Abstract
The coding region of influenza A virus RNA segment 7 from the 1918 pandemic virus, consisting of the open reading frames of the two matrix genes M1 and M2, has been sequenced. While this segment is highly conserved among influenza virus strains, the 1918 sequence does not match any previously sequenced influenza virus strains. The 1918 sequence matches the consensus over the M1 RNA-binding domains and nuclear localization signal and the highly conserved transmembrane domain of M2. Amino acid changes that correlate with high yield and pathogenicity in animal models were not found in the 1918 strain. Phylogenetic analyses suggest that both genes were mammalian adapted and that the 1918 sequence is very similar to the common ancestor of all subsequent human and classical swine matrix segments. The 1918 sequence matches other mammalian strains at 4 amino acids in the extracellular domain of M2 that differ consistently between avian and mammalian strains, suggesting that the matrix segment may have been circulating in human strains for at least several years before 1918.
The influenza virus pandemic of 1918 resulted in the deaths of up to 50 million people worldwide (23, 35). Recently, samples of the 1918 virus RNA have been obtained from several widely separated geographical sites (39). Analyses of viral hemagglutinin (HA), neuraminidase, and nonstructural genes suggest that the 1918 virus was genetically uniform and adapted to its human host (3, 39, 40). The sequences of these genes, however, did not reveal features that could account for the virulence of the virus, although several hypotheses could be discounted. Explanations for the lethality of the virus might therefore lie in the other five viral gene segments.
Influenza A virus RNA segment 7 encodes two proteins, the matrix proteins M1 and M2. M1 mRNA is colinear with the viral RNA, while M2 mRNA is encoded by a spliced transcript (29, 52). The proteins encoded by these mRNAs share their initial 9 amino acids and also have a stretch of 14 amino acids in overlapping reading frames. The M1 protein is a highly conserved 252-amino acid protein. It is the most abundant protein in the viral particle, lining the inner layer of the viral membrane and contacting the ribonucleoprotein core. M1 has been shown to have several functions (28), including regulation of the nuclear export of viral ribonucleoproteins (vRNPs), both permitting the transport of vRNP particles into the nucleus upon infection and preventing newly exported vRNP particles from reentering the nucleus (8, 32). M1 may also be involved in the inhibition of viral transcription in the late stages of infection and regulation of the switch from replication to viral assembly (36, 59, 60). M1 binds RNA (41, 49, 57, 59), vRNPs (34, 38, 41, 57), and lipids (7, 15); dimerizes with other M1 molecules (16, 41); and interacts with both the HA and neuraminidase proteins (1, 11). It is also involved in export to the cytoplasmic membrane, virus assembly, and budding (12, 19, 31).
The 97-amino acid M2 protein is a homotetrameric integral membrane protein that exhibits ion channel activity (4, 20, 21, 30, 37, 46, 50) and is the target of the drug amantadine (18). The ion channel activity of M2 is important both during virion uncoating and during viral budding (14, 37, 46, 47; reviewed in reference 28).
The disease course and pathological damage caused by the 1918 virus are consistent with a virus capable of replicating to titers higher than those of other strains (53, 54). The matrix segment has been shown in several experimental systems to have a significant effect on virulence and viral replication rate (6, 13, 43, 44, 51, 55). In some of these studies, matrix proteins differing by as little as 1 or 2 amino acids conferred striking differences in phenotype. Certain mutations in the M2 gene lead to viruses that are resistant to antiviral drugs (17, 18). It is possible that unique characteristics of either M1 or M2 contributed to the lethality of the 1918 virus.
MATERIALS AND METHODS
Isolation of A/Brevig Mission/1/18 RNA and amplification of the matrix segment.
RNA was isolated by methods described in other publications (27, 48). The 982-nucleotide M1 and M2 complete coding region of the 1918 influenza virus segment 7 was amplified in 15 overlapping fragments varying in length from 97 to 142 nucleotides. Each fragment was reverse transcribed, amplified, and sequenced at least twice. Reverse transcription-PCR, isolation of products, and sequencing have been described previously (39, 40, 48). A list of the primers used is available upon request.
Phylogenetic analyses.
Nucleotide sequences of the M1 and M2 genes were aligned with the genes from other influenza virus isolates by using Lasergene software (DNAStar, Madison, Wis.). Phylogenetic analyses were carried out by use of the neighbor-joining algorithm with distance parameter (p-distance) (Molecular Evolutionary Genetics Analysis, version 1.01, 1993). All analyses were bootstrapped 100 times.
Viral strains.
Matrix segment sequences used in this analysis were obtained from GenBank. For a list of the strains and abbreviations used, see Table 1.
