Abstract
Continued use of antiretroviral therapy despite the emergence of drug-resistant human immunodeficiency virus (HIV) has been associated with the durable maintenance of plasma HIV RNA levels below pretherapy levels. The factors that may account for this partial control of viral replication were assessed in a longitudinal observational study of 20 HIV-infected adults who remained on a stable protease inhibitor-based regimen despite ongoing viral replication (plasma HIV RNA levels consistently >500 copies/ml). Longitudinal plasma samples (n = 248) were assayed for drug susceptibility and viral replication capacity (measured by using a single-cycle recombinant-virus assay). The initial treatment-mediated decrease in plasma viremia was directly proportional to the reduction in replicative capacity (P = 0.01). Early virologic rebound was associated the emergence of a virus population exhibiting increased protease inhibitor phenotypic resistance, while replicative capacity remained low. During long-term virologic failure, plasma HIV RNA levels often remained stable or increased slowly, while phenotypic resistance continued to increase and replicative capacity decreased slowly. The emergence of primary genotypic mutations within protease (particularly V82A, I84V, and L90M) was temporally associated with increasing phenotypic resistance and decreasing replicative capacity, while the emergence of secondary mutations within protease was associated with more-gradual changes in both phenotypic resistance and replicative capacity. We conclude that HIV may be constrained in its ability to become both highly resistant and highly fit and that this may contribute to the continued partial suppression of plasma HIV RNA levels that is observed in some patients with drug-resistant viremia.
All currently available antiretroviral agents select for genotypic mutations that confer reduced phenotypic drug susceptibility (24). Theoretically, the selection of drug-resistant variants in vivo will result in higher levels of viral replication, decreased CD4+ T-cell counts, and a greater risk of disease progression. However, there are limited longitudinal data relating the emergence of drug resistance with subsequent treatment failure. Indeed, observational data from our group and others suggest that the level of viremia remains suppressed below the off-treatment set point even after the emergence of highly resistant human immunodeficiency virus (HIV) and that this partial viral suppression is associated with durable CD4+ T-cell gains, reduced T-cell activation, and reduced T-cell turnover (2, 8, 11, 20, 26, 35).
There is substantial in vitro and in vivo evidence that antiretroviral therapy selects for mutations that impair the inherent ability of HIV to replicate (15). Goudsmit and colleagues, for example, noted that the zidovudine-related T215F/Y mutation is not stable in the absence of the drug and that this mutation therefore confers a significant negative effect on viral replication (19). Similar observations have been made with the M184V mutation associated with lamivudine (3TC) resistance. Patients experiencing virologic failure with 3TC monotherapy often have persistent partial viral suppression (16). Since M184V confers very-high-level phenotypic resistance to 3TC, continued drug activity is unlikely to account for this partial suppression of viral replication (27). Rather, reduced replicative capacity is believed to be the primary cause, perhaps because M184V reduces the processivity of reverse transcriptase (1, 18, 31). Primary protease inhibitor-associated mutations also appear to decrease the enzymatic efficiency of HIV protease (7, 39). The D30N and L90M mutations, for example, confer drug resistance but reduce the ability of HIV to replicate in vitro (32).
We previously studied the replicative capacity of drug-resistant HIV in the setting of a prospective treatment interruption study (12). Replicative capacity was measured in vitro by using recombinant vectors containing patient-derived protease and reverse transcriptase sequences. At study entry, when high-level drug resistance was present, replication capacity was markedly diminished (compared to a wild-type reference). After antiretroviral therapy was discontinued, phenotypic drug resistance waned and the relative capacity of recombinant vectors to replicate increased. This increased replication capacity was temporally associated with an increase of plasma HIV RNA to a new and higher steady-state level. There was a strong correlation between the increase in replicative capacity and the increase in plasma HIV RNA levels, suggesting that the recombinant-vector replication capacity assay provides a direct measurement of in vivo fitness differences between drug-resistant and wild-type variants. These data also suggested that reduced viral fitness is an important factor in persistent partial suppression of viral replication during long-term virologic failure.
The evolution of drug resistance and replicative capacity has not been carefully assessed in patients who remain on long-term combination antiretroviral therapy despite incomplete viral suppression. Current models of viral evolution predict that additional mutations will lead to the emergence of a virus with reduced drug susceptibility, increased replicative capacity, or both and that such evolution will invariably lead to higher levels of viral replication and accelerated loss of peripheral CD4+ T-cell counts (3-6, 14, 17, 29, 33). We therefore performed a longitudinal observational study to determine the evolution of viral characteristics during long-term treatment failure (defined as persistent plasma HIV RNA levels above 500 copies/ml), focusing on the relative contributions of drug susceptibility and replicative capacity. Since we enrolled patients who chose to remain on stable therapy despite incomplete viral suppression, our data may not be generalizable to all patients experiencing virologic failure.
MATERIALS AND METHODS
Design.
This is a longitudinal observational study of 20 patients who, in consultation with their primary-care provider, chose to remain on a stable protease inhibitor-based regimen despite detectable plasma viremia (plasma HIV RNA viremia > 500 copies/ml). Patients were seen every 3 to 6 months up to the time therapy was modified or discontinued. Plasma was archived at each study visit for future analysis. This study was approved by the University of California, San Francisco, Committee on Human Research. All patients provided signed informed consent.
Stored pretreatment plasma samples, obtained prior to initiation of the protease inhibitor-based regimen, were available for 11 patients. All 11 patients had been previously enrolled in clinical studies: 5 in a prospective clinical trial of nelfinavir-saquinavir salvage therapy, 3 in an observational study, and 3 in a clinical trial evaluating indinavir (9, 10). For each patient, plasma samples were obtained from the date protease inhibitor therapy was initiated through the date that virologic failure was confirmed.
Measurements.
Circulating plasma HIV RNA levels were measured by using the branched-chain DNA methodology (Quantiplex version 3.0; limit of quantification, 50 to 500,000 copies of RNA/ml). Plasma HIV RNA levels obtained during clinical practice or as part of an ongoing clinical trial were also used in these analyses. Genotypic resistance testing was performed by population-based sequencing. Phenotypic resistance testing was performed by using a recombinant-virus-based assay (Phenosense; ViroLogic, Inc., South San Francisco, Calif.) (34). Drug susceptibility is reported as the fold change between the 50% inhibitory concentration (IC50) for the patient virus and the IC50 for a drug-sensitive reference virus (NL4-3).
