Abstract
The human immunodeficiency virus type 1 (HIV-1) viral setpoint during the disease-free interval has been strongly associated with future risk of disease progression. An awareness of the correlation between viral setpoint and HIV-1 genetic evolution over time is important in the understanding of viral dynamics and infection. We examined genetic diversity in HIV-1 CRF02_A/G-IbNG-infected seroincident women in Dakar, Senegal; determined whether a viral setpoint kinetic pattern existed for CRF02_A/G-IbNG during the disease-free interval; and correlated viral load level and diversity. Samples were drawn during the disease-free interval from consenting CRF02_A/G-IbNG-infected, antiretroviral therapy-naïve female commercial sex workers in Dakar, Senegal. Based on sequential plasma RNA values, low and high viral setpoint groups were established. Intrapatient diversity and divergence over time was determined from earlier and later time point DNA samples from each person. Most individuals followed the viral setpoint paradigm. For each 1|−|log10 copy/ml of plasma increase in viral load, intrapatient diversity increased by 1.4% (P = 0.028). A greater diversification rate was observed in the high viral setpoint group than in the low viral setpoint group (P = 0.01). Greater nucleotide (P = 0.015) and amino acid (P = 0.048) divergences and a greater nucleotide divergence rate (P = 0.03) were found in the high viral setpoint group. There was no difference between the groups in the ratio of the number of nonsynonymous substitutions per nonsynonymous site to the number of synonymous substitutions per synonymous site. The greater intrapatient diversity, divergence, and diversification rates observed in the high viral setpoint group supports the notion that diversity is driven by cycles of viral replication resulting in accumulated mutations. Recognizing diversity potential based on viral load levels in individuals may inform the design of vaccines and therapies.
A comprehension of human immunodeficiency virus type 1 (HIV-1) diversity is critically important to the study of viral evolution, virulence, disease, and pathogenesis, as well as therapy and effective vaccine design (23). Diversity may impact such processes by influencing viral tropism, immunologic responses to HIV infection, and immune system impairment (39). Most studies evaluating viral diversity and heterogeneity have examined HIV-1 subtype B viruses, and few have looked at diversity within a single non-B subtype or -sub-subtype (38, 46), though the preponderance of HIV-infected individuals worldwide are infected with non-B subtypes. Thus, an understanding of the pattern of non-subtype B diversity within a single HIV-1-infected individual and its association with disease progression may provide insight into viral pathogenesis. For HIV-1 subtype B-infected individuals, progression to HIV disease has been associated with the level of viral load (the viral setpoint is the level at which the viral load remains stable) (13, 26). The stability in viral load over much of the course of infection is thought to result from the struggle between viral replication and host immune responses. This viral setpoint is theoretically maintained for years, is predictive of future clinical outcome, and can be utilized as a prognostic indicator of the rate of disease progression (26). Few studies have evaluated viral load and viral setpoint and their correlation with different HIV-1 subtypes, although there is some indication that viral type and genotype may be associated with viral load levels (14, 29, 32, 33). This is particularly the case with HIV-2 and HIV-1 infections, for HIV-2 viral loads appear to be significantly lower than HIV-1 viral loads over the course of infection. (33)
Most previous studies have sought to examine the correlation between disease progression and intrapatient diversity in subtype B-infected individuals, both adults (1, 18, 20, 22, 24, 25, 40, 41, 47) and children (7, 11, 15, 43), excepting a study examining viral evolution in nonrecombinant subtype A-infected women from Kenya (34) and a study by Sutthent et al. which revealed a correlation between poorer clinical status and increased diversity in subtype E-infected mothers and their infants (44). The majority of studies have evaluated regions of the env gene, a rapidly evolving and functionally important region. Markham et al. examined 285 base pairs (V3 and part of C2/C3) of DNA of the env gene drawn from seroconvertor intravenous drug users from the ALIVE study in Baltimore, Maryland (22). The greatest diversity and divergence was noted in rapid progressors, followed by moderate progressors and finally by nonprogressors, and diversity had a negative correlation with CD4+ cell counts. This suggested that greater diversification was proportional to the rate of progression. McNearney et al. noted similar results for the env V3 region in a smaller study (25). These data supported the Nowak model, which assumes that viral replication, with the resulting accumulation of mutations, drives diversity and therefore antigenic variation, with the immune system controlling disease progression as long as it is able to recognize and control viral replication and emerging variants (30). This model suggests that host immune selection pressures are not driving diversity but rather that the diversity generated by viral replication is driving the disease. Many rounds of viral replication (and therefore high viral load levels) would eventually overwhelm the immune system with new viral variants and result in rapid progression to disease.
Wolinsky et al. studied the V3-V5 region of the env gene in men from the MACS cohort (47). They concluded that, for those in the rapid progressor group, there was little selective pressure for change and that therefore less diversity was evident in this group than in either the moderate or the slow progressor group. Quasispecies diversification and divergence were strongly correlated with a favorable clinical outcome, and this suggested that diversity driven by an effective host immune response resulted in significant pressure on the virus to evolve. Similar results were obtained for HIV-1-infected children in a study by Ganeshan et al. (11).
Multiple intersubtype recombinant viruses have been identified in HIV-1-infected individuals, and several exist as major circulating recombinant forms (23). It is estimated that, worldwide, at least 9 million HIV-1 infections are due to CRF02_A/G-IbNG (23). CRF02_A/G-IbNG was first characterized from a sample collected in Ibadan, Nigeria (31). It is subtype A in the gag gene and the majority of the env gene, subtype G in the long terminal repeats, and a mosaic of A and G in the pol gene. It is epidemic in West and West Central Africa (3, 27) and is the predominant virus circulating in Senegal (38). The CRF02_A/G-IbNG-infected individuals included in our study were antiretroviral therapy-naïve at the time of sampling. We sought to directly examine the genetic diversity of HIV-1 CRF02_A/G-IbNG in seroincident women during the disease-free interval in order to identify patterns of diversity at the viral setpoint, as opposed to during AIDS or primary infection. In this study, we examined intrapatient diversity and divergence in 12 HIV-1 CRF02_A/G-IbNG-infected seroincident women categorized into high and low viral load groups in Dakar, Senegal.
MATERIALS AND METHODS
Study population.
Since 1970, the government of Senegal has required that commercial sex workers register and receive periodic testing and treatment for sexually transmitted diseases. A cohort of consenting commercial sex workers attending the Institut Hygiene Social clinic in Dakar, Senegal, has been followed and studied since 1985, as described previously (38). For the purposes of this study, a subset from this cohort consisting of seroincident CRF02_A/G-IbNG HIV-1-infected individuals was identified. Nucleotide sequencing and subtype characterization were performed as previously described (38). Individuals were included in the study if (i) at least two plasma RNA measurements with a total observation time of at least 6 months were available, (ii) if the individuals were not dually infected with HIV-1 and HIV-2, and (iii) if the samples were not collected after the individual had been diagnosed with AIDS. To ensure that samples were collected after the acute stage of infection had been reached, all initial RNA viral load samples were required to be drawn at least 2 months after the estimated seroconversion date, calculated as the midpoint between the date when the last negative blood sample was collected and the date when the first positive blood sample was collected. The mean of all plasma viral load determinations was calculated for each person. A mean viral load less of than 4,000 RNA copies/ml of plasma was considered low, and one of more than 30,000 RNA copies/ml of plasma was considered high. Individuals were thereby stratified into two groups, the high viral load group (n = 7) and the low viral load group (n = 5), and of course none of the individuals had progressed to AIDS at the time of sampling. Two DNA samples from each individual in the high and low viral load groups were selected, one sample collected at an earlier time and one collected at a later time, with a required observation time of more than 6 months, the purpose being to maximize the observation time (Table 1). There were no significant differences in observation period between the high viral load (median observation period, 2.4 years) and the low viral load (median observation period, 3.2 years) groups (P = 0.64 [Wilcoxon rank sum test]).
