Abstract
The common species C adenoviruses (serotypes Ad1, Ad2, Ad5, and Ad6) infect more than 80% of the human population early in life. Following primary infection, the virus can establish an asymptomatic persistent infection in which infectious virions are shed in feces for several years. The probable source of persistent virus is mucosa-associated lymphoid tissue, although the molecular details of persistence or latency of adenovirus are currently unknown. In this study, a sensitive real-time PCR assay was developed to quantitate species C adenovirus DNA in human tissues removed for routine tonsillectomy or adenoidectomy. Using this assay, species C DNA was detected in Ficoll-purified lymphocytes from 33 of 42 tissue specimens tested (79%). The levels varied from fewer than 10 to greater than 2 × 106 copies of the adenovirus genome/107 cells, depending on the donor. DNA from serotypes Ad1, Ad2, and Ad5 was detected, while the rarer serotype Ad6 was not. When analyzed as a function of donor age, the highest levels of adenovirus genomes were found among the youngest donors. Antibody-coated magnetic beads were used to purify lymphocytes into subpopulations and determine whether viral DNA could be enriched within any purified subpopulations. Separation of T cells (CD4/8- expressing and/or CD3-expressing cells) enriched viral DNA in each of nine donors tested. In contrast, B-cell purification (CD19-expressing cells) invariably depleted or eliminated viral DNA. Despite the frequent finding of significant quantities of adenovirus DNA in tonsil and adenoid tissues, infectious virus was rarely present, as measured by coculture with permissive cells. These findings suggest that human mucosal T lymphocytes may harbor species C adenoviruses in a quiescent, perhaps latent form.
The common adenovirus serotypes of species C (Ad1, Ad2, Ad5, and Ad6) cause roughly 5% of symptomatic upper respiratory tract (5) and 15% of lower respiratory tract (3) infections in children younger than 5 years. In addition to acute disease, the species C adenoviruses establish persistent infections characterized by intermittent viral excretion from immunocompetent hosts (15). Although primary infections are respiratory, species C viruses display prolonged fecal excretion months, and even years, after virus is no longer detected in nasopharyngeal washings (14, 15). Restriction analysis of viruses isolated up to 4 years after initial infection suggested chronic persistent infection rather than reinfection with the same serotype (1).
Numerous early studies documented species C adenovirus isolation following explant of human tonsil and adenoid tissue to culture (11, 18, 28, 32). With few exceptions, tonsil and adenoid tissue yielded no infectious virus immediately after surgical removal. However, infectious virus emerged from these tissues weeks to months after explant. This latter finding, coupled with the observation that tonsils containing adenovirus DNA by in situ hybridization fail to yield infectious virus (25), led investigators to postulate that the virus was latent in these tissues.
Remarkably little is known about the cell type or types that harbor persistent or latent species C adenoviruses. There is some evidence that lymphocytes are one site of virus persistence. There are reliable reports of rare cases of adenovirus and viral DNA in peripheral blood lymphocytes (PBL) during fatal acute infections (2, 12) or from immunosuppressed individuals (7). Species C adenoviruses are rarely found in PBL samples from healthy individuals (7, 12). The only published report of an attempt to identify lymphocytes as the site of virus persistence in tonsils (32), nearly 30 years ago, described the purification of lymphocytes on nylon wool columns prior to bulk culture for virus isolation. In this study, only a small fraction (4%) of the purified lymphocyte samples yielded virus, compared to the majority (62%) of unseparated cultures.
The present study was initiated to apply quantitative PCR in combination with modern cell separation techniques to determine the amounts of species C adenovirus DNA present in tonsil and adenoid lymphocytes and to identify the virus-bearing lymphocyte population(s).
MATERIALS AND METHODS
Tonsil and adenoid cell suspensions.
A total of 42 palatine tonsils or adenoids were obtained from 35 donors (median age, 4 years; range, 1 to 15 years; 43% male, 57% female) undergoing routine tonsillectomies at Egleston Children's Hospital (Atlanta, Ga.). This study received human investigation approval (no. 666-99) from the Internal Review Board of Emory University. Pairs of tonsils from individual donors were pooled, but tonsil and adenoid tissues removed from the same donor were analyzed separately. Surgically removed tissue was placed in HEPES-buffered Hanks' balanced salt solution (HH) containing 5% fetal calf serum, 0.05 mg of gentamicin per ml, and antibiotic-antimycotic mixture (Gibco no. 15240-062) (HH5). Following dissection for pathologic examination, the remaining tissue was immediately available for study. The tissue was pushed through a stainless steel wire screen in HH5 to produce a single-cell suspension. Cells were washed once in the same medium and then stored in 8 to 10 aliquots of 1 × 108 to 2 × 108 cells per ml of 90% fetal calf serum-10% dimethyl sulfoxide in the vapor phase of the liquid nitrogen freezer.
Lymphocyte separations and flow-cytometric analysis.
Lymphocytes were isolated from thawed aliquots by centrifugation over a Ficoll gradient (Ficoll type 400; Sigma). The cells were washed twice in phosphate-buffered saline-2% bovine serum albumin and then resuspended in the appropriate buffer for subsequent magnetic bead separations. Cells were selected using Dynabeads (Dynal) or MACS (Miltenyi Biotech) directly conjugated magnetic beads as specified by the manufacturer. Dynal beads were removed from cells by the addition of the DETACHaBEAD reagent. DETACHaBEAD is a polyclonal anti-Fab antibody specific for the primary antibody on the Dynabead. When it is added to the bead-bound cells, it competes with the antibody-antigen interaction at the cell surface and releases the antibody and bead from the cells, leaving the target cells viable, unstimulated, and without antibody on their surface. MACS beads remained attached to the cell surface when used. To determine the purity of the cell populations before and after magnetic bead separations, the following antibodies were used: pan-T-cell marker CD2 conjugated to phycoerythrin (CD2-PE) (Pharmingen 30055X), pan B-cell marker CD20 conjugated to fluorescein isothiocyanate (CD20-FITC) (Pharmingen 556632), CD4-FITC (Pharmingen 30154X), and CD8-APC (Pharmingen 30329X). Samples were analyzed using a Becton-Dickinson FACSCalibur instrument with CELLQuest software.
Cell digestion and DNA isolation.