TABLE 1.
Matrix gene sequences used in this analysis.
| Abbreviation | Strain name | GenBank accession no. |
|---|---|---|
| BREVIG18 | A/Brevig Mission/1/18 (H1N1) | AY130766 |
| CH/BRESCIA02 | A/chicken/Brescia/1902 (H7N7) | L37795 |
| CH/HK/220/97 | A/chicken/Hong Kong/220/97 (H5N1) | AAF74333 |
| CH/HONGKONG76 | A/chicken/Hong Kong/14/76 (H1N1) | U49119 |
| DK/BAVARJA77 | A/duck/Bavaria/2/77 (H1N1) | M55476 |
| DK/CZECH56 | A/duck/Czechoslovakia/56 (H4N6) | M63537 |
| DK/HONGKONG79 | A/cuck/Hong Kong/698/79 (H5N3) | AF098565 |
| DK/PRIMORJE79 | A/anas acuta/Primorje/695/79 (H3N2) | Z26858 |
| ENGLAND69 | A/England/939/69 (H3N2) | AJ298948 |
| EQ/KENTUCKY81 | A/equine/Kentucky/81 (H3N8) | AF001676 |
| EQ/KENTUCKY86 | A/equine/Kentucky/2/86 (H3N8) | M63540 |
| EQ/MIAMI63 | A/equine/Miami/63 (H3N8) | AF001674 |
| EQ/PRAGUE56 | A/equine/Praque/1/56 (H7N7) | M63533 |
| FIJI83 | A/Fiji/15899/83 (H1N1) | AJ298947 |
| FM47 | A/Fort Monmouth/1/47 (H1N1) | U02084 |
| FPV/WEYBRIDGE34 | A/FPV/Weybridge/34 (H7N7) | L37797 |
| FTWAR50 | A/Fort Warren/1/50 (H1N1) | J02125 |
| FUKUSHIMA96 | A/Fukushima/114/96 (H3N2) | AF038273 |
| GS/GUANGDONG96 | A/goose/Guangdong/1/96 (H5N1) | AAD51929 |
| HK/1073/99 | A/Hong Kong/1073/99 (H9N2) | AAC32081 |
| HK/156/97 | A/Hong Kong/156/97 (H5N1) | AAC32091 |
| HK/482/97 | A/Hong Kong/482/97 (H5N1) | AAC32091 |
| HONGKONG97 | A/Hong Kong/485/97 (H5N1) | AJ278648 |
| MALL/NY78 | A/mallard/New York/6750/78 (H2N2) | M12699 |
| MALL/WISC75 | A/mallard/Wisconsin/169/75 (H5N3) | AF073202 |
| PR34 | A/Puerto Rico/8/34 (H1N1) | NC 002016 |
| QUAIL/ARK93 | A/quail/Arkansas/29209-1/93 (H9N2) | AF156470 |
| SINGAPORE57 | A/Singapore/1/57 (H2N2) | X08093 |
| SW/1937 | A/swine/29/37 (H1N1) | M63517 |
| SW/GERMANY91 | A/swine/Germany/8533/91 (H1N1) | Z26861 |
| SW/HONGKONG94 | A/swine/Hong Kong/273/94 (H1N1) | U49115 |
| SW/IOWA30 | A/swine/Iowa/15/30 (H1N1) | M63534 |
| SW/MARCH52 | A/swine/March/52 (H1N1) | M63518 |
| SW/NETH80 | A/swine/Netherlands/25/80 (H1N1) | Z26862 |
| SW/ONTARIO81 | A/swine/Ontario/2/81 (H1N1) | M63520 |
| SW/WISC61 | A/swine/Wisconsin/1/61 (H1N1) | M63519 |
| TEAL/HONGKONG97 | A/teal/Hong Kong/W312/97 (H6N1) | AF250482 |
| TY/NC88 | A/turkey/North Carolina/1780/88 (H1N1) | M55479 |
| USSR77 | A/USSR/90/77 (H1N1) | M54941 |
| WISC88 | A/Wisconsin/3523/88 (H3N2) | M63521 |
| WISC94 | A/Wisconsin/4755/94 (H3N2) | U53169 |
| WS33 | A/WS/33 (H1N1) | X08089 |
| WSN33 | A/WSN/33 (H1N1) | X08088 |
Nucleotide sequence accession number.
The matrix gene segment coding sequence of A/Brevig Mission/1/18 has been deposited in GenBank under accession no. AY130766.
RESULTS
Sequence of the matrix gene segment.
