Abstract
Herpes simplex virus glycoprotein H (gH) is one of the four virion envelope proteins which are required for virus entry and for cell-cell fusion in a transient system. In this report, the role of the transmembrane and cytoplasmic tail domains of gH in membrane fusion was investigated by generating chimeric constructs in which these regions were replaced with analogous domains from other molecules and by introducing amino acid substitutions within the membrane-spanning sequence. gH molecules which lack the authentic transmembrane domain or cytoplasmic tail were unable to mediate cell-cell fusion when coexpressed with gB, gD, and gL and were unable to rescue the infectivity of a gH-null virus as efficiently as a wild-type gH molecule. Many amino acid substitutions of specific amino acid residues within the transmembrane domain also affected cell-cell fusion, in particular, those introduced at a conserved glycine residue. Some gH mutants that were impaired in cell-cell fusion were nevertheless able to rescue the infectivity of a gH-negative virus, but these pseudotyped virions entered cells more slowly than wild-type virions. These results indicate that the fusion event mediated by the coexpression of gHL, gB, and gD in cells shares common features with the fusion of the virus envelope with the plasma membrane, they point to a likely role for the membrane-spanning and cytoplasmic tail domains of gH in both processes, and they suggest that a conserved glycine residue in the membrane-spanning sequence is crucial for efficient fusion.
Herpes simplex viruses (HSVs) enter cells by fusion of the virus envelope with the host cell plasma membrane at a neutral pH (reviewed by Spear [37]), and four of the HSV envelope glycoproteins, gB, gD, gH, and gL, are not only essential for virus entry (7, 13, 24) but can also induce the fusion of cellular membranes when coexpressed from plasmid vectors in the absence of any other virus components (30, 40). Efficient fusion induction with this system is dependent on the presence of a gD receptor on the plasma membrane of recipient cells (4, 34), but the means by which gB, gD, gH, and gL interact with each other or with other components of the plasma membrane to induce polykaryocyte formation and virus entry remain unclear. It is also uncertain whether the cell-cell fusion induced by the coexpression of gB, gD, gH, and gL mirrors the events which take place when the virion envelope fuses with the plasma membrane during virus entry.
Homologues of HSV type 1 (HSV-1) gB and the gHL heterodimer have been identified in herpesviruses of all subfamilies and are likely to play a direct role in the fusion process. Indeed, studies with pseudorabies virus and human herpesvirus 8 have shown that the expression of gB and gHL is sufficient to induce cell fusion (17, 33) and, in varicella-zoster virus, the expression of gHL alone or gB in combination with gE has been reported to induce fusion (11, 25). Despite the compelling evidence for the activity of gB and gHL as fusion proteins, these molecules have no homology with the fusion proteins of other virus families and no obvious counterparts to the hydrophobic fusion peptides of influenza virus hemagglutinin (HA) or human immunodeficiency virus g41 have been identified. It is also unclear whether these molecules interact directly with plasma membrane components as part of the fusion process. gB contains a GAG-binding domain, but cell surface GAGs are not necessary for cell fusion induced by the expression of HSV-1 glycoproteins (4, 34). The gHL homologue of Epstein-Barr virus associates with a further glycoprotein, g42, which interacts with major histocompatibility complex class II on B cells (22, 23), and, in combination with the gB homologue, this complex mediates cell-cell fusion (17). A putative receptor for human cytomegalovirus gH has been identified (1, 2), though this molecule has yet to be characterized as a cell surface component.
There have been many studies on the effects of site-directed mutagenesis on the function of herpesvirus gB molecules. Cai et al. (7) identified mutations in both the extracellular domain and the cytoplasmic domains of HSV-1 gB which reduced the fusogenic capacity of a syncytial virus. Studies of Epstein-Barr virus gB, pseudorabies virus gB, and HSV gB have identified regions of the cytoplasmic domains that promote and regulate fusion (14, 15, 17, 20, 31), and Wanas et al. (41) showed that specific sequences within the membrane-spanning domain of HSV gB are required for membrane fusion. Studies of gH are much more limited. Modifications to the extracellular and cytoplasmic domains of HSV-1 gH have been reported to abolish membrane fusion and virus infectivity (5, 16, 43), but there are no reports on the role of the membrane-spanning domain. We addressed this issue by constructing chimeric molecules in which the transmembrane or cytoplasmic domains of gH were replaced by equivalent sequences from other transmembrane glycoproteins and also made specific changes to conserved and nonconserved residues within the transmembrane domain. These molecules were examined for their ability to function in cell-cell fusion and in virus entry. Our results confirm the importance of the cytoplasmic tail in the function of gH and show that specific sequences within the transmembrane domain are required for membrane fusion and virus entry.
MATERIALS AND METHODS
Cells and viruses.
Vero cells, COS-7 cells, and CR1 (a gH-positive helper cell line) cells were grown and maintained as described previously (43). 293T cells were grown in Glasgow's minimal essential medium containing 10% fetal calf serum (FCS). A gH-negative mutant of HSV-1, STZgH− (36), was propagated and assayed on CR1 cells.
Mutagenesis of gH and replacement of the transmembrane and cytoplasmic tail domains.
The parental plasmid into which all mutations in gH were introduced was pCDNA3gH. This plasmid contains the gH coding sequence of HSV-1 strain HFEM isolated from pSMH3gH (40) as a HindIII-XbaI fragment. Modified versions of this construct were also generated by site-directed mutagenesis (21) through the introduction of an HpaI site at nucleotide 46141, to give pCDNAgH HpaI, and through that of an AflII site at nucleotide 46180, to give pCDNA3gHAflI1. These sites lie immediately 5′ and 3′, respectively, of the predicted transmembrane coding sequence. Plasmid pS84, encoding the CD8 molecule, was a gift from S. Munro. This construct contains EcoRV and AflII sites immediately 5′ and 3′, respectively, of the predicted transmembrane coding sequence. The coding sequence of HSV-1 gD (nucleotides 138419 to 139601) was cloned into pCDNA3 from the parental plasmid pING-HincIIgD (42). HpaI and AflII sites were introduced 5′ and 3′, respectively, of the predicted transmembrane coding sequence (nucleotides 139439 to 139508) by site-directed mutagenesis.
