Abstract
The human cyclin T1 (hCycT1) protein from the positive transcription elongation factor b (P-TEFb) binds the transactivator Tat and the transactivation response (TAR) RNA stem loop from human immunodeficiency virus type 1 (HIV). This complex activates the elongation of viral transcription. To create effective inhibitors of Tat and thus HIV replication, we constructed mutant hCycT1 proteins that are defective in binding its kinase partner, Cdk9, or TAR. Although these mutant hCycT1 proteins did not increase Tat transactivation in murine cells, their dominant-negative effects were small in human cells. Higher inhibitory effects were obtained when hCycT1 was fused with the mutant Cdk9 protein. Since the autophosphorylation of the C terminus of Cdk9 is required for the formation of the stable complex between P-TEFb, Tat, and TAR, these serines and threonines were changed to glutamate in a kinase-inactive Cdk9 protein. This chimera inhibited Tat transactivation and HIV gene expression in human cells. Therefore, this dominant-negative kinase-inactive mutant Cdk9.hCycT1 chimera could be used for antiviral gene therapy.
The expression of human immunodeficiency virus type 1 (HIV) is regulated by the viral transcriptional transactivator Tat (22). Unlike other DNA-bound activators, Tat binds RNA and effects the elongation step of transcription (41). Without Tat, RNA polymerase II terminates prematurely, and short transcripts accumulate. By binding the transactivation response (TAR) element, which forms an RNA stem loop at the 5′ end of all viral transcripts, Tat relieves this transcriptional block. Tat also requires a cellular cofactor, the positive transcription elongation factor b (P-TEFb) (28, 29, 47). P-TEFb is a heterodimer composed of the cyclin-dependent kinase 9 (Cdk9) and cyclin T1, T2a, T2b, or K (38). Among these C-type cyclins, only cyclin T1 (CycT1) can support Tat transactivation (37, 45). CycT1 but not CycT2 or CycK binds Tat and the central loop of TAR, resulting in the recruitment of Cdk9 to the stalled RNA polymerase II. Cdk9 phosphorylates the C-terminal domain of RNA polymerase II, thus converting the initiating RNA polymerase IIa to its elongating form (RNA polymerase IIo).
CycT1 (726 amino acids) contains two conserved cyclin boxes (from positions 31 to 250), a coiled-coil sequence (from positions 379 to 430), a histidine-rich region (from positions 506 to 530), and a PEST sequence (from positions 709 to 726) (37, 45). N-terminal cyclin boxes are important for binding and activating Cdk9. Early studies revealed that residues from positions 251 to 272 are essential for the binding between Tat and TAR (16). Since murine CycT1, which cannot support Tat transactivation in rodent cells, has a tyrosine at position 261, the cysteine at position 261 in human CycT1 (hCycT1) was found to be essential for this interaction (3, 14, 16, 21). Indeed, the change of this tyrosine to cysteine allows murine CycT1 to support Tat transactivation (14, 16, 21). This region also contains six positively charged residues that serve as an arginine-rich RNA binding motif and a nuclear localization signal. Thus, this sequence is called the Tat-TAR recognition motif. Although the C-terminal region of CycT1 might play a role in TAR binding and binds the C-terminal domain, the N-terminal 272 amino acids of CycT1 are sufficient for Tat transactivation in vitro and in vivo (3, 11, 14-16, 21, 25, 42; K. Fujinaga, D. Irwin, M. Geyer, R. Taube, and B. M. Peterlin, submitted for publication).
The interaction between Tat and CycT1 is dependent on zinc (16). A Zn2+-mediated bond is formed between two cysteines and one histidine in the activation domain (from positions 1 to 48) in Tat and the cysteine at position 261 in hCycT1 (3, 14, 16, 21). No other cysteine and/or histidine is required for this interaction (Fujinaga et al., submitted). In addition, hCycT1 alone cannot bind TAR (45). Although the interaction between CycT1, Tat, and TAR can be detected in the absence of Cdk9 and Cdk9 alone cannot bind TAR, Cdk9 still plays a major role in stabilizing this RNA-protein complex (11, 15). Thus, adding Cdk9 to the electrophoretic mobility shift assay enhances binding between Tat, TAR, and hCycT1 in an ATP-dependent manner (11). Moreover, the autophosphorylation of Cdk9 in its C terminus is required for this binding (15). Mimicking the phosphorylation by mutating three serines (at positions 347, 353, and 357) and two threonines (at positions 350 and 354) to glutamates in the C terminus of Cdk9 (Cdk9-5E) restored this binding. These data imply that the negative charges in Cdk9 are important for the interaction between hCycT1, Tat, and TAR.
Since P-TEFb is essential for Tat transactivation, it is a potential target for anti-HIV therapeutics. To date, a kinase-negative mutant Cdk9 protein [Cdk9(D167N)], an inhibitor that is specific for Cdk9 (flavopiridol), and compounds that bind TAR (phenothiazines) inhibit Tat transactivation (5, 10, 27, 44). However, since this kinase is important for the transcriptional elongation of other cellular genes, inhibiting P-TEFb might have pleiotropic effects. Thus, it is important to develop inactive mutant P-TEFb complexes that are highly specific for Tat.
In this study, we constructed mutant hCycT1 proteins that were deficient in their interactions with Cdk9 or TAR and measured their inhibitory effects on HIV transcription. However, expression studies revealed that these mutant hCycT1 proteins had relatively modest effects, presumably because of the high stability of the endogenous P-TEFb. Stronger inhibitory effects were obtained when the mutant Cdk9 proteins were fused with hCycT1. The chimera between a kinase-inactive Cdk9 that contained glutamates rather than serines and threonines in its C terminus and hCycT1 [Cdk9(D167N,361-5E).hCycT280] especially inhibited Tat transactivation as well as HIV gene expression. Therefore, this fusion protein could provide an effective and specific anti-HIV therapy.
MATERIALS AND METHODS
Plasmid constructions.
The plasmid reporter pHIVSCAT and the plasmid effector pcDNA3-Tat (pTat) have been described elsewhere (13). Mutant hCycT1 proteins were constructed by introducing point mutations into phCycT280 (Fujinaga et al., submitted) by using a transformer site-directed mutagenesis kit (Clontech, Palo Alto, Calif.). The plasmids for mutant Cdk9.hCycT1.Tat chimeras (pCdk9.hCycT280.Tat) were made by inserting the PCR fragments of Cdk9 or the kinase-negative Cdk9(D167N) into phCycT280.Tat (Fujinaga et al., submitted) at BamHI sites.