TABLE 1.
Profiles of two groups in study populationa
| Group and patient identification no. | Mean raw viral load (RNA copies/ml) | Time interval between DNA samples (yr) | Sero- conversion date (mo/yr) | No. of viral load determinations |
|---|---|---|---|---|
| Low viral load | ||||
| 2723 | 616 | 3.3 | 1/96 | 7 |
| 668 | 733 | 2.6 | 12/93 | 3 |
| 225 | 1,521 | 6.5 | 6/91 | 4 |
| 3065 | 2,845 | 2.3 | 3/97 | 3 |
| 1067 | 3,394 | 3.2 | 10/94 | 2 |
| High viral load | ||||
| 1196 | 35,692 | 5.4 | 4/93 | 4 |
| 915 | 43,028 | 7.1 | 1/92 | 7 |
| 394 | 68,440 | 0.75 | 4/96 | 2 |
| 2587 | 92,730 | 4.2 | 1/95 | 4 |
| 2909 | 126,423 | 2.4 | 4/98 | 3 |
| 2997 | 31,738b | 1.4 | 9/96 | 4 |
| 2425 | 216,422b | 1.3 | 3/96 | 4 |
Unless otherwise indicated, viral load was determined by Quantiplex version 2.0 bDNA assay.
Viral load determined by Quantiplex version 3.0 assay.
Viral load assays.
The HIV-1 plasma RNA viral load was quantified by two branched DNA (bDNA) assays (Quantiplex version 2.0 and Quantiplex version 3.0; Bayer Diagnostics, Tarrytown, N.Y.). The viral RNA loads of 10 subjects were quantified by the Quantiplex version 2.0 assay (lower limit of detection, 500 RNA copies/ml) and those of the two remaining subjects were quantified by the Quantiplex version 3.0 assay (lower limit of detection, 50 RNA copies/ml). EDTA-preserved plasma samples were stored at −70oC prior to quantification. Studies have shown that both bDNA assay versions are strongly and significantly correlated with one another and demonstrate excellent linearity, precision, and concordance (6, 21) in studies with duplicate plasma samples. The Quantiplex version 3.0 assay appears to quantify with a level of accuracy that is approximately twofold higher than that provided by the Quantiplex version 2.0 assay (6, 21), though this difference is greater at lower viral load levels; that is, the differences between the assays seems to become less pronounced with increasing viral load levels. This suggests that the Quantiplex version 3.0 assay might overestimate or that the Quantiplex version 2.0 assay might underestimate at lower viral load levels (21). The two samples quantified by the Quantiplex version 3.0 assay in our study were from two individuals in the high viral load group, and the difference in the accuracy of quantification between the two versions would be minimal at that level.
DNA amplification and sequencing by PCR.
Proviral DNA was extracted from peripheral blood mononuclear cells with the Qiagen QIAmp Blood Midi kit (Qiagen, Inc., Chatsworth, Calif.). The DNA was resuspended in Tris-EDTA buffer, and its concentration was determined by measurement of the optical density at 260 nm. One microgram of DNA was used for each PCR. For each individual, three independent PCRs were performed under identical conditions for each sample at both time points to minimize resampling bias (2). To amplify the HIV-1 C2V3 env region, a nested PCR was performed with two sets of primers, WT1-WT2 for the first round and KK30-KK40 for the second round, as previously described (38, 48). Additionally, a single gag fragment was sequenced from each individual to ensure subtype concordance between the gag and the env genes, further verifying the classification of CRF02_A/G-IbNG. A nested PCR with two primer sets (p108-p109 for the first round and p91-p92 for the second round) was performed to amplify the HIV-1 gag 3′-p24/5′-p7 region, as previously described (35). Negative controls were included in each amplification reaction. The PCR products were purified by agarose gel electrophoresis and by using purification columns (gel extraction kit; Qiagen, Inc.). All purified products were subsequently cloned into the pCR2.1 vector (Topo-TA Cloning; Invitrogen, Carlsbad, Calif.). Several clones were processed for each PCR. The plasmid preparation for the double-stranded DNA sequencing was performed by alkaline lysis with silica columns (S.N.A.P. Mini-Prep kit; Invitrogen). Samples were sequenced by dye terminator cycle sequencing with Taq polymerase (Perkin-Elmer Applied Biosystem Division, Foster City, Calif.) and an automatic sequencer (ABI 373; Perkin-Elmer Applied Biosystem Division). Each earlier and later time point sample was processed separately, from DNA extraction to sequence analysis.
Sequence analysis and statistical methods.
For all generated C2V3 viral sequences, a multiple alignment was performed with the Clustal package of multiple alignment programs (Clustal X 1.64), with minor manual adjustments when necessary and complete removal of positions which contained gaps (45). Reference sequences from the HIV sequence database of the Los Alamos National Laboratory were included in certain analyses. Phylogenetic analyses were performed by the neighbor-joining method for both env and gag regions. The alignments were corrected for multiple substitutions, and reliability was estimated by 1,000 bootstrap resamplings (37). Consensus sequences were generated for each individual's earlier and later sequences by using the SeqPublish program (HIV sequence database; Los Alamos National Laboratory). DNA and protein distance analyses were executed with the DNAdist and Prodist programs by using the Phylip 3.572 package (9). Statistical analyses were performed with S-Plus version 6.0 software (Insightful Corporation, Seattle, Wash.) and another software program (version 4.0; Stata Corporation, College Station, Tex.). Viral load values for each individual were log10 transformed for the purpose of statistical analysis.
Nucleotide sequence accession numbers.
All nucleotide sequences were submitted to GenBank and provided with accession numbers (AF526650 to AF526876).
RESULTS
HIV-1 viral load assays in RNA plasma.
All twelve women evaluated in this study had viruses that clustered with CRF02_A/G-IbNG in both the env and the gag fragments. For the low viral load group, the mean viral load ± the standard deviation was 1,822 ± 1,250 RNA copies/ml of plasma (3.02 ± 0.283 log10 RNA copies/ml), and for the high viral load group, the mean viral load ± the standard deviation was 87,782 ± 66,203 RNA copies/ml of plasma (4.78 ± 0.304 log10 RNA copies/ml). Interestingly, 11 of the 12 individuals had a mean fluctuation in viral load measurement of <0.5 log10 RNA copies/ml, which is indicative of a setpoint pattern in viral kinetics (5, 16). A single individual had a viral load measurement that fluctuated by 0.85 log10 RNA copies/ml. These results suggested that our study subjects could be characterized by a stable viral setpoint kinetic pattern, as described earlier.
Earlier and later intrapatient diversity.
