Abstract
The binding of hepatitis C virus glycoprotein E2 to the large extracellular loop (LEL) of CD81 has been shown to modulate human T-cell and NK cell activity in vitro. Using random mutagenesis of a chimera of maltose-binding protein and LEL residues 113 to 201, we have determined that the E2-binding site on CD81 comprises residues Ile182, Phe186, Asn184, and Leu162. These findings reveal an E2-binding surface of approximately 806 Å2 and potential target sites for the development of small-molecule inhibitors of E2 binding.
CD81 is a member of the tetraspanin membrane protein superfamily, characterized by the presence of four transmembrane domains, three intracellular loops, and two extracellular domains, which in the case of CD81 are referred to as the small extracellular loop and the large extracellular loop (LEL). Tetraspanins appear to have an essential function in organizing signaling molecules at the cell surface by associating with other tetraspanins, lineage-specific proteins, integrins, major histocompatibility complex molecules, and signaling proteins in a cell type-dependent manner (for a review, see reference 8). The recent elucidation of the CD81 LEL crystal structure revealed that it is a homodimer, with each monomer being composed of five α-helices (A to E) arranged in a head subdomain (consisting of the last two turns of the A helix, the B, C, and D helices, and their interconnecting loops) atop a stalk subdomain (comprising antiparallel A and E helices) (6). Four cysteine residues, conserved in all tetraspanin sequences, participate in the formation of two disulfide bonds, Cys156-Cys190 and Cys157-Cys175, which stabilize the head subdomain.
Hepatitis C virus (HCV) encodes a ∼3,300-amino-acid polyprotein from which the E1 (polyprotein residues 191 to 383) and E2 (residues 384 to 746) glycoproteins are cleaved cotranslationally. The mature forms of E1 and E2 are noncovalently associated, and each contains an N-terminal ectodomain and a C-terminal transmembrane domain. Recently, it was shown that recombinant soluble E2, E1-E2 complex, and HCV-like particles (12, 14) as well as HCV particles from infectious plasma (14) bind to the CD81 LEL with nanomole-level affinity (13, 14). Furthermore, the LEL can inhibit the binding of E2 to liver sections (13, 14) and of HCV-like particles to MOLT-4 T cells (21). The available evidence suggests that CD81 is unlikely to play a role in HCV entry (1, 4, 13, 16, 17, 21, 22). However, E2-CD81 ligation was recently found to induce in naive and antigen-experienced T cells in vitro a costimulatory signal leading to the production of the proinflammatory cytokine gamma interferon (20). As HCV-associated liver damage is primarily due to a massive infiltration by activated proinflammatory lymphocytes (for a review, see reference 19), these findings raise the possibility that the CD81-E2 interaction plays a role in T-cell-mediated liver inflammation and pathology.
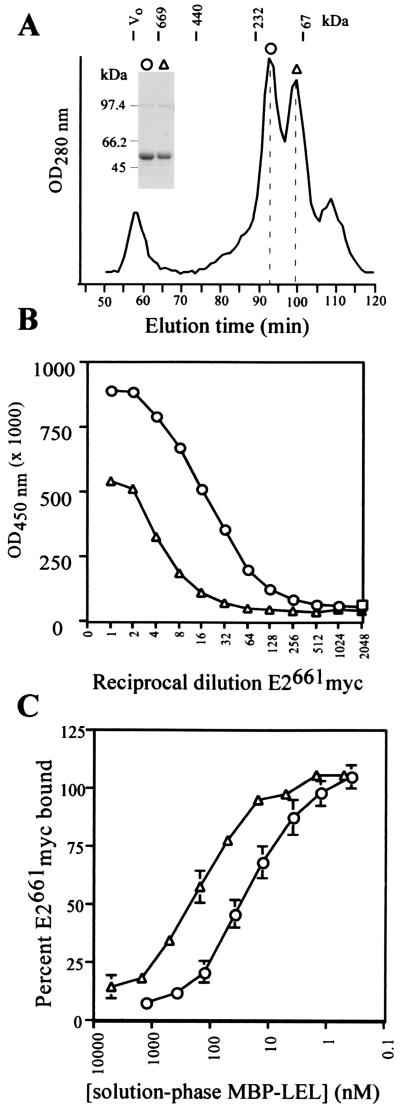
To further characterize the E2-binding residues on the LEL of CD81, we produced a protein chimera consisting of maltose-binding protein (MBP) fused to the N terminus of LEL residues 113 to 201 (MBP-LEL113-201) via a trialanine linker. MBP is monomeric and serves as an ideal scaffold for studying oligomerization determinants of heterologous proteins (2, 7). Superdex 200 gel filtration chromatography of amylose-agarose-purified MBP-LEL113-201 reveals two major species, corresponding approximately in molecular mass to the dimer (eluting at 92 min) and the monomer (100 min) (Fig. 1A). Nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of the MBP-LEL113-201 dimer and monomer peak fractions revealed an ∼48-kDa major species and a trace amount (<5%) of an ∼97-kDa band (Fig. 1A, inset), consistent with the virtual absence of disulfide-linked dimers. The ∼97-kDa trace species is likely to be an artifact induced by boiling of samples in the absence of β-mercaptoethanol, as it was obtained from both the dimer and monomer peaks and was not detected by electrospray mass spectrometry (see below).
