Abstract
The UL24 gene of herpes simplex virus type 1 is conserved across many herpesviruses, but its protein product has not been identified. We expressed the UL24 gene in insect cells from a recombinant baculovirus and used the resulting protein to raise a rat antiserum. In immunoblotting experiments, this antiserum recognized a 30-kDa protein in lysates from infected cells. The identity of this species as UL24 was confirmed by using a virus encoding a truncated form of UL24. On the basis of biochemical fractionation of infected cells, UL24 appeared to be predominantly nucleus associated, especially at later times in infection. Although certain UL24 transcripts exhibit early kinetics, UL24 protein accumulated at later times in infection and levels were decreased sixfold in the presence of the viral DNA synthesis inhibitor phosphonoacetic acid; thus, UL24 was expressed with leaky-late kinetics. The entire UL24 open reading frame is encoded by mRNAs with two different 5′ ends. A mutation that eliminates the more abundant transcripts that originate at the first transcription start site resulted in a 10-fold reduction in the level of UL24 expressed but did not eliminate expression. Thus, the less abundant transcripts originating at the second transcription start site can evidently be translated, although transcripts originating at the first start site appear to be the major contributors to the expression of UL24. We conclude that UL24 is a bona fide herpes simplex virus type 1 protein that associates primarily with nuclei and whose expression is regulated at multiple levels.
The genome of herpes simplex virus type 1 (HSV-1) contains approximately 80 different genes (26), including one known as UL24. UL24 is evolutionarily conserved among many diverse herpesviruses and is considered to be a core herpesvirus gene (12). This conservation suggests an important role for UL24 in the life cycle of these viruses. Indeed, UL24 null mutations or mutations in the conserved regions of UL24 result in decreased viral yields in cultured cells (21). They also can confer a cell type-dependent, syncytial phenotype whereby infected cells fuse together at elevated temperatures (21, 33, 36). In mice, mutations in UL24 cause especially severe defects in viral replication in and reactivation from sensory ganglia (20). UL24 transcripts were identified many years ago (10, 22, 30, 31, 39), but the HSV-1 UL24 protein has not been identified. Although the syncytial phenotype observed in some UL24 mutant viruses has led to the suggestion that UL24 may encode a membrane protein (36, 38), there has been no definitive evidence to support this hypothesis.
Upon HSV-1 infection, immediate-early (α), early (β), and late (γ) viral genes are expressed in a temporal cascade, with expression of late genes being dependent on viral DNA synthesis (reviewed in reference 32). Several viral genes produce multiple transcripts, although little is known about the function of these apparently redundant transcripts. The pattern of transcription of the UL24 gene is quite complex, with six UL24 transcripts expressed during infection with HSV-1 (Fig. 1). The four transcripts that arise from the first two transcription start sites for UL24 (5.6, 5.4, 1.4, and 1.2 kb) contain the entire open reading frame (ORF), and it is not known which, if any, of these transcripts are translated. The remaining two transcripts (5.2 and 0.9 kb) originate within the ORF and have the potential to encode only a truncated form of the protein. The very long (336-base), GC-rich 5′ untranslated region (UTR) contained in the more abundant transcripts originating at the first start site might be expected to inhibit translation by adopting a secondary structure (discussed in reference 23). In addition, this region contains a small upstream ORF that, although its initiation codon's context does not conform to the Kozak consensus (23), could also inhibit (or conceivably boost) expression (reviewed in reference 15). The 5.4- and 1.2-kb transcripts that originate at the second start site, which have a short 5′ UTR, are less abundant. It is not known which transcription start site is more important for expression of UL24 protein. The 1.4-, 1.2-, and 0.9-kb transcripts terminate at the UL24 polyadenylation (polyA) signal and exhibit early kinetics, while the 5.6-, 5.4-, and 5.2-kb transcripts belong to the leaky-late kinetic class and terminate at the UL26 polyA signal (10, 16). However, the kinetics of expression of the putative UL24 protein are not known.
FIG. 1.
The UL24 transcription unit. (A) The top line represents the prototype organization of the HSV-1 genome; the shaded boxes represent the repeat segments, and the thin lines represent the unique regions. The UL24 gene lies at 0.3 map unit. Below is an expanded view of the locus containing UL24. The UL24 ORF is shown as a filled box with an arrow indicating the direction of transcription. The tk and UL25-UL26.5 ORFs are shown as open boxes with arrows. The six UL24 transcripts (thin lines with arrows) originate from three different transcription start sites and terminate at either the UL24 or the UL26 polyA signal. The transcripts that arise from the first two start sites (5.6, 5.4, 1.4, and 1.2 kb) contain the entire UL24 ORF. The other two transcripts (5.2 and 0.9 kb) originate from a start site within the ORF. The three transcripts that utilize the UL24 polyA signal (1.4, 1.2, and 0.9 kb) exhibit early kinetics (β), while the three that terminate at the UL26 signal (5.6, 5.4, and 5.2 kb) exhibit leaky-late kinetics (γ1). These longer transcripts contain sequences corresponding to the UL24, UL25, UL26 and UL26.5 genes. Transcripts arising from the second start site are the least abundant, as indicated by the thinner lines. The UL24 and tk genes are transcribed divergently, and the 5′ ends of the genes overlap. (B) 5′ UTR of UL24. The sequence from HSV-1 strain KOS of the region upstream of the UL24 ORF is shown with the ATG for the initiation codon in bold (19), with the same numbering system as that used by McGeoch et al. (26). Arrows above the sequence indicate the two transcription start sites for UL24 mRNAs that could encode a full-length protein. The short upstream ORF is boxed. This feature is also present in HSV-1 strain 17 and HSV-2 strains G and HG52.