To construct the plasmids for C-terminal deletion mutagenesis, the indicated fragments of Cdk9 or Cdk9(D167N) were amplified with appropriate primers with BamHI sites. Amplified fragments were inserted into the BamHI sites of phCycT280.Tat. Mutant Cdk9(D167N,361-5E).hCycT280.Tat was constructed by inserting a PCR fragment amplified with primers that contain five mismatching nucleotides corresponding to the three serines (at positions 347, 353, and 357) and two threonines (at positions 350 and 354) into phCycT280.Tat at BamHI sites. Mutants Cdk9(D167N,361-5E).hCycT280 and Cdk9(328).hCycT280 were constructed by removing the XbaI fragment from pCdk9(D167N,361-5E).hCycT280.Tat and pCdk9(328).hCycT280.Tat, respectively. All the constructs contain the Myc epitope tag. The expression of the mutant and fusion proteins was confirmed by Western blotting with anti-Myc antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.). Sequences of oligonucleotide primers are available upon request.
Transient transfection and CAT assay.
NIH 3T3 cells were cotransfected with pHIVCAT (0.1 μg) and the indicated phCycT1 and its mutant constructs in the presence of pTat (0.1 μg) (Fig. 1 and 2) or pCdk9.hCycT280.Tat and its mutant constructs (0.5 μg) in the absence of pTat (Fig. 3) with Lipofectamine (Gibco-BRL, Gaithersburg, Md.). Forty-eight hours after transfection, cells were lysed, and the chloramphenicol acetyltransferase (CAT) activities were measured as described before (13). For measuring the inhibitory effects of mutant hCycT1 proteins and mutant Cdk9.hCycT280 chimeras, HeLa cells were cotransfected with pHIVCAT (0.1 μg), pTat (5 ng), and increasing amount of mutant phCycT1 constructs or pCdk9.hCycT280 by using Lipofectamine (Gibco-BRL). Forty-eight hours after transfection, CAT activities in the cell lysates were measured.
FIG. 1.
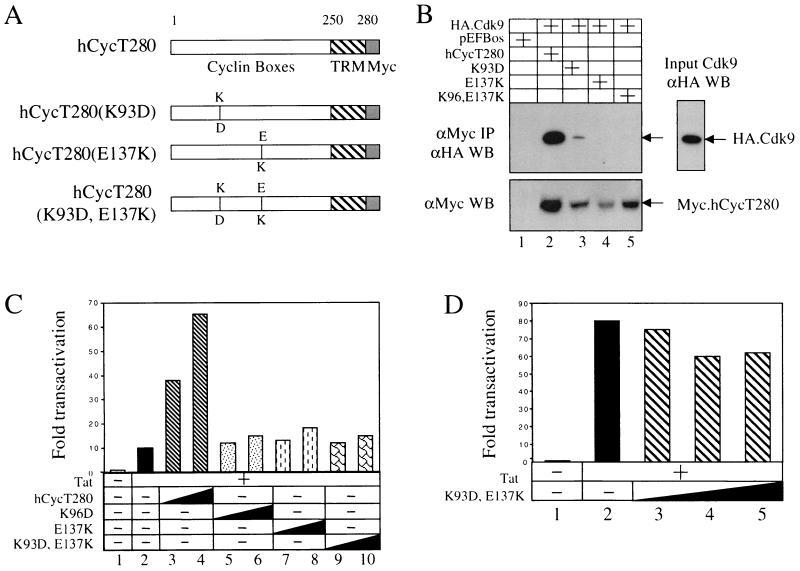
Mutant hCycT1 proteins defective in binding to Cdk9 weakly inhibit Tat transactivation. (A) Schematic presentation of the mutant proteins used in this study. The mutation sites (K93D and E137K) are indicated. TRM, Tat-TAR recognition motif. (B) The mutant hCycT1 proteins do not bind to Cdk9 in vivo. Myc epitope-tagged wild-type hCycT280 protein and mutant hCycT1 proteins were coexpressed with HA epitope-tagged Cdk9 protein in COS cells. Forty-eight hours after transfection, Myc epitope-tagged hCycT1 proteins were immunoprecipitated with the anti-Myc antibody from cell lysates. Bound Cdk9 was visualized by Western blotting with the anti-HA antibody (upper panel). The expression of hCycT1 proteins in the cell lysates was detected by Western blotting with the anti-Myc antibody (lower panel). (C) Mutant hCycT1 proteins do not support Tat transactivation in murine cells. Increasing amounts (0.3 to 0.6 μg) of hCycT280 and its mutant counterparts were coexpressed with Tat (0.1 μg) and pHIVCAT (0.1 μg) in NIH 3T3 cells. Forty-eight hours after the transfection, CAT activities in the cell lysates were measured. The results are presented as fold activation over the CAT activity obtained with pHIVCAT and 0.6 μg of the empty plasmid vector (lane 1). (D) The dominant-negative activity of mutant hCycT280(K93D, E137K) protein in human cells. Increasing amounts (0.2 to 0.8 μg) of hCycT280(K93D, E137K) were coexpressed with Tat (5 ng) in HeLa cells. Forty-eight hours after transfection, CAT activities in the cell lysates were measured. Results are shown as fold activation over the value obtained without Tat. All experiments were performed in triplicate and repeated at least two independent times, and the standard errors of the mean were less than 20%.
FIG. 2.
