Abstract
The Mason-Pfizer monkey virus (M-PMV) Gag protein possesses the ability to assemble into an immature capsid when synthesized in a reticulocyte lysate translation system. In contrast, the human immunodeficiency virus (HIV) Gag protein is incapable of assembly in parallel assays. To enable the assembly of HIV Gag, we have combined or inserted regions of M-PMV Gag into HIV Gag. By both biochemical and morphological criteria, several of these chimeric Gag molecules are capable of assembly into immature capsid-like structures in this in vitro system. Chimeric species containing large regions of M-PMV Gag fused to HIV Gag sequences failed to assemble, while species consisting of only the M-PMV p12 region, and its internal scaffold domain (ISD), fused to HIV Gag were capable of assembly, albeit at reduced kinetics compared to M-PMV Gag. The ability of the ISD to induce assembly of HIV Gag, which normally assembles at the plasma membrane, suggests a common requirement for a concentrating factor in retrovirus assembly. Despite the dramatic effect of the ISD on chimera assembly, the function of HIV Gag domains in that process was found to remain essential, since an assembly-defective mutant of HIV CA, M185A, abolished assembly when introduced into the chimera. This continued requirement for HIV Gag domain function in the assembly of chimeric molecules will allow this in vitro system to be used for the analysis of potential inhibitors of HIV immature particle assembly.
For most retroviruses, including human immunodeficiency virus (HIV), assembly of the immature particle occurs in association with the plasma membrane. However, members of the Betaretrovirus genus (24) preassemble their immature capsids within the cytoplasm. These fully formed immature particles subsequently migrate to the plasma membrane, where envelopment and budding occur (reviewed in references 37 and 43). We have established an in vitro synthesis and assembly system for one member of this genus, Mason-Pfizer monkey virus (M-PMV), and have used that system to more definitively characterize the assembly versus transport functions of the p12 and matrix (MA) domains, respectively, of M-PMV Gag (38, 39). This analysis, together with data from studies in cells (41), established the existence within p12 of an internal scaffold domain (ISD), which functions to increase the efficiency with which M-PMV Gag can assemble into immature particles. A similar synthesis and assembly system, using a wheat germ extract, has been developed for HIV Gag, although this latter system requires the addition of membranes for detectable assembly of HIV immature capsids (31). More-defined in vitro systems using purified Gag or fragments of Gag proteins produced in bacteria have also been developed, and the production of spherical particles, analogous to the immature capsid, and tubular particles, analogous to the mature core, have been reported. Both the composition of the Gag protein or fragment and the specific conditions employed can influence shape determination during assembly in these systems (5-8, 14, 15, 19, 22, 23, 26, 27, 45, 47).
A number of investigators have analyzed Gag domain function in assembly by the creation of chimeric Gag proteins, and they have shown that several domains of Gag can be substituted by the equivalent domains from another retrovirus without blocking particle release (2, 9, 11-13, 35). In one study, chimeric gag genes were constructed by recombining the sequences for the major domains of HIV and murine leukemia virus (MuLV) according to the retroviral protease cleavage sites (12). While the Gag molecules resulting from these precise substitutions could be found assembled into released particles, this could occur only in the context of rescue by a wild-type Gag. In contrast, two studies that examined less-precise substitutions of domains were successful in producing assembly-competent chimeric Gag proteins. In the first, the MuLV NC zinc finger region internal to the protease cleavage sites was substituted into the equivalent region of Rous sarcoma virus (RSV) Gag (13). Not only could this molecule assemble into particles, but it also preferentially packaged RNA containing the MuLV Ψ sequence. In the second, the carboxyl-terminal approximate half of HIV Gag was fused to the amino-terminal approximate half of RSV Gag (2). This RSV/HIV chimera not only efficiently produced particles but it did so with similar kinetics to the parental RSV Gag. These studies, as well as others employing mutagenesis and deletion analysis, have highlighted the remarkable plasticity of the retroviral Gag protein. Indeed, more recent studies have demonstrated the acceptance of heterologous protein interaction domains as substitutes for the functionally similar Gag I domain (1, 29, 49).
To provide a simplified assembly assay in the context of a mammalian cell extract, we constructed a panel of chimeric HIV/M-PMV gag genes in order to determine whether fusion of various regions of M-PMV Gag, including the p12 domain, with regions of HIV Gag would endow the chimeric Gag proteins with the ability to assemble efficiently in the in vitro translation-assembly system. Not only were assembly-competent chimeric molecules produced but also the participation of HIV CA function, within a chimeric molecule, in assembly was established. Furthermore, by using a small-molecule inhibitor of virus assembly, we have demonstrated the utility of the chimeric Gag gene and the assembly assay for the analysis of potential inhibitors of immature HIV capsid assembly.