DNA was prepared from 2 × 106 to 1 × 107 purified lymphocytes essentially as described by Babcock et al. (4). Cells were placed directly in lysis buffer (0.45% NP-40, 0.45% Tween 20, 2 mM MgCl 2, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 0.5 mg of proteinase K per ml) and incubated at 55°C overnight (107 cells/200 μl of lysis buffer). Following incubation at 55°C, the proteinase K was inactivated at 95°C for 10 min. The samples were then vortexed vigorously, and 5-μl volumes were used directly in the PCRs as described below.
Real-time quantitative PCR for adenovirus hexon DNA.
Quantitative analysis of species C adenovirus hexon DNA in tonsillar and adenoidal lymphocytes was performed using real-time PCR (16). Briefly, PCR amplification was carried out in 50-μl reaction mixtures consisting of Qiagen 1× PCR buffer, 2.25 mM MgCl2, 0.2 mM each deoxynucleoside triphosphate (Roche), 2.5 U of Qiagen Hotstart Taq polymerase, 0.5 μM each primer, and 0.3 μM TaqMan probe. Primers were modified from those originally described by Pring-Akerblom et al. (26) to facilitate amplification of a conserved region of the species C adenovirus hexon gene (nucleotides 21049 to 21334 of Ad5; GenBank accession number NC_001406). For the primer and TaqMan probe sequences, see Fig. 1A. Serial 10-fold dilutions (from 5 × 107 to 1 copy) of Ad2 DNA (Gibco) were included in each run to generate a standard curve for quantitative assessment of donor adenovirus DNA. This PCR amplification yielded a 285-bp product. All samples were also tested for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) to normalize variation between samples due to differences in cell counts or input DNA. GAPDH was amplified using real-time PCR with the following primers and probe: sense primer (5′ AAATGAATGGGCAGCCGTTA 3′), antisense primer (5′ TAGCCTCGCTCCACCTGACT 3′), and TaqMan probe (5′-FAM CCTGCCGGTGACTAACCCTGCGCTCCT QSY7-3′). This reaction produced a 105-bp PCR product. Thermocycling profiles for real-time PCR consisted of 1 cycle of 95°C for 15 min followed by 45 cycles of 95°C for 15 s, 53°C for 35 s and 72°C for 30 s in a BioRad I-cycler. All standard dilutions and samples were run in triplicate or duplicate for adenovirus hexon or human GAPDH, respectively.
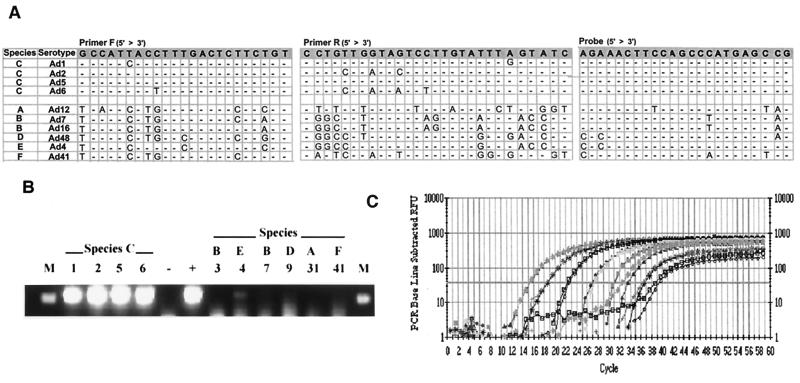
FIG. 1.
Real-time PCR detection of human species C adenovirus DNA. (A) Nucleotide sequences of the primers and TaqMan probe are to the hexon region of Ad5. (B) The species C viruses as well as representatives from each of the other human adenovirus species were amplified by real-time PCR using the hexon primers, and the products were run on an ethidium bromide-stained 1.8% agarose gel. Numbers indicate the adenovirus serotype tested. M, marker; −, negative water control; +, Ad2 positive control DNA. (C) Purified Ad2 DNA was serially 10-fold diluted, and duplicates of each dilution (from 5 × 107 to 5 copies) were tested. The fluorescence intensity (RFU = relative fluorescent units) collected in real time for each sample was plotted against the number of PCR cycles. The horizontal line indicates the fluorescence threshold setting, which is set at 10 standard deviations above the baseline emission.
Nested PCR for adenovirus E1A and fiber DNA.
The adenovirus fiber and E1A genes were detected in tonsil and adenoid lymphocytes by using nested or semi-nested PCR. The PCR amplifications were carried out in a final volume of 50 μl. The reaction mix consisted of Roche 1× PCR buffer, 2.25 mM MgCl 2, 0.2 mM each deoxynucleoside triphosphate (Roche), 2.5 U of Roche Taq polymerase, and 150 nM each primer.
The following set of nested E1A primers, as published by Flomenberg et al. (12), were used: forward outer primer, 5′ GAGTGAACTTTGACCGTYTACGTG 3′; reverse outer primer, 5′ TCCACCTACAAATCATACAGWTCGT 3′; forward inner primer, 5′ TCCGCGTACCGTGTCAAAGT 3′; and reverse inner primer, 5′ GGAACGCGAAGGTGTCTCATT 3′. PCR amplification was carried out with 1 cycle at 95°C for 4 min, 35 cycles of 95°C for 45 s, 56°C for 45 s, and 72°C for 45 s, followed by 1 cycle at 72°C for 3 min in a Hybaid gradient thermocycler. Following the initial round of PCR, 1 μl of primary PCR product was added to fresh PCR mixture and amplified in a second 50-μl nested PCR. The secondary PCR amplification was conducted under the same conditions with the exception of 25 cycles instead of 35.
For detection of the fiber gene, the following seminested primers were used: forward primer, 5′ ACCTTCAACCCCGTGTATCC-3′; outer reverse primer, 5′ GCAATGCTWAGTTTGGAGTC 3′; inner reverse primer, 5′ TGCCCATTTRAGCGCAAGCAT. PCR amplification was carried out in a Hybaid gradient PCR machine. For fiber DNA, PCR amplification was carried out with 1 cycle at 95°C for 4 min, 35 cycles of 95°C for 45 s, 56°C for 45 s, and 72°C for 45 s, followed by 1 cycle at 72°C for 3 min in a Hybaid gradient thermocycler. Following the initial cycle of PCR, 1 μl of primary PCR product was used in a second nested PCR amplification. Again, PCR amplification was done for only 25 cycles in the secondary PCR reaction.