To begin addressing questions regarding UL24 function and regulation, we have raised antiserum that recognizes the UL24 protein and have studied its localization and kinetics of expression and the contribution of various UL24 transcripts to its synthesis.
MATERIALS AND METHODS
Construction of recombinant baculovirus.
The UL24 ORF was excised from pKG3.6 (19) from 15 bp upstream of the initiating ATG, with Pfl23II, to 92 bp downstream of the termination codon, with BamHI. The fragment was blunt ended with Klenow and ligated into prokaryotic expression vector pGEX6P3 (Amersham/Pharmacia), which had been linearized with SmaI, to generate the vector pGEX6P3-UL24. In order to express GST-UL24 in insect cells, the glutathione S-transferase (GST) coding sequence from baculovirus expression vector pAcG3x (Pharmingen) was removed by digestion with SmaI and EcoNI and replaced with a DNA fragment encoding GST-UL24 that had been excised from pGEX6P3-UL24 with SalI and EcoNI, the SalI-generated end being blunted with Klenow. By using the BaculoGold transfection system (Pharmingen) and following the manufacturer's instructions, the resulting transfer vector, pAcG-GST-UL24, was used to generate a baculovirus that expressed GST-UL24 (BvGST-UL24). Baculovirus was propagated on SF9 insect cells in Grace's insect medium supplemented with 10% fetal calf serum.
Protein expression and generation of UL24 antiserum.
To express recombinant GST-UL24, insect cells were grown in spinner flasks to a density of 1 × 106 to 2 × 106 cells per ml and infected with BvGST-UL24 at a multiplicity of infection (MOI) of 1 to 5. At 72 h postinfection (p.i.), the cells were harvested. Cells were resuspended in 20 mM Tris (pH 7.9)-1 mM EDTA-1 M NaCl-1 mM dithiothreitol containing the protease inhibitor cocktail Complete (Roche) and lysed by sonication. GST-UL24 was affinity purified with glutathione-Sepharose, and the GST moiety was cleaved with PreScission Protease (Amersham/Pharmacia); however, despite being cleaved from its GST fusion partner, UL24 was retained on the matrix. The protein was eluted from the matrix in 100-fold-diluted sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) loading dye (containing 0.01% SDS), concentrated with a Speed-Vac (Savant), and then reloaded on an SDS-13% polyacrylamide gel. Gel slices containing recombinant UL24 were used to immunize two rats (procedure carried out by COVANCE Research Products Inc.). A 300-μg antigen sample per animal was used for the initial immunization, and 150 μg was provided for each of the three requested boosts; however, only two boosts were carried out. The antisera used in this study were derived from a production bleed following the second boost.
Protein microsequencing.
A 20-μg sample of GST-UL24 was proteolytically cleaved, and the products were resolved on an SDS-15% polyacrylamide gel in 3-[cyclohexylamino]-1-propanesulfonic acid (CAPS) gel running buffer (24). The proteins were transferred to a polyvinylidene difluoride membrane by electrotransfer in CAPS buffer containing 10% methanol. Protein microsequencing was carried out by the Boston University DNA and Protein Core Facility.
Viruses and cells.
HSV-1 strains KOS and 17 and KOS-derived mutants PAAr5 (7), dl−197/−116 (5), dlsptk (9), and ΔT143 (19) were propagated on Vero cells (American Type Culture Collection) as described previously (8).
Immunoblotting.
Cells in 60-mm-diameter dishes were mock infected or infected with HSV-1 at an MOI of 5 to 10 and harvested at various times p.i. The dishes were placed on ice, and cells were washed in ice-cold phosphate-buffered saline prior to being lysed on the dishes in 500 μl of buffer containing 50 mM Tris (pH 7.9)-1% Triton X-100-0.5% sodium deoxycholate-0.1% SDS-500 mM NaCl. Lysates were cleared by microcentrifugation at 18,000 × g and 4°C for 30 min. Unless otherwise indicated, 200 μl of each of the lysates was concentrated 10-fold with a Speed-Vac concentrator prior to being resolved on an SDS-12.5% polyacrylamide gel. Proteins were transferred to a polyvinylidene difluoride membrane by electrotransfer. Immunoblotting and detection by enhanced chemiluminescence were carried out essentially as described by the manufacturer (Amersham). Antibodies against gB and ICP0 were obtained from Fitzgerald. A mitochondrion-specific antibody was obtained from Chemicon International. Where quantification of signals is reported, the values were determined by running known amounts of whole-cell lysate made from HSV-1-infected cells on the same gel as the experimental samples to generate a standard curve, similar to what is shown in Fig. 4. The signals were scanned and then quantified with NIH Image 1.61.
FIG. 4.
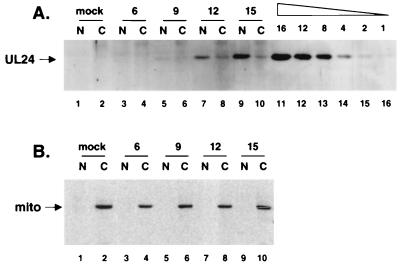
UL24 fractionates primarily with nuclei of infected cells. (A) Mock-infected or HSV-1 strain KOS-infected cells were harvested at 6, 9, 12, and 15 h p.i., as indicated above the panels. Cytoplasmic (C lanes) and nuclear (N lanes) fractions were resolved by SDS-PAGE and analyzed by immunoblotting with anti-UL24 serum. In lanes 11 to 16, the volumes, in microliters, of lysate from HSV-1-infected cells loaded on the gel are indicated above the lanes; these samples served as standards allowing relative quantification of the UL24 signal, the position of which is indicated on the left. (B) As a control, the membrane was subsequently stripped and incubated with an antibody directed against a mitochondrion-specific protein (mito), whose position is indicated on the left.