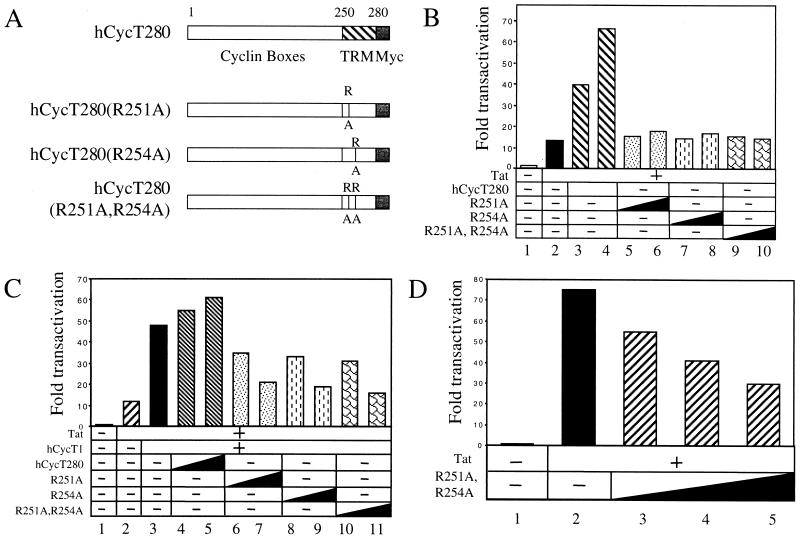
Mutant hCycT1 proteins defective for TAR binding are more potent inhibitors of Tat. (A) Schematic presentation of the mutant proteins used in this study. The mutation sites (R251A and R254A) are indicated. TRM, Tat-TAR recognition motif. (B) Mutant hCycT1 proteins do not support Tat transactivation in murine cells. Increasing amounts (0.3 to 0.6 μg) of hCycT280 and its mutant counterparts were coexpressed with Tat (0.1 μg) and pHIVCAT (0.1 μg) in NIH 3T3 cells. Forty-eight hours after transfection, CAT activities in the cell lysates were measured as for Fig. 1. (C) Mutant hCycT1 proteins can compete with the wild-type hCycT1 protein for Tat transactivation in murine cells. Increasing amounts (0.3 μg to 0.6 μg) of hCycT280 and its mutant counterparts were coexpressed with hCycT1 (0.1 μg), Tat (0.1 μg), and pHIVSCAT (0.1 μg) in NIH 3T3 cells. Forty-eight hours after transfection, CAT activities in the cell lysates were measured. (D) The dominant-negative activity of the mutant hCycT280(R251A,R254A) protein in human cells. Increasing amounts (0.2 μg to 0.8 μg) of hCycT(R251A,R254A) were coexpressed with Tat (5 ng) and pHIVCAT in HeLa cells. Forty-eight hours after transfection, CAT activities in the cell lysates were measured. Results are shown as fold activation over the value obtained without pTat. Variability between experiments was the same as in Fig. 1.
FIG. 3.
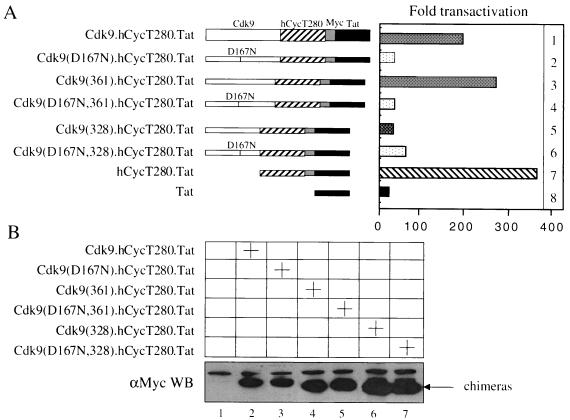
C terminus of Cdk9 is required for Tat transactivation. (A) Cdk9.hCycT280.Tat chimera and its C-terminal truncation mutant counterpart (0.5 μg) were coexpressed with pHIVCAT in NIH 3T3 cells. Forty-eight hours after transfection, CAT activities in the cell lysates were measured. (B) Expression of the fusion proteins in the cell was measured by Western blotting (WB) with the anti-Myc antibody. Variability between experiments was as in Fig. 1.
Coimmunoprecipitation.
COS cells were cotransfected with phCycT280 and its mutant constructs and hemagglutinin (HA) epitope-tagged pCdk9 (43) by using Lipofectamine (Gibco-BRL). Forty-eight hours after transfection, coimmunoprecipitation was performed as described before (42). Briefly, cells were lysed with ELB buffer (50 mM HEPES-KOH [pH 7.9], 0.1% Triton X-100, 5 mM dithiothreitol, 5 mM EDTA, 150 mM NaCl) in the presence of protease inhibitors (Sigma, St. Louis, Mo.) and immunoprecipitated with anti-Myc antibody (Santa Cruz Biotechnology). Following binding to the antibody, reaction mixtures were incubated with protein A-Sepharose beads (Pharmacia, Piscataway, N.J.) at 4°C. Immunoprecipitate reaction products were washed extensively with ELB buffer and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blotting with anti-HA antibody (Santa Cruz Biotechnology).
HIV-1 production assay.
293T cells (105) were cotransfected with 0.4 μg of pNL4-3, which contains the entire HIV-1NL4-3 provirus, and increasing amounts of pCdk9.hCycT.Tat and its mutant constructs by using Lipofectamine (Gibco-BRL). The total amount of DNA was kept constant at 1.2 μg by the addition of empty pEF-Bos. Forty-eight hours after transfection, the viral reverse transcriptase activities in the supernatants from these cells were measured by a standard protocol (5).
RESULTS
Mutant hCycT1 proteins defective in Cdk9 binding weakly inhibit Tat transactivation.
hCycT1 is a C-type cyclin. Among the other C-type cyclins, CycH also plays a role in the initiation of transcription (7, 8, 17, 20, 32, 36). The crystal structure of CycH has been solved (2). The region in CycH that binds its kinase partner, Cdk7, has been mapped (2). To determine the sequences of hCycT1 that bind Cdk9, we first compared residues between CycH and hCycT1. The superimposition of the putative structure of hCycT1 on that of CycH revealed surfaces that should bind Cdk9 (41). In this region, the lysine at position 93 (K93), which is conserved between these two cyclins, is involved in binding Cdk9 (4, 12). Besides K93, a glutamate at position 137 (E137) is also conserved between these two cyclins. Therefore, we constructed first mutant hCycT1 proteins whose charges at these positions were reversed [hCycT280(K93D), hCycT280(E137K), and hCycT280(K93D, E137K)] by site-directed mutagenesis (Fig. 1A).
Since the N-terminal 280 residues in hCycT1 are sufficient for Tat transactivation in vitro and in vivo, all our point mutations were made in this truncated hCycT1 protein (hCycT280) (14). The Myc epitope-tagged wild-type and mutant hCycT proteins were coexpressed with HA epitope-tagged Cdk9 protein in COS cells, followed by immunoprecipitation with anti-Myc antibodies. The interaction between Cdk9 and hCycT280 and mutant CycT1 proteins was visualized by Western blotting with anti-HA antibodies. As presented in Fig. 1B, although the expression levels were much lower, all mutant proteins [hCycT280(K93D), hCycT280(E137K), and hCycT280(K93D,E137K)] failed to interact with Cdk9 in vivo (Fig. 1B, lanes 3 to 5).
Next, we measured the ability of these mutant hCycT1 proteins to support Tat transactivation in murine NIH 3T3 cells (Fig. 1C). Since the endogenous murine CycT1 does not support the formation of the RNA-protein complex with Tat and TAR, Tat activated the HIV-1 long terminal repeat (LTR) (pHIVCAT) weakly in these cells (Fig. 1C, lane 2). However, in the presence of hCycT280, Tat transactivation was 70-fold (Fig. 1C, lane 4). Therefore, the activity of mutant hCycT1 proteins could be measured easily by their coexpression with Tat and pHIVCAT in NIH 3T3 cells.