The entire matrix coding sequence (982 nucleotides) from the shared start codon at nucleotide 26 of the matrix segment to the stop codon of the M2 open reading frame (ORF) at nucleotide 1007 was determined from the frozen sample obtained from Brevig Mission, Alaska (A/Brevig Mission/1/18 [H1N1]), Brevig18. The 1918 sequence and a map of the two overlapping reading frames of an influenza A virus matrix segment are shown in Fig. 1. The theoretical translations of the two ORFs, M1 and M2, are indicated.
FIG. 1.
Sequence of the 1918 matrix segment. (A) Diagram of the matrix gene segment of an influenza A virus showing the positions of the two overlapping ORFs. Nucleotides 1 to 25 and 1008 to 1027 are noncoding. The M1 and M2 ORFs share the first nine codons. (B) Sequence of the A/Brevig Mission/1/18 (H1N1) matrix gene segment coding region (GenBank accession no. AY130766). The coding sequences start at nucleotide 26; M1 and M2 share the initial nine codons. The M1 theoretical translation is shown above the line; that of M2 is shown below the line. The M2 transmembrane is boxed. The M2 ectodomain consists of the first 24 M2 protein amino acids (see also Fig. 3). The numbering of the gene segment is aligned to A/USSR/90/77 (GenBank accession no. M54941) and refers to the sequence in the sense (mRNA) orientation.
M1 gene.
The 1918 matrix segment has an M1 ORF of 756 nucleotides, with a predicted 252-amino acid M1 protein. Several functional domains have been mapped to the M1 protein. The theoretical translation of the Brevig18 M1 sequence predicts that the strain contains the consensus RNA-binding and nuclear localization signal (RKLKR) at residues 101 to 105 (10, 41, 58). The Brevig18 M1 sequence also matches the consensus at residues 148 to 162 (CATCEQIADSQHRSH), where a putative zinc-binding motif has been identified (9, 49). Two residues, those at positions 115 and 137, may be of potential interest. They are V (position 115) and T (position 137) in the 1918 strain (and in all avian and swine strains) and I and A, respectively, in other human viruses. Neither of these residues is located in a known functional domain of the protein.
M2 gene.
The 1918 matrix segment has a spliced M2 ORF of 291 nucleotides, with a predicted 97-amino acid M2 protein. Like other influenza A viruses, the Brevig18 M2 protein has the potentially palmitoylated C at position 50 (45) and the potentially phosphorylated S at position 64 (22) within the cytoplasmic domain. Within the transmembrane domain, residue 28 is I in Brevig18 but V in most other human viruses.
Amantadine resistance region.
Five amino acid sites that have been identified in the transmembrane region of the M2 protein are involved in resistance to the antiviral drug amantadine: sites 26, 27, 30, 31, and 34 (5, 18, 21). The Brevig18 sequence is identical at these positions to that of the amantadine-sensitive strain Singapore57. Thus, it is predicted that the M2 protein of Brevig18 would be sensitive to amantadine.
Phylogenetic analyses.
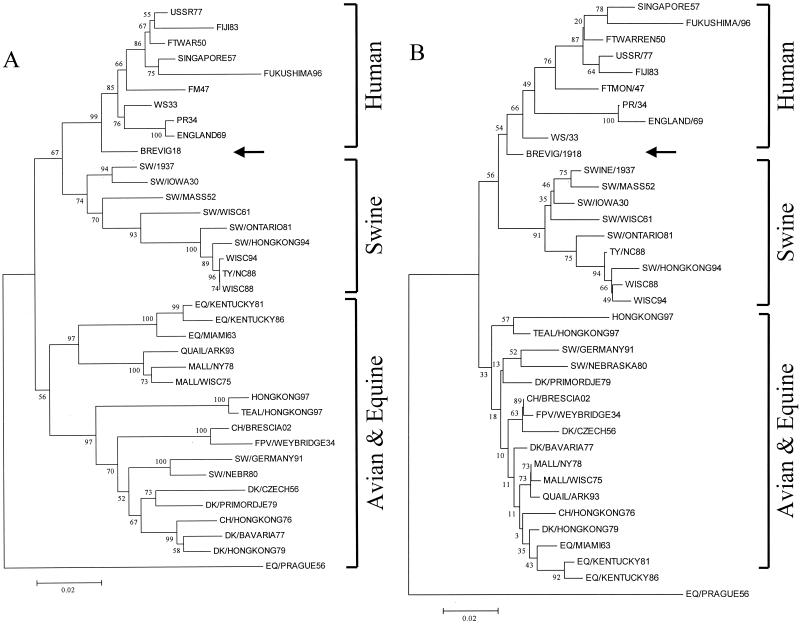
Thirty-seven M1 nucleotide sequences were analyzed by neighbor joining, with the proportion of differences as the p-distance. Four clades were differentiated: human, swine, Eurasian avian, and equine plus North American avian. Brevig18 was located within and near the root of the human clade (Fig. 2A). Synonymous and nonsynonymous changes were analyzed separately. Synonymous changes gave a result essentially identical to that shown in Fig. 2A. Little information was obtained by analyzing nonsynonymous changes, which produced a tree with virtually no horizontal depth for most branches. This was expected given the high degree of conservation of the M1 protein.