Nuclear versus cytoplasmic fractionation.
Vero cells in 100-mm-diameter dishes were infected at an MOI of 5. Cell fractionation was carried out, following Dounce homogenization, on the basis of a protocol described previously (34), with the following modifications: the pH of the cell lysis buffer was 7.9, and the nonionic detergent that it contained was Triton X-100. This buffer also contained the protease inhibitor cocktail Complete-EDTA (Roche). The cells were lysed in a volume of 0.5 ml per dish. The nuclei were pelleted by microcentrifugation at 2,000 × g and 4°C for 5 min; and the cytoplasmic fraction was retained. After the nuclei were washed once in the original lysis buffer, they were lysed in 500 μl of 50 mM Tris (pH 7.9)-1% Triton X-100-0.5% sodium deoxycholate-0.1% SDS containing 500 mM NaCl. Nuclear debris was removed by microcentrifugation at 18,000 × g for 30 min at 4°C. Samples were concentrated fivefold in a Speed-Vac concentrator prior to analysis by SDS-PAGE and immunoblotting.
Mitochondrial fractionation.
Isolation of mitochondria from cells grown in culture was carried out on the basis of the one-step protocol described by Bogenhagen and Clayton (4) and the method described by Marchenko et al. (25). In brief, cells were lysed by Dounce homogenization and mitochondria were removed from the cell lysate by centrifugation and purified on a sucrose gradient. The clearly visible band of mitochondria was collected by lateral suction in a volume of approximately 550 μl. As a control, a similar volume was collected from the top of the gradient.
RESULTS
Identification of the UL24 protein.
In an attempt to identify and study the putative UL24 protein of HSV-1, we set out to generate an antiserum to this protein. The UL24 ORF encodes a predicted protein of 269 amino acids corresponding to a molecular mass of approximately 29.5 kDa. UL24 was expressed as a fusion to GST with a baculovirus expression system in insect cells. The fusion protein produced had an apparent molecular mass of 45 kDa, approximately 13 kDa smaller than the expected size. The GST moiety was removed by proteolytic cleavage, and the 14-kDa protein was gel purified. Given that both the GST fusion protein and the cleaved UL24 protein were smaller than expected, amino-terminal protein microsequencing was carried out on the cleaved product, which confirmed that the protein contained the expected N-terminal UL24 amino acid sequence. We assume that the C-terminal 16 kDa of UL24 was removed by proteolysis within the insect cells or during the subsequent purification. The purified UL24 protein was used to immunize two rats.
The antisera raised were used in immunoblotting analyses of lysates made from HSV-1-infected or mock-infected Vero cells, which were harvested at various times p.i. As controls, the corresponding preimmune sera were tested in parallel. The serum from one of the immunized rats did not recognize a virus-specific protein (data not shown). The immune serum from the second rat recognized a protein with an apparent molecular mass of 30 kDa in the lysates of infected cells (Fig. 2A, lanes 7 and 8), while the corresponding preimmune serum did not (lanes 2 to 4), and this protein was absent from mock-infected cells (lane 5). We detected the 30-kDa protein starting at 11 h p.i., with more protein present at 15 h p.i. Both membranes were subsequently stripped and incubated with an antibody directed against viral protein gB. The gB protein was present in similar amounts in infected cell lysates on both membranes (Fig. 2B); thus, the absence of the 30-kDa protein in lanes 2 to 4 of Fig. 2A was not due to a lack of viral protein.
FIG. 2.
Serum from a rat immunized with recombinant UL24 reacts with a 30-kDa protein present in lysates of HSV-1-infected cells. Lysates from either mock-infected cells (lanes 1 and 5) or HSV-1-infected cells (lanes 2 to 4 and 6 to 8) were harvested at the indicated numbers of hours p.i. (hpi). (A) The lysates were analyzed by immunoblotting with either preimmune serum (lanes 1 to 4) or serum raised against recombinant UL24 (α UL24) (lanes 5 to 8). The positions of molecular weight standards are indicated to the right of each blot. The position of a 30-kDa protein recognized specifically by immune serum in lysates from infected cells is denoted by an asterisk. (B) As a control, each membrane was subsequently stripped and analyzed with a monoclonal antibody to viral protein gB, the position of which is indicated on the right.