FIG. 1.
Characterization of the MBP-LEL113-201 chimera. (A) Separation of MBP-LEL113-201 oligomeric forms by Superdex 200 gel filtration chromatography. Elution positions for protein standards are indicated above the profile; the dotted lines indicate the elution positions for the putative MBP-LEL monomer (98 min) and dimer (92 min). The symbols above gel filtration peaks have been used to identify corresponding MBP-LEL species in panels B and C. The inset shows the results of nonreducing SDS-PAGE analysis of peak fractions (putative MBP-LEL113-201 dimer [circles] and MBP-LEL113-201 monomer [triangles]) collected from Superdex 200 gel filtration experiments. Vo, void volume. (B) Binding of E2661myc to solid-phase MBP-LEL oligomeric forms in EIA. E2661myc binding was detected with MAb 9E10 and a horseradish peroxidase-conjugated anti-mouse antibody. MBP-LEL113-201 dimer, circles; MBP-LEL113-201 monomer, triangles. (C) Competitive EIA. Serial dilutions of MBP-LEL113-201 dimer (circles) and monomer (triangles) were used to compete with the binding of a subsaturating amount of E2661myc to solid-phase MBP-LEL113-201 dimer. Results are means ± standard errors of data from three independent experiments.
Sedimentation equilibrium analysis confirmed the molecular mass of the MBP-LEL113-201 putative dimer. The experimental data, plotted as the normal log of the optical density at 280 nm (OD280) versus the squared radius, results in a straight line that is consistent with the presence of a stable single solute (data not shown). The slope of the line and the observed molecular mass (± standard deviation) of 96,300 ± 4,500 Da closely approximate those of the dimer, whose theoretical mass is 100,518 Da. Next, the disulfide bond statuses of the MBP-LEL113-201 dimer and monomer were examined by treatment with the alkylating agent 4-vinylpyridine (4-VP) followed by mass spectrometry to detect the covalent modification of free sulfhydryl groups (2). The molecular masses of the 4-VP-treated MBP-LEL113-201 dimer (50,264 Da) and monomer (50,260 Da) were unchanged relative to those of their untreated counterparts (50,265 and 50,262 Da, respectively), confirming that two intramolecular disulfide bonds had formed.
To examine the ability of the MBP-LEL113-201 dimer and monomer to bind HCV E2, we developed a solid-phase enzyme immunoassay (EIA) in which plate-bound MBP-LEL chimeras capture E2661myc secreted from transfected 293T cells. Binding is detected by using the anti-myc monoclonal antibody (MAb) 9E10 and peroxidase-conjugated anti-mouse immunoglobulin G. Figure 1B shows that E2661myc binds optimally to the MBP-LEL113-201 dimer; a substantially lower level of binding is observed for the MBP-LEL113-201 monomer. The dimeric form of MBP-LEL113-201 also captured noncovalently associated complexes of E1-E2 from transfected-cell lysates, as determined with the conformation-dependent MAb H53 (data not shown). The ability of the chimeras to inhibit the binding of solution-phase E2661myc to plate-bound MBP-LEL113-201 dimer was used to rule out the possibility that the attachment of MBP chimeras to the solid phase had induced the formation of an E2661myc binding site (Fig. 1C). The 50% inhibitory concentration (IC50) for the MBP-LEL113-201 dimer is approximately 10-fold lower than the IC50 for monomeric MBP-LEL113-201 (30 ± 10 nM versus 283 ± 29 nM), suggesting that dimerization of the LEL enhances binding to E2 by approximately 10-fold.
