Abstract
The neurotrophin receptor (p75NTR) serves as a receptor for rabies virus (RV). We expressed and purified a soluble chimera consisting of the p75NTR ectodomain fused to the human immunoglobulin G1 (IgG1) Fc fragment (p75-Fc). Although p75-Fc interacts with RV, the infectivity of RV did not decrease significantly when it was incubated in the presence of the soluble receptor alone. However, when it was subsequently incubated with an antihuman IgG directed against the Fc fragment of p75-Fc, the infectivity of RV was significantly lowered (>90%), whereas incubation with antihuman IgG alone had no effect. We then selected eight independent RV mutants that were not neutralized by p75-Fc and antihuman IgG (srr [soluble receptor resistant] mutants). Each mutant carried a single mutation in the glycoprotein gene leading to one amino acid substitution in the protein. A total of four different substitutions were found. Two of the mutations were located at position 318 (phenylalanine replaced by a serine or a valine residue), and two were located at position 352 (histidine replaced by a tyrosine or an arginine residue). All of the mutations prevented the interaction with p75NTR as either a soluble or a membrane-anchored form. Two mutants (F318S) and (H352R) resulted in the formation of small plaques on BSR cells, probably due to the slower maturation of the glycoprotein. Immunoprecipitation, immunofluorescence, and neutralization assays showed that the four mutated glycoproteins still interacted with representative anti-RV glycoprotein monoclonal antibodies (MAbs), indicating that p75NTR binds outside of the known RV glycoprotein antigenic sites.
Rabies virus glycoprotein (RVG) is the only surface-exposed viral coat protein. It is organized in trimers (21) that protrude from the lipid envelope. RVG has three major functions in the RV infection process. First, RVG mediates cell binding through interaction with cellular receptors and is the first determinant of Lyssavirus neurotropism (3). Second, RVG mediates fusion between viral and cell membranes through a pH-dependent process (22, 39). Third, RVG, the main viral antigen, induces both humoral (neutralizing antibodies) (66) and cellular immune responses.
RVG is a 505-amino-acid type I membrane glycoprotein (1, 70) with three potential N-glycosylation sites: the first one, located at amino acid 37, is very inefficiently glycosylated (50); the second one, at position 204 or 247, is glycosylated in some strains, but not in others; and the third one, located at amino acid 319, is always glycosylated (67). RVG folding, intracellular transport, and oligomerization have been analyzed for Pasteur virus (PV) and Evelyn Rokitnicki-Abelseth (ERA) virus, and it has been shown that RVG acquired its trimeric organization in the endoplasmic reticulum (18, 65). The putative fusion peptide has also been localized between amino acids 102 and 179 (17).
RVG has been extensively mapped with specific anti-RVG monoclonal antibodies (MAbs) by different laboratories. Our group has selected about 50 RVG mutants from the challenge virus strain (CVS) that are resistant to neutralization by anti-RVG MAbs. Several independent antigenic sites have been defined, depending on their interaction with neutralizing anti-RVG MAbs and on the effect of mutations (6, 29). Site II is the major antigenic site (70% of the MAbs recognize this region). It contains two stretches of amino acid residues located at positions 34 to 42 and 184 to 210 (43) joined by a disulfide bridge between cysteines 35 and 207 (15, 17, 62). Site III is a conformational site (amino acids 330 to 340) and plays a major role in RV propagation in the nervous system and pathogenicity (12, 16, 49, 59). Two minor sites (site a at amino acids 342 and 343 [6] and a linear site at amino acids 251 to 264 [14, 28, 60]) and several other epitopes have also been described (28). RVG can adopt at least two conformations (20): N (for neutral) and I (for fusion inactive). In the I state, RVG is no longer recognized by site II-specific MAbs, but the virus can be neutralized by MAbs that do not neutralize the virus when RVG is in the N conformation (44). This property has been used to select RAIN (resistant to acid-induced neutralization) mutants (44). The corresponding mutations are located either at the N terminus of the RVG sequence (between amino acids 10 and 15) or at amino acid positions 44, 282, 392, and 396, the last ones controlling the conformational changes (19).
The identification of specific neuronal receptors is important for our understanding of the RV neurotropism. Although, wild-type RV strains show a tropism restricted to neurons, laboratory strains such as CVS or PV have a broader tropism and are also able to infect nonneuronal cells in vitro (48, 68). Several proteins have been proposed to mediate virus binding for RV laboratory and/or wild-type strains. The first one is the nicotinic acetyl-choline receptor (nAChR) (32). Sequence homology between RVG (site II) and snake toxins (α-bungarotoxin and d-tubocurarin) suggests that these molecules contain a common binding domain (34). This specific interaction is inhibited either by natural ligands or by a MAb directed against the receptor (33). The neuronal cell adhesion molecule (NCAM) has been proposed to serve as a receptor for RV laboratory strains (55). This molecule is expressed by different types of cells (neurons, lymphocytes, and fibroblasts). We have shown that the neurotrophin receptor (p75NTR) is a specific neuronal receptor for RV. A soluble form of RVG was used as a ligand in an expression cloning strategy with an NG108 cDNA library, which led to the identification of p75NTR (57). Fibroblasts expressing p75NTR can be infected by a wild-type RV isolate, whereas control cells are not susceptible. We recently showed that the glycoprotein of another member of the Lyssavirus genus (EBL2) interacts with p75NTR (58). This receptor is a type I glycoprotein and belongs to the tumor necrosis factor receptor family, characterized by several cysteine-rich domains (CRD) in the ectodomain and a death domain in the cytoplasmic region (5, 9). A few other viral receptors, such as HVEM (herpesvirus entry mediator) (40) and TvB, a receptor for the avian leukosis-sarcoma virus receptor subgroups B and D (7), also belong to this family. Deletion mutants of p75NTR have been engineered to map the RVG binding site. NGF binds to the second and third CRDs with low affinity (4, 63, 69), whereas soluble RVG binds to CRD1 with high affinity (29a).
A reciprocal strategy cannot be used to map the p75 binding site, because most of the deletions in the RVG ectodomain would affect its folding, intracellular processing, and oligomeric structure. Alternative approaches have been developed to map receptor binding sites in viral glycoprotein. Competition experiments between soluble receptors and antiviral glycoprotein MAbs have been the most frequently used strategy. The HVEM-binding region on HSV1 glycoprotein D (gD) was located by this approach (42, 64), but the interacting chains were only identified after determination of the three-dimensional structure of the HVEM-gD complex (8). The most powerful approach for the localization of the receptor binding site is to select mutants that escape neutralization by a soluble form of a viral receptor: they are named “srr (soluble receptor resistant) mutants.” Although neutralizing activity of soluble receptors has been reported for many viruses, such as poliovirus (11, 26), rhinovirus (23, 36), coronavirus (71, 72), human immunodeficiency virus (13, 24, 30, 52, 56), measles virus (10), and hepatitis A viruses (51), characterization of srr mutants has only been described in the cases of poliovirus (11, 26) and coronavirus (37, 38, 45, 46).