To provide genetic tests showing that the 30-kDa protein recognized by the antiserum was, indeed, encoded by the UL24 ORF, strains of HSV-1 with defined lesions in UL24 were exploited. Vero cells were either mock infected or infected with UL24-null virus ΔT143 (19) or parental strain PAAr5 (7). Lysates were harvested at various times p.i., and the lysates were analyzed by immunoblotting with immune serum. In lysates from cells infected with PAAr5, the 30-kDa protein was detected faintly at 10 h p.i. and more intensely at 15 h p.i. (Fig. 3A, lanes 3 and 4), similar to the result obtained with KOS (Fig. 2A). However, in lysates from cells infected with the UL24-null virus, this protein was not detected (lanes 5 to 7), which was consistent with the 30-kDa protein being encoded by UL24. We obtained even stronger evidence in support of this conclusion by using mutant virus dl−197/−116 (5). This virus contains an in-frame deletion in UL24 that results in the removal of amino acids 98 through 124 such that the resulting protein is predicted to be approximately 3 kDa smaller than the wild-type UL24 protein. Vero cells were either mock infected or infected with mutant virus dl−197/−116 or parental strain PAAr5 and harvested at various times p.i. In lysates from cells infected with PAAr5, the 30-kDa protein was detected faintly at 8 h p.i. and more intensely at 12 and 16 h p.i. (Fig. 3C, lanes 3 to 5). In contrast, in lysates from cells infected with dl−197/−116, this 30-kDa protein was absent and a protein of the expected size for the deletion-containing form of the UL24 protein was detected faintly at 8 h p.i. and more intensely at 12 and 16 h p.i. (lanes 7 to 9). These results demonstrate that the absence of the 30-kDa protein in the lysates from cells infected with the UL24-null virus was not simply an indirect effect of the lack of UL24 expression, and thus, the 30-kDa protein is the product of the UL24 ORF.
FIG. 3.
Genetic tests identify UL24 protein in lysates of infected cells. (A) Lysates from cells either mock infected or infected with either PAAr5 or the UL24-null virus ΔT143 were analyzed at the indicated numbers of hours p.i. (hpi) by immunoblotting with serum raised against recombinant UL24. The position of the UL24 protein is indicated on the right. (B) As a control, the membrane was stripped and incubated with a monoclonal antibody directed against gB, the position of which is indicated on the right. (C) Lysates from mock-infected and infected cells were analyzed at the indicated times p.i. by immunoblotting with serum raised against recombinant UL24. The positions of molecular weight standards are indicated on the left. The bracket on the right indicates the positions of UL24 proteins from either parental strain PAAr5 (lanes 3 to 5) or mutant strain dl−197/−116 (lanes 7 to 9) that specifically reacted with the immune serum. (D) As a control, the membrane was stripped and incubated with a monoclonal antibody against viral protein gB, whose position is indicated on the right.
Subcellular location of UL24.
Analyses of the primary structure of UL24 by certain computer programs predict a potential mitochondrial targeting sequence at the N terminus of the protein and suggest that UL24 might localize to mitochondria (IPSORT [3], TargetP [13], MitoProt II [6], and Predotar [http://www.inra.fr/predotar]). A different program suggests that UL24 might localize to the nuclei of cells (PSORT) (28, 29). To investigate the subcellular location of UL24 in infected cells, we initially used each of the UL24 immune sera that were raised in indirect immunofluorescence experiments. However, the various patterns of perinuclear and nuclear staining that we obtained with either of the immune sera were similar to those obtained with the respective preimmune sera (data not shown). We therefore turned to biochemical fractionation. Vero cells infected with HSV-1 at an MOI of 5 were harvested at various times p.i., and the nuclear and cytoplasmic fractions were isolated. The fractions were resolved by SDS-PAGE and analyzed by immunoblotting (Fig. 4). In this experiment, we detected UL24 faintly at 9 h p.i., at which time the amounts of UL24 were similar in the cytoplasmic and nuclear fractions (Fig. 4A, lanes 5 and 6). However, at later times in infection, UL24 appeared to associate primarily with nuclei such that at 12 h p.i., more than half of the UL24 was in the nuclear fraction (lanes 7 and 8) and at 15 h p.i., approximately 70% of the UL24 was present in the nuclear fraction (lanes 9 and 10). To rule out the possibility that the apparent nuclear localization of UL24 was simply a consequence of inefficient lysis of the cells, the membrane was stripped and incubated with antiserum directed against a mitochondrial marker (Fig. 4B). This marker was only detected in the cytoplasmic fractions, indicating that the cells had been lysed efficiently.
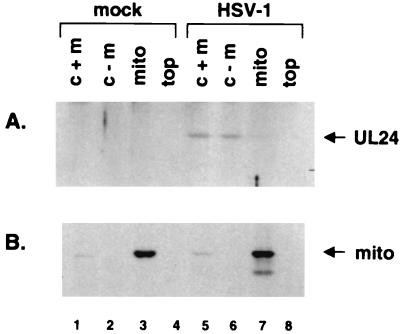
Because it appeared that not all of the UL24 fractionated with nuclei, we tested whether or not the remaining protein localized to the mitochondria of infected cells, as was suggested by certain of the sequence analyses. Cells were either mock infected or infected with HSV-1 at an MOI of 10. At 10.5 h p.i., when a substantial amount of UL24 localized to the cytoplasm, the cytoplasmic fractions were collected and mitochondria were purified on sucrose gradients. Samples from the initial cytoplasmic fractions that contained mitochondria (c + m), the cytoplasmic fractions obtained after the mitochondria had been removed via centrifugation (c − m), gradient-purified mitochondria (mito), and the top fraction of the gradients (top) were resolved by SDS-PAGE and subjected to immunoblotting with anti-UL24 serum. It must be noted that the purified mitochondria were collected in a volume 10 times smaller than the volume of the cell lysates from which they were purified. There was no apparent difference in the amounts of UL24 detected in the cytoplasmic fraction before and after the centrifugation step to remove the mitochondria (Fig. 5A, lanes 5 and 6). Consistent with this observation, very little UL24 was observed in the lane corresponding to the purified mitochondria (lane 7). No UL24 was detected in the fraction taken from the top of the gradient (lane 8). As a control, the blots were subsequently stripped and incubated with an antiserum that recognizes a 62-kDa mitochondrion-specific protein. This protein was detected in the initial cytoplasmic fractions (Fig. 5B, lanes 1 and 5) but not in the fractions depleted of mitochondria (lanes 2 and 6), and in contrast to the results obtained with UL24, this protein was clearly concentrated in the fractions containing the gradient-purified mitochondria (lanes 3 and 7). These results indicate that the isolation of mitochondria was successful and show that very little, if any, of the cytoplasmic portion of UL24 fractionated with mitochondria under these conditions.
