Abstract
A direct comparison of the inhibitory effects of alpha, beta, and gamma interferons (IFNs) on replication of a hepatitis C virus subgenomic replicon in a hepatoma cell line revealed similarities in antiviral potency. However, alternate IFN-induced antiviral mechanisms were suggested following observations of striking differences between IFN-γ and IFN-α/β with respect to strength and durability of the antiviral response and the magnitude and pattern of IFN-mediated gene expression.
Interferons (IFNs) are a family of cytokines that share the property of inducing proteins that inhibit virus replication (23). IFNs are not constitutively synthesized by cells but respond to external stimuli, such as virus infections, that trigger their transient synthesis and secretion. Binding of IFN to its receptor elicits signals that induce the transcription of a specific set of genes. These genes encode proteins that carry out the diverse functions of the IFN system including mediating antiviral activity. IFNs are classified as either alpha/beta IFN (IFN-α/β) or IFN-γ. IFN-α and -β are readily induced by double-stranded viral RNA, primarily in leukocytes and fibroblasts. These IFNs then induce some of the known IFN-responsive antiviral effector mechanisms, including (i) 2′,5′oligoadenylate synthetase (2′,5′-OAS) (which then activates the constitutively expressed RNase L to degrade viral RNAs), (ii) double-stranded RNA-activated protein kinase (which inhibits viral protein synthesis), and (iii) MxA (a GTPase that blocks transport of viral ribonucleoproteins to the nucleus) (23). The IFN-α/β-regulated genes induce direct antiviral activity as part of the early or innate host immune response to limit viral infection. The host adaptive antiviral response mediated by CD8+ cytotoxic T lymphocytes and natural killer cells and by virus-specific neutralizing antibodies then eradicates virus and virus-infected cells. IFN-γ is induced as part of the adaptive immune response by a wide variety of stimuli and is restricted to T cells and natural killer cells (4). Little is known about the direct antiviral activity of IFN-γ, particularly with respect to the mechanism, magnitude, and kinetics of its antiviral activity in comparison with IFN-α and -β.
Hepatitis C virus (HCV), a member of the Flaviviridae family, has a single-stranded positive-sense RNA genome encoding a polyprotein of at least 10 distinct gene products (5). Infections caused by HCV are a major medical concern since as much as 3% of the worldwide population is estimated to be HCV seropositive (1). At present, the most widely prescribed HCV antiviral therapy is the combination of polyethylene glycol-modified IFN-α2b (PEG-Intron) and ribavirin (18), a treatment capable of inducing sustained virologic responses in 54 to 56% of chronically infected patients (11a, 18). Although this is a significant improvement from the sustained response rate of IFN-α monotherapy (6 to 16%) (7, 21, 24) and IFN-α plus ribavirin (40%) (8, 19), improvement in sustained response rates still remains the major goal in developing new HCV treatment modalities. Further elucidation of the mechanisms by which IFNs inhibit HCV replication in the liver could enhance the design of more effective therapeutics. Clinical data from current HCV therapies has provided evidence to suggest that sustained response rates are associated with the profile of the decline of viremia. Response to IFN-α treatment (in combination with ribavirin) in patients with chronic HCV infection is biphasic and is represented by a rapid, initial decline (innate IFN response in the first few days) followed by a second, prolonged response (adaptive IFN response in the first 4 to 12 weeks) (14). More in-depth analyses of these treatment response rates for IFN therapy with patients with chronic HCV infection revealed that a steeper decline in viremia in the second phase of viral clearance correlated well with increased sustained virologic response rates to IFN treatment (25). Such analyses clearly implicate possible roles for two distinct IFN-mediated antiviral responses and, moreover, suggest that the IFN-induced adaptive antiviral response is key to the long-lasting host response to HCV infection. This raises the following questions. (i) What mechanisms are responsible for IFN-mediated viral clearance? (ii) Since IFN-α is primarily involved in the innate or early response to virus, what other IFN-mediated antiviral responses are involved in the adaptive host anti-HCV response? Recent understanding of the critical role of IFN-γ-mediated noncytolytic antiviral activity in the clearance of hepatitis B virus (HBV) in both murine and primate models (reviewed in reference 12) has encouraged further investigation into the direct antiviral activities of IFN-γ, particularly in viral hepatitis infections.