Replicative capacity was measured by using a modification of the phenotypic drug susceptibility assay (12). Briefly, patient-derived reverse transcriptase and protease gene sequences were inserted into a viral vector containing a luciferase gene. Following a single round of viral replication in the absence of drug, luciferase activity was measured and compared to that for a reference virus containing the reverse transcriptase and protease sequences derived from the NL4-3 strain of HIV type 1 (HIV-1). Replicative-capacity measures for each patient were calculated by comparing the luciferase activity generated by recombinant viruses containing patient-derived sequences to that generated by the reference virus, after adjusting for minor differences in transfection efficiencies. Replicative-capacity values were expressed as percentages and reflect the level of replication for patient-derived virus compared to that for the wild-type reference (values less than 100% imply reduced replicative fitness).
Data analysis.
The baseline date for this analysis is defined as the earliest date during virologic failure (defined as a viral load of >500 copies of RNA/ml) at which a plasma sample was available for analysis. Data were censored at the time antiretroviral therapy was modified or discontinued. The following measurements were considered in these analysis: (i) plasma HIV RNA levels, (ii) peripheral CD4+ T-cell counts, (iii) fold change in drug susceptibility to the protease inhibitor administered to the patient, (iv) fold change in drug susceptibility to the nucleoside reverse transcriptase inhibitor, and (v) replicative capacity. Plasma HIV RNA levels and phenotypic susceptibility values were log10 transformed prior to multivariate analyses. Most patients were on a regimen containing two nucleoside reverse transcriptase inhibitors and at least one protease inhibitor. Zidovudine resistance was used as a surrogate for nucleoside reverse transcriptase inhibitor resistance in order to simplify the analysis. For patients on a dual-protease inhibitor-based regimen, the fold change in susceptibility to the pharmacokinetically enhanced protease inhibitor was used. Set point viral load is defined as the viral load prior to the initiation of any therapy or the viral load achieved after at least 16 weeks of a treatment interruption. (23)
The primary analysis focused on all data from study baseline through the time treatment was modified or discontinued or until follow-up for this study was completed. A secondary analysis was performed for the 11 patients for whom plasma samples were available prior to the initiation of the study-related protease inhibitor-based regimen and during the initial virologic response (prior to virologic failure). Changes in viral load, drug susceptibility, and replicative capacity were modeled on data for the 6-month period after therapy was modified.
The temporal relationship between the emergence of genotypic resistance mutations within the protease and replicative capacity was also determined. Only amino acid residue changes known to be associated with protease inhibitor resistance were considered. Mutations were defined as primary or secondary mutations based on published guidelines (D30N, M46I, G48V, I50V, V82A, I84V, and L90M were considered primary mutations; all other resistance-associated mutations were classified secondary) (24).
Statistical analysis.
Analyses were performed with SAS system 8.2 for Windows (SAS Institute, Inc., Cary, N.C.). Mixed effects were assessed with Proc Mixed in the SAS System (R. C. Littell, G. A. Milliken, W. W. Stroup, and R. D. Wolfinger, SAS system for mixed models, SAS Institute, Inc., 1996). Nonparametric statistics were employed for univariate correlations and comparison tests (Spearman rank test and Wilcoxon two-sample test, respectively). Midpoint tendency values are reported as medians and interquartile ranges (IQR), except as noted. All rate estimates and multivariable models were generated in mixed-effect model analyses, with a random effect specified for the individual (intercept) (13). In the analysis of cross-resistance within a drug class, a mixed-effect model with a random effect for the individual (intercept) was employed to account for the contribution of multiple time points for each subject to the analysis.
RESULTS
Baseline values.
Twenty subjects remaining on a stable protease inhibitor-based regimen despite incomplete viral suppression contributed longitudinal data to this analysis (Table 1). Eight patients were experiencing virologic failure of their initial protease inhibitor-based regimen, while the remainder were on their second (n = 12) or third (n = 1) protease inhibitor-based regimen. Subjects were observed for a median of 26.2 months (IQR, 16 to 44 months) (Table 2). Twelve patients switched to a new regimen or interrupted therapy during the study observation (after a median duration on study of 16.5 months).
TABLE 1.
Patient data at study entrya
| Patient no. | Treatment regimenb | Nadir CD4 T-cell count (cells/μl) | Viral load (log copies of RNA/ml)
|
Baseline CD4 T-cell count (cells/μl) | Fold change in susceptibilityd to:
|
Replicative capacity (%)e | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Off therapyc | Baseline | ZDV | NVP | IDV | NFV | SQV | Regimen protease inhibitor | |||||
| 1 | d4T/3TC/RTV/SQV | 169 | 5.19 | 3.73 | 167 | 0.6 | 0.5 | 71 | 91 | 344 | 344 | 23 |
| 2 | d4T/3TC/RTV/SQV | 210 | 5.34 | 3.30 | 331 | 8.1 | 0.3 | 8.7 | 8.7 | 14 | 13.6 | 47 |
| 3 | d4T/3TC/IDV | 10 | 5.11 | 3.27 | 143 | 18 | 0.3 | 4.0 | 2.9 | 1.0 | 4.0 | 5 |
| 4 | ZDV/3TC/RTV | 159 | 4.93 | 3.30 | 145 | 8.5 | 0.3 | 3.7 | 3.6 | 1.1 | 3.7 | 24 |
| 5 | d4T/ddI/RTV/SQV | 97 | 5.70 | 5.51 | 382 | 230 | 0.6 | 11 | 58 | 61 | 61 | 32 |
| 6 | d4T/3TC/RTV/SQV | 102 | 4.84 | 4.56 | 182 | 5.5 | 0.5 | 17 | 25 | 4.1 | 4.1 | 34 |
| 7 | ZDV/3TC/IDV | 346 | 5.12 | 3.81 | 299 | 0.5 | 0.7 | 11 | 16 | 10 | 11 | 41 |
| 8 | 3TC/NVP/NFV/SQV | 148 | 2.70 | 224 | 0.5 | 0.5 | 2.2 | 3.6 | 0.7 | 0.7 | 25 | |
| 9 | d4T/EFV/IDV | 93 | 5.80 | 3.44 | 524 | 0.6 | 222 | 0.9 | 1.2 | 0.8 | 0.9 | 81 |
| 10 | ABC/NVP/NFV/SQV | 200 | 4.35 | 2.74 | 295 | 5.1 | 0.4 | 3.3 | 4.4 | 0.8 | 0.8 | 20 |
| 11 | ABC/NVP/NFV/SQV | 30 | 5.29 | 2.94 | 271 | 3.0 | 0.4 | 40 | 47 | 7.4 | 7.4 | 38 |
| 12 | d4T/ABC/NFV/SQV | 132 | 5.42 | 3.06 | 231 | 0.6 | 0.6 | 1.6 | 4.0 | 0.9 | 0.9 | 6 |
| 13 | d4T/IDV | 237 | 5.85 | 3.40 | 318 | 2.7 | 1.8 | 0.5 | 0.6 | 0.5 | 0.5 | 37 |
| 14 | ZDV/3TC/IDV | 36 | 5.19 | 3.93 | 86 | 87 | 0.5 | 4.4 | 11 | 5.0 | 5.0 | 23 |
| 15 | ABC/NVP/NFV/SQV | 21 | 3.11 | 240 | 0.6 | 39 | 45 | 35 | 3.1 | 3.1 | 17 | |
| 16 | d4T/3TC/NVP/NFV | 121 | 5.49 | 3.31 | 295 | 1.1 | 34 | 8.0 | 28 | 28 | 28 | 23 |
| 17 | d4T/ABC/NFV/SQV | 196 | 5.18 | 3.65 | 260 | 16 | 0.8 | 2.2 | 1.7 | 1.0 | 1.0 | 5 |
| 18 | 3TC/ABC/NFV/SQV | 141 | 5.22 | 2.44 | 315 | 0.4 | 49 | 5.3 | 6.8 | 3.7 | 3.7 | 47 |
| 19 | ZDV/3TC/RTV | 207 | 5.18 | 4.08 | 362 | 8.5 | 0.5 | 7.2 | 11 | 3.9 | 3.9 | 35 |
| 20 | d4T/3TC/ddl/EFV/RTV/APV | 66 | 5.70 | 3.72 | 183 | 61 | 700 | 13 | 32 | 29 | 12 | 22 |
Values obtained at study baseline (earliest point during virologic failure for which a plasma sample could be evaluated).