FIG. 5.
UL24 does not appear to localize to mitochondria. (A) Mock-infected or HSV-1 strain 17-infected cells were harvested at 10.5 h p.i. Fractions of the original lysates containing cytoplasm and mitochondria (c + m), the lysates that had been depleted of mitochondria (c − m), the concentrated, gradient-purified mitochondria (mito), and the top of the gradient (top) were analyzed by immunoblotting with anti-UL24 serum. The position of UL24 is indicated on the right. (B) As a control, the membrane was subsequently stripped and incubated with an antibody directed against a mitochondrial marker (mito), the position of which is indicated on the right.
A major role for transcripts utilizing the first transcription start site in the expression of the UL24 protein.
As discussed earlier, there are six different transcripts for UL24; however, the function, if any, of this complexity is unclear. Transcripts originating from the first two transcription start sites, whether terminating at either the UL24 or the UL26 polyA signal, contain the entire UL24 ORF. Thus, there are four transcripts that could potentially serve to program the translation of UL24 (Fig. 1). The 5′ UTRs of two of these transcripts (5.6 and 1.4 kb) have features that could affect translation, especially negatively (Fig. 1), while the other two transcripts (5.4 and 1.2 kb) are relatively nonabundant (11). To investigate the question of which transcripts express UL24, we took advantage of a virus, dlsptk (9), containing a deletion in the tk gene that removes the promoter and the transcription start site of the 5.6- and 1.4-kb UL24 transcripts so that these transcripts are not expressed (11). Vero cells were either mock infected or infected at an MOI of 10 with either KOS or dlsptk, and at various times p.i., total cell lysates were harvested and analyzed by immunoblotting with anti-UL24 serum. Levels of UL24 expressed in the dlsptk-infected cells were substantially lower than those in cells infected with the wild-type virus at all of the times at which UL24 was detected (Fig. 6A). At 16 h p.i., the level of UL24 expressed in the dlsptk-infected cells was 10-fold lower than that measured for the wild-type virus, on the basis of dilutions of extracts of virus-infected cells run on the same gel. As a control, the membrane was subsequently stripped and incubated with a monoclonal antibody directed against gB. The levels of gB for KOS and dlsptk were similar (Fig. 6B). These results indicate that the 5.4- and 1.2-kb transcripts originating at the second transcription start site can serve as mRNAs for UL24 but suggest that most of the UL24 expressed results from translation of transcripts originating at the first transcription start site.
FIG. 6.
Loss of the 5.6- and 1.4-kb transcripts decreases the levels of UL24 in infected cells. (A) Levels of UL24 were compared among cells infected with KOS (lanes 2 to 5), cells infected with mutant virus dlsptk (6) (lanes 6 to 9), and mock-infected cells (lane 1). At the indicated numbers of hours p.i. (hpi), cells were harvested for analysis by immunoblotting with anti-UL24 serum. The position of UL24 is indicated on the right. (B) As a control, the membrane was stripped and incubated with an antibody against gB, the position of which is indicated on the right.
The UL24 protein is expressed with leaky-late kinetics.
Three of the UL24 transcripts end at the UL24 polyA signal and exhibit early kinetics, and three end at the UL26 polyA signal and exhibit leaky-late kinetics (Fig. 1) (10, 16). These classifications are based both on the temporal kinetics of expression and on the respective dependencies on viral DNA replication. We wished to determine with what kinetics the UL24 protein was expressed. In the experiments shown in Fig. 2, 3, and 6, UL24 was first detected at the same time as or later than the gB protein, the product of a leaky-late gene, and, like gB, continued to accumulate throughout the infection period. To determine the impact of viral DNA replication on the expression of UL24, Vero cells were infected in duplicate either in the presence or in the absence of 400 μg of phosphonoacetic acid (PAA), an inhibitor of viral DNA replication, per ml. At various times p.i., total cell lysates were harvested and total RNA was harvested from the duplicate samples. The cell lysates were analyzed by immunoblotting with the UL24 antiserum (Fig. 7A). PAA treatment resulted in an approximately sixfold decrease in the levels of UL24 protein expressed on the basis of a dilution series of infected-cell lysate run on the same gel (compare lanes 2 and 3 to lanes 5 and 6). As a control, the membrane was stripped and incubated with antibody directed against ICP0, a product of an immediate-early gene, whose expression is not dependent on viral DNA replication. As expected, the amounts of ICP0 present in the presence and absence of PAA were similar (Fig. 7B). The RNA samples were analyzed by Northern blotting, and as we have observed previously (10), inhibition of viral replication had little effect (less than twofold) on the short, early transcripts but a more pronounced effect (approximately sixfold) on the levels of long UL24 transcripts (data not shown). Thus, the effect of PAA on the levels of UL24 protein is consistent with the effect on the levels of the long UL24 transcripts, and we conclude that UL24 protein expression is largely, but not completely, dependent on viral replication, which is consistent with leaky-late kinetics.
FIG. 7.