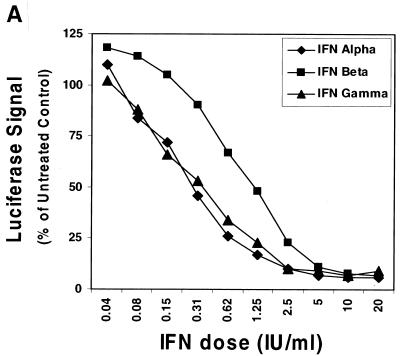
Clearly a comparison of the direct antiviral effects of IFN-α, IFN-β, and IFN-γ on HCV replication could shed light on the existence of distinct IFN-induced anti-HCV antiviral mechanisms. Mechanistic studies of HCV replication have been hindered by the lack of an in vitro cell culture system and a validated small animal model as an alternative to the chimpanzee. Nonetheless, the recent establishment of a selectable HCV subgenomic replicon system (genotype 1b) in Huh-7 hepatoma cells has provided a useful system for studying HCV RNA replication (3, 16, 17, 20). IFN-α/β has antiviral activity in the HCV replicon system (3, 10, 13); however, a comparative analysis of the in vitro antiviral activity of IFN-α/β and IFN-γ has not been demonstrated in this model. The HCV replicon system provides us with a unique opportunity to perform this comparison. First, some of the classical IFN-α/β-induced antiviral effector pathways do not appear to be of importance in this in vitro model (10), and second, the absence of infiltrating IFN-γ-responsive immune cells, such as lymphocytes, in this system allows us to focus on the IFN-γ-induced antiviral effector molecules present in liver cells. We therefore monitored the effects of recombinant IFN-α2b (Intron A; Schering-Plough Corp., Kenilworth, N.J.), IFN-β (Biosource, Camarillo, Calif.), and IFN-γ-1b (Actimmune; InterMune Pharmaceuticals, Palo Alto, Calif.) on viral RNA synthesis in 5-2 cells (a human hepatoma cell line, Huh7, carrying the firefly luciferase reporter HCV subgenomic replicon based on the I389luc-ubi-neo/NS3-3′/5.1 replicon backbone [15]). Cells were maintained in growth medium (Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum and 0.5 to 1 mg of G418/ml). To assess the antiviral activity or potential cytokine-induced toxicity from the IFN treatments, luciferase activity (a direct measurement of reporter replicon RNA synthesis) and cell viability assays were performed. In these assays, replicon cells were incubated for 2 days in the presence of G418 and then treated with 0.04 to 20 IU of IFNs/ml in growth media without phenol red or G418. Luciferase and cellular viability [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt (MTS); Promega, Madison, Wis.] assays were performed at 24 h posttreatment. All three IFNs were capable of suppressing HCV RNA replication as demonstrated by decreased luciferase activity (Fig. 1A) without affecting viability. The 50% effective concentrations (EC50s) for the various IFNs tested were approximately 0.3, 1.2, and 0.3 IU/ml for IFN-α, IFN-β, and IFN-γ, respectively. Interestingly, complete suppression of the replicon-directed luciferase signal was not achieved at IFN concentrations 20- to 60-fold higher than the EC50s. Similar data for IFN-γ has been generated recently with the HCV replicon cell culture model using concentrations ranging from 5 to 5,000 IU/ml (11).
FIG. 1.
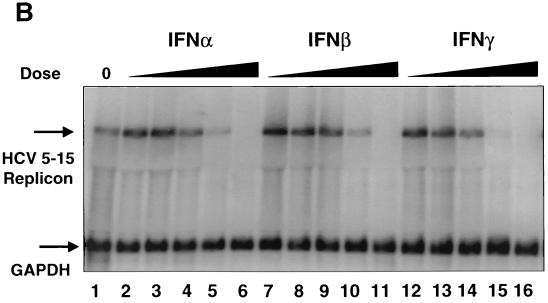
Inhibition of HCV RNA synthesis in the subgenomic replicon cells by IFN-α, -β, and -γ. (A) HCV replicon luciferase reporter cells (I389luc-ubi-neo/NS3-3′/5.1) were incubated in the presence of IFN-α, -β or -γ for 24 h and then assayed for luciferase activity. Luciferase signals were plotted as mean percentages of those for the untreated control cells and are representative of three independently derived experiments. (B) Northern blot analysis of HCV replicon RNA derived from I389neo/NS3-3′/wt replicon (16) cells treated with log increment doses of IFN-α (lanes 2 to 6) or IFN-γ (lanes 12 to 16) (0.01 to 100 IU/ml) or IFN-β (lanes 7 to 11) (0.003 to 33 IU/ml). A 32P-labeled, HCV-specific (NS5A-NS5B) probe was used to detect replicon RNA extracted from the replicon cells that had been treated with various doses of IFN for 72 h. Similarly, a probe specific for glyceraldehyde-3-phosphate dehydrogenase mRNA was used to monitor the level of this cellular RNA.