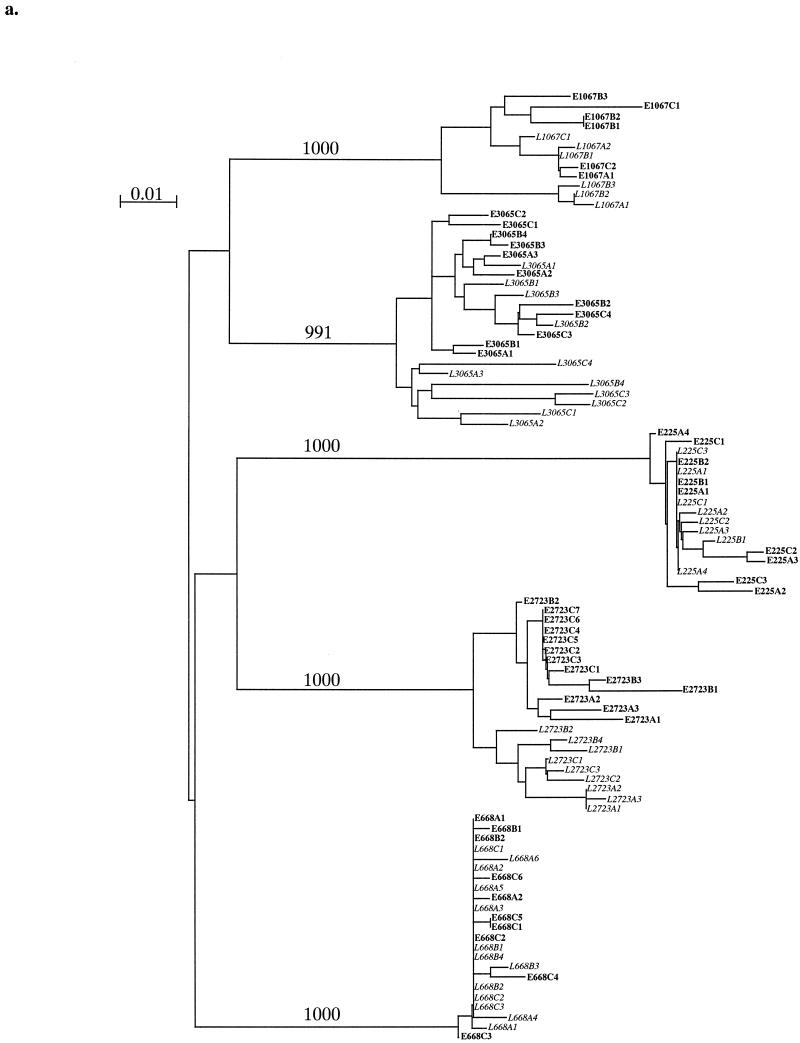
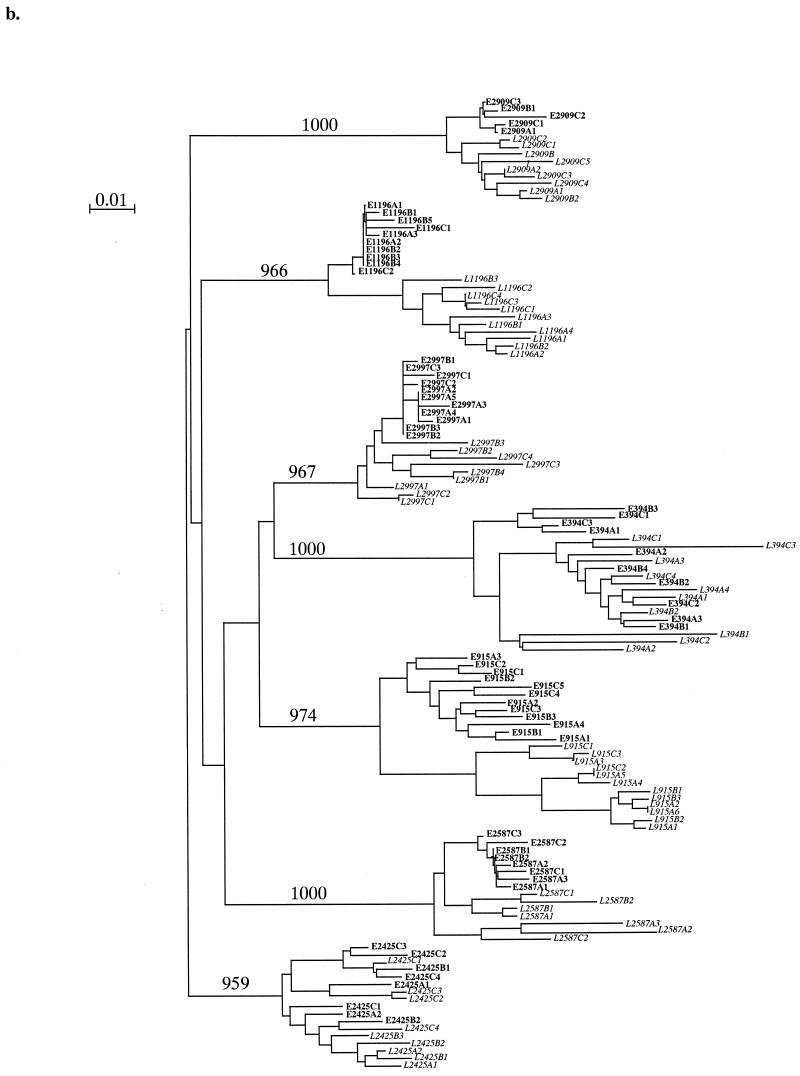
We generated and sequenced multiple C2V3 env clones from each time point. Panels a and b of Fig. 1 show two neighbor-joining phylogenetic trees including all earlier and later time point clones for each sample in the low and high viral load groups, respectively. There was a distinct difference between the high and low viral load groups in the clustering patterns of clones. Two of five low viral load individuals (subjects 2723 and 3065) showed evidence of separate clustering between the times the samples were collected, while the remainder revealed mixed earlier and later clones on branches. Four of five low viral load individuals (subjects 2723, 668, 225, and 1067) displayed horizontal branch lengths for the majority of their later clones that were similar to or shorter than those displayed for earlier clones (Fig. 1a), suggesting that later clones have fewer nucleotide changes (less divergence) than clones do. In the high viral load group, five of seven individuals (subjects 1196, 915, 2587, 2909, and 2997) revealed distinct clustering between all earlier and later clones, and there was evidence of a similar pattern emerging in a sixth individial (subject 2425) (Fig. 1b). Additionally, four of seven individuals (subjects 1196, 915, 2587, and 2997) displayed later clones with horizontal branch lengths that were greater than those of earlier clones, while the remaining individuals (subjects 394, 2909, and 2425) showed evidence of a similar trend in later clones. The most dramatic clustering and the longest clone branch lengths in later samples occurred in the two individuals with the greatest observation times, subjects 1196 and 915. Later clones from members of the high viral load group displayed longer branch lengths and therefore greater divergence those from members of the low viral load group. This phylogenetic analysis suggests that the high viral load group exhibited a higher magnitude of nucleotide divergence over time, a divergence not evident in analyses of earlier samples.
FIG. 1.
Low (a) and high (b) viral setpoint group neighbor-joining phylogenetic trees of multiple clones of HIV-1 env C2V3 sequences. The designations of earlier clones appear in roman type, while those of later clones appear in italics. In the clone designations, the letters E and L respectively indicate early and late time point clones, the letters A, B, and C indicate different PCRs, and the final digit indicates the clone number sequenced. The tree was constructed using 1,000 bootstrap resamplings. The number of resamplings carried out is indicated at each major branch point. Bar, 1% nucleotide divergence.