Restriction digestion and religation of cleaved fragments from these plasmids allowed the assembly of expression constructs in pCDNA3 in which the transmembrane and/or cytoplasmic domain coding sequences of gH were replaced by the equivalent regions from CD8 or gD. In some instances, assembly of the appropriate restriction fragments resulted in a coding change at the ligation site and the resulting plasmids were subsequently modified by site-directed mutagenesis to restore the authentic coding sequence. The coding sequence of the transmembrane domain of influenza virus (strain PR8) HA was constructed by assembling the following synthetic oligonucleotides: 1, 5′ ATTCTGATCTACTCAACTGTCGCCAGTTCACTGGTGCTTTTGGTCTCC 3′; 2, 5′ CTGGGGGCAAATCAGTTTCTGGATGTGTTCTC 3′; 3, 5′ TTAAGAGAACACATCCAGAAACTGATTGCCCCCAGGGAGACCAA 3′; and 4, 5′ AAGCACCAGTGAACTGGCGACAGTTGAGTAGATCGCCAGAAT 3′.
Oligonucleotides 1 and 3 and oligonucleotides 2 and 4 were annealed at 80°C. The HpaI-AflII fragment (corresponding to the transmembrane coding sequence) was excised from pCDNA3gHCD8TM, and the remaining fragment was ligated with the mixture of annealed oligonucleotides to generate pCDNA3gHHATM.
Constructs expressing HSV-1 gH with amino acid substitutions, deletions, and insertions were generated by site-directed mutagenesis of pCDNA3gH, and a construct lacking the cytoplasmic tail was constructed by replacing the codon for leucine 827 with a translational termination codon.
Fluorescence-activated cell sorter (FACS) analysis.
Monolayers of 293T cells seeded to 80% confluence (2 × 106 cells) were cotransfected with plasmids expressing wild-type gL and with a plasmid expressing either wild-type or mutated gH. After 24 h, the transfected cells were harvested with trypsin-EDTA, pelleted, and resuspended in 150 μl of wash buffer (1% rabbit serum in phosphate-buffered saline [PBS]). Cells were divided into three 50-μl aliquots and incubated for 1 h on ice with either 100 μl of the antibody to gH, LP11 (a hybridoma supernatant diluted 1/3 in wash buffer), or wash buffer only. Cells were then washed three times with wash buffer and incubated for 45 min on ice with 100 μl of rabbit anti-mouse fluorescein isothiocyanate (diluted 1/50 in wash buffer) or with wash buffer only. Cells were washed three times with ice-cold PBS before resuspending in 500 μl of PBS. Flow-cytometric analysis was then performed with a Becton Dickinson FACScan cytometer by using the Cellquest program.
Cell fusion assay.
The cell fusion assay was carried out essentially as described by Turner et al. (40) with a few minor modifications. Subconfluent monolayers of 293T cells were cotransfected with plasmids expressing wild-type forms of HSV-1 gB, gD, and gL, together with a plasmid expressing either the wild-type or mutated forms of gH. After 2 days, the transfectants were overlaid with Vero cells, and 24 h later, the number of nuclei that were recruited into syncytia was scored. The outcome of this assay was variable and presumably depended on the state of the cells, the cell density, and the transfection efficiency. Experiments using plasmids expressing wild-type glycoproteins generally resulted in 1,000 to 2,000 nuclei recruited into syncytia, but values as high as 10,000 were obtained on some occasions. Mutant gH molecules were therefore always compared with wild-type molecules in parallel experiments, and results were recorded as a percentage of wild-type fusion. In a previous work (40), transfected COS-7 cells were used as fusion effectors and it was reported that cells transfected with control plasmids gave small, spontaneously appearing syncytia. No background was observed by using 293T cells, and where constructs are described as fusion negative, no syncytia or polykaryocytes were observed.
Virus complementation assay.
A total of 2 × 105 COS-7 cells seeded in six-well dishes were transfected with plasmids expressing either wild-type or mutated forms of gH. Thirty-two hours posttransfection, monolayers were infected with HSV-1 STZgH− at a multiplicity of infection (MOI) of 10 and incubated at 37°C for 1 h. Unpenetrated virions were inactivated by washing once in PBS and once in citrate buffer, pH 3 (135 mM NaCl, 10 mM KCl, 40 mM citric acid), followed by a further two washes with PBS. Monolayers were overlaid with 2 ml of prewarmed Glasgow's minimal essential medium containing 10% FCS. Eighteen hours postinfection, cells were harvested and sonicated and the yield of infectious progeny virions was determined by plaque assay on monolayers of the gH-helper cell line CR1.
Measurement of virus entry rates.
Prechilled monolayers of 106 CR1 cells were inoculated in triplicate with 300 PFU of the progeny virions obtained from the complementation experiments described above and incubated for 1 h at 4°C for virus adsorption to occur. Monolayers were washed with PBS to remove unabsorbed virus, transferred to 37°C, and overlaid with prewarmed medium. After 5, 10, 15, 20, 30, 60, and 90 min at 37°C, virions that had failed to penetrate were inactivated by an acid wash as described above. They were then overlaid with prewarmed medium containing carboxymethyl cellulose. After incubation for 48 h at 37°C, the monolayers were fixed with formal saline and stained with toluidine blue and plaques were counted. In each experiment, one triplicate set of dishes was incubated for 48 h at 37°C without an acid wash to provide an estimate of the total available PFU. Numbers of plaques obtained after acid washing at different times postadsorption were recorded as a percentage of the total number of plaques available.
Measurements of gH incorporation into virions.