To confirm that the inhibitory effect on the luciferase signal was due to suppression of viral RNA synthesis, Northern blot analysis was performed to measure the replicon RNA levels in the IFN-treated cells. Replicon cells that had been plated 24 h previously in G418-containing media were treated with IFNs at different doses and incubated for another 72 h till harvest. Total RNA was extracted and subjected to Northern blotting analysis against an HCV-specific probe. As shown in Fig. 1B, similar inhibitory effects on replicon RNA levels were observed for the three IFNs. This inhibition was specific for HCV RNA, since the level of a cellular mRNA (glyceraldehyde-3-phosphate dehydrogenase) was unaffected. We were unable to correlate temporally the IFN-mediated decreases in HCV replicon RNA levels with suppression of the luciferase reporter replicon because 24 h post-IFN treatment would be insufficient time to detect RNA differences by Northern blotting. Nonetheless, these results clearly demonstrated that both IFN-α/β and IFN-γ can effectively inhibit HCV RNA replication with similar potency in cell culture.
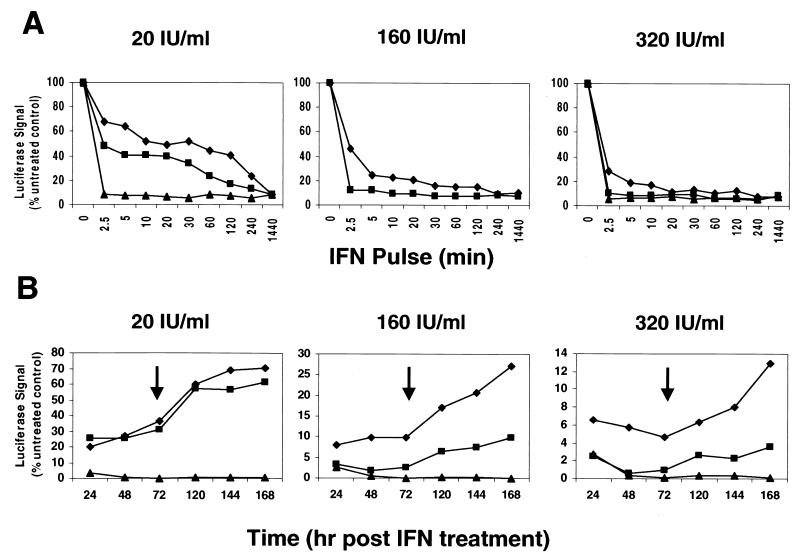
The effect of the IFN exposure time on the antiviral activity of each IFN was further assessed by evaluating the luciferase signal after IFN pulse treatments (20, 160, and 320 IU of IFN-α and -β/ml; 20 and 320 IU of IFN-γ/ml) in 5-2 cells for various times (2.5 min to 24 h). The IFN-containing medium was replaced with IFN-free medium after each treatment period, and luciferase signal was measured 24 h from the start of the IFN incubation. Although IFN pulse times varied, luciferase measurements were not made sooner than 24 h after initiation of the IFN pulse treatment due to the preexisting intracellular pool of replicon RNA and the slow degradation rate of the RNA. The response rate of HCV replication inhibition by each IFN was dose dependent, although IFN-γ was able to achieve maximal suppression of replication with only a 2.5-min pulse at the lowest IFN dose of 20 IU/ml (Fig. 2A). This result indicated that IFN-γ induced a more robust response than IFN-α and -β in the HCV replicon cell culture. Interestingly, a recent report (22) on HBV revealed that IFN-γ induced a slower antiviral effect than IFN-α, suggesting that different temporal mechanisms may be employed by IFN-γ to achieve its antiviral activities against HCV and HBV.