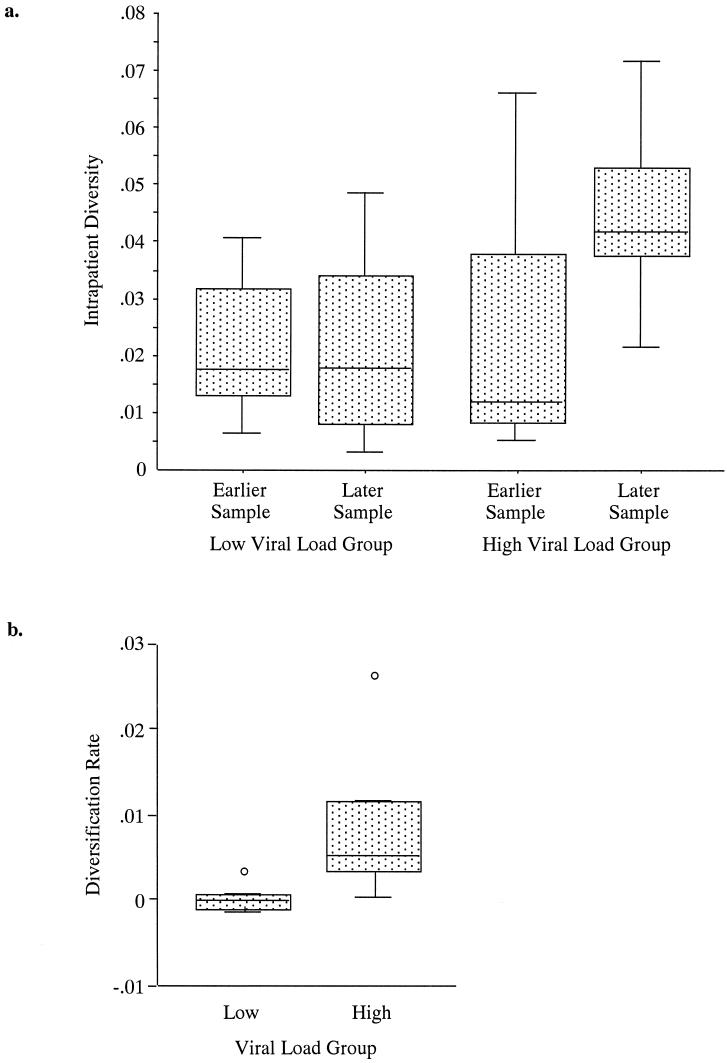
Intrapatient diversity was defined as the mean pairwise nucleotide distance, calculated by measuring the distances between all sequences from a single individual at a single time point, and was calculated at both time points for each individual (Fig. 2a). There was no significant difference in intrapatient diversity between the high (median diversity, 0.018) and low (median diversity, 0.012) viral load groups for the earlier time point (P = 0.88 [Wilcoxon rank sum test]). Therefore, both groups exhibited a similar level of intrapatient diversity at the earlier time point. The difference in intrapatient diversity at the later time point between the high (median diversity, 0.042) and low (median diversity, 0.018) viral load groups was not significant (P = 0.073 [Wilcoxon rank sum test]), although the trend was toward a greater intrapatient diversity in the high viral load group at the later time points, suggesting that high viral load subjects have a greater tendency towards development of a more diverse quasispecies population over time. There was a significant association between the log10-transformed viral load and later intrapatient diversity, with the linear regression model predicting that for each 1|−|log10 copy/ml increase in mean viral load, the diversity increased by 0.014 U (95% confidence interval [CI], 0.0017 to 0.026 U; P = 0.028). Additionally, there was no significant association between the mean log10-transformed viral load and intrapatient diversity at earlier time points (P = 0.54). These models suggest that the intrapatient diversity is positively correlated over the course of infection as a function of the viral load setpoint.
FIG. 2.
Box-and-whisker plots of intrapatient diversity at earlier and later time points (a) and diversification rates (b), stratified by viral load in each case. The line in the middle of the box represents the median and the box extends from the 25th percentile (x[25]) to the 75th percentile (x[75]), which is defined as the interquartile range (IQR). The whiskers extend to the upper and lower adjacent values. The upper adjacent value is less than or equal to x(75) + 1.5(IQR) and the lower adjacent value is greater than or equal to x(25) + 1.5(IQR). Any observed points more extreme than the adjacent values are referred to as outside values or outliers and are plotted individually.
Diversification and diversification rate.
In this study, diversification is defined as the change in intrapatient diversity, or the difference between earlier and later time point intrapatient diversity. The diversification rate is defined as the diversification divided by the observation time (the length of time between collection of earlier and later samples). Examination of the diversification rate revealed a significant difference, almost 200-fold, between the high (median diversification rate, 0.0054 [or 0.54%] per year) and low (median diversification rate, 0.000030 [or 0.003%] per year) viral load groups (P = 0.010 [Wilcoxon rank sum test]). The test was also significant (120-fold difference) when time was excluded (median diversification, 1.2 and 0.01% for high and low groups, respectively; P = 0.018 [Wilcoxon rank sum test]). Thus, the high viral load group displayed a diversification and a diversification rate that were significantly higher than those displayed by the low viral load group (Fig. 2b). However, linear regression models did not demonstrate statistically significant associations (P was 0.075 and 0.12 for the high and low viral load groups, respectively). These differences in the diversification and the diversification rate suggest the possibility that diversity is influenced by the number of cycles of viral replication and the proportional mutation rate.
Nucleotide divergence between all earlier and later viral sequences.
Divergence can be defined as the distance from a founder sequence to a later sequence. The average of nucleotide distance between all earlier and later sequences was calculated for each individual, and this single value approximated the nucleotide divergence from earlier to later virus populations in a single individual. This divergence value was significantly different for the high (median distance, 0.054) and the low (median distance, 0.029) load subjects (P = 0.015 [Wilcoxon rank sum test]), with the high viral load group subjects displaying a greater level of divergence from earlier to later time points (data not shown). Linear regression models identified a positive linear correlation between the log10-transformed viral load level and the nucleotide distance measurement, with a time-adjusted slope of 0.30 (95% CI, 0.083 to 0.52; P = 0.012) and nonadjusted slope of 0.28 (95% CI, 0.058 to 0.50; P = 0.018). A positive correlation between viral load level and the magnitude of nucleotide divergence was identified, with high viral load subjects displaying greater divergence over time.
Amino acid divergence between all earlier and later viral sequences.
The understanding of amino acid diversity and divergence is important inasmuch as the majority of immune responses to infection occur at the protein level. The average amino acid distance between all earlier and later sequences was calculated for each individual, and this single value approximated the amino acid divergence from earlier and later virus populations in a single individual. The amino acid divergence was significantly different for the high and low viral load groups (median divergence, 0.26 versus 0.11; P = 0.048 [Wilcoxon rank sum test]) with the high viral load subjects exhibiting a significantly higher amino acid divergence from earlier to later time points (data not shown). There was a positive but nonsignificant linear correlation between the protein distance and the log10-transformed viral load measurement (P = 0.094). This suggested that the greater nucleotide divergence noted in the high viral load group resulted in amino acid changes and therefore a greater amino acid divergence associated with high viral load levels. The increase in potential nonsynonymous changes in this population may provide evidence of additional selective pressures in individuals carrying high viral loads.