Monolayers of COS-7 cells seeded in six-well dishes at 2 × 105 cells per well were transfected with plasmids expressing either wild-type or mutated forms of gH. Thirty-two hours posttransfection, monolayers were infected with STZgH− at an MOI of 10 and incubated at 37°C for 1 h. Monolayers were overlaid with 2 ml of prewarmed methionine-free minimal essential medium containing 10% FCS and 46 MBq of [35S]methionine (Amersham, Little Chalfont, Buckinghamshire, United Kingdom). Eighteen hours postinfection, the tissue culture supernatant containing released virus was removed and cell debris was removed by centrifugation at 600 × g for 10 min. Virions were pelleted in a Beckman L8-M ultracentrifuge at 55,000 × g for 2 h. Virions were resuspended in and lysed in radioimmunoprecipitation assay buffer (50 mM Tris [pH 7.2], 150 mM NaCl, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 1% Triton X-100) and then immunoprecipitated with 100 μl of the antibody to gH, LP11 (hybridoma supernatant used undiluted). Twenty microliters of a protein A Sepharose beads slurry was added for 1 h at 4°C, and complexes were washed three times in radioimmunoprecipitation assay buffer, heated at 100°C for 5 min, and analyzed by electrophoresis on a 7% denaturing sodium dodecyl sulfate-polyacrylamide gel, followed by autoradiography.
RESULTS
Characterization of gH chimeric constructs in cell-cell fusion.
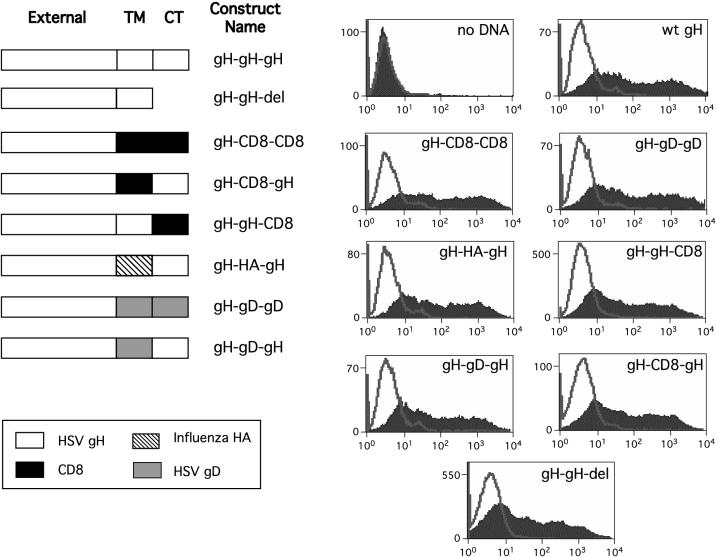
In order to establish whether the transmembrane domain and cytoplasmic tail of HSV-1 gH are required for virus entry and membrane fusion, one or both of these domains were replaced with analogous regions from three other proteins. In addition, the cytoplasmic tail was deleted entirely. Three different membrane-spanning domains from the type 1 membrane glycoproteins were used to replace those of HSV-1 gH. These were those from the human CD8 molecule, influenza virus HA, and HSV-1 gD, and a summary of the panel of constructs produced is shown in Fig. 1.
FIG. 1.
Cell surface expression of chimeric gH constructs. The membrane-spanning domain and/or cytoplasmic domain was replaced with the analogous region from either CD8, influenza virus HA, or HSV-1 gD. The constructs are illustrated, and each is named to indicate its composition with reference to the external domain, transmembrane (TM) domain, and cytoplasmic tail (CT). Thus, gH-gD-gD indicates the external domain of gH fused to the transmembrane and cytoplasmic domains of gD. Each construct was transfected into 293T cells, and cell surface expression was determined by FACS analysis. In each analysis, the filled profile shows cell populations treated with LP11 and the unfilled profile is a control in which treatment with this antibody was omitted. wt, wild type; del, deleted.
Before testing whether replacement of the membrane-spanning domain and/or the cytoplasmic tail of gH with analogous regions from other molecules affects the ability of gH to mediate cell-cell fusion, the levels of cell surface expression of these chimeric molecules in transfected cells were determined by FACS analysis. 293T cells were transfected with plasmids expressing each of the chimeric molecules together with a plasmid expressing gL, and the levels of cell surface gH were measured with monoclonal antibody LP11, an antibody that recognizes the gHL heterodimer. As shown in Fig. 1, all the chimeric gH molecules were expressed at the plasma membrane at levels equivalent to that of wild-type gH, implying that replacement of the gH transmembrane domain and/or cytoplasmic tail with analogous regions from other proteins did not significantly affect the stability of these molecules, their ability to interact with gL, their transport to the cell surface, or their retention in the plasma membrane. All mutants subsequently described in this paper were subjected to this analysis, and all gave results similar to those shown in the graphs in Fig. 1 (right panels).
The effect on cell fusion of replacing the predicted membrane-spanning domain of gH with that of either CD8, HA, or HSV-1 gD and the effect of deleting or replacing the cytoplasmic tail were assessed by using the transient cell-cell fusion assay. Cells were transfected with the chimeric constructs, together with plasmids expressing wild-type forms of gL, gB, and gD. Fusion was assessed after overlaying the transfected cells with Vero cells and counting the number of nuclei that were recruited into syncytia. None of the chimeric molecules induced detectable fusion. This confirms previous reports that modification of the cytoplasmic domain of gH impairs fusion function (5, 43) and suggests that sequences in the transmembrane domain also play a role in this process.
Site-directed mutagenesis of the gH transmembrane domain.