FIG. 2.
Differential response rates of HCV replicon reporter cells to IFN treatment and durability of IFN inhibitory effect. (A) HCV replicon reporter cells (I389luc-ubi-neo/NS3-3′/5.1) were treated with 20, 160, or 320 IU of IFN-α (diamonds) and IFN-β (squares)/ml or with 20 and 320 IU of IFNγ (triangles)/ml for various lengths of time (2.5 min to 24 h) and assayed for luciferase activity at the 24-h time point from the start of IFN treatment. Luciferase signals were plotted as mean percentages of the untreated control cells and are representative of three independently derived experiments. (B) Reporter cells were treated with IFN doses as described above for 4 h and then assayed for the luciferase signal 24, 48, or 72 h posttreatment. In addition, a parallel and equally treated cell culture was trypsinized at the 72-h mark (see arrow), diluted 1:4 into new microtiter plates, and subsequently assayed either 48, 72, or 96 h from this point (120, 144, and 168 total hours post-IFN treatment). Durability of the IFN HCV replication inhibitory effect was plotted over time as a percentage of the luciferase signal derived from mock-treated cells. Data represent means of triplicate values from a typical assay.
The durability of the antiviral response to each of the three IFNs was addressed by following the temporal effect (24 to 168 h) after a 4-h pulse treatment with IFNs at 20, 160, or 320 IU/ml. IFN-pulsed reporter cells were cultured and assayed for the luciferase signal, as described previously, 24, 48, or 72 h post-IFN treatment. In addition, a parallel culture was split 1:4 at the 72-h point to maintain subconfluent growth and then replated onto new microtiter plates for further culturing out to 120, 144, and 168 h post-IFN treatment. IFN-γ produced longer-lasting HCV-inhibitory effects than similar pulses with IFN-α and -β (Fig. 2B). IFN-γ-treated cells were unable to recover HCV replication 72 h following the IFN pulse treatment or even after continuation of the culture (with one culture split) out to 168 h after treatment. In contrast, replicon cells treated with IFN-α and -β appeared to recover slowly following the IFN pulse treatments, as evidenced by a temporal increase in the luciferase signal (Fig. 2B). The dose of IFN was not the determining factor for recovery of HCV replication. The durability of response by an 8- or 16-fold-higher dose of IFN-α and -β, respectively, paralleled the trends seen when comparing the durability of responses at the 20-IU/ml treatment level (Fig. 2B). The maximal inhibition of HCV replicon replication induced by IFN-γ at all doses tested after 24 h was not reversible over a 6-day period. In contrast, the maximal inhibition of HCV replicon replication after 24 h induced by IFN-α and -β was reversible even when the dose of IFN-α and -β was 8 to 16-fold higher than that of IFN-γ. Cell proliferation assays (MTS) performed on parallel and identically treated cell cultures suggested that the replicon cells were viable and continuously growing over the 168-h test period. IFN-mediated cytotoxicity was not a contributing factor for inhibition of HCV replication in all experiments, since the viability of the replicon cells following various doses and exposure times to IFN-α, IFN-β, and IFN-γ was 100% ± 2.4%, 99.6% ± 4.3%, and 98.3% ± 2.4%, respectively. In addition, an experiment addressing the long-term survival of IFN-treated replicon cells in the presence of G418 showed that IFN-γ-pulsed cells (20 and 320 IU/ml) were unable to replicate and survive under G418 selection whereas a significant proportion of IFN-α/β-treated cells grew steadily (data not shown). Collectively, these results showed greater durability of the antiviral response by IFN-γ than of that by IFN-α and -β and suggest a possible alternate mechanism of anti-HCV replication effect by IFN-γ.