FIG. 2.
Phylogenetic trees of the Brevig18 M1 (A) and M2 (B) nucleotide sequences. Both trees are neighbor-joining trees determined by p-distance. The Brevig18 strain is indicated by an arrow. Bootstrap values are given for selected nodes, and distance bars are shown below the trees.
Thirty-seven M2 nucleotide sequences were analyzed by the neighbor-joining method as described above. A tree very similar to that seen with the M1 sequences was obtained (Fig. 2B), except that only three clades were differentiated: human, swine, and avian plus equine. Brevig18 was located within and near the root of the human clade. Analyzing synonymous positions alone gave a similar result. The analysis of nonsynonymous substitutions alone resulted in a tree in which Brevig18 and Sw/Iowa30 were placed within and near the root of the avian-equine clade. However, bootstrap values for this region of the tree were extremely low, suggesting little real support for this topology.
The phylogenetic analyses suggest that the Brevig18 matrix genes, while more avian-like than those of other mammalian influenza viruses, were mammalian adapted. For example, the extracellular domain of the M2 protein contains 4 amino acids that differ consistently between the avian and mammalian clades (M2 residues 14, 16, 18, and 20). Brevig18 matches the mammalian sequence at all 4 of these residues (Fig. 3).
FIG. 3.
Alignment of M2 extracellular domain protein sequences. The first 24 residues of M2 are shown. Amino acids matching Brevig18 are shown as dots. The first 6 residues of Dk/HongKong/79 are unknown and are represented by question marks.
DISCUSSION
Several model systems have indicated that small changes in the matrix proteins can have large effects on phenotype. Replacing the matrix segment of human viruses with that of PR34 consistently results in a higher yield of the strain for vaccine development (2). The PR34 M1 gene has been shown to confer high-growth characteristics in single-gene reassortant strains (55). The PR34 M1 protein, however, differs by fewer than 10 amino acids from modern human strains. Other studies indicate that the matrix segment of WSN33 (like PR34, a repeatedly passaged derivative of an early human strain) can also confer a high-yield phenotype (55). However, between them, the M1 proteins of WSN33 and PR34 share only a few amino acids that differ consistently from modern human strains and each has additional, unique differences. Therefore, it is not clear which amino acid changes are responsible for the high-yield phenotype or even if the same amino acids are responsible in both PR34 and WSN33. At those M1 sites where PR34 and WSN33 share amino acids found in few other strains, the Brevig18 sequence instead matches the human consensus. Only at M1 site 218 does Brevig18 share a characteristic with PR34 and WSN33 that distinguishes them from modern human strains. PR34, WSN33, and Brevig18 all retain the threonine that is found in avian strains, whereas all subsequent human strains have replaced the avian threonine with an alanine or valine.
While the PR34 and WSN33 matrix segments contribute to a high-growth phenotype when reassortant human viruses are grown on chicken eggs, other experiments suggest that minor changes in the matrix proteins also contribute to higher growth in mouse lung. Passage of the FM47 virus through the mouse lung resulted in a single amino acid change in M1, T139A, that was shown in single gene reassortment assays to be responsible for higher yield in mouse lung (42, 43). Adaptation of another human virus (HK68) to mouse resulted in high-yield variants with one amino acid change in M1 (T167A) and one in M2 (D44N) (6). Govorkova et al. adapted one avian (A/black duck/NJ/1580/78 [H2N3]) and one human (A/Japan × Bellamy/57 [H2N1]) virus to the mouse lung and again found that high-growth variants had only 1 or 2 amino acid changes in the matrix proteins (13). Changes making viruses more virulent in the mouse lung also appear to increase growth rates in embryonated chicken eggs and MDCK cells, suggesting that the changes may have functional significance beyond mouse adaptation (13, 56). None of these changes were found in Brevig18.
Several closely related H5N1 viruses were isolated from humans in 1997. When the complete genomes of strains with a high-pathogenicity phenotype were compared to those of low pathogenicity, a total of 5 amino acid differences were found to correlate with the high-pathogenicity phenotype. One change was in M1 (V15I), a change that has also been found in PR34 but not in Brevig18 (24).