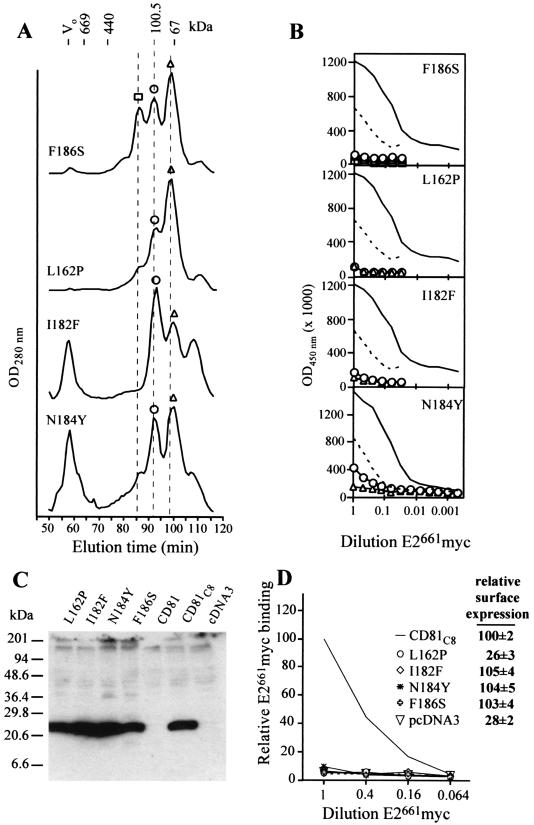
A random mutagenesis approach was used to identify amino acid changes that resulted in the loss of E2-binding function in MBP-LEL113-201 as determined by the method of Lin-Goerke et al. (9). We used the EIA-based E2661myc-binding assay to screen 300 transformants and identified four point mutants (N184Y, L162P, I182F, and F186S) that retained the ability to form dimers (Fig. 2A) but exhibited a decrease in (N184Y) or a complete loss of (L162P, I182F, and F186S) E2661myc-binding function (Fig. 2B). In agreement with the solid-phase assay data, solution-phase N184Y dimer exhibited a lower affinity for E2661myc than its wild-type counterpart, having an IC50 of 766 ± 251 nM, approximately 20-fold higher than the IC50 for the wild-type dimer. Examination of these mutants by 4-VP mass spectrometry confirmed that these mutants had formed two intramolecular disulfide bonds, indicating that the L162P, I182F, N184Y, and F186S mutations are accommodated in the LEL fold (data not shown). Therefore, the ability of these mutations to inhibit LEL E2-binding function implies that Leu162, Ile182, Asn184, and Phe186 are E2 contact residues.
FIG. 2.
