Abstract
DNAs from four wild chimpanzees (Pan troglodytes schweinfurthi) from eastern Africa were screened for 14 DNA viruses and retroviruses. Between two and three viruses were found in each animal. An entire hepatitis B virus (HBV) genome was amplified and sequenced from samples taken from one animal. This indicates that HBV is distributed across the entire range of chimpanzee habitats.
The transfer of microbes between hosts has been occurring from time immemorial. Given their higher mutation rates, microbes are able to adapt rather readily to their hosts. Our closest relative, the chimpanzee, is host to a number of viruses homologous to human pathogens, with simian immunodeficiency virus of chimpanzees (SIVcpz)—which is isogenic with human immunodeficiency virus type 1—being a case in point (6-8, 10, 11, 20, 22). Here, samples from four dead wild chimpanzees in East Africa were screened for a number of DNA viruses and retroviruses.
Small tissue samples (∼1 cm3) were taken in the field from the kidney (animal AK), liver (animal FG), or lung (animals RAS and SAD) and stored in 70% alcohol. Samples were progressively rehydrated in distilled water. DNA was extracted in a laboratory that had never handled any of the viruses listed in Table 1. Total DNA was resuspended in 10 mM Tris-HCl (pH 8) and 1 mM EDTA. Its quality was checked by amplifying the mitochondrial DNA control region (D loop) with a single primer pair. All samples yielded good PCR products. Sequencing showed that all four animals were unambiguously Pan troglodytes schweinfurthi (data not shown), which are found in central and eastern Africa between the Ubangui River and the Great Lakes.
TABLE 1.
Viral test results for chimpanzees
| Virus(es) | Result for chimpanzee:
|
|||
|---|---|---|---|---|
| AK (kidney) | FG (liver) | RAS (lung) | SAD (lung) | |
| SIVcpz | − | − | − | − |
| STLV-1 | − | − | + | − |
| STLV-2 | − | − | − | − |
| Simian foamy virus | − | + | + | + |
| Simian HBV | − | + | − | − |
| TTV circinovirus | + | + | + | + |
| Kaposi's sarcoma-associated herpesvirus | − | − | − | + |
| Adenovirus types 2 and 5 | + | − | − | − |
| Adeno-associated virus | − | − | − | − |
| Simian virus 40 | − | − | − | − |
| JC virus, BK virus | − | − | − | − |
| Parvovirus B19 | − | − | − | − |
| Poxviruses | − | − | − | − |
| Papillomaviruses | − | − | − | − |
With respect to the retroviruses, all four samples proved negative for SIVcpz after extensive analysis with five primer pairs. RAS was positive for simian T-cell leukemia virus type 1 (STLV-1), while FG, RAS, and SAD harbored simian foamy viruses (Table 1). All four animals were infected by the TT circinovirus (TTV), while the liver sample from one animal (FG) was positive by tests with primers specific for hepatitis B virus (HBV). All samples were negative for adeno-associated virus, simian virus 40, JC virus, BK virus, B19 parvovirus, poxviruses, and papillomaviruses (Table 1). When it was tested with degenerate primers specific for the 3′ end of herpesviral DNA polymerase (open reading frame 9), the lung sample from SAD proved to be positive for a virus related to the human Kaposi's sarcoma-associated gammaherpesvirus or human herpesvirus 8, which is not without precedent (5). Overall, between two and three viruses were isolated from each animal. As no sera were available, it was not possible to test for antibodies to any of these viruses or to other viruses.