The amino acid changes found to increase virulence in these studies are distributed over the M1 gene from amino acids 15 to 227 and often fall in areas of no known function. Almost all of the changes resulting in increased yield or virulence are not found in any other strain. Overall, the studies suggest that matrix sequence variation plays an important role in virulence but that virulence cannot be predicted directly from the matrix sequence. The 1918 matrix sequence does not code for any of the single amino acid changes that correlate with virulence in these studies.
While the PR34 matrix segment appears to confer a high-growth phenotype on a wide range of human viruses, other studies suggest that the phenotypic effect of the matrix segment depends on viral context. Murphy et al. showed that replacing the matrix segment of a human H3N2 virus (A/Udorn/307/72 [H3N2]) with that of A/mallard/NY/6750/78 (H2N2) resulted in severe growth restriction in the squirrel monkey respiratory tree, while replacement with the matrix segment of A/pintail/Alberta/119/79 (H4N6) caused no growth restriction (33). The two avian segments differed at only 3 amino acids, 2 in M1 and 1 in M2. Similarly, Subbarao et al. showed that the matrix segment of A/Ann Arbor/6/60 (H2N2) attenuated A/Korea/82 (H3N2) but not A/Udorn/72 (H3N2) (44).
The Brevig18 M1 and M2 sequences do not code for any of the unique amino acid changes that correlate with the high-yield phenotype in different model systems. The amino acids that are unique to the PR34 matrix segment, which may contribute to its ability to confer a high-yield phenotype on many human viruses, are not shared by Brevig18. There is not a single amino acid position in M1 or M2 that is unique to Brevig18, suggesting that if the matrix genes were involved specifically in viral lethality in 1918, a combination of substitutions may have played a role. There is, however, substantial evidence that small changes in matrix sequence, in a particular viral context, can result in a high-yield phenotype. It is possible that the unique Brevig18 matrix sequence would result in a high-yield phenotype in any human strain, but the possibility that it would contribute to lethality only in the context of the other 1918 viral proteins cannot be ruled out. It may well be that a particular constellation of amino acids in several proteins contributed to the virulence of the 1918 strain.
Influenza viruses must have at least a novel HA to cause a pandemic (26). However, they must also be readily transmissible between humans and capable of replicating in the human lung. In the pandemics of 1957 and 1968, all of the gene segments encoding internal proteins (with the exception of PB1) were retained from the strain previously circulating in humans (25). Determining whether the matrix segment was similarly retained in 1918 is complicated because the matrix proteins are very highly conserved. Only about 5% of M1 amino acids differ among the most divergent strains. While almost 20% of M2 amino acids vary, they tend to vary more between species than within them and do not appear to change progressively over time like the surface proteins. Phylogenetic analyses suggest that the matrix proteins are not under continuous positive selective pressure in humans; however, there are certain amino acid residues that distinguish the human clades from those of swine and birds. In M1, 3 residues (115, 121, and 137) distinguish the human clade from the 1930s to the present. Brevig18 matches the human consensus at one of the three, retaining the amino acid characteristic of avian strains at the other two. By contrast, in M2, 7 amino acids are found in all human adapted strains; Brevig18 matches the human consensus at 5 of them (Fig. 3). Of interest is the fact that 4 of these 5 amino acids are coded for by the short stretch of nucleotides that are used in overlapping reading frames in both M1 and M2. Since the corresponding amino acids in M1 are extremely conserved, change in M2 in this area is highly constrained. If Brevig18 was acquired directly from an avian source, these four concerted changes had to have been made very quickly. While this observation might suggest that the 1918 matrix segment was retained from the previously circulating human strain, overall phylogenetic analyses suggest an intermediate position for Brevig18 between avian and subsequent mammalian strains, similar to the phylogenetic position of previously analyzed 1918 gene segments. It will probably not be possible to determine whether or not the 1918 matrix segment shifted immediately before the pandemic without samples of the influenza virus strain circulating in humans before 1918.
Acknowledgments
This work was supported by grants from the National Institute of Allergy and Infectious Diseases, the American Registry of Pathology, and the Department of Veterans Affairs and the intramural funds of the Armed Forces Institute of Pathology.
The opinions contained herein are the private views of the authors and are not to be construed as official or as reflecting the views of the Department of the Army or the Department of Defense.