(A) Superdex 200 gel filtration of F186S, L162P, I182F, and N184Y mutants. Dotted lines represent elution times for the MBP-LEL113-201 monomer (98 min), dimer (92 min), and putative trimer (86 min). Symbols above peaks have been used to identify corresponding MBP-LEL species in panel B. Vo, void volume. (B) EIA of E2661myc binding to solid-phase peak Superdex 200 fractions collected in the experiment whose results are shown in panel A. Binding curves for the MBP-LEL113-201 dimer (solid line) and monomer (dashed line) are included. (C) Synthesis and stability of CD81C8 mutants. Lysates of CD81C8-expressing 293T cells were subjected to reducing SDS-PAGE in 12 to 17% polyacrylamide gradient gels followed by Western blotting with MAb C8, directed to the C-terminal epitope tag. (D) Binding of E2661myc to cell surface-expressed CD81C8 mutants. Transfected CHO-K1 cells were incubated with serial dilutions of concentrated E2661myc tissue culture fluid, washed, and then incubated with radioiodinated MAb 9E10. After being subjected to further washing, the cells were lysed and their radioactivity was quantitated in a Packard Autogamma counter. Relative E2661myc binding is expressed as follows: (c.p.m. bound to cells expressing mutant CD81C8/c.p.m. bound to cells expressing wild-type CD81C8) × 100. The results are representative of two independent transfections. The inset shows the relative cell surface-expression of CD81C8 mutants. Intact CD81C8-expressing CHO-K1 cells were incubated with 125I-labeled MAb 1.3.3.22 for 1 h on ice and then washed prior to quantitation of radioactivity. Relative 125I-labeled MAb 1.3.3.22 binding is expressed as follows: (c.p.m. bound to cells expressing mutant CD81C8/c.p.m. bound to cells expressing wild-type CD81C8) × 100. The means ± standard deviations of data from three independent transfections are shown.
We next confirmed that these four mutations had similar effects on E2-binding function in the context of the entire tetraspanin molecule. We engineered a cytomegalovirus promoter-based expression vector (CD81C8), containing the CD81 open reading frame linked to a C-terminal MAb C8 epitope tag, for transient-transfection experiments. By using the anti-CD81 MAb 1.3.3.22 in flow cytometry, we first confirmed that the C8 epitope tag did not affect the cell surface expression of CD81 (data not shown). Mutations were introduced into CD81C8, and the expression and stability of the mutants in transfected HEK 293T cells were compared with those of the wild type by Western blotting with MAb C8. Figure 2C shows that the mutants and the modified wild-type CD81C8 were expressed in transfected-cell lysates at similar levels, migrating to a position corresponding to a molecular mass of 26 kDa.
The abilities of wild-type and mutated CD81 molecules to bind secreted E2661myc were compared by using a surface binding assay. CD81C8-transfected CHO-K1 cells were incubated with serial dilutions of concentrated tissue culture fluid obtained from E2661myc-transfected HEK 293T cells. E2661myc binding was measured in a gamma counter following incubation with 125I-labeled MAb 9E10 and washing of the cells. Figure 2D indicates that mutations L162P, I182F, N184Y, and F186S in the context of the entire CD81 molecule completely abolished E2661myc-binding ability while cell surface expression of I182F, N184Y, and F186S was unchanged compared to that of the modified wild-type CD81C8, confirming that these residues comprise the E2-binding site. We were unable to detect L162P at the cell surface; therefore, we cannot distinguish whether the L162P mutation results in a translocation-incompetent CD81C8 structure or whether L162P forms part of the MAb 1.3.3.22 epitope. However, given that the MPB-LEL113-201 L162P mutant is able to form dimers and contains two intramolecular disulfide bonds and that its CD81C8 counterpart appears to be as stable as wild-type CD81C8 in transfected cells (Fig. 2C), the latter possibility seems more likely. Flint et al. (4) demonstrated that the binding of MAb 1.3.3.22 to CD81 precludes binding of E2, which suggests that the two binding sites overlap. The epitope of 1.3.3.22 does not appear to involve the Ile182, Asn184, or Phe186 residue but may involve residue Leu162. Together these data suggest that Leu162 may indeed be part of the E2-binding site.
The D helix residues Ile182, Asn184, and Phe186 form part of a solvent-exposed cluster comprising a hydrophobic ridge (Ile181, Ile182, Leu185, and Phe186) adjacent to a hydrophilic base (Asn184 and Thr166) (Fig. 3). The solvent-exposed hydrophobic residues are generally conserved in CD81 sequences, and their presence points to a protein-protein interaction role for this site (6). Consistent with this idea, our data indicate that these residues are involved in E2 binding. Interestingly, the F186L mutation occurs naturally in CD81 of African green monkeys and has been shown previously to eliminate E2 binding when placed in the human CD81 LEL sequence (5, 10). Leu162 occurs within a short 310 helix located between the B and C helices, forming a hydrophobic pocket on the surface of the head subdomain (Fig. 3). A naturally occurring mutation, T163A, at an adjacent residue of the African green monkey CD81 has been shown to enhance E2 binding when placed in the human CD81 sequence (5), suggesting that the Leu162 region modulates E2 binding. The involvement of Leu162 in E2 binding extends the molecular surface of the binding site from 591 Å2 (Thr166, Ile181, Ile182, Asn184, Leu185, and Phe186) to 805 Å2.