As presented in Fig. 2C, the mutant hCycT1 proteins which were defective in Cdk9 binding had little effect on Tat transactivation in murine cells (Fig. 1C, lanes 5 to 10). No differences in the activities were observed among these three mutant proteins (Fig. 1C, lanes 5 to 10). Therefore, we measured the inhibitory effect of hCycT280(K93D,E137K) on Tat transactivation in human cells (Fig. 1D). Increasing amounts of the mutant hCycT280(K93D,E137K) protein were coexpressed with Tat and pHIVCAT in HeLa cells. As presented in Fig. 1D, the mutant hCycT280(K93D,E137K) protein had little to no dominant-negative activity on Tat in HeLa cells (20% reduction at the highest concentration; Fig. 1D, lane 5). This failure reflects low levels of expression of the mutant protein in these cells (Fig. 1B, lower panel). Thus, we conclude that although the mutant hCycT280(K93D,E137K) protein does not bind Cdk9, it also does not inhibit Tat transactivation.
Mutant hCycT1 proteins defective in TAR binding are more potent inhibitors of Tat.
The previous result indicated that the mutant hCycT1 proteins, which were defective in Cdk9 binding, were unstable in cells. This finding is consistent with observations that the unpaired cyclins, i.e., without their kinase partners, are degraded rapidly (33). Therefore, the Cdk9 binding region in hCycT1 is not a good target for effective Tat inhibitors.
Next, we constructed mutant hCycT1 proteins that were defective in TAR binding. The Tat-TAR recognition motif of hCycT1 presumably forms an alpha-helical structure which contains six positively charged residues (41). Among these amino acids, the arginines at positions 251 and 254 are involved in the interaction with TAR (16). Although CycT1 alone cannot bind TAR without Tat, when fused with Tat, the Tat-TAR recognition motif binds TAR (Fujinaga et al., submitted). This implies that no other residues in hCycT1 are required for this interaction. Therefore, we changed these arginines to alanines [hCycT280(R251A), hCycT280(R254A), and hCycT280(R251A,R254A)] (Fig. 2A). These mutant hCycT1 proteins were expressed at similar levels and bound Cdk9 equivalently to the wild-type (hCycT280) protein in COS cells (data not presented). The activity of these proteins was measured by coexpression with Tat and pHIVCAT in NIH 3T3 cells. All of our mutant hCycT1 proteins also failed to rescue Tat transactivation in these cells (Fig. 2B, lanes 5 to 10).
Next, the ability of these mutant hCycT1 proteins to compete with the wild-type hCycT1 protein for Tat transactivation was measured. Increasing amounts of our mutant hCycT1 proteins were coexpressed with Tat, pHIVCAT, and hCycT1 protein in NIH 3T3 cells. As presented in Fig. 2C, all mutant hCycT1 proteins inhibited Tat transactivation in NIH 3T3 cells in the presence of hCycT1 (Fig. 2C, lanes 5 to 11). At the highest concentration, CAT activities were decreased to the basal level (Fig. 2C, compare lanes 7, 9, and 11 to lane 2). These results indicate that the mutant hCycT1 proteins can compete effectively with the wild-type hCycT1 protein for Tat transactivation in murine cells.
Since no difference in activity was observed among these three mutant hCycT1 proteins, the mutant hCycT280(R251A,R254A) was then examined for its dominant-negative effects on Tat transactivation in human cells. Increasing amounts of mutant hCycT280(R251A,R254A) protein were coexpressed with Tat and pHIVCAT in HeLa cells. As presented in Fig. 2D, the mutant hCycT280(R251A,R254A) protein inhibited Tat transactivation up to 50%, which was more potent than the mutant hCycT280(K93D,E137K) protein described above. We conclude that the mutant hCycT1 proteins defective in TAR binding have stronger dominant-negative effects on Tat transactivation than those that do not bind Cdk9.
Fusion proteins between hCycT1 and kinase-inactive Cdk9 proteins result in stronger dominant-negative effects.
Although it competed effectively with the exogenously introduced hCycT1 protein in NIH 3T3 cells, the mutant hCycT280(R251A,R254A) protein exhibited relatively modest dominant-negative effects in HeLa cells. Since the endogenous hCycT1 protein forms a stable, long-lived complex with Cdk9 in cells, the exogenous mutant hCycT1 proteins alone might not be incorporated efficiently into P-TEFb complexes during our assays (35). Therefore, we constructed several fusion proteins between hCycT1and Cdk9 that were inactive for Tat transactivation.
Previously, we demonstrated that hCycT1 containing the N-terminal 280 amino acids (hCycT280) fused with Tat (hybrid hCycT280.Tat protein) binds TAR in vitro and supports Tat transactivation in murine cells (Fujinaga et al., submitted). At first, the full-length wild-type Cdk9 protein was fused with the hybrid hCycT280.Tat (Cdk9.hCycT280.Tat) protein (Fig. 3A). As a negative control, a kinase-inactive Cdk9 [Cdk9(D167N)] protein was fused with the hCycT280.Tat chimera [Cdk9(D167N).hCycT280.Tat] (Fig. 3A). These fusion proteins were examined by coexpression with pHIVCAT in NIH 3T3 cells.
As presented in Fig. 3A, the Cdk9.hCycT280.Tat chimera (lane 1) activated the HIV-1 LTR in the absence of Tat to levels similar to those observed with the hCycT280.Tat chimera (lane 7) or coexpressed hCycT280 and Tat (data not presented). In contrast, the kinase-inactive mutant Cdk9(D167N).hCycT280.Tat fusion protein failed to activate pHIVCAT (Fig. 3A, lane 2). Western blotting demonstrated that these fusion proteins were expressed at similar levels (Fig. 3B, lanes 1 and 2). These results indicate that Cdk9, in the context of the fusion protein, functions as well as the free Cdk9 protein and that the interaction between hCycT1 in the fusion protein and the endogenous Cdk9 is negligible. Therefore, a fusion protein between hCycT1 and the kinase-inactive Cdk9 protein might bypass the problem of the high stability of the endogenous P-TEFb complexes.
Previously, Garber et al. demonstrated that the C-terminal region of Cdk9 is autophosphorylated and that this autophosphorylation enhances the interaction between hCycT1, Tat, and TAR in vitro (15). In addition, they also demonstrated that mutating the serines and threonines (S347, T350, S353, T354, and S357) to glutamates (Cdk9-5E) mimicked this autophosphorylation. With these mutations, the kinase-negative Cdk9 behaved like its wild-type counterpart. Therefore, we constructed a series of C-terminally truncated Cdk9 proteins fused with the hCycT280.Tat chimera (Fig. 3A). As a control, the kinase-inactive Cdk9 [Cdk9(D167N)] protein was also fused with the hybrid hCycT280.Tat protein (Fig. 3A). These fusion proteins were coexpressed with pHIVCAT in NIH 3T3 cells.
