji, U., C. K. Grant, and J. H. Elder. 2000. Feline immunodeficiency virus Vif localizes to the nucleus. J. Virol. 74:2533-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chelbi-Alix, M. K., and H. de The. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene 18:935-941. [DOI] [PubMed] [Google Scholar]
- 12.Chelbi-Alix, M. K., F. Quignon, L. Pelicano, M. H. M. Koken, and H. De Thé. 1998. Resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J. Virol. 72:1043-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Courcoul, M., C. Patience, F. Rey, D. Blanc, A. Harmache, J. Sire, R. Vigne, and B. Spire. 1995. Peripheral blood mononuclear cells produce normal amounts of defective Vif− human immunodeficiency virus type 1 particles which are restricted for the preretrotranscription steps. J. Virol. 69:2068-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniel, R., R. A. Katz, G. Merkel, J. C. Hittle, T. J. Yen, and A. M. Skalka. 2001. Wortmannin potentiates integrase-mediated killing of lymphocytes and reduces the efficiency of stable transduction by retroviruses. Mol. Cell. Biol. 21:1164-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dent, A. L., J. Yewdell, F. Puvion-Dutilleul, M. H. Koken, H. de The, and L. M. Staudt. 1996. LYSP100-associated nuclear domains (LANDs): description of a new class of subnuclear structures and their relationship to PML nuclear bodies. Blood 88:1423-1426. [PubMed] [Google Scholar]
- 16.Desbois, C., R. Rousset, F. Bantignies, and P. Jalinot. 1996. Exclusion of Int-6 from PML nuclear bodies by binding to the HTLV-I Tax oncoprotein. Science 273:951-953. [DOI] [PubMed] [Google Scholar]
- 17.Dettenhofer, M., S. Cen, B. A. Carlson, L. Kleiman, and X.-F. Yu. 2000. Association of human immunodeficiency virus type 1 Vif with RNA and its role in reverse transcription. J. Virol. 74:8938-8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doucas, V., A. M. Ishov, A. Romo, H. Juguilon, M. D. Weitzman, R. M. Evans, and G. G. Maul. 1996. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 10:196-207. [DOI] [PubMed] [Google Scholar]
- 19.Engelhardt, O. G., E. Ullrich, G. Kochs, and O. Haller. 2001. Interferon-induced antiviral Mx1 GTPase is associated with components of the SUMO-1 system and promyelocytic leukemia protein nuclear bodies. Exp. Cell Res. 271:286-295. [DOI] [PubMed] [Google Scholar]
- 20.Everett, R. D., P. Freemont, H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan, L., and K. Peden. 1992. Cell-free transmission of Vif mutants of HIV-1. Virology 190:19-29. [DOI] [PubMed] [Google Scholar]
- 22.Gabuzda, D. H., K. Lawrence, E. Langhoff, E. Terwilliger, T. Dorfman, W. A. Haseltine, and J. Sodroski. 1992. Role of vif in replication of human immunodeficiency virus type 1 in CD4+ T lymphocytes. J. Virol. 66:6489-6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goncalves, J., P. Jallepalli, and D. H. Gabuzda. 1994. Subcellular localization of the Vif protein of human immunodeficiency virus type 1. J. Virol. 68:704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goncalves, J., Y. Korin, J. Zack, and D. Gabuzda. 1996. Role of Vif in human immunodeficiency virus type 1 reverse transcription. J. Virol. 70:8701-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goncalves, J., B. Shi, X. Yang, and D. Gabuzda. 1995. Biological activity of human immunodeficiency virus type 1 Vif requires membrane targeting by C-terminal basic domains. J. Virol. 69:7196-7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollenberg, S. M., R. Sternglanz, P. F. Cheng, and H. Weintraub. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 15:3813-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karczewski, M. K., and K. Strebel. 1996. Cytoskeleton association and virion incorporation of the human immunodeficiency virus type 1 Vif protein. J. Virol. 70:494-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozak, S. L., J. M. Heard, and D. Kabat. 2002. Segregation of CD4 and CXCR4 into distinct lipid microdomains in T lymphocytes suggests a mechanism for membrane destabilization by human immunodeficiency virus. J. Virol. 76:1802-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, L., J. M. Olvera, K. E. Yoder, R. S. Mitchell, S. L. Butler, M. Lieber, S. L. Martin, and F. D. Bushman. 2001. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 20:3272-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madani, N., and D. Kabat. 2000. Cellular and viral specificities of human immunodeficiency virus type 1 Vif protein. J. Virol. 74:5982-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madani, N., and D. Kabat. 1998. An endogenous inhibitor of human immunodeficiency virus in human lymphocytes is overcome by the viral Vif protein. J. Virol. 72:10251-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahajan, R., L. Gerace, and F. Melchior. 1998. Molecular characterization of the SUMO-1 modification of RanGAP1 and its role in nuclear envelope association. J. Cell Biol. 140:259-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marin, M., C. S. Tailor, A. Nouri, and D. Kabat. 2000. Sodium-dependent neutral amino acid transporter type 1 is an auxiliary receptor for baboon endogenous retrovirus. J. Virol. 74:8085-8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Müller, S., and A. Dejean. 1999. Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73:5137-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller, S., M. J. Matunis, and A. Dejean. 1998. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 17:61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ochsenbauer, C., T. Wilk, and V. Bosch. 1997. Analysis of vif-defective human immunodeficiency virus type 1 (HIV-1) virions synthesized in ‘non-permissive' T lymphoid cells stably infected with selectable HIV-1. J. Gen. Virol. 78:627-635. [DOI] [PubMed] [Google Scholar]