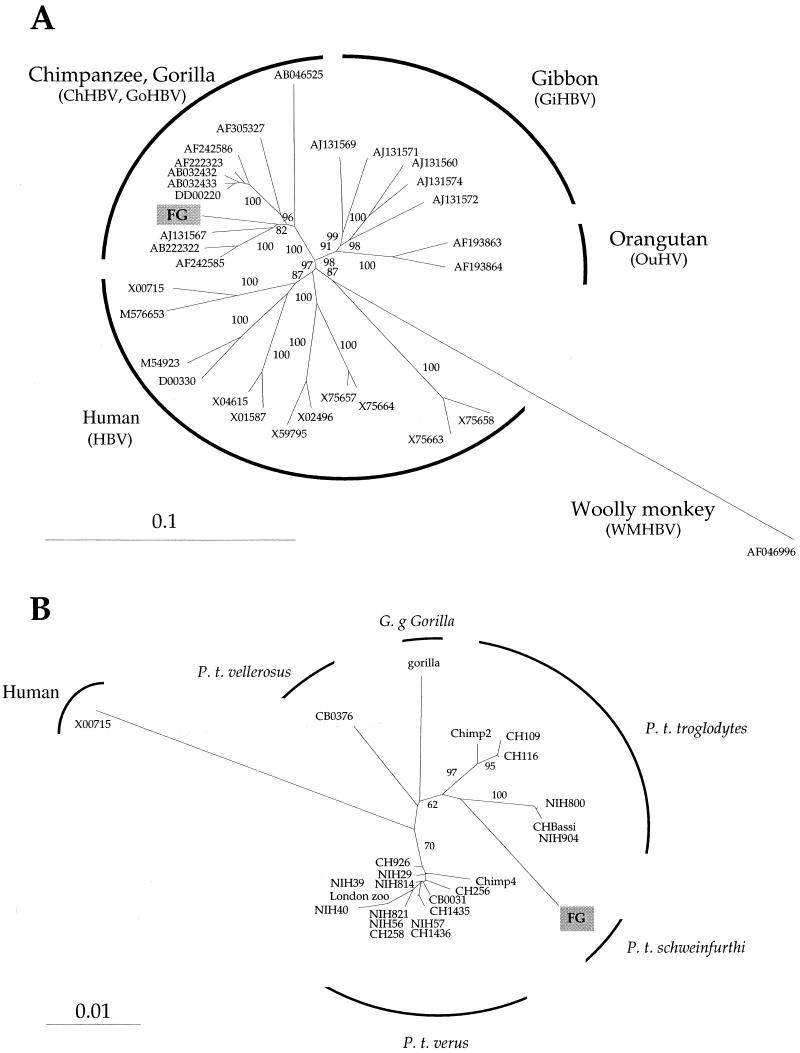
HBV-like viruses are known to infect chimpanzees in West Africa (Pan troglogytes verus) and West Central Africa (Pan troglodytes troglodytes and Pan troglodytes vellerosus) (6, 9, 10, 14, 19, 21) and have been known to infect at least one western lowland gorilla (Gorilla gorilla gorilla) from Cameroon (6). Hence, the virus obtained from FG is apparently the first to be identified from an East African chimpanzee (P. t. schweinfurthi). A complete HBV genome was amplified in six fragments and sequenced. Phylogenetic analysis showed that the complete sequence clustered in a monophyletic group with the chimpanzee and gorilla sequences (Fig. 1A). Analysis of the C, P, S, and X genes generated essentially the same tree, indicating that the virus was not a recombinant. As the database for the major viral surface antigen (HBsAg), encoded by the S gene, is the largest, a tree was constructed from 24 sequences (Fig. 1B). Even though the bootstrap values are not particularly strong, the sequence of the virus obtained from FG clustered more closely with the other viral sequences obtained from the P. t. troglodytes subspecies. This is perhaps not especially surprising, as P. t. troglodytes and P. t. schweinfurthi are the most closely related subspecies (4, 18).
FIG.1.
Phylogenetic analysis of HBV sequences. (A) Phylogenetic comparison of HBV sequences of viruses from chimpanzees, humans, and other nonhuman primates. Sequences were aligned by CLUSTAL W. Nucleotide sequence distances were determined with Dnadist of the Phylip package (version 3.5). Calculated distances were then used by applying the neighbor-joining method to pairwise sequence distances calculated by the Kimura two-parameter method to generate unrooted trees. Horizontal branch lengths are drawn to scale, with the bar indicating 0.1 nucleotide replacement per site. The final output was generated with Treeview. The number at each node represents the percentage of bootstrap replicates (out of 100). Only bootstrap values of ≥80 are given. The sequence of the virus obtained from FG was most closely related to those of chimpanzees and gorilla subtypes (0.5 to 0.7% nucleic acid divergence) and was distinct from the genotypes of human (10 to 15% divergence), gibbon ape (11% divergence), orangutan (11% divergence), and woolly monkey (28% divergence) viruses. (B) Phylogenetic comparison of HBV S-region sequences of viruses from chimpanzee subspecies. Horizontal branch lengths are drawn to scale, with the bar indicating 0.01 nucleotide replacement per site. The number at each node represents the percentage of bootstrap replicates (out of 100). Only bootstrap values of ≥60 are given. Sequences, identified by common names, and their GenBank accession numbers were as follows: CHBassi, AB046525; CB0376, AF305327; chimp4, AF242586; CH926, AF222322; CH256, AB032433; gorilla, AJ131567; CH109, AB222322; chimp2, AF242585; NIH800, AF222318; NIH904, AF222321; CH116, AF305328; NIH814, AF222319; NIH29, AF222312; NIH39, AF222313; NIH821, AF222320; NIH56, AF222316; NIH40, AF222314; CB0031, AF305326; NIH57, AF222317; CH1435, AF305329; CH1436, AF305330; CH258, AB032433; London zoo chimpanzee, DD00220; and the HBV subtype as an outgroup, X00715.