Effect of viral DNA replication on expression of UL24. (A) Mock- or KOS-infected cells were incubated either in the presence (lanes 4 to 6) or in the absence (lanes 1 to 3) of 400 μg of the viral DNA replication inhibitor PAA per ml. At 6 and 12 h p.i. (hpi), lysates were harvested and analyzed by immunoblotting with anti-UL24 serum. The position of UL24 is indicated on the right. (B) As a control, the membrane was stripped and incubated with an antibody directed against immediate-early protein ICP0, the location of which is indicated on the right.
DISCUSSION
Identification of the UL24 protein of HSV-1.
We have identified the UL24 protein of HSV-1 in lysates of cells infected in culture by using antiserum raised against recombinant UL24 and viruses containing mutations that are predicted to affect the size and expression of UL24. As the antiserum generated against UL24 was raised against approximately the first third of the protein, we cannot use this reagent to study the expression of another potential UL24 protein encoded in the 5.2- and 0.9-kb transcripts, which originates within the UL24 ORF (Fig. 1). It will be necessary to generate an antiserum either to the entire UL24 protein or to the C terminus of the protein to address the existence of this alternate product. While this report was in preparation, Hong-Yan et al. (17) reported the identification of the UL24 protein of HSV-2. With antiserum raised against bacterially produced UL24, they detected a 32-kDa protein in infected cells whose expression was dependent on viral DNA replication and which they reported to be associated with virions. This protein has a size similar to that of HSV-1 UL24. Interestingly, despite 74% sequence identity between UL24 of HSV-1 and HSV-2, neither antiserum efficiently recognized the other form of the protein (17; A. Pearson and D. M. Coen, unpublished data).
Localization of UL24.
Mitochondrial targeting sequences typically form amphipathic helices and are rich in arginine, alanine, and serine residues and poor in negatively charged residues (13). Certain computer analyses of UL24 suggested the presence of a possible mitochondrial targeting sequence at the N terminus of the protein. However, upon biochemical fractionation, only trace amounts of UL24 were observed in the mitochondrial fraction, most likely reflecting contamination from other cellular compartments rather than actual localization to mitochondria. It is possible that the very basic nature of the N terminus of UL24 produced a false positive in the sequence analysis.
In the biochemical cell fractionation experiments we conducted, UL24 fractionated primarily with nuclei of infected cells, particularly at later times in infection. In certain of the immunofluorescence experiments that have been reported with antisera against HSV-2 UL24 (17), in addition to perinuclear and cytoplasmic staining, some staining was observed in the nucleus, particularly at later times in infection. In immunofluorescence studies of infected cells with our antisera, we observed variable perinuclear and nuclear staining although similar patterns were observed with preimmune sera. One advantage to the cell fractionation approach we ultimately employed is that it allowed us to quantify the proportion of UL24 associated with the nucleus and in the cytoplasm. In our experiments, it appeared that the ratio of nuclear to cytoplasmic UL24 increased at later times in infection, with most of the UL24 present in the nuclear fraction from at least 12 h p.i., and at the last time point analyzed, 15 h p.i., approximately 70% of the UL24 fractionated with nuclei. Although in these experiments we cannot exclude localization to some other cellular compartment that cofractionated with the nuclei, the association of UL24 with nuclear fractions suggests that UL24 localizes to the nuclei of infected cells.
The main purpose of investigating the subcellular localization of UL24 was to assemble clues to the function of UL24 during infection. The syncytial phenotype associated with UL24 mutations suggests that UL24 is involved, directly or indirectly, in one or several different membrane fusion events. Other genes that, when mutated, cause syncytial phenotypes are those for glycoproteins gK, gB, and gL and UL20, which encodes a virion-associated protein involved in viral transport from the nucleus to the cell membrane (1, 37). It has been suggested that these genes, which all encode proteins that are reported to be virion associated, have a role in modulating the normal fusion of membranes during viral infection (reviewed in references 27 and 35). The observation that UL24 is associated primarily with nuclei is not inconsistent with the syncytial phenotype of UL24 mutants; both gK and UL20 localize primarily to nuclear and perinuclear regions in infected cells, yet the corresponding genes still result in syncytial plaques when mutated (18, 37). Similar to UL20 mutations, UL24 mutations have a cell type-dependent syncytial phenotype (2, 36; discussed in reference 35). Like UL20 and gK, HSV-2 UL24 has been reported to copurify with extracellular virions (17). We were unable to detect UL24 in gradient-purified preparations of extracellular HSV-1 virions (data not shown); however, this may simply have been a question of sensitivity and thus we cannot rule out the possibility that HSV-1 UL24 is a virion protein. Thus, one possibility is that, like UL20 (1) and gK (14), UL24 is involved in egress of HSV from the nucleus to the cell membrane; its nuclear localization could be consistent with participation in early steps of this process. Perhaps when these fusion-related genes are mutated, early steps in virion assembly are altered and cell-cell fusion events that are usually inhibited until the appropriate stage in infection can occur in an aberrant manner. The temperature-dependent nature of the UL24 syncytial phenotype (21) might reflect destabilization of protein-protein interactions in the absence of UL24, although there is no information about any UL24-interacting proteins that might help to elucidate this issue. The antibody reagent described here may help us to gain such information.
Relative contributions of the various UL24 transcripts.