We selected srr mutants to map the precise p75 binding site on the RVG protein and to determine how srr mutations alter the antigenic structure of RVG. We hypothesize that srr mutants could be different from antigenic mutants, and they could subsequently be used to investigate the role of p75 in RV infection. We created a dimeric molecule containing the ectodomain of p75 fused to an Fc fragment of human immunoglobulin G1 (IgG1), which allows purification of the recombinant protein on a protein A-Sepharose column. In this chimera, the IgG hinge that separates and confers flexibility between the Fc fragment and the p75 ectodomain was conserved. Furthermore, the presence of the Fc fragment, inducing dimerization of the molecule, could increase the avidity of the receptor for the virus. First, we showed that p75-Fc and RVG interact directly, indicating that the binding of RV does not require any cellular cofactors. The RV infectivity only decreased significantly when incubation with an anti-Fc antibody was added. Eight independent srr mutants were selected from the RV CVS and characterized. Only four different mutations were found in the glycoprotein. We then studied the interaction between the mutant glycoproteins and specific anti-RVG MAbs.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
BSR cells, a clone of the BHK-21 (baby hamster kidney) line (47), were grown at 37°C in Glasgow's modified minimal essential medium (MEM) (Gibco-BRL) supplemented with 10% calf serum in a humidified incubator in a 5% CO2 atmosphere. COS-7 cells were obtained from Brian Seed (Massachusetts General Hospital, Boston, Mass.). They were grown at 37°C in Dulbecco's modified MEM (DMEM; Gibco-BRL) supplemented with 10% fetal bovine serum (FBS) in a 5% CO2 humidified incubator. Sf21 cells from Spodoptera frugiperda (Invitrogen) were grown at 28°C in TC100 medium (Gibco-BRL) supplemented with 10% FBS.
The RV CVS strain was amplified and titrated by plaque assay as previously described (44) onto BSR cells. Stocks from individual CVS plaques (10 clones) were amplified and used in this selection process. These stocks have been used previously for selecting mutants that escaped neutralization at acidic pH (44).
A recombinant baculovirus expressing the human p75NTR protein was constructed (58) and used to infect Sf21 cells. A rabbit polyclonal anti-p75NTR antibody (Rex) was obtained from Louis Reichardt (University of California, San Francisco), and a hybridoma producing the anti-p75NTR MAb ME-204 was obtained from the American Type Culture Collection. Polyclonal antihuman IgG was obtained from Jackson Laboratories.
Five neutralizing antiglycoprotein MAbs were used: 30AA5 and 40DC2 bind to RVG antigenic site II (43); 50AD1 and 41BC2 bind to site III (59); 9B4 binds to minor site a (6); and two nonneutralizing MAbs,17D2 and 21H8, respectively, bind to the linear epitope (28) and recognize all glycoprotein conformations (35). These antibodies were prepared and characterized in our laboratory.
Expression of a soluble form of receptor, p75-Fc.
The ectodomain of the human p75NTR receptor (amino acids 1 to 222) was cloned in fusion with the cDNA from the human IgG1 heavy chain Fc domain (2, 30). A plasmid (CD4-Fc) was obtained from Brian Seed (Massachusetts General Hospital); the CD4 coding sequence was excised with XhoI and BamHI restriction enzymes and replaced by the human p75NTR coding sequence (kindly provided by Barbara Hempstead, Cornell University, Ithaca, N.Y.). The p75NTR cDNA sequence was amplified by PCR with Tfu polymerase (Promega). The forward primer was complementary to the 5′ signal peptide sequence and contained an XhoI restriction site (underlined) (5′CGCGGGCTCGAGATGGACGGGCCGCGC3′), and the reverse primer corresponded to the 3′ end of the O-glycosylated stalk of p75NTR and contained a BamHI site (underlined) (5′GCGGGATCCCATCGGCCAGGGATC3′). The resulting fragment was digested with BamHI and XhoI and inserted into the linearized plasmid containing the human IgG1 cDNA sequence in frame. The insert was confirmed by sequencing.
COS-7 cells were transiently transfected with the p75-Fc constructs by the DEAE-dextran method (57). The cell supernatants were collected 72 h after transfection, clarified by low-speed centrifugation, and applied to a protein A-Sepharose column (Sigma). The p75-Fc protein was eluted with 0.1 M glycine [pH 3] and neutralized with 1/20 volume of 1 M Tris-HCl (pH 8). The protein was then desalted over FastDialyzer Interbiotech (Interchim) in phosphate-buffered saline (PBS) (10 mM phosphate buffer [pH 7.0], 137 mM NaCl, 2.6 mM KCl) and stored at −80°C. The protein concentration was determined by the Bradford method (Pierce). The protein was also analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE [12% polyacrylamide]) followed by Coomassie blue staining or Western blotting.
Purified recombinant HVEM-Fc, produced in mammalian cells, was purchased from R&D Systems. It contains the HVEM ectodomain fused to the human IgG1 Fc domain (40).
ELISA.
For the enzyme-linked immunosorbent assay (ELISA), purified RV (CVS) was diluted with TD buffer (137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, 25 mM Tris-HCl [pH 7.5]), dispensed into a 96-well plate (200 ng in 50 μl per well), and dried overnight at room temperature. The plate was saturated with 10% skim milk in PBS. Serial dilutions of purified p75-Fc or human IgG (Jackson) were added (50 μl per well) and incubated for 2 h at 37°C in a humidified incubator. The plate was washed three times with PBS containing 1% skim milk. Alkaline phosphatase-conjugated antihuman IgG (Sigma) diluted 1:1,000 in PBS containing 1% skim milk was added (50 μl per well), and the plate was incubated at 37°C for 1 h. Wells were washed three times with PBS. Finally, we added 200 μl of 1.33 mM p-nitrophenyl phosphate (Sigma) in 1 M Tris-HCl (pH 8) to each well. The A405 was measured with a spectrophotometer (Mini Reader; Dynatech Laboratories, Santa Monica, Calif.).
Neutralization by anti-RVG MAb or p75-Fc.
The virus inoculum (103 or 102 PFU) was incubated with ascitic fluids (diluted 100-fold) or with different amounts of p75-Fc, HVEM-Fc, or human IgG for 1 h at 37°C. When specified, 10 μg of antihuman IgG (Pierce) was added for 1 h at 37°C. The neutralization efficiency was estimated by means of a plaque assay with BSR cells as described previously (44).
Selection of srr mutants and sequencing.
Ten independent stocks of CVS (106 PFU each) were incubated with 5 μg of p75-Fc for 1 h at 37°C and then with 10 μg of anti-human IgG for 1 h at 37°C. Nonneutralized virus was plated onto BSR cell monolayers in 24-well plates. After 3 days at 37°C, the cell supernatants were collected and resubmitted to another cycle of neutralization and amplification. Each viral population was amplified three or four times in the presence of p75-Fc and antihuman IgG, and plaques were assayed on BSR cells to select for mutants. Plaques were picked, amplified in BSR cells, and submitted to a neutralization test with p75-Fc plus antihuman IgG. Only mutants in which more than 90% escaped neutralization by p75-Fc were further amplified and characterized. Total RNA was extracted from infected BSR cells with the RNAgents total RNA isolation kit (Promega). The RVG gene cDNA was generated by reverse transcription-PCR (RT-PCR)—first with avian myeloblastosis virus (AMV) reverse transcriptase (Promega) and second with Taq polymerase (Promega)—by using RVG-specific primers. The cDNA fragment was then purified and sequenced with a T7 sequencing kit (Amersham).
RVG immunoprecipitation by anti-RVG MAbs or p75-Fc with virus-extracted RVG.
Viral particles (50 μg) from CVS were solubilized in 1 ml of TD buffer plus 1% CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) (Sigma) for 30 min at 4°C and then spun for 5 min at 10,000 × g. Viral extract (200 μl) was incubated for 2 h at 4°C with 3 μg of either p75-Fc, HVEM-Fc, or human IgG preadsorbed onto protein A-Sepharose. Immune complexes were analyzed by SDS-PAGE (10% polyacrylamide), and RVG was detected by Western blotting with anti-RVG MAb 17D2.
RVG immunoprecipitation by anti-RVG MAbs or p75-Fc with cell-extracted RVG.