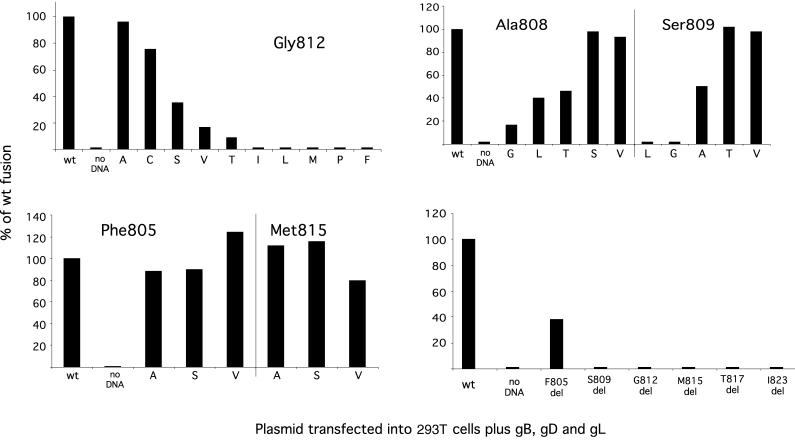
Having found that replacing the gH transmembrane domain with a membrane anchor from a heterologous molecule abolished the ability of gH to mediate fusion, we attempted to determine whether specific amino acids within the predicted transmembrane region of gH are critical for membrane fusion. To this end, we generated a panel of constructs in which some of the amino acids of the transmembrane domain were replaced with different residues. The predicted 21-residue membrane-spanning domains of the gH molecules from a number of different alphaherpesviruses are shown in Fig. 2A. Only 3 residues (marked in bold) are entirely conserved: a conserved pair comprised of alanine at position 808 in the gH sequence and serine at position 809 and a glycine residue at position 812. We therefore changed each of these three residues and two nonconserved residues (methionine 815 and phenylalanine 805) to a number of different amino acids. A summary of all the mutations that were introduced is shown in Fig. 2B.
FIG. 2.
Summary of mutations introduced into the membrane-spanning domain of gH. (A) Alignment of alphaherpesvirus transmembrane domains with conserved residues shown in bold. HSV2, HSV type 2; EHV1, equine herpesvirus 1; EHV4, equine herpesvirus 4; PRV, pseudorabies virus; BHV1, bovine herpesvirus 1; VZV, varicella-zoster virus. (B) Amino acid substitutions. Asterisks identify residues that were deleted.
The effect on cell fusion of introducing these single amino acid changes into the transmembrane domain of gH was measured by using the cell-cell fusion assay described earlier. The results of these experiments are shown in Fig. 3 and are expressed as a percentage of the level of fusion induced by a wild-type gH molecule. Some of the mutations which were introduced at both conserved and nonconserved residues within the gH transmembrane domain had little or no effect on cell fusion. However, six of the mutants at the conserved pair of amino acids A808 and S809 were severely compromised in fusion and all but two of the changes introduced at G812 gave rise to gH molecules which exhibited either reduced fusion phenotypes or were negative in this assay.
FIG. 3.
Fusion activity of gH molecules with alterations to the transmembrane domain. Mutant gH constructs were transfected into 293T cells together with plasmids expressing gD, gB, and gL. After 48 h, untransfected target cells were added and fusion was assessed after a further 24 h by counting nuclei recruited into syncytia. The fusion activity of mutant constructs is given as a percentage of the value obtained using a wild-type (wt) gH expression plasmid in parallel cultures. del, deleted.
Although the experiments described above do not represent an exhaustive mutagenic analysis, the results suggest that the conserved glycine is particularly sensitive to substitution. A role in the fusion process for a centrally conserved glycine in the transmembrane domain of other virus fusion proteins, including that for the vesicular stomatitis virus (VSV) G glycoprotein and influenza virus HA, has been proposed (9, 28). This led us to investigate the effect of repositioning this residue within the transmembrane domain. Constructs were generated in which G812 was shifted by one position in the N-terminal direction (G811, L812) or the C-terminal direction (V812, G813). Neither of these molecules was able to mediate fusion, providing further evidence of the importance of the glycine residue at position 812.
We also constructed a mutant in which G812 was deleted, together with a series of mutants in which other residues in the transmembrane domain were deleted, namely, F805, S809, M815, T817, and I823. Surprisingly, as shown in Fig. 3, all these mutants were nonfunctional in fusion except for F805 deleted, which exhibited 38% of wild-type activity, suggesting that the length of the transmembrane domain may be critical for fusion activity.
Ability of mutant gH molecules to function in virus entry.
The results reported above demonstrate the importance of sequences in the transmembrane and cytoplasmic domains of gH for the function of the molecule in mediating cell-cell fusion. It is reasonable to suppose, but not certain, that this function of gH is similar to its essential role in the fusion of the virus envelope with the plasma membrane during virus entry. We therefore predicted that mutants impaired in the cell-cell fusion assay would also be impaired in virus entry. To test this prediction, we conducted a complementation assay in which cells were transfected with wild-type or mutant expression constructs and subsequently infected with a gH-negative HSV-1 mutant. Pseudotyped progeny virions were then assayed by using a gH-positive helper cell line, and the results were expressed as the yield of infectious virions as a percentage of the yield obtained by using wild-type gH. The results of these experiments did not show a clear correlation between fusion phenotype and virus rescue phenotype. All mutants that were positive in the fusion assay were indistinguishable from wild-type gH in their ability to rescue infectious virus. Fusion-negative constructs fell into two groups. Some, in particular, the chimeric molecules and those containing deletions in the transmembrane sequence, were deficient or impaired in their ability to rescue gH-negative virions. In contrast, all substitution mutants were capable of rescuing infectivity, regardless of their phenotype in the fusion assay. Some of these data are summarized in Table 1.
TABLE 1.