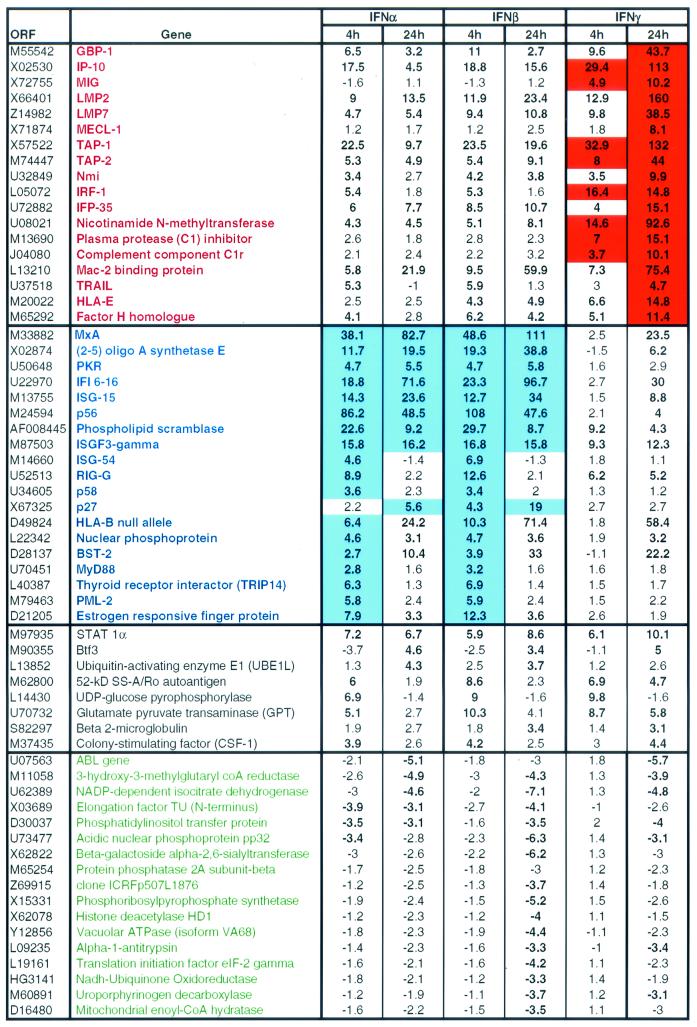
Correlation of the differential IFN-induced gene expression profiles between IFN-α/β and IFN-γ treatments and the strength and durability of the IFN-induced inhibition of HCV replicon replication could provide insight into previously unrecognized IFN-induced anti-HCV pathways and antiviral effector molecules present in the liver. To address this, DNA microarray analyses were performed on RNA samples isolated from untreated or IFN-treated HCV replicon cells that contained the replicon construct I389neo/NS3-3′/wt (16). The cells (2 × 106 to 4 × 106) were plated in 10-cm tissue culture plates, incubated for 2 days in the presence of G418, and then treated for 4 or 24 h with 20 IU of IFN-α2b, IFN-β, or IFN-γ/ml (in growth media without G418) prior to isolation of total cellular RNA. The synthesis of double-stranded cDNA from total cellular RNA (10 μg) and the generation of biotin-labeled cRNA from the double-stranded cDNA template were performed according to Affymetrix (Santa Clara, Calif.) protocols. IFN-induced gene expression changes were determined following comparison with the untreated cells and represented as increased or decreased fold changes. Only genes that had a fold change in expression of >3 were considered significant. The comparisons of the genes upregulated or downregulated by IFN-α, IFN-β, or IFN-γ in the HCV replicon cell culture model are detailed in Table 1. Distinct sets of genes were upregulated at 4 and 24 h by both IFN-α/β and IFN-γ. However, the magnitude of induced gene expression could be clearly categorized into three classes: (i) 18 genes that were preferentially upregulated by IFN-γ, (ii) 19 genes that were preferentially upregulated by IFN-α and IFN-β, and (iii) 8 genes that were upregulated by all three IFNs. The downregulated genes, however, showed no similar trend (Table 1). The differential effect on gene expression by IFN-α/β versus IFN-γ, coupled with similarity in magnitude but differences in durability of the anti-HCV response by IFN-α/β and IFN-γ in the HCV replicon system, suggests that distinct, IFN-type-specific host anti-HCV pathways exist in liver cells. One possible explanation for the difference in durability of the IFN-γ antiviral response from that of IFN-α/β was provided by examining the kinetics of the gene expression induced by IFN-α/β and IFN-γ. The magnitude of antiviral activity (decrease in luciferase signal) of each IFN was similar after either a 4-h pulse or continuous exposure for 24 h. Nevertheless, all of the IFN-γ-induced genes continued to increase expression between 4 and 24 h whereas expression of IFN-α/β-induced genes was maximally induced at 4 h in 10 of the 19 genes. Therefore, we can presume that the differential magnitude and kinetics of gene expression have little impact on the induction of an antiviral state by the IFN-stimulated genes but more likely influence the strength and durability of the antiviral response in the HCV replicon system.