FIG. 3.
Molecular surface of the human CD81 LEL dimer, showing locations of mutations that affect E2 binding (Leu162, Ile182, Asn184, and Phe186). The figure was drawn using GRASP (11) software.
An inspection of E2-binding sites in the context of the human CD81-LEL dimeric structure reveals that one site is rotated 90° relative to the other. The orientation of the LEL dimer such that an E2-binding site projects out from the plane of the membrane (Fig. 3; see also Fig. 4 in reference 6) indicates that the E2-binding site of the partner monomer faces the membrane, making it unavailable for E2 ligation. Such a relationship between binding sites in native CD81 would preclude bivalent binding of CD81 to E2, particularly if virion-associated E2 molecules are related by dyad symmetry, as is the case with the flavivirus counterpart, the E glycoprotein (15, 23). However, such a relationship between binding sites may allow asymmetric binding of E2 and other members of the tetraspanin web to LELs.
The results of recent in vitro studies indicate that the E2-CD81 interaction has immunomodulatory implications including enhanced production of the proinflammatory cytokines gamma interferon and interleukin-4 (20), down-regulation of T-cell receptors (20), and suppression of NK cell activity (3, 18). These findings raise the possibility that E2-CD81 ligation contributes to an HCV-induced liver immunopathology that is characterized by large numbers of liver-infiltrating activated T cells. Our identification of the E2-binding footprint in the context of the human CD81-LEL crystal structure provides new targets for the design of small-molecule inhibitors of the E2-CD81 interaction.
Acknowledgments
We thank Anne Maerz for assistance with expression vector construction, Bruce Kemp for use of the PE Sciex III+ mass spectrometer and the Pharmacia Smart System, and G. J. Howlett, Department of Biochemistry and Molecular Biology, The University of Melbourne, for assistance with sedimentation equilibrium analyses.
This study was supported by NHMRC project grants 156714 and 991153.
REFERENCES
- 1.Agnello, V., G. Abel, M. Elfahal, G. B. Knight, and Q. X. Zhang. 1999. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 96:12766-12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Center, R. J., B. Kobe, K. A. Wilson, T. Teh, G. J. Howlett, B. E. Kemp, and P. Poumbourios. 1998. Crystallization of a trimeric human T cell leukemia virus type 1 gp21 ectodomain fragment as a chimera with maltose-binding protein. Protein Sci. 7:1612-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crotta, S., A. Stilla, A. Wack, A. D'Andrea, S. Nuti, U. D'Oro, M. Mosca, F. Filliponi, R. M. Brunetto, F. Bonino, S. Abrignani, and N. M. Valiante. 2002. Inhibition of natural killer cells through engagement of CD81 by the major hepatitis C virus envelope protein. J. Exp. Med. 195:35-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flint, M., C. Maidens, L. D. Loomis-Price, C. Shotton, J. Dubuisson, P. Monk, A. Higginbottom, S. Levy, and J. A. McKeating. 1999. Characterization of hepatitis C virus E2 glycoprotein interaction with a putative cellular receptor, CD81. J. Virol. 73:6235-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Higginbottom, A., E. R. Quinn, C. C. Kuo, M. Flint, L. H. Wilson, E. Bianchi, A. Nicosia, P. N. Monk, J. A. McKeating, and S. Levy. 2000. Identification of amino acid residues in CD81 critical for interaction with hepatitis C virus envelope glycoprotein E2. J. Virol. 74:3642-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kitadokoro, K., D. Bordo, G. Galli, R. Petracca, F. Falugi, S. Abrignani, G. Grandi, and M. Bolognesi. 2001. CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs. EMBO J. 20:12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobe, B., R. J. Center, B. E. Kemp, and P. Poumbourios. 1999. Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose-binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc. Natl. Acad. Sci. USA 96:4319-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy, S., S. C. Todd, and H. T. Maecker. 1998. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu. Rev. Immunol. 16:89-109. [DOI] [PubMed] [Google Scholar]