As presented in Fig. 3A, an 11-amino-acid truncation from the C terminus [Cdk9(361).hCycT280.Tat] had little effect on pHIVCAT (Fig. 3A, lane 3). In contrast, a 44-amino-acid truncation [Cdk9(328).hCycT280.Tat] from the C terminus abolished this activity on the HIV LTR (Fig. 3A, lane 5). This finding is consistent with previous observations that the C terminus of Cdk9 is required for stable interactions between hCycT1, Tat, and TAR in vitro (15). The kinase-negative version of these fusion proteins [mutants Cdk9(D167N,361).hCycT280.Tat and Cdk9(D167N,328).hCycT280.Tat] also had little to no activity on pHIVCAT, which again indicates that no interaction occurred between hCycT1 in the fusion protein and the endogenous Cdk9 (Fig. 3A, lanes 4 and 6). These fusion proteins were expressed at similar levels in Western blotting (Fig. 3B).
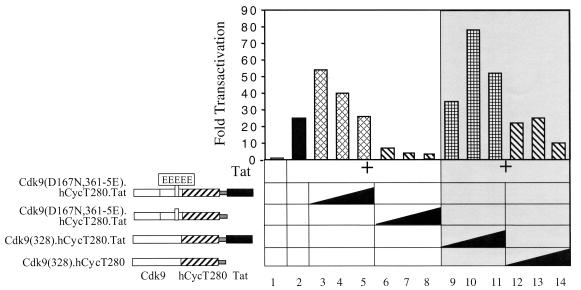
Next, these fusion proteins were examined for their inhibitory effects on Tat transactivation. We also constructed a fusion protein with the kinase-inactive Cdk9 protein containing 361 amino acids and mutations changing S347, T350, S353, T354, and S357 to glutamates [Cdk9(D167N,361-5E).hCycT1.Tat]. Increasing amounts of these mutant Cdk9(328).hCycT280.Tat and Cdk9(D167N,361-5E).hCycT1.Tat chimeras were coexpressed with Tat and pHIVCAT in HeLa cells. As presented in Fig. 4, none of the fusion proteins that contained Tat had significant inhibitory effects on Tat transactivation (lanes 3 though 5 and 9 though 11). In contrast, the fusion proteins without Tat showed a strong dominant-negative effect on Tat transactivation. Indeed, the mutant Cdk9(328).hCycT280 chimera inhibited Tat transactivation by 70% (Fig. 4, lanes 2 and 14, 25-fold versus 10-fold). The mutant Cdk9(D167N,361-5E).hCycT280 chimera showed the strongest inhibitory effect and almost completely blocked Tat transactivation in HeLa cells (Fig. 4, lanes 6 through 8).
FIG. 4.
Kinase-inactive mutant Cdk9(D167N,361-5E).hCycT280 chimera inhibits Tat transactivation. Increasing amounts (0.2 μg to 0.8 μg) of Cdk9.hCycT280.Tat chimera and its mutant counterparts (illustrated on the left of the graph) were coexpressed with Tat (5 ng) and pHIVCAT in HeLa cells. Forty-eight hours after transfection, CAT activities in the cell lysates were measured. Results are shown as fold activation over the value obtained without Tat. Variability between experiments was as in Fig. 1.
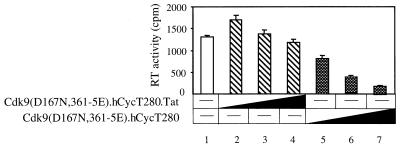
The previous result indicated that the dominant-negative kinase-inactive mutant Cdk9(D167N,361-5E).hCycT280 chimera can inhibit Tat transactivation. Thus, it was examined for its inhibitory effects on HIV gene expression. Increasing amounts of this mutant chimera were coexpressed with the HIV-1 provirus (pNL4-3) in 293T cells. Forty-eight hours later, HIV gene expression was quantified by measuring the viral reverse transcriptase activities released into the supernatant. As presented in Fig. 5, the kinase-inactive mutant Cdk9(D167N,361-5E).hCycT280 chimera inhibited HIV gene expression (Fig. 5, lanes 5 to 7), whereas the mutant Cdk9(D167N,361-5E).hCycT280.Tat chimera had no effects (Fig. 5, lanes 2 to 4). From these results, we conclude that the fusion protein between hCycT1 and inactive Cdk9 protein can inhibit Tat transactivation and HIV-1 gene expression in vivo.
FIG. 5.
Kinase-inactive mutant Cdk9(D167N,361-5E).hCycT280 chimera blocks HIV-1 gene expression. Increasing amounts (0.3 μg to 0.8 μg) of the kinase-negative mutant Cdk9(D167N,361-5E).hCycT280 and Cdk9(D167N,361-5E).hCycT280.Tat chimeras were coexpressed with 0.4 μg of HIV-1 provirus (pNL4-3) in 293T cells. Forty-eight hours after transfection, cell culture supernatants were collected, and the viral reverse transcriptase (RT) activity was measured as described in Materials and Methods.
DISCUSSION
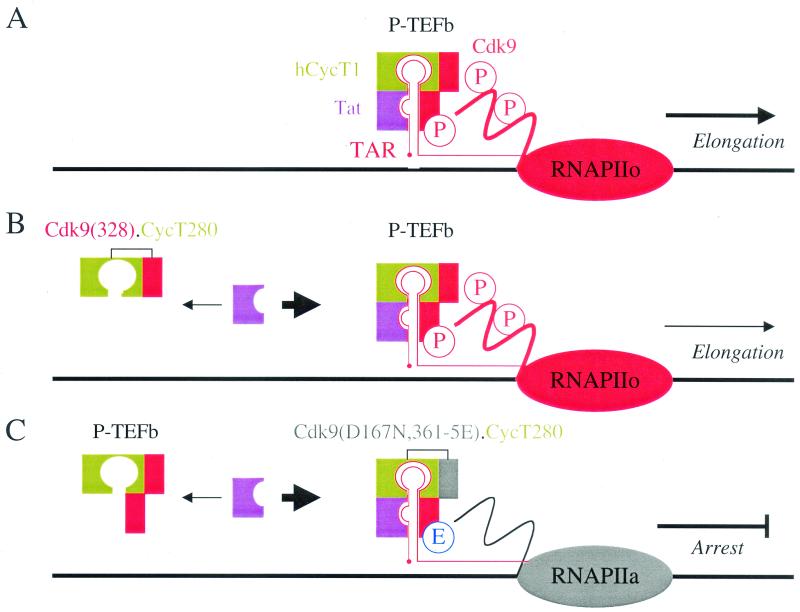
In this study, we defined sequences in hCycT1 that are involved in binding Cdk9 and TAR. However, disrupting these regions in hCycT1 resulted in only modest inhibitory effects on Tat transactivation. Higher inhibitory effects were obtained when hCycT1 was fused with a mutant Cdk9 that was defective for Tat transactivation. Indeed, the kinase-inactive mutant Cdk9(D167N,361-5E).hCycT280 chimera inhibited Tat transactivation as well as HIV gene expression. This chimera effectively forms a protective shield on TAR, which blocks the recruitment of the functional P-TEFb complex. Our results are presented schematically in Fig. 6.