Antiretroviral agents: zidovudine (ZDV), abacavir (ABC), didanosine (ddl), 3TC, stavudine (d4T), efavirenz (EFV), nevirapine (NVP), amprenavir (APV), indinavir (IDV), nelfinavir (NFV), ritonavir (RTV), and saquinavir (SQV).
Load obtained prior to initiation of any therapy or during a treatment interruption lasting more than 16 weeks.
Defined as IC50 for the patient-derived virus/IC50 for the wild-type reference strain.
Defined as the amount of luciferase activity (replication) for the patient-derived vector containing mutant pol and gag gene sequences divided by the luciferase activity of the wild-type reference strain (NL4-3).
TABLE 2.
Rate of change per month for virologic and immunologic measurements during long-term virologic failure of a stable protease inhibitor-based regimena
| Variable | Estimate | P |
|---|---|---|
| (n = 20) | ||
| Plasma HIV RNA level (log copies/ml/mo) | 0.008 | <0.0001 |
| CD4+ T cells (cells/μl/mo) | 1.63 | <0.0001 |
| Protease inhibitor susceptibility (fold change/mo, log transformed) | 0.02 | <0.0001 |
| Zidovudine susceptibility (fold change/mo, log transformed) | 0.01 | <0.0001 |
| Replicative capacity (% change/mo) | −0.17 | 0.009 |
Rate-of-change estimates were calculated from the earliest date during virologic failure for which a specimen was available until therapy was modified or discontinued (median duration of observation, 26.2 months per individual).
At study baseline (the earliest date during confirmed virologic failure for which a plasma sample was available for analysis), patients had been on their study regimens for a median of 3.6 months (IQR, 2.2 to 8.5 months). The median plasma HIV RNA level was 3.65 log10 copies of RNA/ml (IQR, 3.1 to 3.7 log10 copies of RNA/ml), and the median CD4+ T-cell count was 266 cells/ml (IQR, 183 to 316 cells/ml). These levels reflected a median decrease in plasma HIV RNA levels of 1.74 log10 copies of RNA/ml from the off-therapy set point (IQR, decrease of 2.3 to 1.3 log10 copies of RNA/ml) and a median CD4+ T-cell count increase from a pretherapy nadir of 108 cells/ml (IQR, 70 to 174 cells/ml).
The baseline patient-derived virus had a median 4.0-fold decrease in drug susceptibility to the study regimen protease inhibitor (IQR, 1- to 11-fold) and a median 5.6-fold decrease in drug susceptibility to zidovudine (IQR, 1.7- to 8.5-fold). The median replicative capacity at baseline was 25% (IQR, 22 to 37%). This level was significantly lower than the replicative capacities previously reported for untreated patients with wild-type HIV (median of 62%, P < 0.001; data not shown) (11). Replicative capacity was lower among patients with high-level 3TC resistance (median, 23%) than among patients with low-level 3TC resistance (median, 42%) (P = 0.02). There was no association between baseline replicative capacity and baseline resistance to drugs other than 3TC.
Virologic changes during long-term virologic failure.
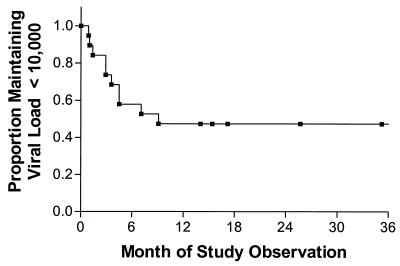
Most patients exhibited significant increases in plasma HIV RNA levels during the first several months of virologic failure, while the remainder maintained a low but detectable level of viremia throughout the study. Of the 19 patients who had a viral load <10,000 copies of RNA/ml at the study baseline, 10 eventually had a confirmed increase in viremia to >10,000 copies of RNA/ml (Fig. 1).
FIG. 1.
Time to virologic rebound to greater than 10,000 copies of RNA/ml. The proportion of patients maintaining plasma HIV RNA levels below 10,000 copies/ml is shown by using the Kaplan-Meier method. All 19 patients who had a plasma HIV RNA levels of <10,000 at the study baseline date are included. The first plasma HIV RNA level greater than 10,000 copies of RNA/ml was used as the failure end point.