BSR cells (106cells in 35-mm-diameter dishes) were infected with each virus at a multiplicity of infection (MOI) of 2. Twenty-four hours postinfection, cells were radiolabeled for 1 h at 37°C with 50 μCi of [35S]methionine and [35S]cysteine (Promix; Amersham) in methionine- and cysteine-free MEM (Gibco-BRL). Cell extracts were obtained from TD buffer plus 1% CHAPS and a protease inhibitor cocktail (2 μg each of chymostatin, leupeptin, antipain, and pepstatin per ml and 16 μg of aprotinin per ml). After incubation for 30 min on ice, cellular debris was removed by centrifugation at 12,000 × g for 15 min. Cell extracts were incubated for 1 h at 4°C with the different anti-RVG MAbs (2 μl of ascitic fluids) or p75-Fc (4 μg) preadsorbed onto protein A-Sepharose. Immune complexes were then analyzed by SDS-PAGE (12% polyacrylamide). Immunoprecipitated RVG was detected by autoradiography with Biomax Kodak films.
Pulse-chase experiments.
Pules-chase experiments were performed as described by Gaudin (18). BSR cell monolayers were infected with CVS or srr mutants at an MOI of 5. After 24 h, the cells were preincubated in methionine- and cysteine-free medium for 30 min. The cells were then labeled with 100 μCi of [35S]methionine and [35S]cysteine (Promix; Amersham) for 3 min. Prewarmed DMEM containing 5 mM methionine and cysteine was then added for the indicated chase periods. Cells were lysed, and RVG was immunoprecipitated with MAb 17D2 as described above.
SDS-PAGE and immunoblot analysis.
Protein samples were boiled for 5 min in Laemmli buffer (27). Electrophoresis was carried out in a discontinuous SDS-PAGE gel. Proteins were either stained with Coomassie blue or transferred onto nitrocellulose membranes (BA85; Schleicher & Schuell). Membranes were saturated with 5% skim milk in Tris-buffered saline(TBS) (50 mM Tris-HCl [pH 8], 150 mM NaCl) containing 0.05% Tween 20 for 30 min and then incubated overnight with the appropriate antibodies. Membranes were washed twice in TBS-Tween and incubated for 1 h with a peroxidase-conjugated secondary antibody (Sigma). The immune complex was detected after extensive washing in TBS-Tween by enhanced chemiluminescence (ECL Western blotting detection reagent; Amersham).
Binding of p75-expressing Sf21 cells to RV-infected BSR cells.
Binding assays were carried out between BSR cells infected with the different mutants and insect cells (Sf21) infected by a recombinant baculovirus and expressing p75NTR at their surface (p75-Sf21) as previously described (58). Briefly, a suspension of radiolabeled p75-Sf21 cells was added to BSR cell monolayers infected with the different mutants (MOI of 2) for 30 h and incubated for 30 min at room temperature. Unbound p75-Sf21 cells were removed by being washed three times with PBS, and specific binding was measured by counting the radioactivity associated with the infected BSR cells.
In parallel experiments, the expression of RVG at the surface of RV-infected BSR cells was monitored by surface immunoprecipitation with anti-RVG MAbs (30AA5 and 21H8) as described previously (58).
RESULTS
Expression of a soluble form of the receptor (p75-Fc).
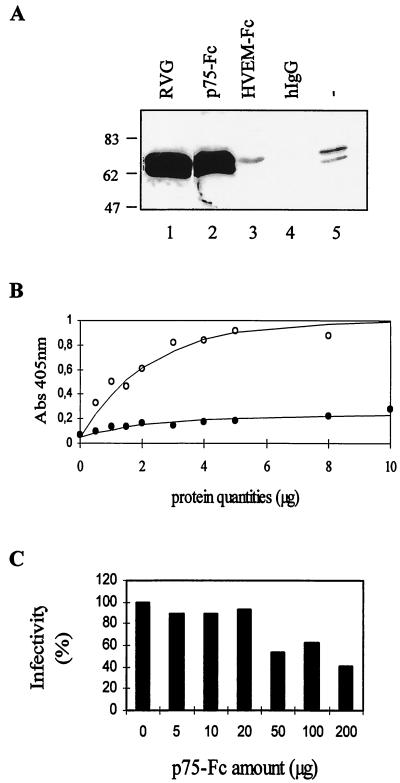
We expressed a soluble form of human p75NTR (p75-Fc) consisting of the whole ectodomain (amino acids 1 to 222, containing the four cysteine-rich domains and the O-glycosylated stalk) fused to the Fc portion of the human IgG1 heavy chain (Fig. 1A). The secreted chimeric receptor was produced in the supernatant of transiently transfected COS-7 cells. An aliquot of the supernatant was run on a denaturing gel under reducing conditions and analyzed by Western blotting. Two bands were detected with a polyclonal anti-p75 antibody: the major band corresponded to the p75-Fc monomer and migrated according to its predicted molecular weight, and the smaller band (indicated by an asterisk in Fig. 1B) likely corresponded to the calf IgG heavy chain from the serum detected by cross-reaction or to a degradation product of p75-Fc. The chimeric molecule (p75-Fc) was also detected by a conformational anti-p75 MAb (ME20.4, binding to CRDs 3 and 4) as well as by a polyclonal anti-human IgG (data not shown). The p75-Fc protein (about 5 μg/ml in the supernatant) was purified on a protein A-Sepharose column. The purity of the soluble receptor was estimated to be ≥90% (Fig. 1C); in some preparations, a small amount of calf IgG from the cell medium was also copurified on the protein A column. Under nonreducing conditions, p75-Fc migrated as a dimer due to the formation of interchain disulfide bridges (Fig. 1C), confirming that the Fc fragment induces dimerization of the recombinant protein. In the subsequent experiments, we used the soluble chimeric recombinant receptor (p75-Fc), which kept its antigenicity.
FIG. 1.
Expression and purification of recombinant p75-Fc protein in COS-7 cells. (A) Structural organization of the p75-Fc chimeric receptor, containing the four cysteine-rich domains and the O-glycosylated stalk from p75NTR fused to the Fc fragment of human IgG1. The chimera receptor was produced in the supernatant of transiently transfected COS-7 cells and analyzed by Western blotting with a rabbit polyclonal anti-p75NTR antibody (B). After purification on a protein A-Sepharose column, the proteins were analyzed under reducing (R) and nonreducing conditions (NR) by SDS-PAGE and Coomassie blue staining of the gel (C).
Interaction between p75-Fc and RVG.
First, we checked if the chimeric receptor was able to interact with RVG. Coprecipitation of trimeric RVG extracted from CVS virus particles was carried out (Fig. 2A). Preadsorbed p75-Fc efficiently pulled down RVG, but not an irrelevant Fc chimeric molecule (HVEM-Fc) or human IgG. Similar results were obtained when the RVG was solubilized under a monomeric conformation obtained by using Triton X-100 extraction (data not shown). We then used an ELISA to investigate the binding of the soluble receptor p75-Fc to RVG at the surface of the virus. A plate was coated with purified RV (CVS strain) and incubated with different amounts of purified p75-Fc or human IgG. A representative experiment is shown in Fig. 2B. p75-Fc bound to the virus in a concentration-dependent manner, with an estimated Kd of 400 nM. Human IgG did not bind to RV under the same conditions. We then investigated the ability of p75-Fc to neutralize RV infectivity. In a first series of experiments, 103 PFU of CVS was incubated with various amounts of p75-Fc, and virus titers were determined by plaque assay on BSR cell monolayers. Titers did not decrease in the presence of up to 20 μg of p75-Fc and were roughly halved in the presence of higher concentrations of the molecule. Therefore, although p75-Fc interacts with RVG either in a soluble form or at the surface of the virus, as shown by ELISA, it did not efficiently neutralize RV infectivity (Fig. 2C).