FIG. 6.
Working hypothesis for the fusion proteins. (A) Wild-type Cdk9 protein (red rectangle) is phosphorylated in its C terminus (indicated by P) and capable of stabilizing the interaction between hCycT1 (green rectangle), Tat (purple rectangle), and TAR (red line), which is required for the hyperphosphorylation of the C-terminal domain of RNA polymerase II (RNAPIIo, red oval) and the elongation of transcription in vivo. (B) Mutant Cdk9(328).hCycT280 chimera that interacts with Tat but not with TAR can squelch Tat transactivation. (C) The kinase-inactive mutant Cdk9(D167N,361-5E).hCycT280 chimera can bind TAR in the presence of Tat as a transcriptionally inactive complex. This complex effectively forms a protective shield on TAR, which blocks the recruitment of the functional P-TEFb complex. RNA polymerase II remains unphosphorylated (RNAPIIa, gray oval).
Activation of transcriptional elongation by Tat is one of the key regulatory steps of HIV replication. Thus, inhibition of this step forms a powerful target for anti-HIV therapy. Several inhibitors such as TAR decoys and dominant-negative Tat protein have been demonstrated to inhibit Tat transactivation (18, 31, 40). Likewise, cellular Tat cofactors such as P-TEFb have been targeted. Until now, the kinase activity of Cdk9 had been the main focus of these efforts (5, 10, 28, 44). However, since P-TEFb is essential for the expression of many cellular genes (6, 9, 23, 26, 38, 39), inhibiting all P-TEFb by overexpression of a kinase-inactive Cdk9 [Cdk9(D167N)] protein or kinase inhibitors should cause growth arrest or cell death (5). Indeed, our attempts to establish stable cell lines expressing the mutant hCycT280(R251A,R254A) protein have been unsuccessful, which implies that mutations in the Tat-TAR recognition motif also disrupt the nuclear localization signal of hCycT1 that is essential for the normal functions of hCycT1 (data not presented).
In addition, the endogenous hCycT1 proteins form a very stable complex with Cdk9 in cells (35). Therefore, the exchange rate between the exogenous mutant hCycT1 proteins and the hCycT1 protein in P-TEFb is expected to be slow, which would delay the onset of dominant-negative effects for HIV replication. For example, although the mutant hCycT1(R251A,R254A) could compete with the exogenous hCycT1 proteins for Tat transactivation in murine cells, the inhibitory effect of this mutant hCycT1 protein in human cells was relatively low (Fig. 2C and D). On the other hand, the free hCycT1 proteins that are unpaired with Cdk9 are unstable and degraded rapidly. Indeed, levels of expression of the mutant hCycT1 proteins that were defective in Cdk9 binding [hCycT280(K93D), hCycT280(E137K), and hCycT280(K93D,E137K)] were much lower than that of the wild-type hCycT280 protein (Fig. 1).
To overcome these difficulties, we focused on Cdk9.hCycT1 fusion proteins. First, they do not interfere with the function of the endogenous P-TEFb complex. Indeed, the mutant Cdk9(D167N-5E).hCycT280 protein had no effect on the basal expression from the HIV LTR or other heterologous promoters, such as cytomegalovirus and Rous sarcoma virus (data not presented). Second, they form a separate and stable P-TEFb complex in cells. It is also to be noted that the Cdk9.hCycT280.Tat chimera but not the kinase-inactive mutant Cdk9(D167N).hCycT280.Tat chimera activated the HIV LTR in murine cells. This observation indicates that the fusion proteins fold correctly to form new P-TEFb complexes and that there is little to no binding between endogenous Cdk9 protein and hCycT280 in the chimeras (Fig. 3).
Since the autophosphorylation in the C terminus of Cdk9 stabilizes the complex between hCycT1, Tat, and TAR, two different forms of Cdk9 [Cdk9(328) and Cdk9(D167N,361-5E)] that cannot support Tat transactivation were fused with hCycT1 (15). However, Tat in the mutant Cdk9(328).hCycT280.Tat and Cdk9(D167N-5E).hCycT280.Tat chimeras could still interact with the wild-type hCycT1 in the endogenous P-TEFb (Fujinaga et al., submitted for publication). Also, the exogenous Tat bound to the endogenous P-TEFb could compete with the fusion proteins for TAR. Thus, although these tripartite fusion proteins did not activate the HIV LTR in murine cells, they did not inhibit Tat transactivation in human cells (Fig. 4).
In contrast, when Tat was removed from this tripartite fusion protein, the mutant Cdk9(328).hCycT280 chimera could inhibit Tat transactivation by squelching the free Tat (Fig. 4; also illustrated in Fig. 6). On the other hand, the kinase-inactive mutant Cdk9(D167N,361-5E).hCycT280 chimera, which mimicked the phosphorylation in the C-terminal region of Cdk9, bound TAR in the presence of Tat but could not phosphorylate the C-terminal domain of RNA polymerase II (15). Since it could trap the exogenous Tat into this inactive complex with TAR, it inhibited HIV transactivation more efficiently in human cells (Fig. 4; also illustrated in Fig. 6). Also, since the phosphorylation of the C-terminal region of Cdk9 is important for TAR binding, its effect is highly specific for Tat and TAR. In this scenario, the bipartite mutant Cdk9(D167N,361-5E).hCycT280 chimera could inhibit Tat transactivation at relatively low concentrations without disturbing the function of the endogenous P-TEFb complex.
Although levels of expression of P-TEFb in cells appear constant at different stages of the cell cycle (19, 30), the activity of P-TEFb is regulated by posttranscriptional modifications such as phosphorylation (11, 15), ubiquitination (24), and its association with 7SK RNA (34, 46). The short HIV transcripts that represent a lack of Tat phenotype are found predominantly in latently infected cells, which implies that the activity of P-TEFb could be low in these cells (1). Cdk9 might be autophosphorylated upon cellular activation, which could render P-TEFb fully capable of supporting Tat transactivation. Since the mutant Cdk9(D167N,361-5E) chimera mimics this activated form of P-TEFb, it would also block Tat transactivation when expressed constitutively in these latently infected cells. Importantly, it would appreciably raise the threshold at which Tat could activate viral replication.