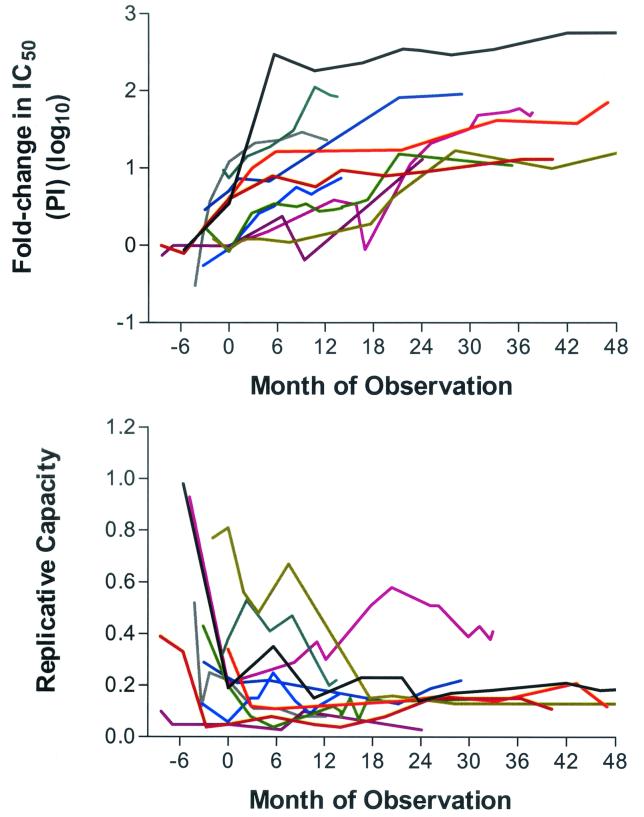
The rate at which plasma HIV RNA levels increased was greater during early virologic failure (0.05 log unit per month during first 9 months) than during later periods. Most patients eventually achieved a relatively steady-state level of plasma viremia (median change of 0.006 log copies/ml per month after month 9). Phenotypic resistance to protease inhibitor and zidovudine therapy increased continuously during prolonged virologic failure for most patients (0.02 [P < 0.0001] and 0.01 log10 fold change in IC50 per month [P < 0.0001] for protease inhibitor and zidovudine susceptibilities, respectively) (Fig. 2 shows the longitudinal phenotypic data from the 11 patients for whom pretreatment values were available). Viral replicative capacity declined slowly or remained stable during virologic failure (minus 0.2% replicative capacity per month; P = 0.009) (Fig. 2). CD4+ T-cell counts rose from study baseline at a rate of 1.63 cells/μl per month (P < 0.0001).
FIG. 2.
Change in protease inhibitor susceptibility and replicative capacity in 11 patients who initiated a new protease inhibitor (PI) regimen and remained on that regimen despite incomplete viral suppression. Day 0 is defined by the study baseline date (earliest available sample during virologic failure of the study regimen). Patient data were censored at the time therapy was discontinued or modified. Replicative capacity is expressed as the ratio of the luciferase activity from vectors containing patient-derived sequences to the luciferase activity from vectors containing wild-type sequences.
Multivariable modeling revealed that the change in protease inhibitor resistance was the best predictor of plasma HIV RNA levels during long-term virologic failure (0.25 higher log10 HIV RNA level per 1 log10-fold increase in IC50; P = 0.03). Furthermore, increasing protease inhibitor resistance was associated with decreasing viral replication capacity over time (0.005 higher log10 fold change in IC50 per 1% decrease in replicative capacity; P = 0.04). There was no association between change in replicative capacity and change in plasma HIV RNA levels during long-term virologic failure.
Longitudinal relationship between genotype, phenotype, and replicative capacity.
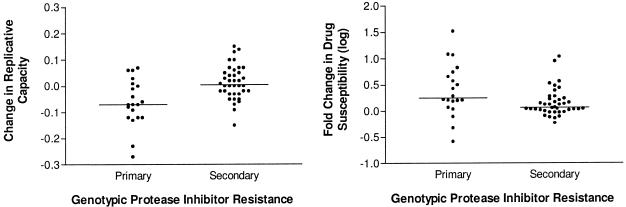
Protease gene sequences from 141 time points were available. At 20 of these time points at least one new primary-site mutation within protease was observed (19 of these changes involved V82A, I84V, and/or L90M). The emergence of a primary-site mutation was temporally associated with a median change in replicative capacity of −7% (IQR, −11 to +1%), while the emergence of secondary mutations was associated with a median increase in replicative capacity of +1% (IQR, −3 to + 6%) (P = 0.003 for primary versus secondary mutations) (Fig. 3). Furthermore, the emergence of a new primary mutation was associated with a large increase in phenotypic resistance (0.26 log10-fold change in IC50; IQR, 0.10 to 0.72 log10-fold change in IC50), while the emergence of a secondary mutation was associated with small increases in phenotypic resistance (0.07 log10-fold change in IC50; IQR, 0.01 to 0.23 log10-fold change in IC50) (P = 0.01 for primary versus secondary mutations).
FIG. 3.
The effect of primary versus secondary mutations within HIV protease on drug susceptibility and replicative capacity. Genotypic resistance sequences, phenotypic resistance levels, and replicative-capacity levels were available for 141 samples. The influence of a new primary or secondary protease inhibitor-associated mutation on change in drug susceptibility and replicative capacity is shown. A primary mutation within protease was defined based on published guidelines (most of the new mutations which emerged in this cohort were V82A, I84V, and L90M) (24). The changes in replicative capacity and phenotypic susceptibility were defined as the differences between the values obtained at the study visit immediately preceding the emergence of the new mutation and those at the study visit at which the new mutation was first observed. Replicative capacity is expressed as the ratio of the luciferase activity from vectors containing patient-derived sequences to the luciferase activity from vectors containing wild-type sequences.
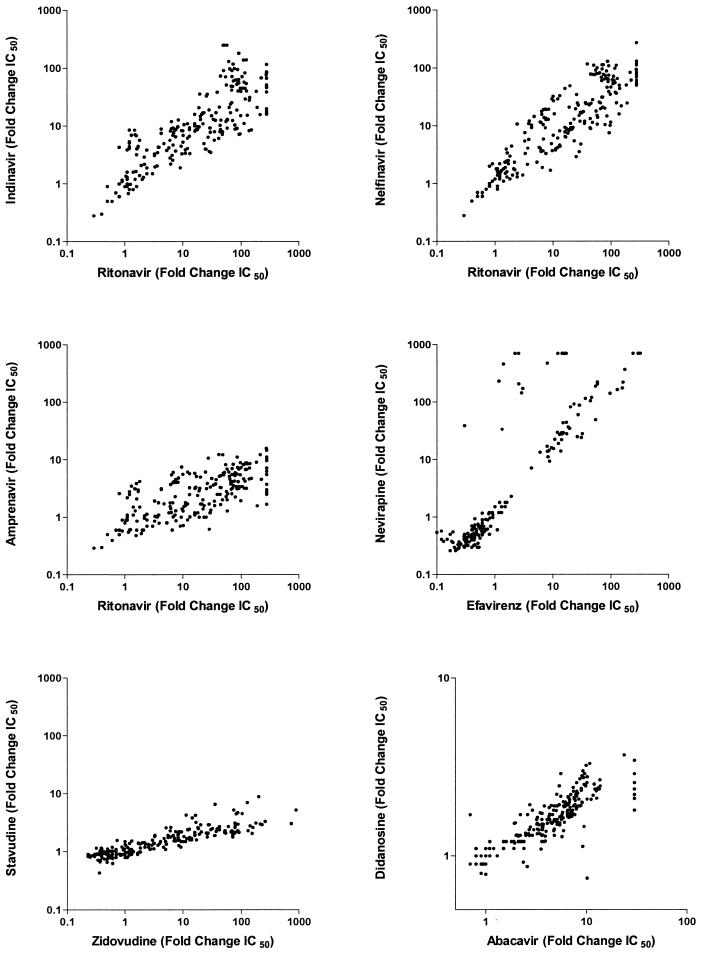
Cross-resistance is common in patients remaining on a stable regimen.