Comparison of the fusion and rescue phenotypes of modified gH moleculesa
| Construct | gH molecules | Fusion | Virus rescue (% of WT) |
|---|---|---|---|
| WT | WT | Positive | 100 |
| Chimeric molecules | gH-CD8-CD8 | Negative | <0.1 |
| gH-gD-gD | Negative | <0.1 | |
| gH-CD8-gH | Negative | <0.1 | |
| gH-gD-gH | Negative | 0.6 | |
| gH-gH-CD8 | Negative | 0.1 | |
| Amino acid substitutions | M815→S | Positive | >50 |
| F805→V | Positive | >50 | |
| F805→S | Positive | >50 | |
| G812→I | Negative | >50 | |
| G812→L | Negative | >50 | |
| S809→L | Negative | >50 | |
| Deletions | G812 del | Negative | 3 |
| M815 dcl | Negative | 3 | |
| I823 del | Negative | 9 |
The ability of each construct to rescue a gH-negative virus was determined as detailed in Materials and Methods. In this assay, the yield of rescued virions using the wild-type gH expression plasmid (WT) varied from 106 to 107 PFU. Background values obtained in the absence of a gH molecule (i.e., residual inoculum) varied from 103 to 5 × 103 PFU. The rescue competence of each mutant molecule was compared to that of the WT molecule in a parallel experiment, and the result is expressed as the percentage of the yield obtained relative to WT rescue. Values below 0.1% are too close to background levels to be reliable. Values above 50% are indistinguishable from WT yields.
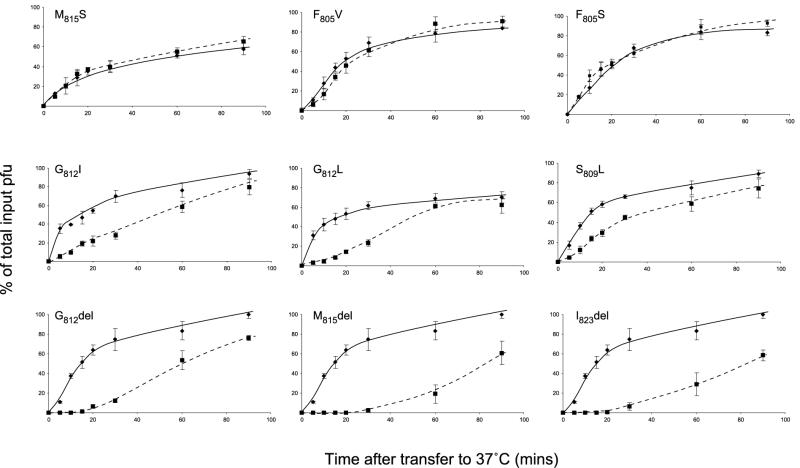
The observation that some gH mutants that are impaired in fusion function but are nevertheless capable of rescuing the infectivity of gH-negative virus was unexpected and suggested that the role of gH in cell-cell fusion might be distinguishable from its role in mediating fusion of the virus envelope with the plasma membrane during virus entry. It is apparent, however, that direct comparison of the results of fusion and rescue assays might be misleading. In the rescue assay, the infectivity of pseudotyped virions was measured by plaque assay on a helper cell line. In this assay, entry was not rate limiting because a virion, once bound, is present for the duration of the assay and will form a plaque regardless of whether it takes minutes or hours to enter the cell. Thus, pseudotyped virions with greatly impaired entry rates would nevertheless be scored as “rescued.” To investigate further the competence of mutant gH molecules in virus entry, we therefore measured the entry rates of pseudotyped virions. Progeny virions were adsorbed to monolayers on ice and shifted to 37°C, and, at various times, bound virions which had yet to enter were inactivated by an acid wash. Entry rate assays of this type were performed with all mutants that gave sufficient yields of infectious pseudotypes in rescue experiments, and representative examples are shown in Fig. 4. Fusion-positive mutants (represented by M815S, F805S, and F805V) gave pseudotyped virions whose entry rates were indistinguishable from those of virions rescued with wild-type gH. All substitution mutants that were fusion negative (represented by G812I, G812L, and S809L) yielded pseudotyped virions with reduced entry rates. Fusion-negative deletion mutants gave reduced yields of pseudotyped virions (Table 1), and this was accompanied by a dramatic reduction in entry rates. Although the data in Fig. 4 are inadequate for detailed kinetic analysis, the behavior of the most impaired mutants indicates a lag before entry begins, perhaps suggesting that gH is responsible for a rate-limiting first stage in the fusion process. The results of these entry assays emphasize the point that infectivity assays may fail to detect entry defects. The data indicate a consistent relationship between the function of gH in cell-cell fusion and in virus entry: all fusion-positive gH constructs gave wild-type levels of infectious virions, and these virions entered cells at the same rate as wild-type virions; fusion-negative constructs yielded pseudotyped virions with reduced entry rates, and, where entry rates were greatly impaired, this was reflected in a reduced infectious yield as judged by plaque assay.
FIG. 4.
Entry rates of pseudotyped virions. Mutant or wild-type gH expression plasmids were transfected into cells which were then infected with a gH-negative mutant of HSV-1. The entry rate of pseudotyped infectious progeny was assessed by adsorbing approximately 300 PFU to monolayers of CR1 cells at 4°C, transferring cultures to 37°C, and inactivating residual inoculum by acid washing at various times after transfer. Plaques were then counted after incubation for a further 2 days. An estimate of the total available infectivity was obtained from cultures that received no acid treatment, and the number of plaques obtained following acid treatment at a particular time was recorded as a percentage of the total available. The entry rate of virions pseudotyped with mutant gH molecules  was compared in parallel cultures with the entry rate of virions pseudotyped with a wild-type gH molecule
was compared in parallel cultures with the entry rate of virions pseudotyped with a wild-type gH molecule  . The means and standard deviations of triplicate cultures are given. del, deleted.
. The means and standard deviations of triplicate cultures are given. del, deleted.