Consensus sequence analysis of nucleotide and amino acid divergence.
Consensus nucleotide and amino acid sequences were generated for all earlier and later time point samples from each subject. These alternate divergence measures were evaluated to confirm the previous findings. The nucleotide divergence between earlier and later consensus sequences was significantly greater in the high viral load group (median divergence, 0.027 [high viral load group] versus 0.0092 [low viral load group]; P = 0.023 [Wilcoxon rank sum test] [data not shown]). Additionally, the amino acid divergence between earlier and later consensus sequences was significantly greater in the high viral load group (median divergence, 0.13 [high viral load group] versus 0.044 [low viral load group]; P = 0.023 [Wilcoxon rank sum test] [data not shown]).
dn/ds ratios as a measure of positive selection pressure.
Characterization of nucleotide substitutions in a gene or genome involves several main forces, including mutation generation, random genetic drift, purifying or negative selection, and occasionally positive selection (12). The ratio of nonsynonymous substitutions per nonsynonymous site to synonymous substitutions per synonymous site (dn/ds ratio) is a measure of positive selection and is frequently used in the HIV-1 system. A dn/ds ratio of greater than 1 may be considered indicative of positive selective pressure. We calculated the dn/ds ratios for our groups of of high and low viral load individuals through the SNAP program (Los Alamos Database), based on the method employed by Nei and Gojobori (28). The calculated dn/ds ratios were not normally distributed; therefore, analysis by the nonparametric Wilcoxon rank sum test was conducted on both direct and loge-transformed values. There was no significant difference between the high and low viral load groups with regard to dn/ds ratios (median dn/ds ratio, 1.20 [high viral load group] versus 0.435 [low viral load group]; median loge dn/ds values, 0.19 [high viral load group] versus 0.83 [low viral load group]; P = 0.168 [Wilcoxon rank sum test]), although the trend was towards a higher median dn/ds ratio in the high viral load individuals. Synonymous changes per synonymous site were similar for individuals carrying high and low viral loads (median ds value, 0.0484 [high viral load group] versus 0.0419 [low viral load group]; median loge ds value, −3.03 [high viral load group] versus −3.17 [low viral load group]; P = 0.465 [Wilcoxon rank sum test]), but nonsynonymous changes per nonsynonymous site were significantly greater in the high viral load group than in the low viral load group (median dn value, 0.0567 [high viral load group] versus 0.0232 [low viral load group]; median loge dn value, −2.87 [high viral load group] versus −3.76 [low viral load group]; P = 0.028 [Wilcoxon rank sum test]). This argues against strong positive selection on one of the two populations but indicates that there may have been a greater number of nonsynonymous substitutions in the high viral load group. It is possible that selective pressures are constant across all individuals regardless of viral load category, and the greater number of nonsynonymous changes in the high viral load group might have occurred secondary to the higher viral replication rate, although this would predict an equivalent increase in the number of synonymous changes. Alternatively, it is possible that the high viral load population is subject to positive selective pressures, which could include diversifying selection for amino acid changes. This could select for a more cytopathic, pathogenic, or robust viral variant, as has been suggested previously (22).
Interaction between the effects of viral load and time and its effect on nucleotide and amino acid divergence.
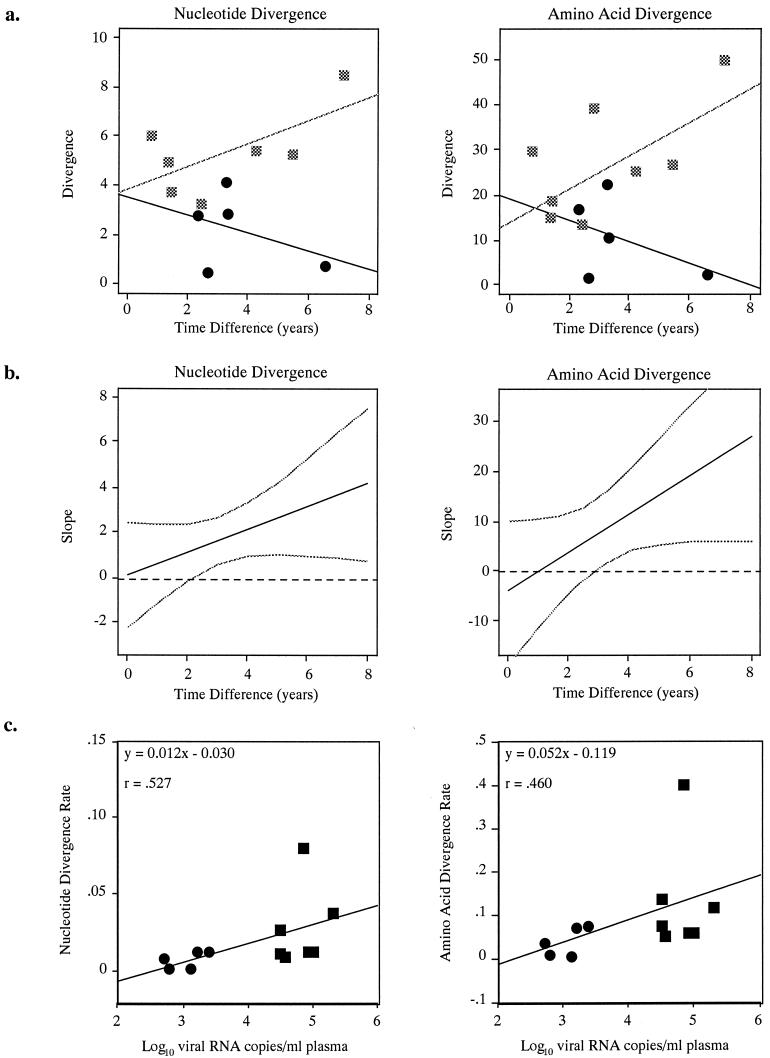
The rate of viral divergence over time appeared to differ by viral load level. This interaction was noted for viral load measured as a binary variable (Fig. 3a) and as a continuous quantitative variable (Fig. 3b.) Fig. 3a shows a plot of the nucleotide and amino acid divergence (mean nucleotide distance and mean protein distance between all earlier and later clones) versus the observation time separately by viral load group. In each plot, the best-fit regression lines for the low and high viral load groups are substantially nonparallel, illustrating the interaction. Statistical tests for interaction in the linear regression model gave P values of 0.148 and 0.089 for mean nucleotide and mean protein distances, respectively. Statistical tests for interaction in additional linear regression models for consensus nucleotide distances (P = 0.033) and consensus protein distances (P = 0.052) were performed (data not shown). The results were sensitive to the individual with low viral load in the bottom right corner of each plot; this and the small sample size imply that formal inferences should be made cautiously. Figure 3b illustrates the interaction when viral load is considered quantitatively for nucleotide and amino acid divergence. In each plot, the black line represents the estimated increase in mean divergence (100%) per 1|−|log10 increase in viral load as a function of time and the gray lines are 95% pointwise confidence intervals. The steepness of the (positive) slope indicates the strength of interaction, with a zero slope indicating no interaction. Statistical tests for interaction in the linear regression model gave P values of 0.175 and 0.102 for mean nucleotide and mean protein distances, respectively. Statistical tests for interaction in additional linear regression models for consensus nucleotide distances (P = 0.069) and consensus protein distances (P = 0.097) were performed (data not shown). In sum, the data suggest that the rate of viral divergence over time was modified by viral load, with a higher viral load leading to a considerably higher divergence rate. The relationship between divergence and time was different between high and low viral load levels, with evidence of a strong interaction between observation time and viral load level.