We hypothesized that the major UL24 transcripts containing the entire UL24 ORF (5.6 and 1.4 kb), which arise from the first transcription start site, might not be translated because of features of their long 5′ UTR (Fig. 1) (10). Alternatively, given the low abundance of the transcripts originating at the second start site (5.4 and 1.2 kb), it was reasonable to question whether these minor transcripts contributed at all to the expression of the UL24 protein. In fact, a virus with the first transcription start of UL24 deleted still expressed UL24 protein, indicating that the minor transcripts that arise from use of the second transcription start site are, indeed, translated. Even so, the level of UL24 expression by this mutant may be artificially high, as previous results suggest that it overexpresses the 5.4- and 1.2-kb transcripts (11). Regardless, these results suggest that the transcripts originating at the first start site are the main contributors to UL24 expression. It is not clear what role, if any, the second transcription start site serves. Construction and analysis of a virus with this second start site deleted would help address this question.
In the presence of the viral DNA replication inhibitor PAA, the levels of UL24 protein were decreased approximately sixfold. Thus, the UL24 protein is expressed with leaky-late kinetics. In the report describing the identification of the UL24 protein of HSV-2, no UL24 protein was detected in the presence of PAA (17). Whether this result reflects a difference between HSV-1 and HSV-2 or differences in the sensitivities of the assays used in the different experiments remains to be determined. In the presence of PAA, there is an approximately sixfold decrease in the levels of the long, leaky-late transcripts but little or no effect (twofold or less) on the short, early transcripts. The effect on the long transcripts is similar to the effect on the protein levels and could therefore account in full for the decrease in UL24 protein that we observed. These results support a hypothesis that the long, leaky-late UL24 transcripts are most important for the overall levels of UL24 protein in cells infected in culture. In fact, we have no evidence that the short transcripts are translated. Thus, there remains the question of what function, if any, is served by having two sets of UL24 transcripts that have the same 5′ end. Unlike the long transcripts that have the potential to encode the UL24, UL25, UL26, and UL26.5 proteins, the short UL24 transcripts appear to be able to function solely as templates for the synthesis of the UL24 polypeptide, and yet it appears that the short transcripts may play only a small, if any, role in the translation of UL24. One possibility is that the translation of the short and long transcripts is differentially regulated in a cell type-dependent manner.
The regulation of UL24 transcripts is highly complex, entailing not only multiple transcription start sites and polyA signals but also antisense regulation by tk mRNA and regulation at a step after transcription initiation by ICP27 (10, 11, 16). It is not known why the expression of UL24 is so complicated. The antiserum described here should allow further study of the relationships between regulation of UL24 transcripts and expression of UL24 protein.
Acknowledgments
We thank A. Griffiths for helpful discussions.
This work was supported by grant R01 AI26126 from the National Institutes of Health to D.M.C.
REFERENCES
- 1.Avitabile, E., P. L. Ward, C. Di Lazzaro, M. R. Torrisi, B. Roizman, and G. Campadelli-Fiume. 1994. The herpes simplex virus UL20 protein compensates for the differential disruption of exocytosis of virions and viral membrane glycoproteins associated with fragmentation of the Golgi apparatus. J. Virol. 68:7397-7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines, J. D., P. L. Ward, G. Campadelli-Fiume, and B. Roizman. 1991. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J. Virol. 65:6414-6424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannai, H., Y. Tamada, O. Maruyama, K. Nakai, and S. Miyano. 2001. Views: fundamental building blocks in the process of knowledge discovery, p. 233-238. In I. Russell and J. Kolen (ed.), Proceedings of the 14th International Florida Artificial Intelligence Research Society Conference. AAAI Press, Menlo Park, Calif.
- 4.Bogenhagen, D., and D. A. Clayton. 1974. The number of mitochondrial deoxyribonucleic acid genomes in mouse L and human HeLa cells. Quantitative isolation of mitochondrial deoxyribonucleic acid. J. Biol. Chem. 249:7991-7995. [PubMed] [Google Scholar]
- 5.Böni, J., and D. M. Coen. 1989. Examination of the roles of transcription factor Sp1-binding sites and an octamer motif in trans induction of the herpes simplex virus thymidine kinase gene. J. Virol. 63:4088-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Claros, M. G., and P. Vincens. 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241:779-786. [DOI] [PubMed] [Google Scholar]
- 7.Coen, D. M., D. P. Aschman, P. T. Gelep, M. J. Retondo, S. K. Weller, and P. A. Schaffer. 1984. Fine mapping and molecular cloning of mutations in the herpes simplex virus DNA polymerase locus. J. Virol. 49:236-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coen, D. M., H. E. Fleming, Jr., L. K. Leslie, and M. J. Retondo. 1985. Sensitivity of arabinosyladenine-resistant mutants of herpes simplex virus to other antiviral drugs and mapping of drug hypersensitivity mutations to the DNA polymerase locus. J. Virol. 53:477-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coen, D. M., M. Kosz-Vnenchak, J. G. Jacobson, D. A. Leib, C. L. Bogard, P. A. Schaffer, K. L. Tyler, and D. M. Knipe. 1989. Thymidine kinase-negative herpes simplex virus mutants establish latency in mouse trigeminal ganglia but do not reactivate. Proc. Natl. Acad. Sci. USA 86:4736-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook, W. J., and D. M. Coen. 1996. Temporal regulation of herpes simplex virus type 1 UL24 mRNA expression via differential polyadenylation. Virology 218:204-213. [DOI] [PubMed] [Google Scholar]