TABLE 1.
Genes upregulated and downregulated by 4-h and 24-h treatment with IFN-α, IFN-β, and IFN-γa
Microarray analysis of gene expression changes induced by 20 IU/ml of IFN-α and IFN-β or IFN-γ/ml in HCV replicon cells (I389neo/NS3-3′/wt). Total cellular RNA (10 μg) was used to synthesize double-stranded cDNA, which was subsequently used to generate biotin-labeled cRNA. Fragmented cRNA (20 μg) target was hybridized to a Hu6800 full-length GeneChip array (Affymetrix) at 45°C overnight. The chips were washed (Fluidics Station 400; Affymetrix) and stained with phycoerythrin-labeled streptavidin (Molecular Probes, Eugene, Oreg.) and then scanned on a Hewlett Packard/Agilent GeneArray scanner. Data was analyzed using the Affymetrix Data Mining Tool (version 3.0). The manufacturer recommended analyses to assess variability in array hybridization (efficiency and normalization), and cDNA synthesis efficiency were performed as previously described (9). Genes are listed by open reading frame (ORF) GenBank accession numbers or TIGR database HT number). Genes preferentially upregulated by IFN-γ are listed in red with fold changes highlighted in red, whereas genes preferentially upregulated by IFN-α and IFN-β are listed in blue with fold changes highlighted in blue. Genes not preferentially upregulated by all three IFNs are listed in black. Downregulated genes are listed in green. Bold numbers represent fold changes of >3.
Our gene expression data generated in the HCV replicon-containing hepatoma cell line (Huh-7) showed some striking similarity to data generated in previous comparative gene profiling experiments with IFNs. IFN-α/β or IFN-γ (1,000 IU/ml, 6-h exposure)-stimulated gene expression profiles were demonstrated in a human fibrosarcoma cell line, HT1080, in the absence of viral replication, by DNA microarray analysis (9). Of the 18 genes preferentially upregulated by IFN-γ in the HCV replicon-containing cell line shown in Table 1, 10 of those genes were upregulated in the IFN-stimulated HT1080 cells, and 7 of those 10 genes (encoding IRF-1, TAP-1, LMP7, LMP2, MECL-1, C1r, and HLA-E) showed preferential upregulation by IFN-γ. In addition, of the 19 genes preferentially upregulated by IFN-α and -β in the HCV replicon-containing cell line (Table 1), 16 were present in the IFN-stimulated HT1080 cells and 12 of 16 showed preferential upregulation by IFN-α and IFN-β. These data show that in the absence of viral replication, the differential gene profiles induced by IFN-α/β and IFN-γ are strikingly similar to our observations with the HCV replicon-containing Huh7 cells.