FIG. 3.
(a) Nucleotide and amino acid divergence (y axis) plotted against observation time in years (x axis) with regression lines. Squares, high viral load subjects; circles, low load subjects. (b) Slope of nucleotide and amino acid divergence (y axis, represented by mean distances) plotted against observation time (x axis) for all subjects. Solid line, estimated (percent) increase in mean divergence per 1|−|log10 increase in viral load as a function of time interval; broken line, 95% pointwise confidence interval; dashed horizontal line, zero line. (c) Number (log10 transformed) of viral RNA copies/ml of plasma (y axis) for each subject plotted against nucleotide and amino acid divergence rates (x axis, mean distance divided by observation time.).
An alternate method to discern the aforementioned interaction was to substitute a divergence rate (divergence divided by observation time) for the divergence measures. There was a significant positive correlation between the nucleotide and amino acid divergence rates and the log10-transformed viral load measurement (by both the Wilcoxon rank sum test and Spearman's correlation coefficient) in most of the divergence measures. Nucleotide and amino acid divergence rates are plotted against viral load in Fig. 3c, and the positive correlation between viral load level and divergence is evident.
DISCUSSION
Biologically relevant changes in viral replication occur at >0.5 log10 RNA copies/ml, and it has therefore been suggested that changes of <0.5 log10 RNA copies/ml over time are indicative of the viral setpoint (5, 16). The data for the majority of individuals evaluated in this cohort appear to corroborate the viral setpoint paradigm. The viral load measurements of 11 of 12 of our subjects fluctuated by <0.5 log10 RNA copies/ml, while a single individual had a variability of 0.85 log10 RNA copies/ml. All RNA measurements were taken during the disease-free interval. Our subjects provided a unique opportunity to evaluate viral loads during the disease-free interval for a non-B subtype. While the majority of setpoint studies have described subtype B-infected individuals undergoing antiretroviral therapy at some point during sampling, our samples were collected from antiretroviral therapy-naïve individuals, providing an opportunity to elucidate the natural history of viral kinetics in the absence of additional selective pressures.
Many studies have evaluated HIV-1 subtype B diversity at single and sequential time points and its correlation with disease progression (1, 7, 11, 15, 18, 20, 22, 24, 25, 40, 41, 43, 47). These studies included individuals treated with antiretroviral drugs and sampled both during the disease-free interval and once clinical AIDS was established. Some studies evaluated both HIV-1 seroincident and seroprevalent individuals. A few studies did not directly examine sequence when calculating diversity, while others did not control for resampling bias. To our knowledge, this is the first study to examine the relationship between viral load level and genetic diversity in a non-B subtype during the disease-free interval. We attempted to characterize the relationship between genetic diversity and the HIV-1 RNA viral load and also sought to determine whether the relationship was linear and positively or negatively correlated. Additionally, this design provided an opportunity to examine diversity during the disease-free interval, unaltered by the viral dynamics of clinical AIDS or acute infection. Not surprisingly, the magnitude of diversity was positively correlated to viral load level, such that individuals carrying high viral loads demonstrated a greater intrapatient diversity, diversification, diversification rate, and divergence over time in their nucleotide and protein sequences than did individuals carrying low viral loads. Further, the greater the observation time, the greater the generated divergence between the earlier and the later samples in the high viral load group. Accordingly, intrapatient diversity was similar between individuals carrying high and low viral loads earlier in infection, with a trend towards a greater intrapatient diversity in the high viral load group later in infection. Much of this may be attributable to mutations generated by increasing rounds of viral replication in the individuals carrying high viral loads as well as to rapid virus turnover. Reverse transcriptase has a high error rate and an inadequate proofreading and repair mechanism. Hence, the mutation rate of HIV-1 has a profound effect on genetic variability (4), and if diversity generated by mutation rate is directly proportional to rounds of replication, this diversity will be positively compounded by time. Not surprisingly, the high levels of nucleotide diversity and divergence found in the high viral load group was magnified in individuals with the greatest observation time, a finding similar to what has been observed for subtype B infections (17). It therefore seems quite likely that viral replication rate and the resultant level of circulating virus is the primary driving force in the generation of nucleotide diversity in this population of CRF02_A/G-IbNG-infected individuals.
All individuals generated diversity and divergence proportional to their viral load level: individuals with high viral load were characterized by high diversity and divergence and individuals with low viral load were characterized by lower diversity and divergence, as has been described previously in studies of HIV-1 subtype B (22, 25). Increased nucleotide diversity and divergence which resulted in concordant amino acid divergence was noted in high viral setpoint individuals. Accordingly, individuals carrying high viral loads possessed a greater number of nonsynonymous nucleotide changes. This might be consistent with initial strong immune pressures on the virus population until rapidly replicating variants predominated to push the individual to AIDS (30) or on individual virulent variants resulting due to the accumulation of nonsynonymous changes during the progression to AIDS (40). There was no significant difference in the dn/ds ratio between high and low viral load individuals. However, one cannot rule out the possibility of positive selective forces acting on the high viral load population, or of the absence of positive selection on the low viral load population. Even the presence of small selective forces, whether positive or negative in nature, could influence variation in the population (4). The magnitude of sequence divergence noted in the high viral setpoint individuals in this study is most likely a direct function of a higher viral replication rate and duration of infection (time) to compound resulting error-associated mutations. Alternatively, one cannot rule out new, unique, and virulent viral variants that emerge as a result of high replication rate in the individuals with high viral setpoints and continue to reestablish new populations as infection time progresses.
Unlike many of the published studies of diversity, which evaluated subtype B-infected homosexual men, this study was an evaluation of a cohort composed solely of women infected with non-subtype B virus through a heterosexual mode of transmission. The role of gender-specific differences in the biology of HIV disease pathogenesis and transmission remains to be fully characterized, although the data suggest that such differences may exist in viral load (8, 10, 36, 42) and pathogenesis (8, 10). There is also evidence that HIV-1-infected women from Mombasa, Kenya, harbor viruses of greater heterogeneity than do HIV-1-infected men of the same population (19). Such gender-specific differences in biology and transmission may affect the interpretation of diversity in this population as well as the general applicability of any findings to other HIV-1-infected populations.
It is also possible that diversity seen in the env C2V3 region may not be fully representative of diversity across the entire genome and that selective forces towards positive selection in this region may not be applicable to forces across the entire genome. However, the V3 region is an important functional region for virus tropism and pathogenicity, cytotoxic T-lymphocyte responses, humoral antibody responses, and coreceptor usage and is also a region of frequent mutations (22). Therefore, examination of mutations and diversity in this region may represent genetic diversity in functionally important domains more susceptible to selection. Future studies to further characterize the relationship between genetic diversity and viral load are clearly needed.