FIG. 2.
p75-Fc interacts with RVG. (A) Coprecipitation. RVG, solubilized from CVS particles with 1% CHAPS, was incubated with preadsorbed p75-Fc (lane 2), HVEM-Fc (lane 3), human IgG (lane 4), or protein A-Sepharose (lane 5). The immune complexes were analyzed by SDS-PAGE, and the presence of RVG was detected by Western blotting with anti-RVG 17 D2. In lane 1, solubilized RVG was loaded. (B) ELISA. Various amounts of purified p75-Fc (open circles) or human IgG1 (solid circles) were applied to a 96-well plate that had been coated with 200 ng of purified virus per well. Bound recombinant proteins were detected with an antihuman IgG alkaline phosphatase-conjugated antibody, with p-nitrophenyl phosphate as a substrate. A405 was measured. The data are the means from duplicate wells. (C) Neutralization. CVS (103 PFU) was incubated with different amounts of purified p75-Fc. A plaque assay was then performed with BSR cell monolayers. The residual infectivity was calculated as the ratio of virus titers obtained in the presence and absence of p75-Fc.
Selection of mutants resistant to neutralization by a soluble form of the receptor (srr mutants).
Our goal was to map the RVG domain involved in p75NTR binding by selecting independent srr mutants that escaped neutralization by a soluble form of the receptor. We hypothesized that the binding of p75-Fc to the virus was insufficient to prevent RV from interacting with cells and therefore infecting them. Thus, we tried to increase the neutralization efficiency of the p75-Fc molecule by adding an incubation step with an antihuman IgG specific to the Fc part of the chimera, expecting that it would increase the avidity of the p75-Fc-anti-Fc complex for RV or would increase the steric hindrance. This antibody should not interfere with the interaction between p75-Fc and RVG, which occurs at the N-terminal part of p75NTR (29a).
Efficient neutralization with p75-Fc was achieved when an additional incubation with an antihuman IgG antibody was performed (Fig. 3). No neutralization was observed when HVEM-Fc or human IgG was used alone or in combination with an antihuman IgG antibody (Fig. 3). When we increased the viral inoculum up to 106 PFU, the neutralization rate was only about 90% when both p75-Fc and anti-Fc antibody were used (data not shown) and never reached the efficiency observed with an anti-RVG specific neutralizing MAb (>99.99%), conditions required for selection of mutants in a single step. To increase the proportion of srr mutants in the viral populations, three consecutive passages in the presence of 5 μg of p75-Fc and 10 μg of antihuman IgG antibody were performed with 10 independent stocks of CVS. At each step, the survivors were amplified on BSR cell monolayers, which do not express the p75NTR receptor, but do express alternative receptors (53, 55, 68), allowing cell infection and amplification of srr mutants.
FIG. 3.
Neutralization of CVS by p75-Fc and antihuman IgG. The CVS inoculum (about 100 PFU) was incubated with different amounts (1 to 10 μg) of p75-Fc, HVEM-Fc, and human IgG for 1 h at 37°C alone (open bars) or with 10 μg of antihuman IgG (solid bars) for 1 additional h at 37°C. Virus was then titrated onto BSR cells by plaque assay. The data are the means of two neutralizations.
After three or four passages under neutralization conditions, each viral population was titrated by plaque assay in the presence of p75-Fc and anti-Fc antibody, and individual plaques were picked, amplified, and confronted again with p75-Fc plus anti-Fc antibody. We did not isolate mutants from two viral stock populations (CVS-1 and CVS-3). In total, eight independent mutants from the other eight CVS stocks were isolated and fully characterized. All of them except one, 4-1, showed a plating efficiency in presence of p75-Fc ≥1 (Table 1). The entire gene of the glycoprotein was sequenced for four of them, and partial sequences were obtained for the others (Table 2). Only four different substitutions were identified among the eight independent mutants. Two were located at position 318 of the viral glycoprotein (F318V and F318S) next to the N-glycosylation site, and two were located at position 352 (H352R and H352Y). The F318S and H352R mutants resulted in the formation of small plaques on BSR cells. The other two mutants gave bigger plaques than CVS (Table 2).
TABLE 1.
Selection of mutants resistant to the neutralization by p75-Fc
| Name of mutant | Parent virus | Titer
|
Plating efficiencya | |
|---|---|---|---|---|
| − anti-Fc Ab | p75-Fc + anti-Fc Ab | |||
| 2-4 | CVS-2 | 5 × 107 | 5 × 107 | 1 |
| 4-1 | CVS-4 | 7.5 × 107 | 5 × 107 | 0.7 |
| 5-4 | CVS-5 | 1 × 107 | 1 × 107 | 1 |
| 6-5 | CVS-6 | 5.5 × 107 | 7 × 107 | 1.3 |
| 7-5 | CVS-7 | 4.7 × 107 | 8.3 × 107 | 1.8 |
| 8-2 | CVS-8 | 5 × 107 | 6.5 × 107 | 1.3 |
| 9-1 | CVS-9 | 2.3 × 107 | 4 × 107 | 1.7 |
| 10-5 | CVS-10 | 1.5 × 107 | 1.5 × 107 | 1 |
The plating efficiency of the mutants was determined by dividing the titer obtained in presence of p75-Fc by the titer obtained in its absence.
TABLE 2.
Characterization of eight independent mutants resistant to neutralization by p75-Fca
| Name of mutant | Plaque phenotype | Mutation in glycoprotein gene
|
||
|---|---|---|---|---|
| Amino acid no. | Nature of substitution | Change in codon | ||
| 2-4 | small | 352 | H to R | CAT to CGT |
| 4-1 | small | 352 | H to R | CAT to CGT |
| 5-4∗ | large | 318 | F to V | TTC to GTC |
| 6-5∗ | small | 318 | F to S | TTC to TCC |
| 7-5∗ | large | 352 | H to Y | CAT to TAT |
| 8-2 | large | 318 | F to V | TTC to GTC |
| 9-1∗ | small | 352 | H to R | CAT to CGT |
| 10-5 | small | 318 | F to S | TTC to TCC |
One mutant per CVS stock was amplified and analyzed for the plaque phenotype on BSR cells. The entire gene of the glycoprotein was sequenced for the mutants noted by an asterisk, and partial sequence (amino acids 254 to 429) was obtained for the others.
Characterization of the srr mutants. (i) Interaction with anti-RVG MAbs.
Until now, no mutations had been described at positions 318 and 352. In order to determine if these mutations alter the antigenicity of the glycoprotein, we studied the interaction between the mutants and anti-RVG MAbs directed against site II (30AA5 and 40DC2), site III (50AD1 and 41BC2), site a (9B4), the linear epitope (17D2), and the acid-specific site (21H8). Five of these MAbs are neutralizing towards CVS, and all neutralized the srr mutants (Table 3). RVG from the different srr mutants was immunoprecipitated by all of the antibodies tested, but not by p75-Fc, whereas the CVS glycoprotein was immunoprecipitated by all of the MAbs and by p75-Fc (Fig. 4A). Because 50AD1 (site III specific) is an IgM and cannot be monitored by immunoprecipitation, we used immunofluorescence to investigate its binding to RV mutants, which was found to be normal (Fig. 4B). These results suggest that mutations at positions 318 and 352, located outside the previously characterized antigenic sites, do not alter the RVG antigenic structure (Table 3).
TABLE 3.
Interaction of srr mutants with anti-RVG MAbsa
| Virus | Characterization ofa:
|
||||||
|---|---|---|---|---|---|---|---|
| 21H8 (IP) | Site II, 30AA5-40DC2 (IP, IF, and NT) | Site III
|
Site a, 9B4 (IP and NT) | Linear site, 17D2 (IP) | P75-Fc (IP and NT) | ||
| 41BC2 (IP and NT) | 50AD1 (IF and NT) | ||||||
| CVS | + | + | + | + | + | + | + |
| F318V | + | + | + | + | + | + | − |
| F318S | + | + | + | + | + | + | − |
| H352Y | + | + | + | + | + | + | − |
| H352R | + | + | + | + | + | + | − |
Mutants were characterized for their interaction with anti-RVG MAbs by immunoprecipitation (IP), immunofluorescence (IF), or neutralization (NT).