Our data correlating in vitro antiviral activity by the three IFNs and gene expression profiles identified by microarray analysis suggest that some of these genes may initiate or mediate the antiviral action needed to eliminate HCV replication. Which of these genes are functionally relevant can be determined only with an in vivo antiviral response to HCV. Interestingly, DNA microarray analyses have been performed to determine changes in liver gene expression during the course of an acute-resolving HCV infection in a chimpanzee (2). Clearance of viremia in serum and liver occurred between weeks 6 and 8. The most notable changes in gene expression occurred in the 33 IFN-responsive genes which were grouped in one of three different expression patterns: (i) genes which peaked early (day 7) and then declined, (ii) genes which peaked late (week 6), and (iii) genes which peaked early and whose levels were sustained until clearance of viremia. Interestingly, of the 18 genes preferentially upregulated by IFN-γ in the HCV replicon-containing cells, expression of 8 of these genes was found in the liver biopsies from the HCV-infected chimpanzee. More importantly, all eight of these genes (genes for GBP-1, IP-10, TAP-1, LMP7, LMP2, MECL-1, IFP35, and Mac-2 binding protein) were expressed maximally at week 6, concurrent with the initiation of viral clearance. In addition, 9 of the 19 IFN-α/β-induced genes were also found in the HCV-infected liver biopsies. In contrast to the eight IFN-γ-induced genes, only one IFN-α-induced gene, that for MxA, had a peak expression at week 6. The other seven genes peaked early, although five of these maintained high expression levels at week 6 (genes for 2′,5′-OAS, ISG-15, p56, RIG-G, and p27). These data imply that the IFN-γ-stimulated genes have more functional relevance than IFN-α/β-induced genes in initiating a successful host adaptive immune response to HCV infection. Alternatively, expression of IFN-γ-stimulated genes might need to occur before IFN-α/β-induced genes can have a functional impact on viral clearance in HCV infections. Evidence to support these hypotheses comes from a recent study with 25 patients with chronic HCV infection. Patients were treated with the combination IFN-α-ribavirin or IFN-α alone (6), and the HCV-specific T-cell reactivity was monitored during treatment, at the end of treatment, and at 24-week follow-up. Similar to the chimpanzee data, this study showed that T-cell reactivity increased at around treatment weeks 4 to 8. Resolution of HCV viremia (sustained antiviral response) in patients treated with the combination IFN-α-ribavirin or IFN-α alone was more likely for patients who developed HCV-specific T-cell proliferation with increased IFN-γ production.
Using DNA microarray analyses, Robek and colleagues compared changes in cellular gene expression that accompany the IFN-mediated inhibition of replication in the livers of the 1.3-HBV transgenic mice and in the HBV-Met hepatocyte cell line in vitro (22). A number of genes were common to all experiments. Six of those genes were common also to our list of genes preferentially upregulated by IFN-γ, namely, the MIG, IP10, LMP2, LMP7, MECL-1, and GBP-1 genes. Collectively these data suggest that a common IFN-induced antiviral mechanism exists in HBV and HCV that can occur in host nonlymphocyte cells. This antiviral activity is presumably part of the host adaptive immune response to virus infection and is independent of the classical IFN-α/β-induced innate host antiviral mechanisms (MxA, PKR, and 2′,5′-OAS).
The remarkable consistency of the profile of IFN-γ-induced gene expression, irrespective of cell type and presence of replicating virus, coupled with their peak expression at the initiation of viral clearance (HCV) in vivo provides strong evidence that these IFN-induced genes may have direct antiviral activity. The question remaining is which of the key IFN-γ-regulated genes mediate these direct antiviral effects. Our preliminary data suggest that the IFN-induced chemokines IP10 and MIG and proteasome subunits LMP2, LMP7, and MECL-1 do not have direct antiviral activity using the HCV subgenomic replicon system (data not shown) or may require more than one component to trigger the antiviral state.
In summary, we have shown that IFN-α/β and IFN-γ mediate antiviral effects of similar magnitude in HCV replicon-containing Huh-7 cells. However, kinetic analyses showed that the antiviral effect of IFN-γ was strong and durable, whereas IFN-α and IFN-β induced a less robust and more transient antiviral response against HCV. Indeed, IFN-γ-treated HCV replicon cells under drug selection failed to recover and form colonies after long-term culture, suggesting either extended suppression or potential eradication of HCV replicons. Studies are ongoing to distinguish between these two hypotheses. Furthermore, two classes of IFN-regulated genes were also identified. The class of genes that were preferentially upregulated by IFN-γ correlated with some of the genes shown by others to be (i) expressed highly at the peak of an acute-resolving host response to HCV in a chimpanzee and (ii) important in the noncytolytic cytokine-mediated antiviral effects of the host adaptive immune response. This study along with other previous observations suggests that IFN-α/β and IFN-γ induce distinct direct antiviral effector molecules and alternate antiviral pathways in the IFN-mediated suppression of HCV replication. The contribution of these distinct IFN-induced pathways to host innate and adaptive direct antiviral responses warrants further investigation.
Acknowledgments
We thank Ralf Bartenschlager and colleagues for supplying the HCV subgenomic replicon system.