It is also necessary to address and better clarify the role of immunologic correlates and their associations with viral load. We have recognized an interaction between genetic diversity, viral setpoint, and duration of infection. Therefore, elucidation of the etiology and directionality of these variables and their influences on one another may provide additional prognostic markers for disease progression in regions where non-B subtypes predominate. In particular, accounting for such diversity during vaccine design might allow for selection of more broadly neutralizing epitopes for use in negative partners in discordant couples, babies of HIV-1-infected mothers, or in other individuals at risk for transmission by viruses that establish high viral setpoints. Elucidation of the interactions between diversity and markers of disease progression will be pivotal for vaccine design, therapy development, and disease pathogenesis studies.
Acknowledgments
We thank Mary Fran McLane, Christopher Mullins, Donald Hamel, Abdoulaye Dieng-Sarr, and Aissatou Gueye-Ndiaye for consistent support and assistance and Wei-Kung Wang for critical review and suggestions.
This work was supported in part by U.S. Army grant DAMD 17-95-C-5005 as well as by NIH grants AI 43879 and AI 467274.
REFERENCES
- 1.Bagnarelli, P., F. Mazzola, S. Menzo, M. Montroni, L. Butini, and M. Clementi. 1999. Host-specific modulation of the selective constraints driving human immunodeficiency virus type 1 env gene evolution. J. Virol. 73:3764-3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker-Pergola, G., J. L. Mellquist, J. R. Eshleman, J. B. Jackson, and S. H. Eshleman. 1999. Improved detection of human immunodeficiency virus type 1 variants by analysis of replicate amplification reactions: relevance to studies of human immunodeficiency virus type 1 vertical transmission. Mol. Diagn. 4:261-268. [DOI] [PubMed] [Google Scholar]
- 3.Carr, J. K., T. Laukkanen, M. O. Salminen, J. Albert, A. Alaeus, B. Kim, E. Sanders-Buell, D. L. Birx, and F. E. McCutchan. 1999. Characterization of subtype A HIV-1 from Africa by full genome sequencing. AIDS 13:1819-1826. [DOI] [PubMed] [Google Scholar]
- 4.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 5.de Wolf, F., I. Spijkerman, P. T. Schellekens, M. Langendam, C. Kuiken, M. Bakker, M. Roos, R. Coutinho, F. Miedema, and J. Goudsmit. 1997. AIDS prognosis based on HIV-1 RNA, CD4+ T-cell count and function: markers with reciprocal predictive value over time after seroconversion. AIDS 11:1799-1806. [DOI] [PubMed] [Google Scholar]
- 6.Elbeik, T., E. Charlebois, P. Nassos, J. Kahn, F. M. Hecht, D. Yajko, V. Ng, and K. Hadley. 2000. Quantitative and cost comparison of ultrasensitive human immunodeficiency virus type RNA viral load assays: Bayer bDNA Quantiplex versions 3.0 and 2.0 and Roche PCR Amplicor Monitor version 1.5. J. Clin. Microbiol. 38:1113-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Essajee, S. M., H. Pollack, G. Rochford, I. Oransky, K. Krasinski, and W. Borkowsky. 2000. Early changes in quasispecies repertoire in HIV-infected infants: correlation with disease progression. AIDS Res. Hum. Retrovir. 16:1949-1957. [DOI] [PubMed] [Google Scholar]
- 8.Farzadegan, H., D. R. Hoover, J. Astemborski, C. M. Lyles, J. Margolick, R. Markham, T. Quinn, and D. Vlahov. 1998. Sex differences in HIV-1 viral load and progression to AIDS. Lancet 352:1510-1514. [DOI] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1989. PHYLIP: phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 10.Fideli, U. S., S. A. Allen, R. Musonda, S. Trask, B. H. Hahn, H. Weiss, J. Mulenga, F. Kasolo, S. H. Vermund, and G. M. Aldrovandi. 2001. Virologic and immunologic determinants of heterosexual transmission of human immunodeficiency virus type 1 in Africa. AIDS Res. Hum. Retrovir. 17:901-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ganeshan, S., R. E. Dickover, B. T. M. Korber, Y. J. Bryson, and S. M. Wolinsky. 1997. Human immunodeficiency virus type 1 genetic evolution in children with different rates of development of disease. J. Virol. 71:663-677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graur, D., and W. H. Li. 2000. Fundamentals of molecular evolution, 2nd ed. Sinauer Associates, Sunderland, Mass.
- 13.Ho, D. D. 1996. Viral counts count in HIV infection. Science 272:1124-1125. [DOI] [PubMed] [Google Scholar]
- 14.Hu, D. J., S. Vanichseni, T. D. Mastro, S. Raktham, N. L. Young, P. A. Mock, S. Subbarao, B. S. Parekh, L. Srisuwanvilai, R. Sutthent, C. Wasi, W. Heneine, and K. Choopanya. 2001. Viral load differences in early infection with two HIV-1 subtypes. AIDS 15:683-691. [DOI] [PubMed] [Google Scholar]
- 15.Hutto, C., Y. Zhou, J. He, R. Geffin, M. Hill, W. Scott, and C. Wood. 1996. Longitudinal studies of viral sequence, viral phenotype, and immunologic parameters of human immunodeficiency virus type 1 infection in perinatally infected twins with discordant disease courses. J. Virol. 70:3589-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaufmann, G. R., P. Cunningham, A. D. Kelleher, J. Zaunders, A. Carr, J. Vizzard, M. Law, and D. A. Cooper, et al. 1998. Patterns of viral dynamics during primary human immunodeficiency virus type 1 infection. J. Infect. Dis. 178:1812-1815. [DOI] [PubMed] [Google Scholar]
- 17.Kils-Hutten, L., R. Cheynier, S. Wain-Hobson, and A. Meyerhans. 2001. Phylogenetic reconstruction of intrapatient evolution of human immunodeficiency virus type 1: predominance of drift and purifying selection. J. Gen. Virol. 82:1621-1627. [DOI] [PubMed] [Google Scholar]
- 18.Liu, S. L., T. Schacker, L. Musey, D. Shriner, M. J. McElrath, L. Corey, and J. I. Mullins. 1997. Divergent patterns of progression to AIDS after infection from the same source - human immunodeficiency virus type 1 evolution and antiviral responses. J. Virol. 71:4284-4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long, E. M., H. L. Martin, J. K. Kreiss, S. M. J. Rainwater, L. Lavreys, D. J. Jackson, J. Rakwar, K. Mandaliya, and J. Overbaugh. 2000. Gender differences in HIV-1 diversity at time of infection. Nature Med. 6:71-75. [DOI] [PubMed] [Google Scholar]
- 20.Lukashov, V. V., C. L. Kuiken, and J. Goudsmit. 1995. Intrahost human immunodeficiency virus type 1 evolution is related to length of the immunocompetent period. J. Virol. 69:6911-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manegold, C., C. Krempe, H. Jablonowski, L. Kajala, M. Dietrich, and O. Adams. 2000. Comparative evaluation of two branched-DNA human immunodeficiency virus type 1 RNA quantification assays with lower detection limits of 50 and 500 copies per milliliter. J. Clin. Microbiol. 38:914-917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Markham, R. B., W. C. Wang, A. E. Weisstein, Z. Wang, A. Munoz, A. Templeton, J. Margolick, D. Vlahov, T. Quinn, H. Farzadegan, and X. F. Yu. 1998. Patterns of HIV-1 evolution in individuals with differing rates of CD4+ T-cell decline. Proc. Natl. Acad. Sci. USA 95:12568-12573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCutchan, F. E. 2000. Understanding the genetic diversity of HIV-1. AIDS 14:S31-S44. [PubMed] [Google Scholar]