FIG. 4.
Interaction between the srr mutant glycoprotein and p75-Fc or anti-RVG MAbs. BSR cells were infected with the different srr mutants at MOIs of 2 to 5. (A) After 24 h, the cells were radiolabeled for 1 h and then solubilized in TD buffer containing 1% CHAPS. Cell extracts were immunoprecipitated with anti-RVG MAbs or with p75-Fc preadsorbed onto protein A-Sepharose (protA) and analyzed by SDS-PAGE before autoradiography. (B) The interaction was also analyzed by immunofluorescence of RV-infected cells 18 h postinfection. Cells were incubated with MAb 50AD1, fixed with paraformaldehyde, and then incubated with a fluorescein isothiocyanate-conjugated antimouse antibody. Magnification, ×50.
(ii) Interaction with membrane-anchored p75NTR.
We analyzed the binding of srr mutant glycoproteins to a membrane-anchored form of p75NTR. Although poliovirus receptor srr mutants were selected for their resistance to a soluble form of the receptor, they were still able to bind to this receptor when expressed at the cell surface (11). We developed a specific binding assay between RV-infected BSR cells and radiolabeled Sf21 cells infected by a recombinant virus and expressing p75NTR at the surface (p75-Sf21) (58). The expression of srr-mutated RVG was monitored by cell surface immunoprecipitation with two anti-RVG MAbs (Fig. 5A). Cells infected by mutants F318V and H352Y expressed twice as much glycoprotein as CVS-infected cells, whereas cells infected by mutants F318S and H352R expressed the same amount of glycoprotein as CVS-infected cells. No radiolabeled p75-Sf21 cells bound to cell monolayers that have been infected with any of the srr mutants, even though they contained normal amounts of glycoprotein at their surface. Conversely, considerable amounts of p75-Sf21 cells bound when the BSR monolayer was infected with CVS (Fig. 5B). Both substitutions at amino acid 318 or 352 in RVG completely abolished the interaction with p75NTR as either a soluble or membrane-anchored receptor.
FIG. 5.
Binding of srr mutant glycoproteins to a membrane-anchored form of p75NTR. (A) RVG cell surface expression. BSR cells were infected with CVS or srr mutants, radiolabeled for 24 h, and incubated with MAbs 30AA5 and 21H8 at 4°C for 2 h. Cells were lysed, and immune complexes were immunoprecipitated on protein-A Sepharose before SDS-PAGE analysis. (B) Binding of radiolabeled p75-Sf21 cells to BSR cell monolayers infected with CVS or srr mutants. After washing, BSR cell-associated radioactivity was counted. Experiments were carried out in triplicate. MI, mock infected.
Folding of srr mutant RVG.
The migration profiles of glycoprotein were modified for two mutants (F318S and H352R) (Fig. 4A and 5A), but for the other two (F318V and H352Y), RVG migrated as a doublet in CVS (two of three of the molecules bearing two sugar chains and one of three of the molecules bearing a single sugar chain), whereas the srr glycoproteins (F318S and H352R) migrated as a triplet. This difference correlated with the plaque phenotype: F318S and H352R resulted in smaller plaques than CVS. This suggests that the transport or folding of the glycoprotein could be affected in these two mutants.
We used pulse-chase experiments (18) to compare the levels of maturation of RVG in the srr mutants (F318S and H352R) and in CVS. Immediately after the pulse, newly synthesized CVS-RVG, immunoprecipitated by MAb 17D2, migrated as a doublet, with one band corresponding to the addition of one high-mannose sugar chain and one corresponding to the addition of two sugar chains (Fig. 6). The addition of complex sugars in the Golgi apparatus increased the molecular weight, resulting in the presence of additional bands of high molecular weight and a decrease or disappearance of the smaller bands. No major differences were observed between CVS and the mutants in the first 20 min of chase. The larger band appears more slowly in the mutants than in CVS. The immature glycoprotein (the smallest band) was still present after 120 min of chase for the srr mutants (F318S and H352R). Immunofluorescence with an anti-RVG MAb showed an increased accumulation of the glycoprotein in the Golgi apparatus when BSR cells were infected by these mutants (data not shown). These data are in agreement with a slower transport of the glycoprotein correlated with a lower viral production and smaller plaques.
FIG. 6.
Maturation of the srr mutant glycoproteins. BSR cells were infected with CVS or srr mutants (F318S and H352R) at an MOI of 5 for 20 h and then pulse-labeled for 3 min and chased for the indicated times. Cells were solubilized with TD buffer containing 1% CHAPS, and glycoproteins were immunoprecipitated with anti RVG MAb 17D2 before SDS-PAGE analysis and autoradiography.
DISCUSSION
We report the selection and characterization of eight independent RV mutants (CVS) resistant to neutralization by a soluble form of p75NTR (srr mutants). We have shown that two substitutions, (phenylalanine 318 substituted for by a serine or a valine and histidine 352 substituted for by a tyrosine or an arginine) prevent RVG from interacting with p75NTR (soluble or membrane-anchored form) and confer resistance to neutralization by p75-Fc plus antihuman IgG.
Our selection protocol, run on 10 independent CVS stocks, led to the selection of independent srr mutants in 8 of them. The mutants were selected on fibroblasts (BSR) that do not express p75NTR. This selection process relies on the capability of the RV CVS laboratory strain to use alternative receptors on the surface of BSR cells, such as NCAM (55) or gangliosides (53, 54). Only four different substitutions, located in codons 318 and 352 of the viral glycoprotein gene, were found. Phenylalanine 318 was replaced by a valine (two mutants) or a serine (two mutants), and histidine 352 was replaced by a tyrosine (one mutant) or an arginine (three mutants) (Table 2). Mutants with the same substitution were selected in independent CVS stocks, suggesting that our selection protocol was fairly exhaustive and that most of the substitutions that were abrogating interaction with p75NTR and compatible with viral viability have been selected. Furthermore, a nucleotide transversion (T→G), believed to be less frequent than transition events, was independently found twice. The number (4) of srr mutations was similar to the number of mutants selected for their resistance to the neutralization by a single MAb (6, 28, 43, 49) (Fig. 7).
FIG. 7.
Localization of the srr mutations (indicated by asterisks) in the RVG antigenic structure. The abscissa shows the number of different amino acid substitutions found at each amino acid position. Most of the mutations have already been described previously (6, 28, 43, 44, 49, 59), with the exception of the mutations at positions 203, 210 in site II, 331 in site III, and 342 and 354 in minor site a (F. Lafay and C. Tuffereau, unpublished results).
The soluble p75-Fc receptor efficiently bound to RVG either in a soluble form or at the surface of the virus, confirming that no cellular coreceptors are required for this interaction, as previously demonstrated (58). p75-Fc alone did not significantly neutralize RV infectivity. This cannot be attributed to the dimeric nature of p75-Fc, because another soluble form (monomeric and containing only the CRD region) was also unable to neutralize RV infectivity, although it efficiently bound the virus (M. Mousli, unpublished results). The most likely explanation for this poor neutralization is that the interaction between p75-Fc and the virus was too weak. Indeed, the apparent Kd between the virus and p75-Fc was much higher (about 2 logs) than that between isolated RVG trimers and p75NTR (29a; M. Mousli and P. England, unpublished observations). This could suggest that the conformation of the glycoprotein at the surface of the virus is different than in soluble RVG and/or that the accessibility of the p75NTR binding site is restricted. Further experiments are required to distinguish between these two hypotheses. However, soluble viral receptors often bind without neutralizing. For instance, immunoadhesin (HVEM-Fc) binds to herpes simplex virus type 1 (HSV-1), but poorly neutralizes virus infectivity (40), and no mutants have yet been selected. Under our conditions, the addition of an antibody directed against a domain distinct from the binding domain led to neutralization. The antihuman antibody increased either the steric hindrance due to receptor binding or the avidity of the complex for RVG, thus preventing cell interaction and subsequent infection. Alternative methods to increase the neutralization have been proposed; for example, although monomeric soluble CD46 does not neutralize measles virus infectivity, octamerization of the soluble receptor increases the neutralization by up to 99% (10). Both the addition of an antibody and multimerization of the soluble receptor could improve viral neutralization.