REFERENCES
- 1.Ali, A., R. T. Avalos, E. Ponimaskin, and D. P. Nayak. 2000. Influenza virus assembly: effect of influenza virus glycoproteins on the membrane association of M1 protein. J. Virol. 74:8709-8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baez, M., P. Palese, and E. D. Kilbourne. 1980. Gene composition of high-yielding influenza vaccine strains obtained by recombination. J. Infect. Dis. 141:362-365. [DOI] [PubMed] [Google Scholar]
- 3.Basler, C. F., A. H. Reid, J. K. Dybing, T. A. Janczewski, T. G. Fanning, H. Zheng, M. Salvatore, M. L. Perdue, D. E. Swayne, A. Garcia-Sastre, P. Palese, and J. K. Taubenberger. 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl. Acad. Sci. USA 98:2746-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, C. M., L. H. Pinto, T. A. Cross, and R. A. Lamb. 1999. The influenza virus M2 ion channel protein: probing the structure of the transmembrane domain in intact cells by using engineered disulfide cross-linking. Virology 254:196-209. [DOI] [PubMed] [Google Scholar]
- 5.Belshe, R., M. Smith, C. Hall, R. Betts, and A. Hay. 1988. Genetic basis of resistance to rimantadine emerging during treatment of influenza virus infection. J. Virol. 62:1508-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, E., H. Liu, L. Kit, S. Baird, and M. Nesrallah. 2001. Pattern of mutation in the genome of influenza A virus on adaptation to increased virulence in the mouse lung: identification of functional themes. Proc. Natl. Acad. Sci. USA 98:6883-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bucher, D. J., I. G. Kharitonenkov, J. A. Zakomirdin, V. B. Grigoriev, S. M. Klimenko, and J. F. Davis. 1980. Incorporation of influenza virus M-protein into liposomes. J. Virol. 36:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bui, M., E. G. Wills, A. Helenius, and G. R. Whittaker. 2000. Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. J. Virol. 74:1781-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elster, C., E. Fourest, F. Baudin, K. Larsen, S. Cusack, and R. W. Ruigrok. 1994. A small percentage of influenza virus M1 protein contains zinc but zinc does not influence in vitro M1-RNA interaction. J. Gen. Virol. 75:37-42. [DOI] [PubMed] [Google Scholar]
- 10.Elster, C., K. Larsen, J. Gagnon, R. W. Ruigrok, and F. Baudin. 1997. Influenza virus M1 protein binds to RNA through its nuclear localization signal. J. Gen. Virol. 78:1589-1596. [DOI] [PubMed] [Google Scholar]
- 11.Enami, M., and K. Enami. 1996. Influenza virus hemagglutinin and neuraminidase glycoproteins stimulate the membrane association of the matrix protein. J. Virol. 70:6653-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez-Puertas, P., C. Albo, E. Perez-Pastrana, A. Vivo, and A. Portela. 2000. Influenza virus matrix protein is the major driving force in virus budding. J. Virol. 74:11538-11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Govorkova, E. A., A. S. Gambaryan, E. C. Claas, and Y. A. Smirnov. 2000. Amino acid changes in the hemagglutinin and matrix proteins of influenza A (H2) viruses adapted to mice. Acta Virol. 44:241-248. [PubMed] [Google Scholar]
- 14.Grambas, S., and A. J. Hay. 1992. Maturation of influenza A virus hemagglutinin-estimates of the pH encountered during transport and its regulation by the M2 protein. Virology 190:11-18. [DOI] [PubMed] [Google Scholar]
- 15.Gregoriades, A., and B. Frangione. 1981. Insertion of influenza M protein into the viral lipid bilayer and localization of site of insertion. J. Virol. 40:323-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harris, A., F. Forouhar, S. Qiu, B. Sha, and M. Luo. 2001. The crystal structure of the influenza matrix protein M1 at neutral pH: M1-M1 protein interfaces can rotate in the oligomeric structures of M1. Virology 289:34-44. [DOI] [PubMed] [Google Scholar]
- 17.Hay, A. J., N. C. Kennedy, J. J. Skehel, and G. Appleyard. 1979. The matrix protein gene determines amantadine-sensitivity of influenza viruses. J. Gen. Virol. 42:189-191. [DOI] [PubMed] [Google Scholar]
- 18.Hay, A. J., A. J. Wolstenholme, J. J. Skehel, and M. H. Smith. 1985. The molecular basis of the specific anti-influenza action of amantadine. EMBO J. 4:3021-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helenius, A. 1992. Unpacking the incoming influenza virus. Cell 69:577-578. [DOI] [PubMed] [Google Scholar]