- 24.McDonald, R. A., D. L. Mayers, R. C. Y. Chung, K. F. Wagner, S. Rattokim, D. L. Birx, and N. L. Michael. 1997. Evolution of human immunodeficiency virus type 1 env sequence variation in patients with diverse rates of disease progression and T-cell function. J. Virol. 71:1871-1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNearney, T., Z. Hornickova, R. Markham, A. Birdwell, M. Arens, A. Saah, and L. Ratner. 1992. Relationship of human immunodeficiency virus type 1 sequence heterogeneity to stage of disease. Proc. Natl. Acad. Sci. USA 89:10247-10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mellors, J. W., C. R. Rinaldo, P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 27.Montavon, C., C. Toure-Kane, F. Liegeois, E. Mpoudi, A. Bourgeois, L. Vergne, J. L. Perret, A. Boumah, E. Saman, S. Mboup, E. Delaporte, and M. Peeters. 2000. Most env and gag subtype A HIV-1 viruses circulating in West and West Central Africa are similar to the prototype A/G recombinant virus IbNG. J. Acquir. Immune Defic. Syndr. 23:363-374. [DOI] [PubMed] [Google Scholar]
- 28.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 29.Neilson, J. R., G. C. John, J. K. Carr, P. Lewis, J. K. Kreiss, S. Jackson, R. W. Nduati, D. Mbori-Ngacha, D. D. Panteleeff, S. Bodrug, C. Giachetti, M. A. Bott, B. A. Richardson, J. Bwayo, J. Ndinya-Achola, and J. Overbaugh. 1999. Subtypes of human immunodeficiency virus type 1 and disease stage among women in Nairobi, Kenya. J. Virol. 73:4393-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nowak, M. A., R. M. Anderson, A. R. McLean, T. F. Wolfs, J. Goudsmit, and R. M. May. 1991. Antigenic diversity thresholds and the development of AIDS. Science 254:963-969. [DOI] [PubMed] [Google Scholar]
- 31.Olaleye, D. O., Z. J. Sheng, T. M. Howard, and S. Rasheed. 1996. Isolation and characterization of a new subtype A variant of human immunodeficiency virus type I from Nigeria. Trop. Med. Int. Health 1:97-106. [DOI] [PubMed] [Google Scholar]
- 32.Popper, S. J., A. D. Sarr, A. Gueye-Ndiaye, S. Mboup, M. E. Essex, and P. J. Kanki. 2000. Low plasma human immunodeficiency virus type 2 viral load is independent of proviral load: low virus production in vivo. J. Virol. 74:1554-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popper, S. J., A. D. Sarr, K. U. Travers, A. Gueye-Ndiaye, S. Mboup, M. E. Essex, and P. J. Kanki. 1999. Lower human immunodeficiency virus (HIV) type 2 viral load reflects the difference in pathogenicity of HIV-1 and HIV-2. J. Infect. Dis. 180:1116-1121. [DOI] [PubMed] [Google Scholar]
- 34.Poss, M., and J. Overbaugh. 1999. Variants from the diverse virus population identified at seroconversion of a clade A human immunodeficiency virus type 1-infected woman have distinct biological properties. J. Virol. 73:5255-5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renjifo, B., W. Fawzi, D. Mwakagile, D. Hunter, G. Msamanga, D. Spiegelman, M. Garland, C. Kagoma, A. Kim, B. Chaplin, E. Hertzmark, and M. Essex. 2001. Differences in perinatal transmission among human immunodeficiency virus type 1 genotypes. J. Hum. Virol. 4:16-25. [PubMed] [Google Scholar]
- 36.Rezza, G., A. C. Lepri, A. D. Monforte, P. Pezzotti, F. Castelli, F. Dianzani, A. Lazzarin, A. DeLuca, M. Arlotti, F. Leoncini, P. E. Manconi, G. Rizzardini, L. Minoli, A. Poggio, G. Ippolito, A. N. Phillips, M. Moroni, et al. 2000. Plasma viral load concentrations in women and men from different exposure categories and with known duration of HIV infection. J. Acquir. Immune Defic. Syndr. 25:56-62. [DOI] [PubMed] [Google Scholar]
- 37.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 38.Sankale, J. L., D. Hamel, A. Woolsey, T. Traore, M. C. Dia, A. Gueye-Ndiaye, M. Essex, S. Mboup, and P. Kanki. 2000. Molecular evolution of human immunodeficiency virus type 1 subtype A in Senegal: 1988-1997. J. Hum. Virol. 3:157-164. [PubMed] [Google Scholar]
- 39.Sarr, A. D., J. L. Sankale, D. J. Hamel, K. U. Travers, A. Gueye-Ndiaye, M. Essex, S. Mboup, and P. J. Kanki. 2000. Interaction with human immunodeficiency virus (HIV) type 2 predicts HIV type 1 genotype. Virology 268:402-410. [DOI] [PubMed] [Google Scholar]
- 40.Shankarappa, R., P. Gupta, G. H. Learn, A. G. Rodrigo, C. R. Rinaldo, M. C. Gorry, J. I. Mullins, P. L. Nara, and G. D. Ehrlich. 1998. Evolution of human immunodeficiency virus type 1 envelope sequences in infected individuals with differing disease progression profiles. Virology 241:251-259. [DOI] [PubMed] [Google Scholar]
- 41.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489-10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sterling, T. R., C. M. Lyles, D. Vlahov, J. Astemborski, J. Margolick, and T. Quinn. 1999. Sex differences in longitudinal human immunodeficiency virus type 1 RNA levels among seroconvertors. J. Infect. Dis. 180:666-672. [DOI] [PubMed] [Google Scholar]
- 43.Strunnikova, N., S. C. Ray, R. A. Livingston, E. Rubalcaba, and R. P. Viscidi. 1995. Convergent evolution within the V3 loop domain of human immunodeficiency virus type 1 in association with disease progression. J. Virol. 69:7548-7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sutthent, R., S. Foongladda, S. Chearskul, N. Wanprapa, S. Likanonskul, U. Kositanont, S. Riengrojpitak, S. Sahaphong, and C. Wasi. 1998. V3 sequence diversity of HIV-1 subtype E in infected mothers and their infants. J. Acquir. Immune Defic. Syndr. 18:323-331. [DOI] [PubMed] [Google Scholar]
- 45.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The Clustal-X windows interface—flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, Z., C. M. Lyles, C. Beyrer, D. D. Celentano, D. Vlahov, C. Natpratan, R. Markham, C. Khamboonruang, K. Nelson, and X. F. Yu. 1998. Diversification of subtype E human immunodeficiency virus type 1 env in heterosexual seroconverters from northern Thailand. J. Infect. Dis. 178:1507-1511. [DOI] [PubMed] [Google Scholar]
- 47.Wolinsky, S. M., B. T. M. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whetsell, M. R. Furtado, Y. Z. Cao, D. D. Ho, J. T. Safrit, and R. A. Koup. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537-542. [DOI] [PubMed] [Google Scholar]
- 48.Wolinsky, S. M., C. M. Wike, B. T. Korber, C. Hutto, W. P. Parks, L. L. Rosenblum, K. J. Kunstman, M. R. Furtado, and J. L. Munoz. 1992. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 255:1134-1137. [DOI] [PubMed] [Google Scholar]