Regardless of the method used (immunoprecipitation, immunofluorescence, or neutralization), no difference was observed between CVS and the srr mutants in their interactions with representative anti-RVG MAbs. In agreement with this result, none of the previously identified antigenic mutations were located at amino acid positions 318 and 352. This suggests that srr mutations map outside of the known antigenic sites of the RV glycoprotein (Fig. 4 and Table 3). These mutations are probably located in an external domain of the glycoprotein, which has not yet been identified by other methods. However, the protocols used to select antigenic mutants with neutralizing MAbs were more straightforward, and with the exception of the RAIN mutants (44), the neutralization efficiency was sufficient (>99.99%) to allow mutants to be selected in one step. It could be worth investigating whether any of the nonneutralizing anti-RVG MAbs (28) can be rendered neutralizing by the addition of a secondary antibody (antimouse IgG) and determining where the selected mutations map.
Two substitutions, one (F318S) located next to an N-glycosylation site and the other 34 amino acids downstream (H352R), appeared to alter some of the viral growth characteristics. These mutants resulted in smaller plaques than the parental CVS or the other mutants (Table 2), the viral titers were usually 1 log lower (data not shown), and RVG maturation seemed to be delayed (Fig. 6).
The srr mutations were located on both sides of RVG antigenic site III and minor site a (between amino acids 330 and 343) (Fig. 7). However, as mentioned above, regardless of the method used (immunoprecipitation, immunofluorescence, or neutralization), no differences between CVS and the srr mutants for interaction with site III- and site a-specific MAbs were observed. This suggests that mutations in the p75NTR-binding domain did not drastically modify the conformation of the antigenic site III or of the minor site a. Conversely, the double mutation (K330N+R333M), in site III reduced the interaction between the glycoprotein and a membrane-anchored form of p75 (57) and conferred partial resistance to neutralization by p75-Fc plus anti-IgG (C. Langevin, unpublished observations). It is noteworthy that the K330N+R333M mutant, resulting from two independent mutational events (12), could not have been selected on CVS stocks in this selection process.
The role of p75NTR in RV infection is still unknown, and some previous reports need to be reevaluated in the light of recent data. It has been reported that the p75−/− mice generated by Lee and coworkers (31) are equally as susceptible to RV infection as control mice (25). However, it has recently been shown (61) that these mice express a smaller membrane-anchored form of p75NTR (sp75) containing CRD1. We have shown that this domain is sufficient for RVG binding (29a). The srr mutants will be very useful for further studies on the role of p75NTR in RV infection. Dechant and coworkers recently generated true knockout (KO) mice. These mice have a more severe phenotypic alteration, resulting in a perinatal death rate of about 50% (41, 61). In future studies, we will compare the characteristics of CVS infection in p75−/− mice with those of srr mutants in normal mice (with the same genetic background), hoping that symptoms, lethality, and viral propagation in the central nervous system will be the same.
Acknowledgments
We are very appreciative of the excellent technical assistance provided by Jacqueline Bénéjean and Hélène Raux. We thank Anne Flamand, Florence Lafay, Patrice Coulon, and Patrick England for critical comments and careful reading of the manuscript.
Christelle Langevin is a graduate student supported by a fellowship from the Ministère de Education Nationale et de la Recherche. This work was supported by grants from the PRFMIP and from the 5th Framework Program of the EU (QLRT-1999-00573).
REFERENCES
- 1.Anilionis, A., W. H. Wunner, and P. J. Curtis. 1981. Structure of the glycoprotein gene in rabies virus. Nature (London) 294:275-278. [DOI] [PubMed] [Google Scholar]
- 2.Aruffo, A., I. Stamenkovic, M. Melnick, C. B. Underhill, and B. Seed. 1990. CD44 is the principal cell surface receptor for hyaluronate. Cell 61:1303-1313. [DOI] [PubMed] [Google Scholar]
- 3.Badrane, H., C. Bahloul, P. Perrin, and N. Tordo. 2001. Evidence of two Lyssavirus phylogroups with distinct pathogenicity and immunogenicity. J. Virol. 75:3268-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baldwin, A. N., and E. M. Shooter. 1995. Zone mapping of the binding domain of the rat low affinity nerve growth factor receptor by the introduction of novel N-glycosylation sites. J. Biol. Chem. 270:4594-4602. [DOI] [PubMed] [Google Scholar]
- 5.Barker, P. A. 1998. p75NTR: a study in contrasts. Cell Death Differ. 5:346-356. [DOI] [PubMed] [Google Scholar]
- 6.Benmansour, A., H. Leblois, P. Coulon, C. Tuffereau, Y. Gaudin, A. Flamand, and F. Lafay. 1991. Antigenicity of rabies virus glycoprotein. J. Virol. 65:4198-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brojatsch, J., J. Naughton, M. M. Rolls, K. Zingler, and T. A. Young. 1996. CAR1, a TNFR related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell 87:845-855. [DOI] [PubMed] [Google Scholar]
- 8.Carfi, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169-179. [DOI] [PubMed] [Google Scholar]
- 9.Chao, M. V. 1994. The p75 neurotrophin receptor. J. Neurobiol. 25:1373-1385. [DOI] [PubMed] [Google Scholar]
- 10.Christiansen, D., P. Devaux, B. Réveil, A. Evlashev, B. Horvat, J. Lamy, C. Rabourdin-Combe, J. H. M. Cohen, and D. Gerlier. 2000. Octamerization enables soluble CD46 receptor to neutralize measles virus in vitro and in vivo. J. Virol. 74:4672-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Colston, E., and V. R. Racaniello. 1994. Soluble receptor-resistant poliovirus mutants identify surface and internal capsid residues that control interaction with the cell receptor. EMBO J. 13:5855-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coulon, P., J.-P. Thernaux, A. Flamand, and C. Tuffereau. 1998. An avirulent mutant of rabies virus is unable to infect motoneurons in vivo and in vitro. J. Virol. 72:273-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deen, K., J. S. McDougal, R. Inacker, G. Folena-Wasserman, J. Artos, J. Rosenberg, P. J. Maddon, R. Axel, and R. W. Sweet. 1988. A soluble CD4 (T4) protein inhibits AIDS virus infection. Nature (London) 331:82-85. [DOI] [PubMed] [Google Scholar]