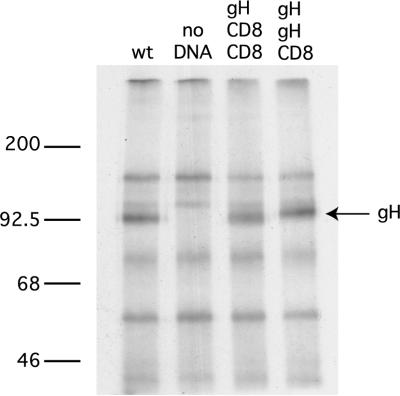
The rescue of infectious pseudotypes by using gH expression constructs requires the incorporation of a functional molecule into the virus envelope, whereas the cell-cell fusion assay requires fusion function alone. However, the behavior of all mutant gH molecules in the infectivity rescue and entry rate assays can be explained by their fusion phenotype: none of the mutants we examined had retained fusion function while losing the ability to rescue infectivity. We attempted to demonstrate that chimeric molecules containing foreign transmembrane and cytoplasmic domains were incorporated into virions. Two constructs (gH-CD8-CD8 and gH-gH-CD8) were transfected into COS-7 cells, and these cells were infected with a gH-negative mutant in the presence of [35S]methionine. Progeny virions were harvested from the culture supernatant by centrifugation, lysed, and gH immunoprecipitated. As shown in Fig. 5, the progeny obtained in the presence of either construct contained levels of gH equivalent to those obtained when cells were transfected with a construct expressing wild-type gH. These results must be interpreted with caution because the purity of these virus pellets cannot be ascertained. Attempts to obtain gradient-purified virions resulted in yields that were too low to allow gH detection in pseudotypes rescued by wild-type or mutant gH expression plasmids. Nevertheless, the results shown in Fig. 5 are consistent with the behavior of mutant gH molecules in fusion and rescue assays and suggest that the assembly of gH into virions is not dependent on specific sequences in the transmembrane or cytoplasmic domains. The same conclusion has been drawn for gD (12, 42).
FIG. 5.
Incorporation of chimeric gH molecules into virus particles. Cells were transfected with plasmids expressing wild-type gH (wt) or the chimeric constructs gH-CD8-CD8 or gH-gH-CD8 and were then infected with a gH-negative mutant at an MOI of 10 in the presence of [35S]methionine. Progeny virions were pelleted from culture supernatants, and lysates were examined for their isotope-labeled gH content by immunoprecipitation. Molecular masses (in kilodaltons) are indicated on the left.
DISCUSSION
Four HSV-1 glycoproteins, gD, gB, and gHL, are required to mediate cell-cell fusion in a transient transfection assay and are essential for virus entry. It is uncertain whether cell-cell fusion induced by the expression of gD, gB, and gHL is analogous to fusion of the virus envelope with the plasma membrane during virus entry. Syncytium formation observed following infection with fusogenic mutants of HSV is clearly distinct from the virus entry process because it requires the expression of several genes that are not required for virus entry (10, 18) and is dependent on a number of alternative syncytium mutations (38). In this sense, the cell fusion observed by coexpression of gD, gB, and gHL is more akin to virus entry than to virus-induced syncytial function because it occurs in the absence of syncytium mutations and in the absence of other gene functions (40). Regardless of the relationship between these different fusion events, the results presented here suggest that the role of gH in cell-cell fusion is similar to its role in virus entry. All mutant gH molecules which were functional in cell fusion assays were able to rescue the infectivity of gH-negative virions, and the pseudotypes entered cells at normal rates. Mutant gH molecules which were nonfunctional in cell fusion either failed to rescue infectivity or produced pseudotypes with reduced entry rates.
Replacement of the transmembrane region or the cytoplasmic tail of gH with analogous sequences from other transmembrane proteins resulted in chimeric molecules that were unable to mediate fusion and were nonfunctional or greatly impaired in their ability to mediate virus entry. Both these domains must therefore contain specific features required for function. The cytoplasmic tail of gH comprises 14 residues, and previous reports have shown that, while deletion of the C-terminal 6 residues has no detectable effect, further deletion dramatically reduces polykaryocyte formation by syncytial HSV strains (5, 43). Our results are entirely consistent with these findings but throw no further light on the mechanism involved.
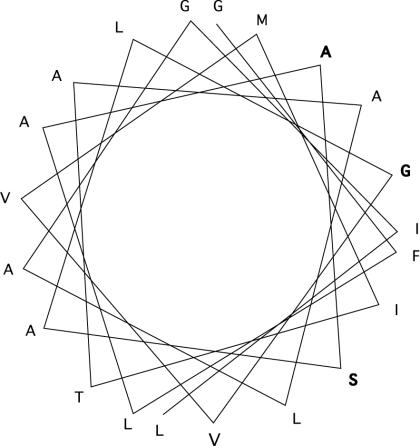
Since replacement of the transmembrane domain of gH with the equivalent domain of CD8, influenza virus HA, or HSV gD abolished fusion function, we examined this region in further detail. The 21-residue predicted transmembrane sequence contains conserved amino acids at three positions in all alphaherpesviruses—namely, ala808, ser809, and gly812. It is notable that an α−helical wheel plot (Fig. 6) of the transmembrane sequence places all three conserved residues on one face of the wheel, suggesting that this face might be involved in interactions with other molecules in the membrane. Analysis of single amino acid substitution mutants confirmed that these three conserved residues are more sensitive to substitution than two nonconserved residues, but a much more exhaustive analysis would be required to demonstrate that one face of the helical wheel is more sensitive to modification than the other.
FIG. 6.
Helical wheel plot of the HSV-1 gH membrane-spanning domain. The wheel illustrates the positions of the membrane-spanning residues when the transmembrane sequence forms an α-helix. Residues conserved in all alphaherpesviruses are shown in bold.
Of the residues examined, gly812 seemed particularly sensitive to substitution and function was also abolished when this residue was moved to position 811 or 813. This is of interest because glycine residues are frequently found in the center of fusion protein transmembrane sequences, and, in the case of the VSV G glycoprotein, a central glycine is essential for membrane fusion (9). It has been proposed that such glycine residues act as helix breakers, thus distorting the bilayer and promoting membrane fusion. This view is supported by the fact that a conserved proline residue (also a helix breaker) is found in the transmembrane domains of foamy virus and murine leukemia virus envelope proteins and is essential for fusion function in both instances (35, 39). It seems unlikely, however, that glycine 812 in the transmembrane domain of gH can act as a helix breaker because the substitution of proline abolishes function, while alanine (which stabilizes the helix) is functional (Fig. 3).