- 11.Cook, W. J., K. K. Wobbe, J. Böni, and D. M. Coen. 1996. Regulation of neighboring gene expression by the herpes simplex virus type 1 thymidine kinase gene. Virology 218:193-203. [DOI] [PubMed] [Google Scholar]
- 12.Davison, A. J. 1993. Herpesvirus genes. Rev. Med. Virol. 3:237-244. [Google Scholar]
- 13.Emanuelsson, O., H. Nielsen, S. Brunak, and G. von Heijne. 2000. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300:1005-1016. [DOI] [PubMed] [Google Scholar]
- 14.Foster, T. P., and K. G. Kousoulas. 1999. Genetic analysis of the role of herpes simplex virus type 1 glycoprotein K in infectious virus production and egress. J. Virol. 73:8457-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geballe, A. P., and M. S. Sachs. 2000. Translational control by upstream open reading frames, p. 595-614. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Hann, L. E., W. J. Cook, S. L. Uprichard, D. M. Knipe, and D. M. Coen. 1998. The role of herpes simplex virus ICP27 in the regulation of UL24 gene expression by differential polyadenylation. J. Virol. 72:7709-7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong-Yan, Z., T. Murata, F. Goshima, H. Takakuwa, T. Koshizuka, Y. Yamauchi, and Y. Nishiyama. 2001. Identification and characterization of the UL24 gene product of herpes simplex virus type 2. Virus Genes 22:321-327. [DOI] [PubMed] [Google Scholar]
- 18.Hutchinson, L., C. Roop-Beauchamp, and D. C. Johnson. 1995. Herpes simplex virus glycoprotein K is known to influence fusion of infected cells, yet is not on the cell surface. J. Virol. 69:4556-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irmiere, A. F., M. M. Manos, J. G. Jacobson, J. S. Gibbs, and D. M. Coen. 1989. Effect of an amber mutation in the herpes simplex virus thymidine kinase gene on polypeptide synthesis and stability. Virology 168:210-220. [DOI] [PubMed] [Google Scholar]
- 20.Jacobson, J. G., S. H. Chen, W. J. Cook, M. F. Kramer, and D. M. Coen. 1998. Importance of the herpes simplex virus UL24 gene for productive ganglionic infection in mice. Virology 242:161-169. [DOI] [PubMed] [Google Scholar]
- 21.Jacobson, J. G., S. L. Martin, and D. M. Coen. 1989. A conserved open reading frame that overlaps the herpes simplex virus thymidine kinase gene is important for viral growth in cell culture. J. Virol. 63:1839-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kibler, P. K., J. Duncan, B. D. Keith, T. Hupel, and J. R. Smiley. 1991. Regulation of herpes simplex virus true late gene expression: sequences downstream from the US11 TATA box inhibit expression from an unreplicated template. J. Virol. 65:6749-6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozak, M. 1999. Initiation of translation in prokaryotes and eukaryotes. Gene 234:187-208. [DOI] [PubMed] [Google Scholar]
- 24.LeGendre, N., and P. Matsudaira. 1988. Direct protein microsequencing from Immobilon-P transfer membrane. BioTechniques 6:154-159. [PubMed] [Google Scholar]
- 25.Marchenko, N. D., A. Zaika, and U. M. Moll. 2000. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J. Biol. Chem. 275:16202-16212. [DOI] [PubMed] [Google Scholar]
- 26.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 27.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakai, K. 2000. Protein sorting signals and prediction of subcellular localization. Adv. Protein Chem. 54:277-344. [DOI] [PubMed] [Google Scholar]
- 29.Nakai, K., and M. Kanehisa. 1992. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14:897-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Read, G. S., J. A. Sharp, and W. C. Summers. 1984. In vitro and in vivo transcription initiation sites on the TK-encoding BamHI Q fragment of HSV-1 DNA. Virology 138:368-372. [DOI] [PubMed] [Google Scholar]
- 31.Read, G. S., and W. C. Summers. 1982. In vitro transcription of the thymidine kinase gene of herpes simplex virus. Proc. Natl. Acad. Sci. USA 79:5215-5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 33.Sanders, P. G., N. M. Wilkie, and A. J. Davison. 1982. Thymidine kinase deletion mutants of herpes simplex virus type 1. J. Gen. Virol. 63:277-295. [DOI] [PubMed] [Google Scholar]
- 34.Sandri-Goldin, R. M. 1998. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 12:868-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spear, P. G. 1993. Membrane fusion induced by herpes simplex virus, p. 201-232. In J. Bentz (ed.), Viral fusion mechanisms. CRC Press, Inc., Boca Raton, Fla.
- 36.Tognon, M., R. Guandalini, M. G. Romanelli, R. Manservigi, and B. Trevisani. 1990. Phenotypic and genotypic characterization of locus Syn 5 in herpes simplex virus 1. Virus Res. 18:135-150. [DOI] [PubMed] [Google Scholar]
- 37.Ward, P. L., G. Campadelli-Fiume, E. Avitabile, and B. Roizman. 1994. Localization and putative function of the UL20 membrane protein in cells infected with herpes simplex virus 1. J. Virol. 68:7406-7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward, P. L., and B. Roizman. 1994. Herpes simplex genes: the blueprint of a successful human pathogen. Trends Genet. 10:267-274. [DOI] [PubMed] [Google Scholar]
- 39.Wilkie, N. M., R. P. Eglin, P. G. Sanders, and J. B. Clements. 1980. The association of herpes simplex virus with squamous carcinoma of the cervix, and studies of the virus thymidine kinase gene. Proc. R. Soc. Lond. B Biol. Sci. 210:411-421. [DOI] [PubMed] [Google Scholar]