- 14.Dietzschold, B., M. Gore, D. Marchadier, H.-S. Niu, H. M. Bunschoten, L. Otvos, Jr., W. H. Wunner, H. C. J. Ertl, A. D. M. E. Osterhaus, and H. Koprowski. 1990. Structural and immunological characterization of a linear virus-neutralizing epitope of the rabies virus glycoprotein and its possible use in a synthetic vaccine. J. Virol. 64:3804-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dietzschold, B., T. J. Wiktor, R. Macfarlan, and A. Varrichio. 1982. Antigenic structure of rabies glycoprotein: ordering and immunological characterization of the large CNBr cleavage fragments. J. Virol. 44:595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dietzschold, B., W. Wunner, T. Wiktor, A. D. Lopes, M. Lafon, A. L. Smith, and H. Koprowski. 1983. Characterization of an antigenic determinant of the glycoprotein that correlates with pathogenicity of rabies virus. Proc. Natl. Acad. Sci. USA 80:70-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durrer, P., Y. Gaudin, R. H. W. Ruigrok, R. Graf, and J. Brunner. 1995. Photolabelling identifies a putative fusion domain in the envelope glycoprotein of rabies and vesicular stomatitis viruses. J. Biol. Chem. 270:17575-17581. [DOI] [PubMed] [Google Scholar]
- 18.Gaudin, Y. 1997. Folding of rabies virus glycoprotein: epitope acquisition and interaction with endoplasmic reticulum chaperones. J. Virol. 71:3742-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaudin, Y., H. Raux, A. Flamand, and R. W. H. Ruigrok. 1996. Identification of amino acids controlling the low-pH-induced conformational changes of rabies virus glycoprotein. J. Virol. 70:7371-7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaudin, Y., R. W. H. Ruigrok, M. Knossow, and A. Flamand. 1993. Low-pH conformational changes of rabies virus glycoprotein and their role in membrane fusion. J. Virol. 67:1365-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaudin, Y., R. W. H. Ruigrok, C. Tuffereau, M. Knossow, and A. Flamand. 1992. Rabies virus glycoprotein is a trimer. Virology 187:627-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gaudin, Y., C. Tuffereau, D. Segretain, M. Knossow, and A. Flamand. 1991. Reversible conformational changes and fusion activity of rabies virus glycoprotein. J. Virol. 65:4853-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greve, J. M., C. P. Forte, C. W. Marlor, A. M. Meyer, H. Hoover-Litty, D. Wunderlich, and A. McClelland. 1991. Mechanism of receptor-mediated rhinovirus neutralization defined by two soluble forms of ICAM-1. J. Virol. 65:6015-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hussey, R. E., N. E. Richardson, M. Kowalski, N. R. Brown, H. C. Chang, R. F. Siliciano, T. Dorfman, B. Walker, J. Sodroski, and E. L. Reinherz. 1988. A soluble CD4 protein selectively inhibits HIV replication and syncytium formation. Nature (London) 331:78-81. [DOI] [PubMed] [Google Scholar]
- 25.Jackson, A. C., and H. Park. 1999. Experimental rabies virus infection of p75 neurotrophin receptor-deficient mice. Acta Neuropathol. 98:641-644. [DOI] [PubMed] [Google Scholar]
- 26.Kaplan, G. G., D. Peters, and V. R. Racaniello. 1990. Poliovirus mutants resistant to neutralization with soluble cell receptors. Science 250:1596-1599. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli, U. K. 1970. Cleavage of stuctural proteins during the assembly of the head of the bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 28.Lafay, F., A. Benmansour, K. Chebli, and A. Flamand. 1996. Immunodominant epitopes defined by a yeast-expressed library of random fragments of the rabies virus glycoprotein map outside major antigenic sites. J. Gen. Virol. 77:339-346. [DOI] [PubMed] [Google Scholar]
- 29.Lafon, M., T. Wiktor, and R. I. Macfarlan. 1983. Antigenic sites on the rabies virus glycoprotein: analysis with monoclonal antibodies. J. Gen. Virol. 64:843-851. [DOI] [PubMed] [Google Scholar]
- 29a.Langevin, C., H. Jaaro, S. Bressanelli, M. Fainzilber, and C. Tuffereau. 2002. Rabies virus glycoprotein (RVG) is a trimeric ligand for the N-terminal cysteine-rich domain of the mammalian p75 neurotrophin receptor. J. Biol. Chem. 277:37655-37662. [DOI] [PubMed] [Google Scholar]
- 30.Langner, K. D., M. Niedrig, P. Fultz, D. Anderson, G. Reiner, H. Repke, H. Gelderblom, B. Seed, J. Hilfenhaus, and G. Zettlmeissl. 1993. Anti-viral effect of different CD4-immunoglobulin constructs against HIV-1 and SIV: immunological characterization, pharmacokinetic data and in vivo experiments. Arch. Virol. 130:157-170. [DOI] [PubMed] [Google Scholar]
- 31.Lee, K. F., H. Li, L. J. Huber, S. C. Landis, A. H. Sharpe, M. V. Chao, and R. Jaenisch. 1992. Targeted mutation of the gene encoding the low affinity NGF receptor p75 leads to deficits in the peripheral sensory nervous system. Cell 69:737-749. [DOI] [PubMed] [Google Scholar]
- 32.Lentz, T. L., T. G. Burrage, A. L. Smith, J. Crick, and G. H. Tignor. 1982. Is the acetylcholine receptor a rabies receptor? Science 215:182-184. [DOI] [PubMed] [Google Scholar]
- 33.Lentz, T. L., E. Hawrot, and P. T. Wilson. 1987. Synthetic peptides corresponding to sequences of snake venom neurotoxins and rabies virus bind to the nicotinic acetylcholine receptor. Proteins 2:298-307. [DOI] [PubMed] [Google Scholar]
- 34.Lentz, T. L., P. T. Wilson, E. Hawrot, and D. W. Speicher. 1984. Amino acid sequence similarity between rabies virus glycoprotein and snake curaremimetic neurotoxins. Science 226:847-848. [DOI] [PubMed] [Google Scholar]
- 35.Maillard, A. P., and Y. Gaudin. 2002. Rabies virus glycoprotein can fold in two alternative, antigenically distinct conformations depending on membrane-anchor type. J. Gen. Virol. 83:1465-1476. [DOI] [PubMed] [Google Scholar]
- 36.Marlin, S. D., D. E. Staunton, T. A. Springer, C. Stratowa, W. Sommergruber, and V. J. Merluzzi. 1990. A soluble form of intercellular adhesion molecule-1 inhibits rhinovirus infection. Nature (London) 344:70-72. [DOI] [PubMed] [Google Scholar]
- 37.Matsuyama, S., R. Watanabe, and F. Taguchi. 2001. Neurovirulence in mice of soluble receptor-resistant (srr) mutants of mouse hepatitis virus: intensive apoptosis caused by less virulent srr mutant. Arch. Virol. 146:1643-1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuyama, S., and F. Taguchi. 2000. Impaired entry of soluble receptor-resistant mutants of mouse hepatitis virus into cells expressing MHVR2 receptor. Virology 273:80-89. [DOI] [PubMed] [Google Scholar]
- 39.Mifune, K., M. Ohuchi, and K. Mannen. 1982. Hemolysis and cell fusion by rhabdoviruses. FEBS Lett. 137:293-297. [DOI] [PubMed] [Google Scholar]
- 40.Montgomery, R. I., M. S. Warner, B. J. Lumb, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells is mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 41.Naumann, T., E. Casademunt, E. Hollerbach, J. Hofmann, G. Dechant, M. Frotscher, and Y.-D. Barde. 2002. Complete deletion of the neurotrophin receptor p75NTR leads to long-lasting increases in the number of basal forebrain cholinergic neurons. J. Neurosci. 22:2409-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nicola, A. V., M. Ponce de Leon, R. Xu, W. Hou, J. C. Whitbeck, C. Krummenacher, R. I. Montgomery, P. G. Spear, R. J. Eisenberg, and G. H. Cohen. 1998. Monoclonal antibodies to distinct sites on herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J. Virol. 72:3595-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prehaud, C., P. Coulon, F. Lafay, C. Thiers, and A. Flamand. 1988. Antigenic site II of the rabies glycoprotein: structure and role in viral virulence. J. Virol. 62:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raux, H., P. Coulon, F. Lafay, and A. Flamand. 1995. Monoclonal antibodies which recognize the acidic configuration of the rabies glycoprotein at the surface of the virion can be neutralizing. Virology 210:400-408. [DOI] [PubMed] [Google Scholar]