MATERIALS AND METHODS
DNA constructs.
All constructs are derivatives of two previously described plasmids, pTFCG.M100A, which expresses M-PMV gag, pro, and pol (38), and pDAB72, which expresses HIV-1 strain BH10 gag plus a truncated pol consisting of a portion of the proteinase (obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases; 16). Expression from pDAB72 produces HIV Gag plus a truncated Gag-Pro. The proteinase domain is not functional. Plasmid pTFCG.M100A was modified by deletion of a BamHI-to-BamHI fragment containing most of pol and a portion of pro to create pTFCG.Bam(−). This modified plasmid produces only M-PMV Gag and a truncated Gag-Pro and thus provides a more appropriate control for the pDAB72 HIV construct than the original pTFCG.M100A.
In addition, each of these plasmids was modified to remove the frameshift signal to ensure that a single Gag translation product was produced. The first of these derivatives, pDAB72.FS(−), is described elsewhere (48). The second, pTFCG.FS(−), was constructed by site-directed mutagenesis of the ribosomal frameshift sequence to introduce silent mutations into the gag reading frame. Using appropriate primers and pTFCG.Bam(−) as the template, the M-PMV gag-pro frameshift signal sequence CAG GGA AAC GGG was altered to CAA GGC AAT GGG (25). The resulting PCR product was cleaved by NcoI and BsgI and ligated into pTFCG.Bam(−), which had been similarly digested with the same enzymes. A fortuitous result of the silent mutations was the creation of an additional BsrDI restriction site that was used for screening of clones. Loss of the production of frameshift products from this construct was confirmed by in vitro synthesis. Selected clones were further confirmed by sequencing.
All chimeric gag genes described in this report (Fig. 1) were constructed by PCR synthesis of gag gene fragments followed by subcloning into one of the four T7 expression vectors, pDAB72, pTFCG.Bam(−), pDAB72.FS(−), or pTFCG.FS(−). The sequences of primers and the specifics of the cloning are available upon request. All constructs were initially screened by restriction analysis, secondarily screened for the synthesis of a chimeric Gag protein of appropriate size, and then finally confirmed by commercial sequencing (Lonestar Labs). The names of these plasmids are as follows: pTFCGChA, pTFCGChB, pTFCGChC, pTFCGChD, pDABCh1.M120A, pDABCh2, pDABCh3, pDABCh3a, pDABCh4, pDABCh5, pDABCh6, pDABCh7, pDABΔp1Δp6, pDABΔp6, and pDAB(−)MA. Plasmid pDABCh1.M120A is so named to reflect the introduction of a methionine-to-alanine substitution within the M-PMV MA coding region in order to prevent efficient internal initiation of translation. The introduction of this substitution into the native M-PMV Gag has no deleterious effect on capsid assembly or virus infectivity (38).
FIG. 1.
Schematic representations of HIV, M-PMV, and chimeric Gag proteins, as well as truncated Gag species used for controls. Each precursor protein is labeled on the left. Within each schematic, sequences derived from HIV are shown in white while sequences derived from M-PMV are shown in black. As indicated in the box at bottom right, the HIV and M-PMV MHR sequences are depicted as cross-marked and striped regions, respectively. Within each schematic, the domains corresponding to mature Gag proteins and peptides are labeled. For HIV, these are p17MA, p24CA, p2, p7NC, p1, and p6. For M-PMV, these are p10MA, pp24, p12, p27CA, p14NC, and p4. The fusion points between amino acid sequences derived from each virus are at the ends of the domains as shown or are indicated above the fusion point by the domain designation and the amino acid number within that domain. For example, CA:185 indicates that the fusion is to residue number 185 of the CA domain.
The specifics of construction for one chimera and its corresponding control are of note. The construction of chimera 3 (Fig. 1) presented a challenge since the point of fusion for M-PMV p12 with p1 of HIV Gag falls within the HIV gag-pol frameshift region. Similarly, the point of deletion for Δp6 also falls within the frameshift region (25). The secondary structure within this region consists of a stem loop that, although it performs its function in the context of mRNA, might also form in the DNA. This would be particularly likely if the sequence, as in this case for the purpose of making the chimera, were part of a single-stranded DNA oligo. Thus, after numerous attempts to perform PCR from a primer in this region, we were forced to modify the DNA sequence to reduce the likelihood of stem-loop formation. The stem-loop sequence downstream of the HIV gag-pol frameshift sequence is TC TGG CCT TCC TAC AAG GGA AGG CCA GG. By the incorporation of silent mutations it was modified to TA TGG CCC TCA TAC AAG GGG AGA CCC GG. These changes removed 6 of the potential 12 bp in the stem (25). In addition, even after modification of the stem sequence, we found it necessary to use Taq polymerase with this primer rather than Pfu Turbo (Stratagene), which failed to produce any product. The final construct was tested for the in vitro synthesis of a protein of the expected size and then was confirmed by commercial sequencing as for the other chimeric constructs.