PCR products were visualized on a 1.8% agarose gel stained with ethidium bromide. The E1A PCR products were 371 and 172 bp from the primary and the nested PCR, respectively. The fiber primary PCR product was 408 bp, and the seminested PCR product was 175 bp.
To avoid sample-to-sample contamination, different rooms and dedicated equipment were used for DNA purification and processing, PCR setup, and gel analysis. The PCR setup hood was treated with a UV light for 15 min prior to setting up any PCR amplification. Positive-displacement pipettes were used for PCR setup, and experimental samples were interspersed with blank or negative samples. In the experiments shown here, no signal was detected in any negative control sample.
Assay for infectious virus.
A total of 106 Ficoll-purified lymphocytes from 16 donors (containing from 0 to 5 × 105 adenovirus genomes per 107 cells) were added to subconfluent monolayers of human ME180 cells, and the cultures were monitored visually for cytopathic effects every 2 or 3 days for 4 weeks.
Adenovirus serotype determination.
PCR products were sequenced by two different methods. Fiber nested PCR products were gel purified, directly ligated into the PGEM-T easy vector (Promega), and transduced into competent bacterial cells for a Qiagen minipreparation of the DNA from two or three colonies. The PGEM-T vector with the cloned in PCR fragment was then sequenced using an upstream T7 primer. Alternatively, real-time PCR hexon products were directly sequenced after gel purification using the Qiagen gel extraction kit. Virus serotyping was performed by sequence comparison between the PCR product and known nucleotide sequences for species C adenoviruses published in GenBank by the National Center for Biotechnology Information. Serotypes were identified by conserved nucleotide base changes.
RESULTS
Specificity, detection limit, and dynamic range of the real-time PCR assay.
To assess the presence of adenovirus species C DNA in human tonsil and adenoid lymphocytes, a sensitive real-time PCR assay was developed using adenovirus hexon-specific primers and a TaqMan probe. Primers were selected from a region highly conserved among species C viruses but significantly divergent among other species (Fig. 1A). As expected, the primers were able to amplify all four of the species C viruses but did not amplify representatives from the other species (Fig. 1B). The faint band seen in the Ad4 lane is nonspecific because the hexon TaqMan probe did not hybridize with this product (data not shown).
To define the sensitivity and efficiency of this primer-probe set in the real-time PCR assay, 10-fold serial dilutions of Ad2 genomic DNA, ranging from 5 × 107 to 5 copies, were tested. This assay was regularly able to detect 5 copies of the genome and, as the Poisson distribution would predict, could inconsistently detect 1 copy of the genome. The range of the assay allowed quantitation over at least 7 orders of magnitude (Fig. 1C). When the threshold cycle values of the standard dilutions were plotted against the log10 of the starting copy number, the correlation coefficient values were usually higher than 0.990 and the slope of the line was greater than −3.8, indicating high amplification efficiency (data not shown).
Lymphocytes from adenoids and tonsils contain species C adenovirus DNA.
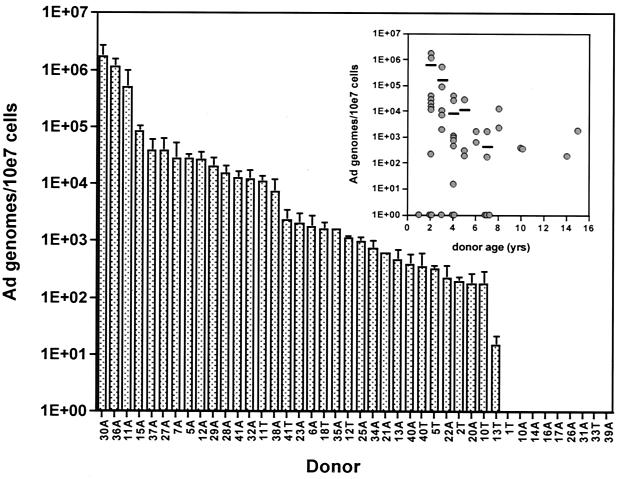
Previous studies have clearly identified adenovirus DNA in tonsil and adenoid tissues, but the cell type(s) harboring the viral genome has never been identified. The presence of adenovirus in lymphoid cells of tonsil and adenoid tissues was approached by preparation of single-cell suspensions followed by Ficoll purification of lymphocytes. Adenovirus DNA from the hexon region of the genome was detectable in 33 of 42 samples tested (79%) (Fig. 2). Adenovirus DNA quantities in positive samples ranged from 20 to 2 × 106 copies per 107 cells. No obvious correlation with the sex of the donor was observed (data not shown). When analyzed as a function of donor age, however, the highest levels of adenovirus genomes were found among the youngest donors (Fig. 2 inset). In seven cases, both adenoids and tonsils were available from the same donor. In two of the seven sets of tissues the number of adenovirus genomes was about the same in tonsils and adenoids, but in the other five sets the number in the adenoids was much greater than that seen in palatine tonsils from the same donor (Fig. 2). Indeed, among all samples analyzed to date, large quantities of adenovirus genomes are more likely to be found in adenoids than in tonsils.
FIG. 2.
Distribution and quantitation of adenovirus DNA per 107 lymphocytes in 42 tonsil and adenoid samples from 35 donors. Real-time PCR was performed on DNA purified from Ficoll-purified lymphocytes from tonsils (T) and adenoids (A). Cellular input DNA amounts were normalized to quantities of GAPDH DNA between samples being compared. Individual donors were tested two to five times, and the results are presented as mean and standard error of the mean for all experiments. (Inset) Number of Ad genomes/107 cells as a function of donor age. Each point represents a single sample. Horizontal lines are the average number of genomes for each age group in which more than two samples were tested.