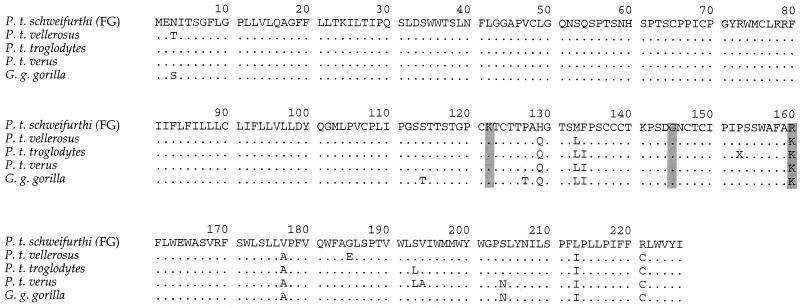
The HBsAg protein sequence of the HBV obtained from chimpanzee FG was aligned with those of a number of chimpanzee and gorilla viruses (Fig. 2). It differed by 3 to 5%. For the human viruses, HBsAg is characterized by the a group-specific antigenic determinant and two sets of mutually exclusive subtype-specific sero-determinants, y/d and w/r (1, 12), resulting from two lysine/arginine (K/R) polymorphisms at positions 122 and 160 (15). Hence, there are four possible serotypes, adw, adr, ayw, and ayr, the last being extremely rare and confined to eastern Asia (3, 25). To date, the chimpanzee HBsAg sequences have been uniform at these two sites, manifesting the equivalent of the adw subtype. The HBV sequence obtained from chimpanzee FG presents an arginine residue at position 160 typical of an adr subtype (Fig. 2). It is possible, therefore, that chimpanzee viruses might have the antigenicities of their human counterparts. Interestingly, orangutan and gibbon HBVs from Asia show the K/R polymorphism at position 160, although they have an invariant arginine at position 122, which is predictive of ayw and ayr serotypes (23). Hence, all four counterparts to the human polymorphisms may be found among great apes.
FIG. 2.
Alignment of the small S protein from chimpanzee HBVs with the sequence of the virus obtained from the animal FG (P. t. schweinfurthi). Only differences are shown. Grey boxes represent amino acid positions involved in determining the human HBV serodeterminants d/y, a, and r/w, respectively. The other sequences shown are from viruses isolated from P. t. vellerosus (sequence CB0376, accession number AF305327), P. t. troglodytes (sequence chimp2, accession number AF242585), P. t. verus from the London zoo (sequence DD00220), and G. g. gorilla (accession number AJ131567).
In terms of transmission of some of these viruses, it is perhaps worth noting that both FG and AK died after having shown influenza-like symptoms. This might bear on the finding of an adenovirus in AK. Among humans, HBV is highly infectious; presumably, therefore, FG would have been infectious also. The case of SAD is somewhat particular; he was killed after having attacked, bitten, and killed three small children. The STLV-1-positive animal, RAS, had been killed by a group of chimpanzees from a neighboring social community (24). His throat and testes had been torn out. Since monkey bites can transmit foamy viruses (8, 16), aggression by chimpanzees might also be conducive to the transmission of some of these viruses. However, as TTV, human T-cell leukemia virus type 1, HBV, and Kaposi's sarcoma-associated herpesvirus or human herpesvirus 8 are widely dispersed in human populations, it is not obvious that these animals represent a health hazard. Quite possibly, a pathologist is more vulnerable to infections than a zookeeper. Nevertheless, there is increasing contact between humans and chimpanzees via the bush meat trade (7). The number of viruses and, by extrapolation, the number of microbes (13) harbored by animals reemphasize the point that animals constitute pools of infectious agents. Given the highly related cellular biochemistries of chimpanzees and humans, the transfer of infections, perhaps in both directions, is probably facile.