The level of phenotypic drug resistance increased over time in most patients, particularly within the protease inhibitor class (Fig. 2). Reduced susceptibility to one drug was often associated with reduced susceptibility to drugs within the same therapeutic class (Fig. 4). This was particularly true among protease inhibitors. For example, reduced susceptibility to ritonavir was strongly associated with reduced susceptibility to indinavir (0.43-unit change in fold change in IC50 for ritonavir per 1-unit increase in fold change in IC50 for indinavir; P < 0.0001), nelfinavir (0.93-unit change in fold change in IC50 for ritonavir per 1-unit increase in fold change in IC50 for nelfinavir; P < 0.0001), and amprenavir (2.7-unit change in fold change in IC50 for ritonavir per 1-unit increase in fold change in IC50 for amprenavir; P = 0.0005). Among the nucleoside reverse transcriptase inhibitors, there was a strong correlation between the zidovudine and stavudine susceptibilities (28.7-unit change in fold change in IC50 for zidovudine per 1-unit increase in fold change in IC50 for stavudine; P = 0.004) and a strong association between abacavir and didanosine (4.56-unit change in fold change in IC50 for abacavir per 1-unit increase in fold change in IC50 for didanosine; P < 0.0001). Finally, in the nonnucleoside reverse transcriptase inhibitor drug class, reduced susceptibility to efavirenz was strongly associated with reduced susceptibility to nevirapine (0.33-unit change in fold change in IC50 for efavirenz per 1-unit increase in fold change in IC50 for nevirapine; P < 0.0001).
FIG. 4.
Cross-resistance patterns during long-term virologic failure of combination antiretroviral therapy. The level of drug susceptibility is defined as IC50 for the patient-derived virus/IC50 of the wild-type reference (fold change).
CD4+ T-cell changes during long-term virologic failure.
Five of 20 patients experienced declining CD4+ T-cell counts during the study and stopped or switched therapy at an earlier date than the balance of the cohort, resulting in less observation time (average of 26 versus 34 months). These patients did not differ from the balance of patients with respect to baseline plasma HIV RNA levels, resistance phenotype, or replicative capacity at study baseline. However, patients with declining CD4+ counts during prolonged virologic failure tended to have a poorer virologic response to therapy than those whose CD4+ counts did not decline (median, 0.32 log10 copies of RNA/ml above baseline versus 0.49 log10 copies of RNA/ml below baseline; P = 0.06).
Initial virologic response to protease inhibitor therapy in a subset of patients.
Eleven of the 20 patients had samples available from before treatment initiation, thus allowing measurement of changes during the initial virologic response to treatment. At date of initiation of a new regimen, median replicative capacity was 35% (IQR, 23 to 42%) and the plasma HIV RNA level was 4.23 log10 copies/ml. Patients had limited evidence of protease inhibitor and zidovudine resistance (median decreases in drug susceptibility of 1.9-fold and 2.9-fold, respectively). The median nadir HIV RNA was 3.15 log10 copies/ml (IQR, 2.75 to 3.73 log10 copies/ml), or 1.3 log10 copies/ml below the pretherapy baseline (IQR, 2.19 to 1.05 log10 copies/ml below the pretherapy baseline). This nadir HIV RNA was achieved a median of 1.9 months (IQR, 1.3 to 4.2 months) after the initiation of a new protease inhibitor-based regimen. Phenotypic resistance to the protease inhibitor regimen increased at a rate of 0.08 log10-fold change in IC50 per month (P < 0.0001). Viral replication capacity decreased at a rate of 2.1% per month (P = 0.009), or an approximately 13% decrease during the first 6 months of response. Phenotypic resistance to zidovudine did not change significantly (0.02 log10-fold change in IC50 per month; P = 0.13). In a multivariate model, change in plasma HIV RNA levels during initial virologic response was significantly associated with change in replication capacity (HIV RNA decreased 0.02 log10 copies/ml per 1% decrease in replicative capacity; P = 0.02) but not with change in protease inhibitor or zidovudine resistance (0.27 log10-fold change in IC50 per month [P = 0.34] and −0.26 log10-fold change in IC50 per month [P = 0.33], respectively).
DISCUSSION
We previously reported that interrupting antiretroviral therapy in patients with highly resistant HIV results in the reemergence of an archived wild-type variant with greater replicative capacity and that this shift in viral phenotype during the treatment interruption was temporally associated with increased levels of viral replication and decreased CD4+ T-cell counts (12). These data provided indirect support for continuing a stable antiretroviral regimen in such patients, particularly if no treatment options that would likely result in complete viral suppression are available. However, the evolution of HIV among patients who remain on a stable but failing regimen has not been carefully studied.
In our present study, we observed the evolution of drug resistance, viral replicative capacity, plasma HIV RNA levels, and CD4+ T-cell counts in patients who remained on a stable protease inhibitor-based regimen despite detectable plasma viremia. A consistent pattern of viral load, phenotype, and replication capacity evolution was observed in most study subjects. First, initial viral-load decreases in response to therapy were directly proportional to decreases in viral replicative capacity during the same period. This is consistent with prior observations made during a structured treatment interruption, where the increase in replicative capacity was strongly correlated with the increase in plasma HIV RNA levels (12). Second, virologic rebound was characterized by an early rise in plasma HIV RNA levels, followed in most patients by a quasi-steady state during which the level of plasma viremia remained relatively stable or increased gradually. This new viral-load steady state remained well below the pretherapy viral-load set point. Third, virologic rebound was temporally associated with the emergence of drug-associated resistance mutations and a viral population with reduced phenotypic drug susceptibility. The rate that protease inhibitor drug susceptibility decreased (or resistance increased) was most rapid during the early phases of virologic failure but was observed to increase in some patients during 2 to 3 years of observation. Finally, the drug-resistant viral population that was established at the beginning of virologic failure exhibited reduced replicative capacity in vitro compared to a wild-type reference virus. Once established, replicative capacity generally remained stable, with decreases observed as primary protease inhibitor mutations emerged and small increases observed as secondary mutations emerged. Thus, ongoing viral evolution in the presence of antiretroviral therapy did not restore replication capacity in any significant manner.