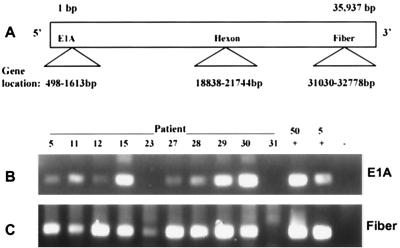
The results described above detect only a single gene, hexon, located near the center of the 36-kb adenovirus genome. Nested PCR was used to detect two additional regions of the viral genome in lymphocytes from hexon-positive donors, the fiber gene (near the 3′ end of the genome) and the E1A gene (near 5′ end of the genome) (Fig. 3A). For this experiment, nine donors, whose lymphocytes contained amounts of hexon DNA ranging from 2 × 103 to 1.7 × 106 copies/107 cells, were chosen. All nine donors were also positive for fiber DNA (Fig. 3C), and eight of the nine were also positive for E1A DNA (Fig. 3B). Notably, donor 23, the single case where E1A was not detected, also contained the smallest quantity of hexon DNA among the nine by real-time PCR. Whether the inability to detect E1A in this donor was a result of the lower sensitivity of those primers or represents a true deletion of that region has not been determined. The fiber and E1A nested PCR assays are a measure of end-point PCR product accumulation and are not quantitative, therefore, no comparisons could be made, based on differences in band intensity, between samples. Neither fiber nor E1A DNA was detected in samples from donor 31, who was also negative for hexon DNA. These findings suggest that intact adenovirus DNA is likely present in human tonsil and adenoid lymphocytes.
FIG. 3.
Detection of proximal (E1A) and distal (Fiber) regions of the adenovirus genome in tonsil and adenoid lymphocytes. (A) Relative location of E1A, hexon, and fiber genes within the adenovirus genome. (B) Nested PCR for adenovirus E1A DNA (172-bp product). (C) Nested PCR for adenovirus fiber DNA (175-bp product). Samples from all patients shown were positive for hexon by real-time PCR, except for the sample from patient 31, which was negative. Nested PCR was performed on DNA purified from Ficoll-purified lymphocytes. +, positive control using either 50 or 5 copies of Ad2 DNA as template; −, negative water control.
Detection of infectious virus in tonsil and adenoid lymphocytes.
To determine whether infectious virus was present in tonsil and adenoid lymphocytes at the time of surgical removal of the tissue, Ficoll-purified lymphocytes from 16 donors were cocultured with permissive human ME180 cells and observed for cytopathic effects for up to 28 days. Of the 16 donor samples tested, 12 contained adenovirus DNA by PCR. Only 1 of the samples (6%) produced infectious virus on coculture with ME180 cells (data not shown). The one positive sample was obtained from donor 15, whose adenoids contained among the highest levels of viral DNA. However, no cytopathic effect was detected in cocultures of lymphocytes from donor 11, whose adenoids contained even higher levels of viral DNA.
CD4+/CD8+ lymphocytes, but not CD19+ B cells, from adenoids contain adenovirus DNA.
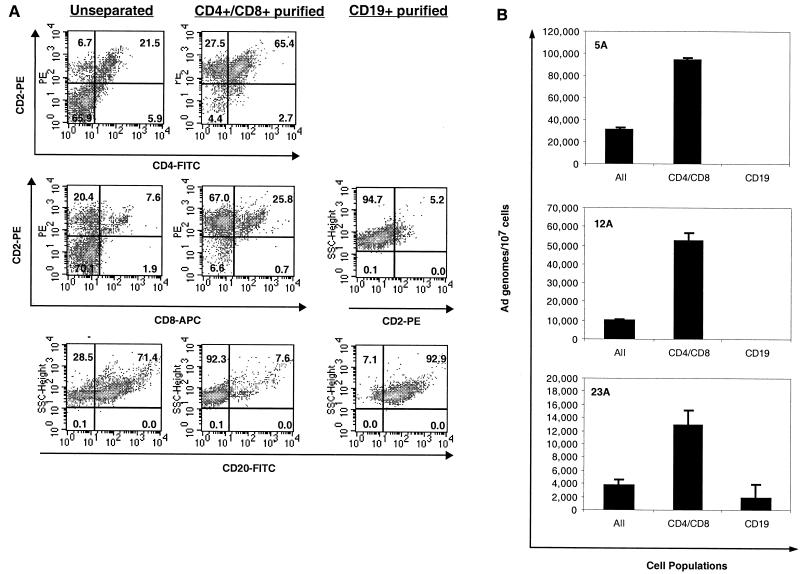
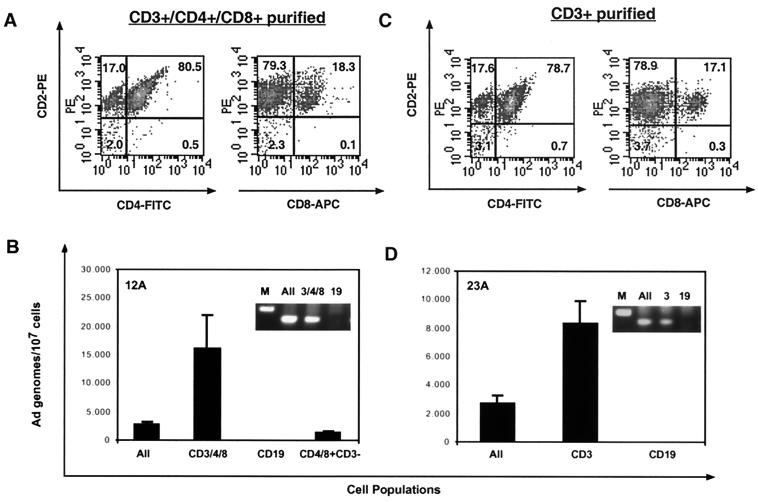
To determine which lymphocyte subpopulations harbor adenovirus DNA, initial separations of cells into CD4+ CD8+ and CD19+ cell fractions were performed using Dynal antibody-coated magnetic beads. Directly conjugated anti-CD4 and anti-CD8 beads were combined and used to enrich T cells. Directly conjugated anti-CD19 beads were used to purify B cells. By using this approach, we obtained populations of cells that were consistently greater than 90% pure and often greater than 95% pure as measured by flow cytometry (Fig. 4A). Unseparated cells contained between 23 and 29% CD2+ cells and between 71 and 77% CD20+ B cells. After magnetic bead purification, the CD4+ CD8+ population contained 91 to 96% CD2+ T cells, and the CD19+ purified cells contained 92 to 97% CD20+ B cells. In the experiment in Fig. 4B, total DNA was isolated from separated and unseparated cells and adenovirus DNA quantitated by real-time PCR for hexon DNA. Adenovirus DNA was detected in the unseparated cells, and in each case, on average a fourfold enrichment was observed in the CD4+ CD8+ population (Fig. 4B). This enrichment is what would be expected if all or most of the adenovirus DNA copurified with a cell population representing one-third to one-quarter of the total lymphocytes. No adenovirus DNA was detected in the B cells of two of the three donors shown. Donor 23A, who did appear to have some viral DNA associated with the B-cell population, still shows a significant depletion in the amount of viral DNA per 107 cells compared to the amount in unseparated cells. The lack of adenovirus DNA enrichment suggests that the viral DNA detected here is likely not from the CD19+ B cells and could be due to residual contaminating CD4+ CD8+ cells, which make up 5% of this CD19-enriched population.