In conclusion, screening for viruses is easy to perform and can extend our knowledge of the microbial floras among the great apes. The finding of an HBV in a P. t. schweinfurthi animal means that the virus is dispersed throughout the entire range of chimpanzee habitats from West Africa to the Great Lakes, unlike SIVcpz, which is found only among central African subspecies (2, 7, 17).
Nucleotide sequence accession number.
The sequences of the complete HBV genome of animal FG can be found in GenBank under accession number AF498266.
Acknowledgments
This work was supported by grants from the Institut Pasteur and the Agence Nationale pour la Recherche sur le SIDA.
Footnotes
This paper is dedicated to the memory of Bill Hamilton.
REFERENCES
- 1.Bancroft, W. H., F. K. Mundon, and P. K. Russell. 1972. Detection of additional antigenic determinants of hepatitis B antigen. J. Immunol. 109:842-848. [PubMed] [Google Scholar]
- 2.Corbet, S., M. C. Müller-Trutwin, P. Versmisse, S. Delarue, A. Ayouba, J. Lewis, S. Brunak, P. Martin, F. Brun-Vezinet, F. Simon, F. Barre-Sinoussi, and P. Mauclere. 2000. env sequences of simian immunodeficiency viruses from chimpanzees in Cameroon are strongly related to those of human immunodeficiency virus group N from the same geographic area. J. Virol. 74:529-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Courouce-Pauty, A. M., A. Plancon, and J. P. Soulier. 1983. Distribution of HBsAg subtypes in the world. Vox Sang. 44:197-211. [DOI] [PubMed] [Google Scholar]
- 4.Gagneux, P., M. K. Gonder, T. L. Goldberg, and P. A. Morin. 2001. Gene flow in wild chimpanzee populations: what genetic data tell us about chimpanzee movement over space and time. Philos. Trans. R. Soc. Lond. B 356:889-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greensill, J., J. A. Sheldon, K. K. Murthy, J. S. Bessonette, B. E. Beer, and T. F. Schulz. 2000. A chimpanzee rhadinovirus sequence related to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8: increased detection after HIV-1 infection in the absence of disease. AIDS 14:F129-F135. [DOI] [PubMed] [Google Scholar]
- 6.Grethe, S., J.-O. Heckel, W. Rietschel, and F. T. Hufert. 2000. Molecular epidemiology of hepatitis B virus variants in nonhuman primates. J. Virol. 74:5377-5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 8.Heneine, W., W. M. Switzer, P. Sandstrom, J. Brown, S. Vedapuri, C. A. Schable, A. S. Khan, N. W. Lerche, M. Schweizer, D. Neumann-Haefelin, L. E. Chapman, and T. M. Folks. 1998. Identification of a human population infected with simian foamy viruses. Nat. Med. 6:403-407. [DOI] [PubMed] [Google Scholar]
- 9.Hu, X., A. Javadian, P. Gagneux, and B. H. Robertson. 2001. Paired chimpanzee hepatitis B virus (ChHBV) and mtDNA sequences suggest different ChHBV genetic variants are found in geographically distinct chimpanzee subspecies. Virus Res. 79:103-108. [DOI] [PubMed] [Google Scholar]
- 10.Hu, X., H. S. Margolis, R. H. Purcell, J. Ebert, and B. H. Robertson. 2000. Identification of hepatitis B virus indigenous to chimpanzees. Proc. Natl. Acad. Sci. USA 97:1661-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huet, T., R. Cheynier, A. Meyerhans, G. Roelants, and S. Wain-Hobson. 1990. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature 345:356-359. [DOI] [PubMed] [Google Scholar]
- 12.Le Bouvier, G. L. 1971. The heterogeneity of Australia antigen. J. Infect. Dis. 123:671-675. [DOI] [PubMed] [Google Scholar]