- 20.Holsinger, L. J., and R. A. Lamb. 1991. Influenza virus M2 integral membrane protein is a homotetramer stabilized by formation of disulfide bonds. Virology 183:32-43. [DOI] [PubMed] [Google Scholar]
- 21.Holsinger, L. J., D. Nichani, L. H. Pinto, and R. A. Lamb. 1994. Influenza A virus M2 ion channel protein: a structure-function analysis. J. Virol. 68:1551-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holsinger, L. J., M. A. Shaughnessy, A. Micko, L. H. Pinto, and R. A. Lamb. 1995. Analysis of the posttranslational modifications of the influenza virus M2 protein. J. Virol. 69:1219-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, N. P., and J. Mueller. 2002. Updating the accounts: global mortality of the 1918-1920 “Spanish” influenza pandemic. Bull. Hist. Med. 76:105-115. [DOI] [PubMed] [Google Scholar]
- 24.Katz, J., X. Lu, T. Tumpey, C. Smith, M. Shaw, and K. Subbarao. 2000. Molecular correlates of influenza A H5N1 virus pathogenesis in mice. J. Virol. 74:10807-10810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawaoka, Y., S. Krauss, and R. G. Webster. 1989. Avian-to-human transmission of the PB1 gene of influenza A viruses in the 1957 and 1968 pandemics. J. Virol. 63:4603-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kilbourne, E. D. 1977. Influenza pandemics in perspective. JAMA 237:1225-1228. [PubMed] [Google Scholar]
- 27.Krafft, A. E., B. W. Duncan, K. E. Bijwaard, J. K. Taubenberger, and J. H. Lichy. 1997. Optimization of the isolation and amplification of RNA from formalin-fixed, paraffin-embedded tissue: the Armed Forces Institute of Pathology Experience and Literature Review. Mol. Diagn. 2:217-230. [DOI] [PubMed] [Google Scholar]
- 28.Lamb, R. A., and R. M. Krug. 2001. Orthomyxoviridae: the viruses and their replication, p. 1487-1531. In D. A. Knipe, P. M. Howley, et al. (ed.), Fields virology, 4th ed., vol. 1. Lippincott Williams and Wilkins, Philadelphia, Pa.
- 29.Lamb, R. A., C. J. Lai, and P. W. Choppin. 1981. Sequences of mRNAs derived from genome RNA segment 7 of influenza virus: colinear and interrupted mRNAs code for overlapping proteins. Proc. Natl. Acad. Sci. USA 78:4170-4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamb, R. A., S. L. Zebedee, and C. D. Richardson. 1985. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 40:627-633. [DOI] [PubMed] [Google Scholar]
- 31.Latham, T., and J. M. Galarza. 2001. Formation of wild-type and chimeric influenza virus-like particles following simultaneous expression of only four structural proteins. J. Virol. 75:6154-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martin, K., and A. Helenius. 1991. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67:117-130. [DOI] [PubMed] [Google Scholar]
- 33.Murphy, B., A. Buckler-White, W. London, and M. Snyder. 1989. Characterization of the M protein and nucleoprotein genes of an avian influenza A virus which are involved in host range restriction in monkeys. Vaccine 7:557-561. [DOI] [PubMed] [Google Scholar]
- 34.Murti, K. G., P. S. Brown, W. J. Bean, Jr., and R. G. Webster. 1992. Composition of the helical internal components of influenza virus as revealed by immunogold labeling/electron microscopy. Virology 186:294-299. [DOI] [PubMed] [Google Scholar]
- 35.Patterson, K. D., and G. F. Pyle. 1991. The geography and mortality of the 1918 influenza pandemic. Bull. Hist. Med. 65:4-21. [PubMed] [Google Scholar]
- 36.Perez, D. R., and R. O. Donis. 1998. The matrix 1 protein of influenza A virus inhibits the transcriptase activity of a model influenza reporter genome in vivo. Virology 249:52-61. [DOI] [PubMed] [Google Scholar]
- 37.Pinto, L. H., L. J. Holsinger, and R. A. Lamb. 1992. Influenza virus M2 protein has ion channel activity. Cell 69:517-528. [DOI] [PubMed] [Google Scholar]
- 38.Rees, P. J., and N. J. Dimmock. 1981. Electrophoretic separation of influenza virus ribonucleoproteins. J. Gen. Virol. 53:125-132. [DOI] [PubMed] [Google Scholar]
- 39.Reid, A. H., T. G. Fanning, J. V. Hultin, and J. K. Taubenberger. 1999. Origin and evolution of the 1918 “Spanish” influenza virus hemagglutinin gene. Proc. Natl. Acad. Sci. USA 96:1651-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reid, A. H., T. G. Fanning, T. A. Janczewski, and J. K. Taubenberger. 2000. Characterization of the 1918 “Spanish” influenza virus neuraminidase gene. Proc. Natl. Acad. Sci. USA 97:6785-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sha, B., and M. Luo. 1997. Structure of a bifunctional membrane-RNA binding protein, influenza virus matrix protein M1. Nat. Struct. Biol. 4:239-244. [DOI] [PubMed] [Google Scholar]