FIG. 4.
Analysis of adenovirus DNA in separated lymphocyte subpopulations. (A) Ficoll-purified lymphocyte populations were purified from adenoid lymphocytes with directly conjugated Dynal magnetic beads to CD4 and CD8 together or CD19 as detailed in Materials and Methods. The resulting cells were then stained with fluorescent antibodies and analyzed on the flow cytometer for purity. CD2 was used as a pan-T-cell marker. CD20 was used as a pan-B-cell marker. (B) Real-time PCR for hexon was performed on 5 μl of DNA prepared from the purified CD4+ CD8+, CD19+, and unseparated (All) cells. Adenovirus DNA values in samples were normalized by referring the viral copy number to the actual amount of input cellular DNA as estimated by quantification of the GAPDH gene in each sorted population.
CD3+ T lymphocytes from adenoids contain adenovirus DNA.
Both dendritic cells and macrophages can express CD4 on their surface (31). Thus, the presence of viral DNA in the CD4+ population could come from dendritic cells or macrophages, as well as from T cells. To distinguish among these possibilities, cells were first purified into CD4+ CD8+ and CD19+ populations. The CD4+ CD8+ population was then further purified with anti-CD3 beads to positively select only the T cells. These CD3+ CD4+ CD8+ T cells were >97% CD2+ (Fig. 5A) and the CD19 B cells were again >95% CD20+ (not shown). Adenovirus DNA was enriched in the CD3+ cell population (Fig. 5B). No viral DNA was detected in the CD19+ population. When the remaining CD4+ CD8+ cells that did not bind to the CD3 beads (CD4+ CD8+ CD3−) were tested, they contained very little viral DNA, suggesting that the viral DNA resides solely in the CD3+ fraction in this donor. The unseparated, CD4+ CD8+ CD3+, and CD19+ cell fractions were also tested by nested PCR for the adenovirus fiber gene, confirming the presence of viral DNA in the T cell fractions (Fig. 5B inset).
FIG. 5.
Analysis of Ad DNA in separated lymphocyte subpopulations. (A) Lymphocyte populations were purified from adenoid lymphocytes with Dynal magnetic beads to CD4 and CD8 together or CD19. The beads were then removed from the CD4+ CD8+-selected cells as detailed in Materials and Methods, and the cells were further purified with MACS magnetic beads to CD3. The purified cell populations were stained with fluorescent antibodies and analyzed on the flow cytometer for purity. (B) Real-time PCR for hexon was performed on DNA prepared from the purified CD3+ CD4+ CD8+, CD19+, and unseparated (All) cells. Cells that were CD4+ CD8+ but CD3− were also tested. Adenovirus DNA levels in samples were normalized by referring the viral copy number to the actual amount of input cellular DNA estimated by quantification of the GAPDH gene in each sorted population. (Inset) Nested PCR for adenovirus fiber DNA was performed on the unseparated (All), CD4+ CD8+ CD3+, and CD19+ samples, and the PCR products were run on a 1.8% agarose gel stained with ethidium bromide. M, marker. (C) Lymphocyte populations were purified from adenoid lymphocytes with Dynal magnetic beads to CD19. T cells were directly obtained with MACS magnetic beads to CD3. The resulting cells were then analyzed for purity by flow cytometry. (D) Real-time PCR for hexon was performed on DNA prepared from the purified CD3+, CD19+, and unseparated cells (All). (Inset) Nested PCR for adenovirus fiber DNA was performed on the same samples, and the PCR products were run on a 1.8% agarose gel stained with ethidium bromide. M, marker.
Identical results were obtained when lymphocytes underwent CD19 fractionation followed directly by CD3 purification without an intervening CD4 CD8 purification (Fig. 5C and D). Using this cell purification protocol, T cells were >95% CD2+. Nested PCR for the fiber gene in these sorted populations again showed adenovirus DNA in the CD3+ T cells and not in the CD19+ B cells (Fig. 5D inset). In all, nine of nine donors examined showed an enrichment of adenovirus DNA in their CD3+ or the CD4+ CD8+ T-cell population.
Characterization of the serotype found in mucosal lymphocytes by fiber and hexon region sequence comparison.
To determine adenovirus serotype, either fiber or hexon PCR-amplified DNA from a total of 22 selected donors (Table 1) was sequenced and compared with published sequences of species C adenoviruses. Serotype identification was obtained for all donor sequences based on conserved serotype-specific nucleotide patterns. Ten donors were sequenced by cloning of the fiber nested PCR product into the PGEM T easy vector followed by sequencing of the miniprepped DNA. This method distinguishes serotypes Ad1, Ad5, and Ad2/6. The sequence of serotypes Ad2 and Ad6 are identical in this region. Of the 10 donors sequenced by this method, 2 donors had PCR products from both their adenoid and tonsil tissue sequenced. Both tissues from a single donor contained identical serotypes by sequence.
TABLE 1.
Characterization of serotype by sequence comparison
| Donora | Serotype from sequence:
|
|
|---|---|---|
| Fiber | Hexon | |
| 2T | Ad5 | |
| 4A | Ad5 | |
| 5A | Ad2/6 | Ad2 |
| 5T | Ad2/6 | Ad2 |
| 6A | Ad5 | |
| 7A | Ad1 | Ad1 |
| 9A | Ad5 | |
| 10T | Ad5 | |
| 11A | Ad1 | |
| 12A | Ad5 | Ad5 |
| 13A | Ad2/6 | |
| 13T | Ad2/6 | |
| 15A | Ad1 | Ad1 |
| 23A | Ad1 | |
| 27A | Ad5 | |
| 28A | Ad1 | |
| 29A | Ad2 | |
| 30A | Ad1 | |
| 32A | Ad1 | |
| 35A | Ad2 | |
| 36A | Ad1 | |
| 37A | Ad5 | |
| 40T | Ad1 | |
| 41A | Ad1 | |
| 41T | Ad1 | |
T, tonsil; A, adenoid.