- 13.Leininger, J. R., K. J. Donham, and M. J. Rubino. 1978. Leprosy in a chimpanzee. Morphology of the skin lesions and characterization of the organism. Vet. Pathol. 15:339-346. [DOI] [PubMed] [Google Scholar]
- 14.MacDonald, D. M., E. C. Holmes, J. C. M. Lewis, and P. Simmonds. 2000. Detection of hepatitis B virus infection in wild-born chimpanzees (Pan troglodytes verus): phylogenetic relationships with human and other primate genotypes. J. Virol. 74:4253-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okamoto, H., M. Imai, F. Tsuda, T. Tanaka, Y. Miyakawa, and M. Mayumi. 1987. Point mutation in the S gene of hepatitis B virus for a d/y or w/r subtypic change in two blood donors carrying a surface antigen of compound subtype adyr or adwr. J. Virol. 61:3030-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sandstrom, P. A., K. Oanh Phan, W. M. Switzer, T. Fredeking, L. Chapman, W. Heneine, and T. M. Folks. 2000. Simian foamy virus infection among zoo keepers. Lancet 355:551-552. [DOI] [PubMed] [Google Scholar]
- 17.Santiago, M. L., C. M. Rodenburg, S. Kamenya, F. Bibollet-Ruche, F. Gao, E. Bailes, S. Meleth, S. J. Soong, J. M. Kilby, Z. Moldoveanu, B. Fahey, M. N. Muller, A. Ayouba, E. Nerrienet, H. M. McClure, J. L. Heeney, A. E. Pusey, D. A. Collins, C. Boesch, R. W. Wrangham, J. Goodall, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2002. SIVcpz in wild chimpanzees. Science 295:465.. [DOI] [PubMed] [Google Scholar]
- 18.Stone, A. C., R. C. Griffiths, S. L. Zegura, and M. F. Hammer. 2002. High levels of Y-chromosome nucleotide diversity in the genus Pan. Proc. Natl. Acad. Sci. USA 99:43-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takahashi, K., B. Brotman, S. Usuda, S. Mishiro, and A. M. Prince. 2000. Full-genome sequence analyses of hepatitis B virus (HBV) strains recovered from chimpanzees infected in the wild: implications for an origin of HBV. Virology 267:58-64. [DOI] [PubMed] [Google Scholar]
- 20.Tobaly-Tapiero, J., J. De Celis-Kosmas, P. Bittoun, J. Lasneret, A. M. Poorters, M. E. Eladari, and R. Emanoil-Ravier. 1996. Isolation and characterization of infectious full-length DNA clones of chimpanzee foamy viruses SFV6 and SFV7: evidence for a Taf-dependent internal promoter. Res. Virol. 147:17-27. [DOI] [PubMed] [Google Scholar]
- 21.Vaudin, M., A. J. Wolstenholme, K. N. Tsiquaye, A. J. Zuckerman, and T. J. Harrison. 1988. The complete nucleotide sequence of the genome of a hepatitis B virus isolated from a naturally infected chimpanzee. J. Gen. Virol. 69:1383-1389. [DOI] [PubMed] [Google Scholar]
- 22.Verschoor, E. J., S. Langenhuijzen, and J. L. Heeney. 1999. TT viruses (TTV) of non-human primates and their relationship to the human TTV genotypes. J. Gen. Virol. 80:2491-2499. [DOI] [PubMed] [Google Scholar]
- 23.Warren, K. S., H. Niphuis, Heriyanto, E. J. Verschoor, R. A. Swan, and J. L. Heeney. 1998. Seroprevalence of specific viral infections in confiscated orangutans (Pongo pygmaeus). J. Med. Primatol. 27:33-37. [DOI] [PubMed] [Google Scholar]
- 24.Wrangham, R. W. 1999. The evolution of coalitionary killing. Yearb. Phys. Anthropol. 42:1-30. [DOI] [PubMed] [Google Scholar]
- 25.Yamishita, Y., S. Kurashina, Y. Miyakawa, and M. Mayumi. 1975. South-to-north gradient in distribution of the r determinant of hepatitis B surface antigen in Japan. J. Infect. Dis. 131:567-569. [DOI] [PubMed] [Google Scholar]