- 9.Lin-Goerke, J. L., D. J. Robbins, and J. D. Burczak. 1997. PCR-based random mutagenesis using manganese and reduced dNTP concentration. BioTechniques 23:409-412. [DOI] [PubMed] [Google Scholar]
- 10.Meola, A., A. Sbardellati, B. Bruni Ercole, M. Cerretani, M. Pezzanera, A. Ceccacci, A. Vitelli, S. Levy, A. Nicosia, C. Traboni, J. McKeating, and E. Scarselli. 2000. Binding of hepatitis C virus E2 glycoprotein to CD81 does not correlate with species permissiveness to infection. J. Virol. 74:5933-5938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nicholls, A., K. A. Sharp, B. Honig, C. T. Tseng, E. Miskovsky, M. Houghton, and G. R. Klimpel. 1991. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11:281-296. [DOI] [PubMed] [Google Scholar]
- 12.Owsianka, A., R. F. Clayton, L. D. Loomis-Price, J. A. McKeating, and A. H. Patel. 2001. Functional analysis of hepatitis C virus E2 glycoproteins and virus-like particles reveals structural dissimilarities between different forms of E2. J. Gen. Virol. 82:1877-1883. [DOI] [PubMed] [Google Scholar]
- 13.Petracca, R., F. Falugi, G. Galli, N. Norais, D. Rosa, S. Campagnoli, V. Burgio, E. Di Stasio, B. Giardina, M. Houghton, S. Abrignani, and G. Grandi. 2000. Structure-function analysis of hepatitis C virus envelope-CD81 binding. J. Virol. 74:4824-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pileri, P., Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani. 1998. Binding of hepatitis C virus to CD81. Science 282:938-941. [DOI] [PubMed] [Google Scholar]
- 15.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 16.Seipp, S., H. M. Mueller, E. Pfaff, W. Stremmel, L. Theilmann, and T. Goeser. 1997. Establishment of persistent hepatitis C virus infection and replication in vitro. J. Gen. Virol. 78:2467-2476. [DOI] [PubMed] [Google Scholar]
- 17.Takikawa, S., K. Ishii, H. Aizaki, T. Suzuki, H. Asakura, Y. Matsuura, and T. Miyamura. 2000. Cell fusion activity of hepatitis C virus envelope proteins. J. Virol. 74:5066-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tseng, C. T., and G. R. Klimpel. 2002. Binding of the hepatitis C virus envelope protein e2 to CD81 inhibits natural killer cell functions. J. Exp. Med. 195:43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valiante, N. M., A. D'Andrea, S. Crotta, F. Lechner, P. Klenerman, S. Nuti, A. Wack, and S. Abrignani. 2000. Life, activation and death of intrahepatic lymphocytes in chronic hepatitis C. Immunol. Rev. 174:77-89. [DOI] [PubMed] [Google Scholar]
- 20.Wack, A., E. Soldaini, C. Tseng, S. Nuti, G. Klimpel, and S. Abrignani. 2001. Binding of the hepatitis C virus envelope protein E2 to CD81 provides a co-stimulatory signal for human T cells. Eur. J. Immunol. 31:166-175. [DOI] [PubMed] [Google Scholar]
- 21.Wellnitz, S., B. Klumpp, H. Barth, S. Ito, E. Depla, J. Dubuisson, H. E. Blum, and T. F. Baumert. 2002. Binding of hepatitis C virus-like particles derived from infectious clone H77C to defined human cell lines. J. Virol. 76:1181-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wunschmann, S., J. D. Medh, D. Klinzmann, W. N. Schmidt, and J. T. Stapleton. 2000. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J. Virol. 74:10055-10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yagnik, A. T., A. Lahm, A. Meola, R. M. Roccasecca, B. B. Ercole, A. Nicosia, and A. Tramontano. 2000. A model for the hepatitis C virus envelope glycoprotein E2. Proteins 40:355-366. [DOI] [PubMed] [Google Scholar]