In addition, the real-time PCR hexon products from 16 donors were gel purified and directly sequenced. Distinctions between serotypes could again be made based on conserved nucleotide base changes within the region. The samples from 4 of these 16 donors were repeats of tissues sequenced by the previous method in the fiber region. All four of these tissues revealed the same serotype by hexon sequencing as they did by fiber sequencing. Again, when the tonsil and adenoid samples from donor 41 were serotyped separately by hexon sequence comparison, both tissues contained the same serotype. As expected, for all 22 donors whose PCR products were serotyped, Ad1, the serotype most commonly isolated from humans (15, 17), was the most common serotype identified in this study group (10 of 22 donors) (Table 1). Ad5 and Ad2 were identified in eight and four donors, respectively. None of the samples appeared to contain more than one serotype. No obvious correlations were observed between adenovirus serotype and donor age, gender, or number of genomes/107 cells. None of the samples was found to contain serotype Ad6, which is the rarest of the species C serotypes (14, 15, 17).
DISCUSSION
The findings reported here support the longstanding but poorly documented suspicion that species C adenoviruses form latent infections in human lymphocytes. Lymphocytes as a site of latency have been postulated in part because of the finding of nonreplicating adenovirus in mucosal lymphoid tissues (11, 18, 25), although the cell types harboring the viral DNA were not identified. In addition, several studies have characterized atypical, even nonlytic infections of human lymphocyte cell lines in vitro. Lavery et al. (21) reported that infection of a series of B- and T-cell lines with Ad2 or Ad5 generally yielded delayed kinetics of viral DNA and mRNA production compared to infection of HeLa cells, and the percentage of infected cells varied from 6 to 60% depending on the cell line. Several groups have reported the establishment of persistently infected lymphocyte cell lines, in which viral DNA is maintained and small quantities of infectious virus are produced while cell growth kinetics remain normal (6, 13, 30). These observations suggest that the species C adenoviruses may be capable of an alternative life-style in a lymphocyte cell population, leading to nonlytic replication and long-term persistence.
At present the number of virus-bearing cells in the lymphocyte population is unknown. Considering that the highest level of viral genome copies among donors tested was 2 × 106 per 107 lymphocytes, it is possible that a relatively large number of cells (up to 20%) carry a single copy of the viral genome in that donor. At the other extreme, given that lytic infection of a fully permissive cell yields 5,000 PFU per cell, 400 cells could account for 2 × 106 viral genomes. Currently, a limiting-dilution approach is being used to distinguish between these extremes. Experiments to date indicate that latently infected T lymphocytes carry an average of 30 copies of the viral genome each (data not shown). Thus, in a donor containing 2 × 106 viral genomes per 107 lymphocytes, 1 in every 150 cells contains adenovirus DNA.
Lymphocyte subpopulations were enriched to determine whether the viral genomes would be similarly enriched in a defined lymphocyte subtype, acknowledging that the virus-containing cell would probably remain rare even in an enriched subpopulation. Nonetheless, enrichment for viral DNA following enrichment of a specific lymphocyte population is strong evidence for a cell-specific viral association. In these experiments, viral DNA is enriched with T lymphocytes in nine of nine donors tested. This was initially accomplished using magnetic beads from Dynal directly coupled with antibodies to CD4 and CD8, both markers of T lymphocyte subpopulations. In subsequent experiments, magnetic beads from another supplier, Miltenyi, coupled with antibody to CD3, a pan-T-cell marker, were used to confirm that viral DNA is enriched in T lymphocytes and to eliminate the possibility that some unknown characteristic of the CD4/8 Dynal beads was responsible for enrichment of viral DNA. Viral DNA is enriched whether cells are first exposed to the CD3 beads and washed and then the remaining cells are exposed to CD19 beads (to enrich B lymphocytes) or whether the protocol is reversed and the CD19 beads are used first. This approach controls for the possibility that virus-bearing cells stick nonselectively to magnetic beads. Finally, in recent experiments, tonsil and adenoid lymphocytes have been separated using fluorescence-activated cell sorting, which has confirmed the enrichment of viral DNA in the CD3+ T-cell population (data not shown). More precise definition of the virus-bearing lymphocyte subpopulation, based on expression of T-lymphocyte cell surface markers, is the subject of ongoing investigation in the laboratory.
In the experiments whose results are shown in Fig. 4, a “depleted” cell population was also analyzed for viral DNA. This population consists of the cells that remain after sequential removal of CD19+ and CD4+ CD8+ cells by magnetic-bead purification. This cell fraction still contains roughly 5 to 25% CD4+ CD8+ cells and 55 to 80% CD19+ cells and therefore cannot be considered a purified cell population. It does, however, contain 5 to 20% of a non-T-cell (CD4− CD8−), non-B-cell (CD19−) type that accounts for 1% or less of the unseparated cell population. Interestingly, three of the nine donors tested in this study show viral DNA enrichment in this “depleted” population in addition to the enrichment of viral DNA seen in their T-cell population. These findings suggest that adenovirus DNA can also be located in a non-T-cell, non-B-cell type in addition to the T cells. Future experiments will focus on determining the identity of this other adenovirus-bearing cell type with new lymphocyte and phagocyte cell surface markers.
In this study, the highest levels of adenovirus DNA are found in 2-year-old donors, with only one tissue sample available from a donor younger than 2 years. Among all donors, there is a decrease in the amount of adenovirus DNA with age after 2 years. In a large prospective study, Edwards et al. (8) found that the vast majority of primary infections with species C adenoviruses occurred within the first 2 years of life. Acute species C infections are found in only 7 of 1,018 clinical respiratory illnesses in older children and young adults (10, 11) and are virtually never seen in military recruits (34). Thus, the quantity of adenovirus DNA remaining in tonsil tissue is highest in the age group most recently infected, with a steady decrease thereafter. This is reminiscent of early studies looking for outgrowth of adenoviruses from explanted tonsils, in which there was an age-related decrease in the percentage of samples yielding virus from roughly 50% among donors younger than 9 years to about 10% from donors older than 19 years (11, 18, 32). The mechanism of loss of adenovirus DNA with age is unknown but could stem either from immune elimination or from depletion of latent stores through reactivation. Nonetheless, infectious virus (11, 18, 32) as well as adenoviral DNA (this study) can still be detected in the oldest donors tested.