Since HIV utilizes the cellular transcriptional machinery for its own replication, it is important to inhibit this step without disturbing cellular functions. Recently, compounds that inhibit the kinase activity of Cdk9 (flavopiridol [5]) or TAR binding activity of hCycT1 and Tat (phenothiazines [27]) were discovered. Flavopiridol inhibits HIV replication at a concentration lower than that at which it causes cytotoxic effects. These compounds were targeted to HIV-specific processes such as the interaction between CycT1, Tat, and TAR. Here, we present a new molecule that can block both of these activities and is a highly specific and effective inhibitor of Tat transactivation. Its use alone or in combination with these other compounds could represent a new therapy for AIDS.
Acknowledgments
We thank Paula Zupanc-Ecimovic for expert secretarial assistance and the members of the laboratory for help with the work and comments on the manuscript.
K.F. is the recipient of an NIH training grant (TG AI2441-11). This work was supported by a grant from the NIH (RO1 AI49104-01).
REFERENCES
- 1.Adams, M., L. Sharmeen, J. Kimpton, J. M. Romeo, J. V. Garcia, B. M. Peterlin, M. Groudine, and M. Emerman. 1994. Cellular latency in human immunodeficiency virus-infected individuals with high CD4 levels can be detected by the presence of promoter-proximal transcripts. Proc. Natl. Acad. Sci. USA 91:3862-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersen, G., A. Poterszman, J. M. Egly, D. Moras, and J. C. Thierry. 1996. The crystal structure of human cyclin H. FEBS Lett. 397:65-69. [DOI] [PubMed] [Google Scholar]
- 3.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1998. Recruitment of a protein complex containing Tat and cyclin T1 to TAR governs the species specificity of HIV-1 Tat. EMBO J. 17:7056-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieniasz, P. D., T. A. Grdina, H. P. Bogerd, and B. R. Cullen. 1999. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc. Natl. Acad. Sci. USA 96:7791-7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chao, S. H., K. Fujinaga, J. E. Marion, R. Taube, E. A. Sausville, A. M. Senderowicz, B. M. Peterlin, and D. H. Price. 2000. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. J. Biol. Chem. 275:28345-28348. [DOI] [PubMed] [Google Scholar]
- 6.Chao, S. H., and D. H. Price. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 276:31793-31799. [DOI] [PubMed] [Google Scholar]
- 7.Cujec, T. P., H. Okamoto, K. Fujinaga, J. Meyer, H. Chamberlin, D. O. Morgan, and B. M. Peterlin. 1997. The HIV transactivator TAT binds to the CDK-activating kinase and activates the phosphorylation of the carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:2645-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drapkin, R., and D. Reinberg. 1994. The multifunctional TFIIH complex and transcriptional control. Trends Biochem. Sci. 19:504-508. [DOI] [PubMed] [Google Scholar]
- 9.Eberhardy, S. R., and P. J. Farnham. 2001. c-Myc mediates activation of the cad promoter via a post-RNA polymerase II recruitment mechanism. J. Biol. Chem. 276:48562-48571. [DOI] [PubMed] [Google Scholar]
- 10.Flores, O., G. Lee, J. Kessler, M. Miller, W. Schlief, J. Tomassini, and D. Hazuda. 1999. Host-cell positive transcription elongation factor b kinase activity is essential and limiting for HIV type 1 replication. Proc. Natl. Acad. Sci. USA 96:7208-7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fong, Y. W., and Q. Zhou. 2000. Relief of two built-in autoinhibitory mechanisms in P-TEFb is required for assembly of a multicomponent transcription elongation complex at the human immunodeficiency virus type 1 promoter. Mol. Cell. Biol. 20:5897-5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraldi, A., P. Licciardo, B. Majello, A. Giordano, and L. Lania. 2001. Distinct regions of cyclinT1 are required for binding to CDK9 and for recruitment to the HIV-1 Tat/TAR complex. J. Cell. Biochem. Suppl. 36:247-253. [DOI] [PubMed] [Google Scholar]
- 13.Fujinaga, K., T. P. Cujec, J. Peng, J. Garriga, D. H. Price, X. Grana, and B. M. Peterlin. 1998. The ability of positive transcription elongation factor B to transactivate human immunodeficiency virus transcription depends on a functional kinase domain, cyclin T1, and Tat. J. Virol. 72:7154-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujinaga, K., R. Taube, J. Wimmer, T. P. Cujec, and B. M. Peterlin. 1999. Interactions between human cyclin T, Tat, and the transactivation response element (TAR) are disrupted by a cysteine to tyrosine substitution found in mouse cyclin T. Proc. Natl. Acad. Sci. USA 96:1285-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garber, M. E., T. P. Mayall, E. M. Suess, J. Meisenhelder, N. E. Thompson, and K. A. Jones. 2000. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 Tat-P-TEFb complex to TAR RNA. Mol. Cell. Biol. 20:6958-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garber, M. E., P. Wei, V. N. KewalRamani, T. P. Mayall, C. H. Herrmann, A. P. Rice, D. R. Littman, and K. A. Jones. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 12:3512-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Martinez, L. F., G. Mavankal, J. M. Neveu, W. S. Lane, D. Ivanov, and R. B. Gaynor. 1997. Purification of a Tat-associated kinase reveals a TFIIH complex that modulates HIV-1 transcription. EMBO J. 16:2836-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham, G. J., and J. J. Maio. 1990. RNA transcripts of the human immunodeficiency virus transactivation response element can inhibit action of the viral transactivator. Proc. Natl. Acad. Sci. USA 87:5817-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrmann, C. H., R. G. Carroll, P. Wei, K. A. Jones, and A. P. Rice. 1998. Tat-associated kinase activity is regulated by distinct mechanisms in peripheral blood lymphocytes and promonocytic cell lines. J. Virol. 72:9881-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoeijmakers, J. H., J. M. Egly, and W. Vermeulen. 1996. TFIIH: a key component in multiple DNA transactions. Curr. Opin. Genet. Dev. 6:26-33. [DOI] [PubMed] [Google Scholar]