REFERENCES
- 1.Alter, M. J., D. Kruszon-Moran, O. V. Nainan, G. M. McQuillan, F. Gao, L. A. Moyer, R. A. Kaslow, and H. S. Margolis. 1999. The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. N. Engl. J. Med. 341:556-562. [DOI] [PubMed] [Google Scholar]
- 2.Bigger, C. B., K. M. Brasky, and R. E. Lanford. 2001. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J. Virol. 75:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 4.Boehm, U., T. Klamp, M. Groot, and J. C. Howard. 1997. Cellular responses to interferon-gamma. Annu. Rev. Immunol. 15:749-795. [DOI] [PubMed] [Google Scholar]
- 5.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 6.Cramp, M. E., S. Rossol, S. Chokshi, P. Carucci, R. Williams, and N. V. Naoumov. 2000. Hepatitis C virus-specific T-cell reactivity during interferon and ribavirin treatment in chronic hepatitis C. Gastroenterology 118:346-355. [DOI] [PubMed] [Google Scholar]
- 7.Davis, G. L., L. A. Balart, E. R. Schiff, K. Lindsay, H. C. Bodenheimer, Jr., R. P. Perrillo, W. Carey, I. M. Jacobson, J. Payne, J. L. Dienstag, et al. 1989. Treatment of chronic hepatitis C with recombinant interferon alfa. A multicenter randomized, controlled trial. N. Engl. J. Med. 321:1501-1506. [DOI] [PubMed] [Google Scholar]
- 8.Davis, G. L., R. Esteban-Mur, V. Rustgi, J. Hoefs, S. C. Gordon, C. Trepo, M. L. Shiffman, S. Zeuzem, A. Craxi, M. H. Ling, J. Albrecht, et al. 1998. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. N. Engl. J. Med. 339:1493-1499. [DOI] [PubMed] [Google Scholar]
- 9.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frese, M., T. Pietschmann, D. Moradpour, O. Haller, and R. Bartenschlager. 2001. Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol. 82:723-733. [DOI] [PubMed] [Google Scholar]
- 11.Frese, M., V. Schwärzle, K. Barth, N. Krieger, V. Lohmann, S. Mihm, O. Haller, and R. Bartenschlager. 2002. Interferon-gamma inhibits replication of subgenomic and genomic hepatitis C virus RNAs. Hepatology 35:694-703. [DOI] [PubMed] [Google Scholar]
- 11a.Fried, M. W., M. L. Shiffman, R. K. Reddy, C. Smith, G. Marino, F. Goncales, D. Haeussinger, M. Diago, G. Carosi, J.-P. Zarski, J. Hoffmann, and J. Yu. 2001. Digestive Disease Week 2001. Gastroenterology 120(Suppl. 1):A55. [Google Scholar]
- 12.Guidotti, L. G., and F. V. Chisari. 2001. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 19:65-91. [DOI] [PubMed] [Google Scholar]
- 13.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrmann, E., A. U. Neumann, J. M. Schmidt, and S. Zeuzem. 2000. Hepatitis C virus kinetics. Antivir. Ther. 5:85-90. [PubMed] [Google Scholar]
- 15.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 17.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manns, M. P., J. G. McHutchinson, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 19.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, J. K. Albrecht, et al. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 20.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poynard, T., V. Leroy, M. Cohard, T. Thevenot, P. Mathurin, P. Opolon, and J. P. Zarski. 1996. Meta-analysis of interferon randomized trials in the treatment of viral hepatitis C: effects of dose and duration. Hepatology 24:778-789. [DOI] [PubMed] [Google Scholar]
- 22.Robek, M. D., S. F. Wieland, and F. V. Chisari. 2002. Inhibition of hepatitis B virus by interferon requires proteasome activity. J. Virol. 76:3570-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tine, F., S. Magrin, A. Craxi, and L. Pagliaro. 1991. Interferon for non-A, non-B chronic hepatitis. A meta-analysis of randomised clinical trials. J. Hepatol. 13:192-199. [DOI] [PubMed] [Google Scholar]
- 25.Zeuzem, S., E. Herrmann, J. H. Lee, J. Fricke, A. U. Neumann, M. Modi, G. Colucci, and W. K. Roth. 2001. Viral kinetics in patients with chronic hepatitis C treated with standard or peginterferon alpha2a. Gastroenterology 120:1438-1447. [DOI] [PubMed] [Google Scholar]