- 37.Parkinson, J., and R. D. Everett. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 74:10006-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reddy, T. R., G. Kraus, O. Yamada, D. J. Looney, M. Suhasini, and F. Wong-Staal. 1995. Comparative analyses of human immunodeficiency virus type 1 (HIV-1) and HIV-2 Vif mutants. J. Virol. 69:3549-3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakai, H., R. Shibata, J.-I. Sakuragi, S. Sakuragi, M. Kawamura, and A. Adachi. 1993. Cell-dependent requirement of human immunodeficiency virus type 1 Vif protein for maturation of virus particles. J. Virol. 67:1663-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646-650. [DOI] [PubMed] [Google Scholar]
- 41.Simon, J. H., D. L. Miller, R. A. Fouchier, M. A. Soares, K. W. Peden, and M. H. Malim. 1998. The regulation of primate immunodeficiency virus infectivity by Vif is cell species restricted: a role for Vif in determining virus host range and cross-species transmission. EMBO J. 17:1259-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon, J. H., N. C. Gaddis, R. A. Fouchier, and M. H. Malim. 1998. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat. Med. 4:1397-1400. [DOI] [PubMed] [Google Scholar]
- 43.Simon, J. H. M., E. A. Carpenter, R. A. M. Fouchier, and M. H. Malim. 1999. Vif and the p55Gag polyprotein of human immunodeficiency virus type 1 are present in colocalizing membrane-free cytoplasmic complexes. J. Virol. 73:2667-2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon, J. H. M., and M. H. Malim. 1996. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J. Virol. 70:5297-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simon, J. H. M., A. M. Sheehy, E. A. Carpenter, R. A. M. Fouchier, and M. H. Malim. 1999. Mutational analysis of the human immunodeficiency virus type 1 Vif protein. J. Virol. 73:2675-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simon, J. H. M., T. E. Southerling, J. C. Peterson, B. E. Meyer, and M. H. Malim. 1995. Complementation of vif-defective human immunodeficiency virus type 1 by primate, but not nonprimate, lentivirus vif genes. J. Virol. 69:4166-4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sova, P., and D. J. Volsky. 1993. Efficiency of viral DNA synthesis during infection of permissive and nonpermissive cells with vif-negative human immunodeficiency virus type 1. J. Virol. 67:6322-6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sternsdorf, T., K. Jensen, B. Reich, and H. Will. 1999. The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J. Biol. Chem. 274:12555-12566. [DOI] [PubMed] [Google Scholar]
- 49.Szekely, L., K. Pokrovskaja, W.-Q. Jiang, H. de The, N. Ringertz, and G. Klein. 1996. The Epstein-Barr virus-encoded nuclear antigen EBNA-5 accumulates in PML-containing bodies. J. Virol. 70:2562-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tailor, C. S., A. Nouri, C. G. Lee, C. Kozak, and D. Kabat. 1999. Cloning and characterization of a cell surface receptor for xenotropic and polytropic murine leukemia viruses. Proc. Natl. Acad. Sci. USA 96:927-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trono, D. 1995. HIV accessory proteins: leading roles for the supporting cast. Cell 82:189-192. [DOI] [PubMed] [Google Scholar]
- 52.Turelli, P., V. Doucas, E. Craig, B. Mangeat, N. Klages, R. Evans, G. Kalpana, and D. Trono. 2001. Cytoplasmic recruitment of INI1 and PML on incoming HIV preintegration complexes: interference with early steps of viral replication. Mol. Cell 7:1245-1254. [DOI] [PubMed] [Google Scholar]
- 53.von Schwedler, U., J. Song, C. Aiken, and D. Trono. 1993. vif is crucial for human immunodeficiency virus type 1 proviral DNA synthesis in infected cells. J. Virol. 67:4945-4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilkinson, G. W., C. Kelly, J. H. Sinclair, and C. Rickards. 1998. Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J. Gen. Virol. 79:1233-1245. [DOI] [PubMed] [Google Scholar]
- 55.Zhang, H., R. J. Pomerantz, G. Dornadula, and Y. Sun. 2000. Human immunodeficiency virus type 1 Vif protein is an integral component of an mRNP complex of viral RNA and could be involved in the viral RNA folding and packaging process. J. Virol. 74:8252-8261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhong, S., S. Muller, S. Ronchetti, P. S. Freemont, A. Dejean, and P. P. Pandolfi. 2000. Role of SUMO-1-modified PML in nuclear body formation. Blood 95:2748-2752. [PubMed] [Google Scholar]
- 57.Zong, R. T., C. Das, and P. W. Tucker. 2000. Regulation of matrix attachment region-dependent, lymphocyte-restricted transcription through differential localization within promyelocytic leukemia nuclear bodies. EMBO J. 19:4123-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]