These longitudinal data indicate that there is a complex relationship between viral replication, drug resistance, and replicative capacity during long-term treatment with an incompletely suppressive regimen. Viral evolution leads to increased drug resistance and, as a result, increased ability of HIV to replicate in the presence of the drug (i.e., plasma HIV RNA levels increase). However, the emergence of drug resistance reduces the inherent ability of HIV to replicate (replicative capacity). Continued drug pressure caused the level of drug resistance to progressively increase, but the replicative capacity remained low and did not increase for most patients. This dynamic between increasing resistance and stable or decreasing replicative capacity likely contributed to the stable maintenance of plasma HIV RNA levels below pretherapy levels, as suggested by others (22). Further support for this hypothesis can be found in the temporal relationship between viral genotype and phenotype. The emergence of primary-site mutations within HIV protease was temporally associated with significant increases in resistance and significant decreases in replicative capacity. In contrast, the emergence of secondary changes was temporally associated with gradual changes in resistance and a small increase in replicative capacity. Thus, it appears that HIV first evolves to preserve its capacity to replicate in the presence of a drug but sacrifices enzymatic and replicative efficiency in the process. Subsequent evolutionary steps to evade the drug's selective pressure generally result in much less dramatic changes in both resistance levels and replicative capacity. In this way, drug-resistant viruses appears to be “painted into a corner” of a viral fitness landscape from which they cannot readily escape without sacrificing the ability to replicate in the presence of the drug. This inability of HIV to fully restore replicative capacity during long-term therapy with a failing regimen likely accounts in part for the durable maintenance of partial viral suppression in some patients with drug-resistant viremia (12).
Although most patients in this cohort did well during the period of observation, it is noteworthy that some patients experienced declining CD4+ T-cell counts. These patients tended to have less-profound treatment-mediated virologic responses, consistent with previous observations by our group and others (8, 30, 38). In addition, despite partial viral suppression, most patients experienced continuous increases in plasma viremia over time. Thus, although treatment-mediated benefits were durable in this cohort, they were not permanent. This critical observation needs to be considered in the context of the observation that drug resistance (and cross-resistance) continually increased over time, even as partial viral suppression was maintained. Thus, delaying a treatment modification in patients with incomplete viral suppression may result in continued treatment benefit for years but at significant costs in terms of the future ability to achieve complete viral suppression.
The replicative-capacity assay used in this study compares an NL4-3 viral construct containing patient-derived reverse transcriptase, protease, and the 3′ end of gag with a wild-type NL4-3 reference stain. Several factors that may affect viral replication in vivo are not measured. For example, the report of very slow disease progression among a cohort of untreated patients infected with a nef-deleted HIV-1 strain provides the most dramatic illustration of the impact of viral fitness on outcome in vivo (28). The contribution of nef to replicative capacity is not measured with the replicative-capacity assay used in this study. Envelope diversity, which may affect viral fitness in vivo (e.g., non-syncytium-inducing/R5 versus syncytium-inducing/CXCR4 phenotypes), is also not measured in this assay (21, 36). Although the assay includes the p7/p1 and p1/p6 Gag cleavage sites, compensatory changes at other Gag cleavage sites may act to partially restore replicative capacity during protease inhibitor therapy but were not measured in this assay (37, 40). Thus, an isolated replicative-capacity measurement may not be expected to correlate with the level of plasma viremia in vivo. However, in longitudinal studies involving serial samples from a single individual, the change in replicative capacity over time may be more meaningful, since most of these unmeasured viral factors remain relatively stable (i.e., most of the evolutionary pressure during treatment is directed at reverse transcriptase and protease). This hypothesis is supported by the consistent association between change in replicative capacity and change in plasma HIV RNA levels observed during the initial virologic response (as shown here) and during treatment interruption (as shown previously) (12).
There are several important limitations to our data that deserve mention. First, patients who did poorly during early virologic failure were unlikely to be eligible for our analysis (selection bias). Some patients experience rapid rebounds in viremia and rapid decreases in CD4+ T-cell counts during early virologic failure; such patients are likely to have patterns of viral evolution very different from those observed here (25). Second, those who continued to do well over time were more likely to remain on a stable regimen (survival bias). Five of 20 patients experienced declining CD4+ T-cell counts during the study and stopped or switched therapy at an earlier date than the balance of the cohort, resulting in less observation time than that for patients who remained on therapy (average of 26 versus 34 months). However, when we truncated the analysis to the first 20 months of follow-up, the study outcomes remained the same (data not shown). Third, our data are limited to patients who were receiving therapy with nucleoside reverse transcriptase inhibitors and protease inhibitors; it is not clear how well our results would translate to patients treated with other treatment regimens. Fourth, many of the patients studied here were heavily pretreated with other agents prior to prospective observation. This was particularly true for the nucleoside analogue class. Further studies of patients failing an initial regimen are needed, but such studies may not feasible given the current standard of therapy, which is to switch therapy during failure of a first or second regimen. Finally, although a relationship between viral load, phenotypic resistance, and replicative capacity was observed in this study, the strength of these associations was relatively modest. It is likely that other factors, such as the host response to the drug-resistant variant, also contribute to the diverse treatment outcomes observed in this patient population.
In conclusion, continued antiretroviral therapy is associated with durable virologic benefit in patients with drug-resistant viremia. This occurs despite the continued selection of drug-associated resistance mutations. Virus evolution in the presence of a drug results in increasing levels of phenotypic resistance and cross-resistance but reduced replicative capacity. As a consequence, the level of viremia remains partially suppressed and CD4+ T-cell counts remain elevated. How to weigh the benefit and risks for a patient remaining on a stable regimen while experiencing a partial virologic response to therapy remains unclear and will likely need to be assessed in a larger randomized clinical study.
Acknowledgments
This work was supported by grant CC99-SF-001 from the California University-wide AIDS Research Program, by the UCSF-Gladstone Institute of Virology and Immunology Center for AIDS Research (P30 MH59037), and by grant 5-MO1-RR00083-37 from the Division of Research Resources, NIH.