- 21.Ivanov, D., Y. T. Kwak, E. Nee, J. Guo, L. F. Garcia-Martinez, and R. B. Gaynor. 1999. Cyclin T1 domains involved in complex formation with Tat and TAR RNA are critical for tat-activation. J. Mol. Biol. 288:41-56. [DOI] [PubMed] [Google Scholar]
- 22.Jones, K. A., and B. M. Peterlin. 1994. Control of RNA initiation and elongation at the HIV-1 promoter. Annu. Rev. Biochem. 63:717-743. [DOI] [PubMed] [Google Scholar]
- 23.Kanazawa, S., T. Okamoto, and B. M. Peterlin. 2000. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity 12:61-70. [DOI] [PubMed] [Google Scholar]
- 24.Kiernan, R. E., S. Emiliani, K. Nakayama, A. Castro, J. C. Labbé, T. Lorca, K.-I. Nakayama, and M. Benkirane. 2001. Interaction between cyclin T1 and SCFSKP2 targets CDK9 for ubiquitination and degradation by the proteasome. Mol. Cell. Biol. 21:7956-7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kwak, Y. T., D. Ivanov, J. Guo, E. Nee, and R. B. Gaynor. 1999. Role of the human and murine cyclin T proteins in regulating HIV-1 tat-activation. J. Mol. Biol. 288:57-69. [DOI] [PubMed] [Google Scholar]
- 26.Lee, D. K., H. O. Duan, and C. Chang. 2001. Androgen receptor interacts with the positive elongation factor P-TEFb and enhances the efficiency of transcriptional elongation. J. Biol. Chem. 276:9978-9984. [DOI] [PubMed] [Google Scholar]
- 27.Lind, K. E., Z. Du, K. Fujinaga, B. M. Peterlin, and T. L. James. 2002. Structure-based computational database screening, in vitro assay, and NMR assessment of compounds that target TAR RNA. Chem. Biol. 9:185-193. [DOI] [PubMed] [Google Scholar]
- 28.Mancebo, H. S., G. Lee, J. Flygare, J. Tomassini, P. Luu, Y. Zhu, J. Peng, C. Blau, D. Hazuda, D. Price, and O. Flores. 1997. P-TEFb kinase is required for HIV Tat transcriptional activation in vivo and in vitro. Genes Dev. 11:2633-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marshall, N. F., J. Peng, Z. Xie, and D. H. Price. 1996. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J. Biol. Chem. 271:27176-27183. [DOI] [PubMed] [Google Scholar]
- 30.Martin-Serrano, J., K. Li, and P. D. Bieniasz. 2002. Cyclin T1 expression is mediated by a complex and constitutively active promoter and does not limit human immunodeficiency virus type 1 Tat function in unstimulated primary lymphocytes. J. Virol. 76:208-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Modesti, N., J. Garcia, C. Debouck, M. Peterlin, and R. Gaynor. 1991. Trans-dominant Tat mutants with alterations in the basic domain inhibit HIV-1 gene expression. New Biol. 3:759-768. [PubMed] [Google Scholar]
- 32.Morgan, D. O. 1997. Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13:261-291. [DOI] [PubMed] [Google Scholar]
- 33.Nakayama, K. I., S. Hatakeyama, and K. Nakayama. 2001. Regulation of the cell cycle at the G1-S transition by proteolysis of cyclin E and p27Kip1. Biochem. Biophys. Res. Commun. 282:853-860. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen, V. T., T. Kiss, A. A. Michels, and O. Bensaude. 2001. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 414:322-325. [DOI] [PubMed] [Google Scholar]
- 35.O'Keeffe, B., Y. Fong, D. Chen, S. Zhou, and Q. Zhou. 2000. Requirement for a kinase-specific chaperone pathway in the production of a Cdk9/cyclin T1 heterodimer responsible for P-TEFb-mediated tat stimulation of HIV-1 transcription. J. Biol. Chem. 275:279-287. [DOI] [PubMed] [Google Scholar]
- 36.Parada, C. A., and R. G. Roeder. 1996. Enhanced processivity of RNA polymerase II triggered by Tat-induced phosphorylation of its carboxy-terminal domain. Nature 384:375-378. [DOI] [PubMed] [Google Scholar]
- 37.Peng, J., Y. Zhu, J. T. Milton, and D. H. Price. 1998. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 12:755-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price, D. H. 2000. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol. Cell. Biol. 20:2629-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Renner, D. B., Y. Yamaguchi, T. Wada, H. Handa, and D. H. Price. 2001. A highly purified RNA polymerase II elongation control system. J. Biol. Chem. 276:42601-42609. [DOI] [PubMed] [Google Scholar]
- 40.Sullenger, B. A., H. F. Gallardo, G. E. Ungers, and E. Gilboa. 1990. Overexpression of TAR sequences renders cells resistant to human immunodeficiency virus replication. Cell 63:601-608. [DOI] [PubMed] [Google Scholar]
- 41.Taube, R., K. Fujinaga, J. Wimmer, M. Barboric, and B. M. Peterlin. 1999. Tat transactivation: a model for the regulation of eukaryotic transcriptional elongation. Virology 264:245-253. [DOI] [PubMed] [Google Scholar]
- 42.Taube, R., X. Lin, D. Irwin, K. Fujinaga, and B. M. Peterlin. 2002. Interaction between P-TEFb and the C-terminal domain of RNA polymerase II activates transcriptional elongation from sites upstream or downstream of target genes. Mol. Cell. Biol. 22:321-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Wijnen, A. J., F. Aziz, X. Grana, A. De Luca, R. K. Desai, K. Jaarsveld, T. J. Last, K. Soprano, A. Giordano, J. B. Lian, et al. 1994. Transcription of histone H4, H3, and H1 cell cycle genes: promoter factor HiNF-D contains CDC2, cyclin A, and an RB-related protein. Proc. Natl. Acad. Sci. USA 91:12882-12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang, D., C. de la Fuente, L. Deng, L. Wang, I. Zilberman, C. Eadie, M. Healey, D. Stein, T. Denny, L. E. Harrison, L. Meijer, and F. Kashanchi. 2001. Inhibition of human immunodeficiency virus type 1 transcription by chemical cyclin-dependent kinase inhibitors. J. Virol. 75:7266-7279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92:451-462. [DOI] [PubMed] [Google Scholar]
- 46.Yang, Z., Q. Zhu, K. Luo, and Q. Zhou. 2001. The 7SK small nuclear RNA inhibits the CDK9/cyclin T1 kinase to control transcription. Nature 414:317-322. [DOI] [PubMed] [Google Scholar]
- 47.Zhu, Y., T. Pe'ery, J. Peng, Y. Ramanathan, N. Marshall, T. Marshall, B. Amendt, M. B. Mathews, and D. H. Price. 1997. Transcription elongation factor P-TEFb is required for HIV-1 tat transactivation in vitro. Genes Dev. 11:2622-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]