- 45.Saeki, K., N. Ohtsuka, and F. Taguchi. 1997. Identification of spike protein residues of murine coronavirus responsible for receptor-binding activity by use of soluble receptor-resistant mutants. J. Virol. 71:9024-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saeki, K., N. Ohtsuka, and F. Taguchi. 1998. Isolation and characterization of murine coronavirus mutants resistant to neutralization by soluble receptors. Adv. Exp. Med. Biol. 440:11-16. [DOI] [PubMed] [Google Scholar]
- 47.Sato, M., N. Maeda, H. Yoshida, M. Urade, and S. Saito. 1977. Plaque formation of herpes virus hominis type 2 and rubella virus in variants isolated from the colonies of BHK21/WI-2 cells formed in soft agar. Arch. Virol. 53:269-273. [DOI] [PubMed] [Google Scholar]
- 48.Schneider, L. G., M. Horzineck, and H. D. Makeda. 1971. Purification of rabies virus from tissue culture. Arch. Gesamte Virusforsch. 34:351-359. [DOI] [PubMed] [Google Scholar]
- 49.Seif, I., P. Coulon, P. E. Rollin, and A. Flamand. 1985. Rabies virulence: effect on pathogenicity and sequence characterization of rabies virus mutations affecting antigenic site III of the glycoprotein. J. Virol. 53:926-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shakin-Eschelman, S. H., A. T. Remaley, A. T. Eshleman, W. H. Wunner, and S. L. Spitalnik. 1992. N-linked glycosylation of rabies virus glycoprotein. J. Biol. Chem. 267:10690-10698. [PubMed] [Google Scholar]
- 51.Silberstein, E., G. Dveksler, and G. G. Kaplan. 2001. Neutralization of hepatitis A virus (HAV) by an immunoadhesin containing the cysteine-rich region of HAV cellular receptor-1. J. Virol. 75:717-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith, D. H., R. A. Byrn, S. A. Marsters, T. Gregory, J. E. Gropman, and D. J. Capon. 1987. Blocking of HIV-1 infectivity by a soluble, secreted form of the CD4 antigen. Science 238:1704-1707. [DOI] [PubMed] [Google Scholar]
- 53.Superti, F., M. Derer, and H. Tsiang. 1984. Mechanism of rabies virus entry into CER cells. J. Gen. Virol. 65:781-789. [DOI] [PubMed] [Google Scholar]
- 54.Superti, F., B. Hauttecoeur, M. J. Morelec, P. Goldoni, B. Bizzini, and H. Tsiang. 1986. Involvement of gangliosides in rabies virus infection. J. Gen. Virol. 67:47-56. [DOI] [PubMed] [Google Scholar]
- 55.Thoulouze, M. I., M. Lafage, M. Schachner, U. Hartmann, H. Cremer, and M. Lafon. 1998. The neural cell adhesion molecule is a receptor for rabies virus. J. Virol. 72:7181-7190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Traunecker, A., W. Like, and K. Karjalaine. 1988. Soluble CD4 molecules neutralize human immunodeficiency virus type 1. Nature (London) 331:84-86. [DOI] [PubMed] [Google Scholar]
- 57.Tuffereau, C., J. Benejean, D. Blondel, B. Kieffer, and A. Flamand. 1998. Low-affinity nerve-growth factor receptor (P75NTR) can serve as a receptor for rabies virus. EMBO J. 17:7250-7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tuffereau, C., E. Desmezieres, J. Benejean, C. Jallet, A. Flamand, N. Tordo, and P. Perrin. 2001. Interaction of lyssaviruses with the low-affinity nerve-growth factor receptor p75(NTR). J. Gen. Virol. 82:2861-2867. [DOI] [PubMed] [Google Scholar]
- 59.Tuffereau, C., H. Leblois, J. Bénéjean, P. Coulon, F. Lafay, and A. Flamand. 1989. Arginine or lysine in position 333 of ERA and CVS glycoprotein is necessary for rabies virulence. Virology 172:206-212. [DOI] [PubMed] [Google Scholar]
- 60.van der Heijden, R. W. J., J. P. M. Langedijk, J. Groen, F. G. C. M. UytdeHaag, R. H. Meloen, and A. D. M. E. Osterhaus. 1993. Structural and functional studies on a unique linear neutralizing antigenic site (G5) of the rabies virus glycoprotein. J. Gen. Virol. 74:1539-1545. [DOI] [PubMed] [Google Scholar]
- 61.von Schack, D., E. Casademunt, R. Schweigreiter, M. Meyer, M. Bibel, and G. Dechant. 2001. Complete ablation of the neurotrophin receptor p75NTR causes defects both in the nervous and the vascular system. Nat. Neurosci. 4:977-978. [DOI] [PubMed] [Google Scholar]
- 62.Walker, P. J., and K. Kongsuwan. 1999. Deduced structural model for animal rhabdovirus. J. Gen. Virol. 80:1211-1220. [DOI] [PubMed] [Google Scholar]
- 63.Welcher, A. A., C. M. Bitler, M. J. Radeke, and E. M. Shooter. 1991. Nerve growth factor binding domain of the nerve growth factor receptor. Proc. Natl. Acad. Sci. USA 88:159-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Whitbeck, J. C., S. A. Conolly, S. H. Willis, W. Hou, C. Krummenacher, M. Ponce de Leon, H. Lou, I. Baribaud, R. J. Eisenberg, and G. H. Cohen. 2001. Localization of the gD-binding region of the human herpes simplex virus receptor, HveA. J. Virol. 75:171-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whitt, M. A., L. Buonocore, C. Préhaud, and J. K. Rose. 1991. Membrane fusion activity, oligomerization, and virus assembly of the rabies virus glycoprotein. Virology 185:681-688. [DOI] [PubMed] [Google Scholar]
- 66.Wiktor, T., E. Gyorgy, D. Schlumberger, F. Sokol, and H. Koprowski. 1973. Antigenic properties of rabies virus components. J. Immunol. 110:269-276. [PubMed] [Google Scholar]
- 67.Wunner, W., B. Dietzschold, C. Smith, M. Lafon, and E. Golub. 1985. Antigenic variants of CVS rabies virus with altered glycosylation sites. Virology 140:1-12. [DOI] [PubMed] [Google Scholar]
- 68.Wunner, W. H., K. J. Reagan, and H. Koprowsky. 1984. Characterization of saturable binding sites for rabies virus. J. Virol. 50:691-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yan, H., and M. V. Chao. 1991. Disruption of cysteine-rich repeats of the p75 nerve growth factor receptor leads to loss of ligand binding. J. Biol. Chem. 266:12099-12104. [PubMed] [Google Scholar]
- 70.Yelverton, E., S. Norton, J. F. Objeski, and D. V. Goeddel. 1983. Rabies virus glycoprotein analogs: biosynthesis in Escherichia coli. Science 219:614-620. [DOI] [PubMed] [Google Scholar]
- 71.Zelus, B. D., D. R. Wessner, G. S. Dvesksler, and K. V. Holmes. 1998. Neutralizationof MHV-A59 by soluble recombinant receptor glycoproteins. Adv. Exp. Med. Biol. 440:3-9. [DOI] [PubMed] [Google Scholar]
- 72.Zelus, B. D., D. R. Wessner, R. K. Williams, M. N. Pensiero, F. T. Phibbs, M. DeSouza, G. S. Dveksler, and K. V. Holmes. 1998. Purified, soluble recombinant mouse hepatitis virus receptor, Bgp1b and Bgp2 murine coronavirus receptors differ in mouse hepatitis virus binding and neutralization activities. J. Virol. 72:7237-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]