REFERENCES
- 1.Back, N. K., M. Nijhuis, W. Keulen, C. A. Boucher, B. O. Oude Essink, A. B. van Kuilenburg, A. H. van Gennip, and B. Berkhout. 1996. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 15:4040-4049. [PMC free article] [PubMed] [Google Scholar]
- 2.Belec, L., C. Piketty, A. Si-Mohamed, C. Goujon, M. C. Hallouin, S. Cotigny, L. Weiss, and M. D. Kazatchkine. 2000. High levels of drug-resistant human immunodeficiency virus variants in patients exhibiting increasing CD4+ T cell counts despite virologic failure of protease inhibitor-containing antiretroviral combination therapy. J. Infect. Dis. 181:1808-1812. [DOI] [PubMed] [Google Scholar]
- 3.Berkhout, B. 1999. HIV-1 evolution under pressure of protease inhibitors: climbing the stairs of viral fitness. J. Biomed. Sci. 6:298-305. [DOI] [PubMed] [Google Scholar]
- 4.Bleiber, G., M. Munoz, A. Ciuffi, P. Meylan, and A. Telenti. 2001. Individual contributions of mutant protease and reverse transcriptase to viral infectivity, replication, and protein maturation of antiretroviral drug-resistant human immunodeficiency virus type 1. J. Virol. 75:3291-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borman, A. M., S. Paulous, and F. Clavel. 1996. Resistance of human immunodeficiency virus type 1 to protease inhibitors: selection of resistance mutations in the presence and absence of the drug. J. Gen. Virol. 77:419-426. [DOI] [PubMed] [Google Scholar]
- 6.Coffin, J. M. 1995. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science 267:483-489. [DOI] [PubMed] [Google Scholar]
- 7.Croteau, G., L. Doyon, D. Thibeault, G. McKercher, L. Pilote, and D. Lamarre. 1997. Impaired fitness of human immunodeficiency virus type 1 variants with high-level resistance to protease inhibitors. J. Virol. 71:1089-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deeks, S. G., J. D. Barbour, R. M. Grant, and J. N. Martin. 2002. Duration and predictors of CD4 T-cell gains in patients who continue combination therapy despite detectable plasma viremia. AIDS 16:201-207. [DOI] [PubMed] [Google Scholar]
- 9.Deeks, S. G., R. M. Grant, G. W. Beatty, C. Horton, J. Detmer, and S. Eastman. 1998. Activity of a ritonavir plus saquinavir-containing regimen in patients with virologic evidence of indinavir or ritonavir failure. AIDS 12:F97-F102. [DOI] [PubMed] [Google Scholar]
- 10.Deeks, S. G., N. S. Hellmann, R. M. Grant, N. T. Parkin, C. J. Petropoulos, M. Becker, W. Symonds, M. Chesney, and P. A. Volberding. 1999. Novel four-drug salvage treatment regimens after failure of a human immunodeficiency virus type 1 protease inhibitor-containing regimen: antiviral activity and correlation of baseline phenotypic drug susceptibility with virologic outcome. J. Infect. Dis. 179:1375-1381. [DOI] [PubMed] [Google Scholar]
- 11.Deeks, S. G., R. Hoh, R. M. Grant, T. Wrin, J. D. Barbour, A. Narvaez, D. Cesar, K. Abe, M. B. Hanley, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, and M. K. Hellerstein. 2002. CD4+ T cell kinetics and activation in human immunodeficiency virus-infected patients who remain viremic despite long-term treatment with protease inhibitor-based therapy. J. Infect. Dis. 185:315-323. [DOI] [PubMed] [Google Scholar]
- 12.Deeks, S. G., T. Wrin, T. Liegler, R. Hoh, M. Hayden, J. D. Barbour, N. S. Hellmann, C. J. Petropoulos, J. M. McCune, M. K. Hellerstein, and R. M. Grant. 2001. Virologic and immunologic consequences of discontinuing combination antiretroviral-drug therapy in HIV-infected patients with detectable viremia. N. Engl. J. Med. 344:472-480. [DOI] [PubMed] [Google Scholar]
- 13.Diggle, P. J., K. Y. Liang, and S. L. Zeger. 1994. Analysis of longitudinal data. Oxford University Press, Oxford, United Kingdom.
- 14.Eastman, P. S., J. Mittler, R. Kelso, C. Gee, E. Boyer, J. Kolberg, M. Urdea, J. M. Leonard, D. W. Norbeck, H. Mo, and M. Markowitz. 1998. Genotypic changes in human immunodeficiency virus type 1 associated with loss of suppression of plasma viral RNA levels in subjects treated with ritonavir (Norvir) monotherapy. J. Virol. 72:5154-5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erickson, J. W., S. V. Gulnik, and M. Markowitz. 1999. Protease inhibitors: resistance, cross-resistance, fitness and the choice of initial and salvage therapies. AIDS 13:S189-S204. [PubMed] [Google Scholar]
- 16.Eron, J. J., S. L. Benoit, J. Jemsek, R. D. MacArthur, J. Santana, J. B. Quinn, D. R. Kuritzkes, M. A. Fallon, M. Rubin, et al. 1995. Treatment with lamivudine, zidovudine, or both in HIV-positive patients with 200 to 500 CD4+ cells per cubic millimeter. N. Engl. J. Med. 333:1662-1669. [DOI] [PubMed] [Google Scholar]
- 17.Frenkel, L. M., and J. I. Mullins. 2001. Should patients with drug-resistant HIV-1 continue to receive antiretroviral therapy? N. Engl. J. Med. 344:520-522. [DOI] [PubMed] [Google Scholar]
- 18.Frost, S. D., M. Nijhuis, R. Schuurman, C. A. Boucher, and A. J. Brown. 2000. Evolution of lamivudine resistance in human immunodeficiency virus type 1-infected individuals: the relative roles of drift and selection. J. Virol. 74:6262-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goudsmit, J., A. de Ronde, E. de Rooij, and R. de Boer. 1997. Broad spectrum of in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J. Virol. 71:4479-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grabar, S., V. Le Moing, C. Goujard, C. Leport, M. D. Kazatchkine, D. Costagliola, and L. Weiss. 2000. Clinical outcome of patients with HIV-1 infection according to immunologic and virologic response after 6 months of highly active antiretroviral therapy. Ann. Intern. Med. 133:401-410. [DOI] [PubMed] [Google Scholar]
- 21.Groenink, M., R. A. Fouchier, S. Broersen, C. H. Baker, M. Koot, A. B. van't Wout, H. G. Huisman, F. Miedema, M. Tersmette, and H. Schuitemaker. 1993. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science 260:1513-1516. [DOI] [PubMed] [Google Scholar]