The ease with which adenovirus DNA is detected in tonsil and adenoid lymphocytes stands in stark contrast to the findings of others looking for adenovirus DNA in PBLs. Using a sensitive nested PCR assay, Flomenberg et al. (12) found adenovirus DNA in PBL from 0 of 33 healthy adults and only 1 of 40 pediatric donors. Another PCR study of >200 donors reported only 1.7% of healthy human adults with adenovirus DNA in their PBLs (7). PBLs and mucosal-derived lymphocytes (tonsil and adenoid) have very different circulation and homing patterns (29). Thus, the virus may be strictly associated with the mucosal-lymphocyte compartment and rarely found circulating in the periphery. Virus shedding in the stool also supports a mucosal association of the virus. The virus may primarily infect mucosal lymphocytes; alternatively, the virus could induce the expression of homing receptors that target infected lymphocytes to the mucosa.
Although early epidemiologic studies concluded that persistent adenovirus infections were benign, recent evidence employing PCR to identify pathogens at sites of disease suggest a role for the species C viruses in a variety of chronic diseases in immunocompetent individuals. A remarkable 80% of children with asthma have adenovirus DNA in their nasopharynx (compared with only 5% of age-matched controls) when tested at a time of disease quiescence (23). An equally high percentage of asthmatic children yield viral capsid protein on bronchoalveolar lavage (22). These findings suggest smoldering virus production at the site of lung inflammation, which may contribute to disease pathogenesis by increasing the local inflammatory response. Persistent adenovirus DNA in the lungs is postulated to be a cofactor in chronic obstructive pulmonary disease (COPD) in adults. Using a semiquantitative PCR method, Matsuse et al. (24) found significant increases in the amount of adenovirus DNA in the lungs of smokers with COPD compared to the lungs of smokers without COPD. Indeed, latent infection with Ad5 exacerbated lung inflammation caused by cigarette smoke in a guinea pig model system (33). The association between adenovirus and cigarette smoking in lung pathogenesis goes even further. PCR screening detects species C adenovirus DNA in one-third of small-cell lung cancers, a disease found virtually only in tobacco smokers (19). Although it is clear that even normal children and adults carry adenovirus DNA in their respiratory tract, adenovirus levels are not increased in all respiratory tract diseases. The level of adenovirus DNA is not elevated in adenocarcinoma or squamous cell carcinoma of the lung (19), idiopathic pulmonary fibrosis (20), cystic fibrosis (9), or chronic sinusitis (27). Thus, elucidation of the mechanisms by which species C adenoviruses persist in mucosal lymphoid tissues may inform attempts to identify their role in chronic inflammatory diseases. The molecular dynamics of long-term adenovirus persistence and latency, such as altered viral DNA replication, transcription, or virion production, are currently the subject of ongoing investigation in the laboratory.
Acknowledgments
We thank Pablo Stolovitzky, Carlos Abramowski, and Catherine South for their help with tissue specimen collection, as well as Guy Beresford for all of his technical assistance with the I-cycler. We also thank Brian Holloway and Karen McCaustland of the Biotechnology Core Facility Branch, CDC.
This work was supported by NIH grant CA-58736 and a grant from the Emory University Research Committee.
REFERENCES
- 1.Adrian, T., G. Schafer, M. K. Cooney, J. P. Fox, and R. Wigand. 1988. Persistent enteral infections with adenovirus types 1 and 2 in infants: no evidence of reinfection. Epidemiol. Infect. 101:503-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andiman, W. A., R. I. Jacobson, and G. Tucker. 1977. Leukocyte-associated viremia with adenovirus type 2 in an infant with lower-respiratory-tract disease. N. Engl. J. Med. 297:100-101. [DOI] [PubMed] [Google Scholar]
- 3.Avila, M. M., G. Carballal, H. Rovaletti, B. Ebekian, M. Cusminsky, and M. Weissenbacher. 1989. Viral etiology in acute lower respiratory infections in children from a closed community. Am. Rev. Respir. Dis. 140:634-637. [DOI] [PubMed] [Google Scholar]
- 4.Babcock, G. J., L. L. Decker, M. Volk, and D. A. Thorley-Lawson. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395-404. [DOI] [PubMed] [Google Scholar]
- 5.Brandt, C. D., H. W. Kim, A. J. Vargosko, B. C. Jeffries, J. O. Arrobio, B. Rindge, R. H. Parrott, and R. M. Chanock. 1969. Infections in 18,000 infants and children in a controlled study of respiratory tract disease. I. Adenovirus pathogenicity in relation to serologic type and illness syndrome. Am. J. Epidemiol. 90:484-500. [DOI] [PubMed] [Google Scholar]
- 6.Chu, Y., K. Sperber, L. Mayer, and M. T. Hsu. 1992. Persistent infection of human adenovirus type 5 in human monocyte cell lines. Virology 188:793-800. [DOI] [PubMed] [Google Scholar]
- 7.Durepaire, N., J. P. Rogez, M. Verdier, S. Rogez, P. Weinbreck, and F. Denis. 1997. Detection of adenovirus DNA by polymerase chain reaction in peripheral blood lymphocytes from HIV-infected patients and a control group: preliminary results. J Acquir. Immune Defic. Syndr. Hum. Retrovirol. 14:189-190. [DOI] [PubMed] [Google Scholar]
- 8.Edwards, K. M., J. Thompson, J. Paolini, and P. F. Wright. 1985. Adenovirus infections in young children. Pediatrics 76:420-424. [PubMed] [Google Scholar]