The other notable set of functionally impaired mutants contained single-residue deletions in the transmembrane sequence. Indeed, most of these mutants, as judged by virus entry assays, were more impaired than any of the substitution mutants. It is difficult to interpret these results unambiguously because single-residue deletions affect the position of other residues in a helix as well as reduce the overall length of the transmembrane region. Nevertheless, at 21 residues, the predicted gH transmembrane domain is close to the lower limit for a membrane-spanning helix, and since a deletion at positions across the domain results in loss of function, it is reasonable to suppose that the gH transmembrane domain is close to its critical minimum length.
There are many reports of the role of transmembrane sequences in the function of viral fusion proteins. No clear picture has emerged from these studies. In the case of influenza virus HA, it is clear that a transmembrane anchor is required for full fusion activity (27, 29) but a variety of transmembrane sequences can suffice (28). In contrast, specific sequence requirements within the transmembrane domain have been identified, for example, in human immunodeficiency virus type 1 (19), murine leukemia virus (39), foamy viruses (35), coronavirus (3, 8), VSV (9), Newcastle disease virus (26), and measles virus (6). In some instances, the results are conflicting: functional VSV G glycoprotein in which the transmembrane domain was replaced with analogous regions of other proteins have been described (32) but site-directed mutagenesis of the transmembrane domain has revealed specific sequence requirements for function (9). It is not, in any case, valid to compare gH with these molecules because gH is clearly not a fusion protein in the same sense. The gHL heterodimer is essential for HSV-mediated membrane fusion but must cooperate with two other transmembrane proteins, gD and gB, to achieve fusion. We have little idea of how these molecules cooperate to induce fusion. It is clear that gD must react with specific receptors, but its membrane anchor and cytoplasmic tail contain no specific sequence requirements. In contrast, although we know nothing of potential receptors for the ectodomain of HSV-1 gH, it is clear that there are specific sequence requirements in the transmembrane and cytoplasmic tail that play important roles, either in interacting with other proteins or acting directly in the fusion process, and it is notable that similar conclusions have been drawn from studies of gB.
Acknowledgments
We thank Duncan Wilson for helpful discussions and Susanne Bell and Birgitte Bruun for invaluable assistance.
This work was supported by the Wellcome Trust, United Kingdom, and we thank the MRC for the award of a cooperative grant.
REFERENCES
- 1.Baldwin, B. R., M. Kleinberg, and S. Keay. 1996. Molecular cloning and expression of receptor peptides that block human cytomegalovirus/cell fusion. Biochem. Biophys. Res. Commun. 219:668-673. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin, B. R., C. O. Zhang, and S. Keay. 2000. Cloning and epitope mapping of a functional partial fusion receptor for human cytomegalovirus gH. J. Gen. Virol. 81:27-35. [DOI] [PubMed] [Google Scholar]
- 3.Bos, E. C., L. Heijnen, W. Luytjes, and W. J. Spaan. 1995. Mutational analysis of the murine coronavirus spike protein: effect on cell-to-cell fusion. Virology 214:453-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Browne, H., B. Bruun, and T. Minson. 2001. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH and gL. J. Gen. Virol. 82:1419-1422. [DOI] [PubMed] [Google Scholar]
- 5.Browne, H. M., B. C. Bruun, and A. C. Minson. 1996. Characterization of herpes simplex virus type 1 recombinants with mutations in the cytoplasmic tail of glycoprotein H. J. Gen. Virol. 77:2569-2573. [DOI] [PubMed] [Google Scholar]
- 6.Caballero, M., J. Carabana, J. Ortego, R. Fernandez-Munoz, and M. L. Celma. 1998. Measles virus fusion protein is palmitoylated on transmembrane-intracytoplasmic cysteine residues which participate in cell fusion. J. Virol. 72:8198-8204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, W., B. Gu, and S. Person. 1988. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J. Virol. 62:2596-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, K. W., Y. Sheng, and J. L. Gombold. 2000. Coronavirus-induced membrane fusion requires the cysteine-rich domain in the spike protein. Virology 269:212-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cleverley, D. Z., and J. Lenard. 1998. The transmembrane domain in viral fusion: essential role for a conserved glycine residue in vesicular stomatitis virus G protein. Proc. Natl. Acad. Sci. USA 95:3425-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis-Poynter, N., S. Bell, T. Minson, and H. Browne. 1994. Analysis of the contributions of herpes simplex virus type 1 membrane proteins to the induction of cell-cell fusion. J. Virol. 68:7586-7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duus, K. M., C. Hatfield, and C. Grose. 1995. Cell surface expression and fusion by the varicella-zoster virus gH:gL glycoprotein complex: analysis by laser scanning confocal microscopy. Virology 210:429-440. [DOI] [PubMed] [Google Scholar]
- 12.Feenstra, V., M. Hodaie, and D. C. Johnson. 1990. Deletions in herpes simplex virus glycoprotein D define nonessential and essential domains. J. Virol. 64:2096-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forrester, A., H. Farrell, G. Wilkinson, J. Kaye, N. Davis-Poynter, and T. Minson. 1992. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J. Virol. 66:341-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster, T. P., J. M. Melancon, and K. G. Kousoulas. 2001. An alpha-helical domain within the carboxyl terminus of herpes simplex virus type 1 (HSV-1) glycoprotein B (gB) is associated with cell fusion and resistance to heparin inhibition of cell fusion. Virology 287:18-29. [DOI] [PubMed] [Google Scholar]