- 22.Hance, A. J., V. Lemiale, J. Izopet, D. Lecossier, V. Joly, P. Massip, F. Mammano, D. Descamps, F. Brun-Vezinet, and F. Clavel. 2001. Changes in human immunodeficiency virus type 1 populations after treatment interruption in patients failing antiretroviral therapy. J. Virol. 75:6410-6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatano, H., S. Vogel, C. Yoder, J. A. Metcalf, R. Dewar, R. T. Davey, Jr., and M. A. Polis. 2000. Pre-HAART HIV burden approximates post-HAART viral levels following interruption of therapy in patients with sustained viral suppression. AIDS 14:1357-1363. [DOI] [PubMed] [Google Scholar]
- 24.Hirsch, M. S., F. Brun-Vezinet, R. T. D'Aquila, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, C. Loveday, J. W. Mellors, B. Clotet, B. Conway, L. M. Demeter, S. Vella, D. M. Jacobsen, and D. D. Richman. 2000. Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA panel. JAMA 283:2417-2426. [DOI] [PubMed] [Google Scholar]
- 25.Huang, W., V. De Gruttola, M. Fischl, S. Hammer, D. Richman, D. Havlir, R. Gulick, K. Squires, and J. Mellors. 2001. Patterns of plasma human immunodeficiency virus type 1 RNA response to antiretroviral therapy. J. Infect. Dis. 183:1455-1465. [DOI] [PubMed] [Google Scholar]
- 26.Kaufmann, D., G. Pantaleo, P. Sudre, and A. Telenti. 1998. CD4-cell count in HIV-1-infected individuals remaining viraemic with highly active antiretroviral therapy (HAART). Swiss HIV Cohort study. Lancet 351:723-724. [DOI] [PubMed] [Google Scholar]
- 27.Larder, B. A., S. D. Kemp, and P. R. Harrigan. 1995. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science 269:696-699. [DOI] [PubMed] [Google Scholar]
- 28.Learmont, J. C., A. F. Geczy, J. Mills, L. J. Ashton, C. H. Raynes-Greenow, R. J. Garsia, W. B. Dyer, L. McIntyre, R. B. Oelrichs, D. I. Rhodes, N. J. Deacon, and J. S. Sullivan. 1999. Immunologic and virologic status after 14 to 18 years of infection with an attenuated strain of HIV-1. A report from the Sydney Blood Bank Cohort. N. Engl. J. Med. 340:1715-1722. [DOI] [PubMed] [Google Scholar]
- 29.Maeda, Y., D. J. Venzon, and H. Mitsuya. 1998. Altered drug sensitivity, fitness, and evolution of human immunodeficiency virus type 1 with pol gene mutations conferring multi-dideoxynucleoside resistance. J. Infect. Dis. 177:1207-1213. [DOI] [PubMed] [Google Scholar]
- 30.Marschner, I. C., A. C. Collier, R. W. Coombs, R. T. D'Aquila, V. DeGruttola, M. A. Fischl, S. M. Hammer, M. D. Hughes, V. A. Johnson, D. A. Katzenstein, D. D. Richman, L. M. Smeaton, S. A. Spector, and M. S. Saag. 1998. Use of changes in plasma levels of human immunodeficiency virus type 1 RNA to assess the clinical benefit of antiretroviral therapy. J. Infect. Dis. 177:40-47. [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Picado, J., K. Morales-Lopetegi, T. Wrin, J. G. Prado, S. D. Frost, C. J. Petropoulos, B. Clotet, and L. Ruiz. 2002. Selection of drug-resistant HIV-1 mutants in response to repeated structured treatment interruptions. AIDS 16:895-899. [DOI] [PubMed] [Google Scholar]
- 32.Martinez-Picado, J., A. V. Savara, L. Sutton, and R. T. D'Aquila. 1999. Replicative fitness of protease inhibitor-resistant mutants of human immunodeficiency virus type 1. J. Virol. 73:3744-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nijhuis, M., R. Schuurman, D. de Jong, J. Erickson, E. Gustchina, J. Albert, P. Schipper, S. Gulnik, and C. A. Boucher. 1999. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS 13:2349-2359. [DOI] [PubMed] [Google Scholar]
- 34.Petropoulos, C. J., N. T. Parkin, K. L. Limoli, Y. S. Lie, T. Wrin, W. Huang, H. Tian, D. Smith, G. A. Winslow, D. J. Capon, and J. M. Whitcomb. 2000. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 44:920-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piketty, C., P. Castiel, L. Belec, D. Batisse, A. Si Mohamed, J. Gilquin, G. Gonzalez-Canali, D. Jayle, M. Karmochkine, L. Weiss, J. P. Aboulker, and M. D. Kazatchkine. 1998. Discrepant responses to triple combination antiretroviral therapy in advanced HIV disease. AIDS 12:745-750. [DOI] [PubMed] [Google Scholar]
- 36.Quinones-Mateu, M. E., S. C. Ball, A. J. Marozsan, V. S. Torre, J. L. Albright, G. Vanham, G. van Der Groen, R. L. Colebunders, and E. J. Arts. 2000. A dual infection/competition assay shows a correlation between ex vivo human immunodeficiency virus type 1 fitness and disease progression. J. Virol. 74:9222-9233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson, L. H., R. E. Myers, B. W. Snowden, M. Tisdale, and E. D. Blair. 2000. HIV type 1 protease cleavage site mutations and viral fitness: implications for drug susceptibility phenotyping assays. AIDS Res. Hum. Retroviruses 16:1149-1156. [DOI] [PubMed] [Google Scholar]
- 38.Staszewski, S., V. Miller, C. Sabin, C. Schlecht, P. Gute, S. Stamm, T. Leder, A. Berger, E. Weidemann, A. Hill, and A. Phillips. 1999. Determinants of sustainable CD4 lymphocyte count increases in response to antiretroviral therapy. AIDS 13:951-956. [DOI] [PubMed] [Google Scholar]
- 39.Zennou, V., F. Mammano, S. Paulous, D. Mathez, and F. Clavel. 1998. Loss of viral fitness associated with multiple Gag and Gag-Pol processing defects in human immunodeficiency virus type 1 variants selected for resistance to protease inhibitors in vivo. J. Virol. 72:3300-3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, Y. M., H. Imamichi, T. Imamichi, H. C. Lane, J. Falloon, M. B. Vasudevachari, and N. P. Salzman. 1997. Drug resistance during indinavir therapy is caused by mutations in the protease gene and in its Gag substrate cleavage sites. J. Virol. 71:6662-6670. [DOI] [PMC free article] [PubMed] [Google Scholar]