- 42.Smeenk, C. A., and E. G. Brown. 1994. The influenza virus variant A/FM/1/47-MA possesses single amino acid replacements in the hemagglutinin, controlling virulence, and in the matrix protein, controlling virulence as well as growth. J. Virol. 68:530-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smeenk, C. A., K. E. Wright, B. F. Burns, A. J. Thaker, and E. G. Brown. 1996. Mutations in the hemagglutinin and matrix genes of a virulent influenza virus variant, A/FM/1/47-MA, control different stages in pathogenesis. Virus Res. 44:79-95. [DOI] [PubMed] [Google Scholar]
- 44.Subbarao, E., M. Perkins, J. Treanor, and B. Murphy. 1992. The attenuation phenotype conferred by the M gene of the influenza A/Ann Arbor/6/60 cold-adapted virus (H2N2) on the A/Korea/82 (H3N2) reassortant virus results from a gene constellation effect. Virus Res. 25:37-50. [DOI] [PubMed] [Google Scholar]
- 45.Sugrue, R. J., R. B. Belshe, and A. J. Hay. 1990. Palmitoylation of the influenza A virus M2 protein. Virology 179:51-56. [DOI] [PubMed] [Google Scholar]
- 46.Sugrue, R. J., and A. J. Hay. 1991. Structural characteristics of the M2 protein of influenza A viruses: evidence that it forms a tetrameric channel. Virology 180:617-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takeuchi, K., and R. A. Lamb. 1994. Influenza virus M2 protein ion channel activity stabilizes the native form of fowl plague virus hemagglutinin during intracellular transport. J. Virol. 68:911-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taubenberger, J. K., A. H. Reid, A. E. Krafft, K. E. Bijwaard, and T. G. Fanning. 1997. Initial genetic characterization of the 1918 “Spanish” influenza virus. Science 275:1793-1796. [DOI] [PubMed] [Google Scholar]
- 49.Wakefield, L., and G. Brownlee. 1989. RNA-binding properties of influenza A virus matrix protein M1. Nucleic Acids Res. 17:8569-8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, C., R. A. Lamb, and L. H. Pinto. 1994. Direct measurement of the influenza A virus M2 protein ion channel activity in mammalian cells. Virology 205:133-140. [DOI] [PubMed] [Google Scholar]
- 51.Ward, A. C. 1997. Virulence of influenza A virus for mouse lung. Virus Genes 14:187-194. [DOI] [PubMed] [Google Scholar]
- 52.Winter, G., and S. Fields. 1980. Cloning of influenza cDNA into M13: the sequence of the RNA segment encoding the A/PR/8/34 matrix protein. Nucleic Acids Res. 8:1965-1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winternitz, M. C., I. M. Wason, and F. P. McNamara. 1920. The pathology of influenza. Yale University Press, New Haven, Conn.
- 54.Wolbach, S. B. 1919. Comments on the pathology and bacteriology of fatal influenza cases, as observed at Camp Devens, Mass. Johns Hopkins Hosp. Bull. 30:104. [Google Scholar]
- 55.Yasuda, J., D. J. Bucher, and A. Ishihama. 1994. Growth control of influenza A virus by M1 protein: analysis of transfectant viruses carrying the chimeric M gene. J. Virol. 68:8141-8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yasuda, J., T. Toyoda, M. Nakayama, and A. Ishihama. 1993. Regulatory effects of matrix protein variations on influenza virus growth. Arch. Virol. 133:283-294. [DOI] [PubMed] [Google Scholar]
- 57.Ye, Z., T. Liu, D. P. Offringa, J. McInnis, and R. A. Levandowski. 1999. Association of influenza virus matrix protein with ribonucleoproteins. J. Virol. 73:7467-7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ye, Z., D. Robinson, and R. Wagner. 1995. Nucleus-targeting domain of the matrix protein (M1) of influenza virus. J. Virol. 69:1964-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ye, Z. P., N. W. Baylor, and R. R. Wagner. 1989. Transcription-inhibition and RNA-binding domains of influenza A virus matrix protein mapped with anti-idiotypic antibodies and synthetic peptides. J. Virol. 63:3586-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zvonarjev, A. Y., and Y. Z. Ghendon. 1980. Influence of membrane (M) protein on influenza A virus virion transcriptase activity in vitro and its susceptibility to rimantadine. J. Virol. 33:583-586. [DOI] [PMC free article] [PubMed] [Google Scholar]