- 15.Gage, P. J., M. Levine, and J. C. Glorioso. 1993. Syncytium-inducing mutations localize to two discrete regions within the cytoplasmic domain of herpes simplex virus type 1 glycoprotein B. J. Virol. 67:2191-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galdiero, M., A. Whiteley, B. Bruun, S. Bell, T. Minson, and H. Browne. 1997. Site-directed and linker insertion mutagenesis of herpes simplex virus type 1 glycoprotein H. J. Virol. 71:2163-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haan, K. M., S. K. Lee, and R. Longnecker. 2001. Different functional domains in the cytoplasmic tail of glycoprotein B are involved in Epstein-Barr virus-induced membrane fusion. Virology 290:106-114. [DOI] [PubMed] [Google Scholar]
- 18.Haanes, E. J., C. M. Nelson, C. L. Soule, and J. L. Goodman. 1994. The UL45 gene product is required for herpes simplex virus type 1 glycoprotein B-induced fusion. J. Virol. 68:5825-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helseth, E., U. Olshevsky, D. Gabuzda, B. Ardman, W. Haseltine, and J. Sodroski. 1990. Changes in the transmembrane region of the human immunodeficiency virus type 1 gp41 envelope glycoprotein affect membrane fusion. J. Virol. 64:6314-6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klupp, B. G., R. Nixdorf, and T. C. Mettenleiter. 2000. Pseudorabies virus glycoprotein M inhibits membrane fusion. J. Virol. 74:6760-6768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kunkel, T. A. 1985. Rapid and efficient site-specific mutagenesis without phenotypic detection. Proc. Natl. Acad. Sci. USA 82:488-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, Q., M. K. Spriggs, S. Kovats, S. M. Turk, M. R. Comeau, B. Nepom, and L. M. Hutt-Fletcher. 1997. Epstein-Barr virus uses HLA class II as a cofactor for infection of B lymphocytes. J. Virol. 71:4657-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, Q., S. M. Turk, and L. M. Hutt-Fletcher. 1995. The Epstein-Barr virus (EBV) BZLF2 gene product associates with the gH and gL homologs of EBV and carries an epitope critical to infection of B cells but not of epithelial cells. J. Virol. 69:3987-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ligas, M. W., and D. C. Johnson. 1988. A herpes simplex virus mutant in which glycoprotein D sequences are replaced by β-galactosidase sequences binds to but is unable to penetrate into cells. J. Virol. 62:1486-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maresova, L., T. J. Pasieka, and C. Grose. 2001. Varicella-zoster virus gB and gE coexpression, but not gB or gE alone, leads to abundant fusion and syncytium formation equivalent to those from gH and gL coexpression. J. Virol. 75:9483-9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGinnes, L., T. Sergel, and T. Morrison. 1993. Mutations in the transmembrane domain of the HN protein of Newcastle disease virus affect the structure and activity of the protein. Virology 196:101-110. [DOI] [PubMed] [Google Scholar]
- 27.Melikyan, G. B., S. A. Brener, D. C. Ok, and F. S. Cohen. 1997. Inner but not outer membrane leaflets control the transition from glycosylphosphatidylinositol-anchored influenza hemagglutinin-induced hemifusion to full fusion. J. Cell. Biol. 136:995-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melikyan, G. B., S. Lin, M. G. Roth, and F. S. Cohen. 1999. Amino acid sequence requirements of the transmembrane and cytoplasmic domains of influenza virus hemagglutinin for viable membrane fusion. Mol. Biol. Cell 10:1821-1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melikyan, G. B., J. M. White, and F. S. Cohen. 1995. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes. J. Cell. Biol. 131:679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muggeridge, M. I. 2000. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J. Gen. Virol. 81:2017-2027. [DOI] [PubMed] [Google Scholar]
- 31.Nixdorf, R., B. G. Klupp, A. Karger, and T. C. Mettenleiter. 2000. Effects of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J. Virol. 74:7137-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Odell, D., E. Wanas, J. Yan, and H. P. Ghosh. 1997. Influence of membrane anchoring and cytoplasmic domains on the fusogenic activity of vesicular stomatitis virus glycoprotein G. J. Virol. 71:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pertel, P. E. 2002. Human herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J. Virol. 76:4390-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pertel, P. E., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 35.Pietschmann, T., H. Zentgraf, A. Rethwilm, and D. Lindemann. 2000. An evolutionarily conserved positively charged amino acid in the putative membrane-spanning domain of the foamy virus envelope protein controls fusion activity. J. Virol. 74:4474-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodger, G., J. Boname, S. Bell, and T. Minson. 2001. The assembly and organization of glycoproteins B, C, D and H in HSV-1 particles lacking individual glycoproteins: no evidence for the formation of a complex of these molecules. J. Virol. 75:710-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spear, P. G. 1993. Entry of alphaherpesviruses into cells. Semin. Virol. 4:167-180. [Google Scholar]
- 38.Spear, P. G. 1993. Membrane fusion induced by herpes simplex virus, p. 201-232. In J. Bentz (ed.), Viral fusion mechanisms. CRC Press Inc., Boca Raton, Fla.
- 39.Taylor, G. M., and D. A. Sanders. 1999. The role of the membrane-spanning domain sequence in glycoprotein-mediated membrane fusion. Mol. Cell. Biol. 10:2803-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wanas, E., S. Efler, K. Ghosh, and H. P. Ghosh. 1999. Mutations in the conserved carboxy-terminal hydrophobic region of glycoprotein gB affect infectivity of herpes simplex virus. J. Gen. Virol. 80:3189-3198. [DOI] [PubMed] [Google Scholar]
- 42.Whiteley, A., B. Bruun, T. Minson, and H. Browne. 1999. The effects of targeting herpes simplex virus type 1 gD to the endoplasmic reticulum and trans-Golgi network. J. Virol. 73:9515-9520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson, D. W., N. Davis-Poynter, and A. C. Minson. 1994. Mutations in the cytoplasmic tail of herpes simplex virus glycoprotein H suppress cell fusion by a syncytial strain. J. Virol. 68:6985-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]