- 9.Eissa, N. T., C. S. Chu, C. Danel, and R. G. Crystal. 1994. Evaluation of the respiratory epithelium of normals and individuals with cystic fibrosis for the presence of adenovirus E1a sequences relevant to the use of E1a− adenovirus vectors for gene therapy for the respiratory manifestations of cystic fibrosis. Hum. Gene Ther. 5:1105-1114. [DOI] [PubMed] [Google Scholar]
- 10.Evans, A. 1957. Endemic respiratory disease. Postgrad. Med. 21:329-338. [DOI] [PubMed] [Google Scholar]
- 11.Evans, A. 1958. Latent adenovirus infections of the human respiratory tract. Am. J. Hyg. 67:256-266. [DOI] [PubMed] [Google Scholar]
- 12.Flomenberg, P., E. Gutierrez, V. Piaskowski, and J. T. Casper. 1997. Detection of adenovirus DNA in peripheral blood mononuclear cells by polymerase chain reaction assay. J. Med. Virol. 51:182-188. [DOI] [PubMed] [Google Scholar]
- 13.Flomenberg, P., V. Piaskowski, J. Harb, A. Segura, and J. T. Casper. 1996. Spontaneous, persistent infection of a B-cell lymphoma with adenovirus. J. Med. Virol. 48:267-272. [DOI] [PubMed] [Google Scholar]
- 14.Fox, J. P., C. D. Brandt, F. E. Wassermann, C. E. Hall, I. Spigland, A. Kogon, and L. R. Elveback. 1969. The virus watch program: a continuing surveillance of viral infections in metropolitan New York families. VI. Observations of adenovirus infections: virus excretion patterns, antibody response, efficiency of surveillance, patterns of infections, and relation to illness. Am. J. Epidemiol. 89:25-50. [DOI] [PubMed] [Google Scholar]
- 15.Fox, J. P., C. E. Hall, and M. K. Cooney. 1977. The Seattle Virus Watch. VII. Observations of adenovirus infections. Am. J. Epidemiol. 105:362-386. [DOI] [PubMed] [Google Scholar]
- 16.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR Genome Res. 6:986-94. [DOI] [PubMed] [Google Scholar]
- 17.Hillis, W. D., M. R. Cooper, and F. B. Bang. 1973. Adenovirus infections in West Bengal. I. Persistence of viruses in infants and young children. Indian J. Med. Res. 61:980-988. [PubMed] [Google Scholar]
- 18.Israel, M. 1962. The viral flora of enlarged tonsils and adenoids. J. Pathol. Bacteriol. 84:169-176. [Google Scholar]
- 19.Kuwano, K., M. Kawasaki, R. Kunitake, N. Hagimoto, Y. Nomoto, T. Matsuba, Y. Nakanishi, and N. Hara. 1997. Detection of group C adenovirus DNA in small-cell lung cancer with the nested polymerase chain reaction. J. Cancer Res. Clin. Oncol. 123:377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuwano, K., Y. Nomoto, R. Kunitake, N. Hagimoto, T. Matsuba, Y. Nakanishi, and N. Hara. 1997. Detection of adenovirus E1A DNA in pulmonary fibrosis using nested polymerase chain reaction. Eur. Respir. J. 10:1445-1449. [DOI] [PubMed] [Google Scholar]
- 21.Lavery, D., S. M. Fu, T. Lufkin, and S. Chen-Kiang. 1987. Productive infection of cultured human lymphoid cells by adenovirus. J. Virol. 61:1466-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Macek, V., J. Sorli, S. Kopriva, and J. Marin. 1994. Persistent adenoviral infection and chronic airway obstruction in children. Am. J. Respir. Crit. Care Med. 150:7-10. [DOI] [PubMed] [Google Scholar]
- 23.Marin, J., D. Jeler-Kacar, V. Levstek, and V. Macek. 2000. Persistence of viruses in upper respiratory tract of children with asthma. J. Infect. 41:69-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsuse, T., S. Hayashi, K. Kuwano, H. Keunecke, W. A. Jefferies, and J. C. Hogg. 1992. Latent adenoviral infection in the pathogenesis of chronic airways obstruction. Am. Rev. Respir. Dis. 146:177-184. [DOI] [PubMed] [Google Scholar]
- 25.Neumann, R., E. Genersch, and H. J. Eggers. 1987. Detection of adenovirus nucleic acid sequences in human tonsils in the absence of infectious virus. Virus Res. 7:93-97. [DOI] [PubMed] [Google Scholar]
- 26.Pring-Akerblom, P., F. E. Trijssenaar, T. Adrian, and H. Hoyer. 1999. Multiplex polymerase chain reaction for subgenus-specific detection of human adenoviruses in clinical samples. J. Med. Virol. 58:87-92. [PubMed] [Google Scholar]
- 27.Ramadan, H. H., R. W. Farr, and S. J. Wetmore. 1997. Adenovirus and respiratory syncytial virus in chronic sinusitis using polymerase chain reaction. Laryngoscope 107:923-925. [DOI] [PubMed] [Google Scholar]
- 28.Rowe, W., R. J. Huebner, and L. K. Gillmore. 1953. Isolation of a cytopathic agent from human adenoids undergoing spontaneous degeneration in tissue culture. Proc. Soc. Exp. Biol. Med. 84:570-573. [DOI] [PubMed] [Google Scholar]
- 29.Salmi, M., D. Adams, and S. Jalkanen. 1998. Cell adhesion and migration. IV. Lymphocyte trafficking in the intestine and liver. Am. J. Physiol. 274:G1-G6. [DOI] [PubMed] [Google Scholar]
- 30.Silver, L., and C. W. Anderson. 1988. Interaction of human adenovirus serotype 2 with human lymphoid cells. Virology 165:377-387. [DOI] [PubMed] [Google Scholar]
- 31.Summers, K. L., B. D. Hock, J. L. McKenzie, and D. N. Hart. 2001. Phenotypic characterization of five dendritic cell subsets in human tonsils. Am. J. Pathol. 159:285-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Veen, J., and M. Lambriex. 1973. Relationship of adenovirus to lymphocytes in naturally infected human tonsils and adenoids. Infect. Immun. 7:604-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vitalis, T. Z., I. Kern, A. Croome, H. Behzad, S. Hayashi, and J. C. Hogg. 1998. The effect of latent adenovirus 5 infection on cigarette smoke-induced lung inflammation. Eur. Respir. J. 11:664-669. [PubMed] [Google Scholar]
- 34.Woolridge, R., J. Grayston, J. Whiteside, C. Loosli, M. Friedman, and W. Pierce. 1956. Studies on acute respiratory illness in Naval recruits with emphasis on the adenovirus (APC-RI). J. Infect. Dis. 99:182-187. [DOI] [PubMed] [Google Scholar]