The introduction of the assembly-defective HIV-1 mutant CA (M185A) into chimera 4 Gag was accomplished by subcloning the mutation from pET11a-CA-M185A, generously provided by Wesley Sundquist (University of Utah, Salt Lake City), into pDABCh4. The region of CA containing the mutation was excised from pET11a-CA-M185A by digestion with PpuMI and SpeI. The resulting fragment was then ligated into the similarly digested pDABCh4 to create pDABCh4.M185A. The presence of the mutation was confirmed by commercial sequencing.
In vitro analysis of immature capsid assembly.
Transcription and translation of wild-type and chimeric Gag protein were performed simultaneously with the TNT-coupled reticulocyte lysate system (Promega) in the presence of [35S]methionine. Products of these synthesis reactions were analyzed on sucrose gradients. Reaction mixtures were diluted to a total volume of 200 μl with 30% (wt/wt) sucrose in gradient buffer containing 20 mM Tris (pH 8.0), 100 mM NaCl, 1 mM EDTA, and 0.1% Triton X-100. Diluted samples were then loaded onto 2.2-ml continuous 30-to-55% (wt/wt) sucrose gradients in gradient buffer. Gradients were centrifuged in a TLS-55 rotor (Beckman Instruments) for 2 h at 55,000 rpm. Approximately 200-μl fractions were taken by hand with a Pipetman (Gilson) from the top of the gradient. The pellet was resuspended in 200 μl of 55% (wt/wt) sucrose in gradient buffer. Aliquots (5 μl) of each fraction were dissolved in sodium dodecyl sulfate (SDS) sample buffer and then loaded onto an SDS-10% polyacrylamide gel. After electrophoresis, radioactive bands were visualized and/or quantified by analysis on a Storm 840 PhosphorImager and ImageQuant software (Molecular Dynamics).
Inhibition assay.
The inhibition assay was performed with 1,1′-bi(4-anilinonaphthaline)-5-sulfonic acid (bis-ANS) and chimera 4, essentially as shown previously with anti-Gag antibodies and M-PMV Gag (39). Briefly, the in vitro synthesis reaction was initiated at 30°C and allowed to continue for 30 min. At that time, aliquots of the reaction mixture were removed and added to prewarmed tubes containing cycloheximide or cycloheximide plus bis-ANS. The final concentrations of cycloheximide and bis-ANS, where present, were 10 μg/ml and 1 mM, respectively.
Electron microscopy.
For analysis of in vitro synthesis products by electron microscopy, reactions were diluted to a total of 1 ml in gradient buffer without detergent and centrifuged at 70,000 rpm for 1 h in a TLA-100.3 rotor with microcentrifuge inserts (Beckman Instruments). The resulting pellets were then fixed overnight in 1% glutaraldehyde in phosphate-buffered saline, pH 7.0, at 4°C. After a rinse in phosphate-buffered saline, pellets were postfixed in 1% buffered osmium tetroxide for 1 h. These pellets were rinsed once more and then dehydrated in a graded series of ethanol solutions beginning with 50% and ending with 100%. After dehydration, pellets were rinsed three times with propylene oxide and then embedded in Polybed. Ultrathin sections were acquired by using a Rechert-Jung Ultra Cut E ultramicrotome. After staining with uranyl acetate and lead citrate, sections were examined and photographed by using a Hitachi-7000 transmission electron microscope.
RESULTS
Construction of initial chimeric gag genes.
We have constructed a panel of chimeric HIV/M-PMV gag genes with the goal of establishing whether regions from M-PMV Gag could confer upon the HIV Gag protein the ability to assemble capsids in an in vitro translation-assembly system. Previously, we demonstrated by both sucrose gradient and thin-section electron microscopy analyses that in this reticulocyte lysate-based translation system, HIV Gag does not assemble into immature capsid-like structures (39).
Initial experiments utilized a series of gross fusions of large portions of each gag sequence (Fig. 1, chimeras [Ch] A to D). The first of these, chimera A, was designed to recreate with M-PMV sequences the assembly-competent Gag fusion protein of Bennett et al. (RHB) that was constructed from RSV sequences (2). The resulting chimera A precursor contains two major homology regions (MHRs). The second CA fusion, chimera B, was designed to remove the M-PMV MHR. Chimera B, as well as chimeras C, D, 1, and 2, were constructed by taking into account the boundaries of the known tertiary elements as described in the high-resolution structures of HIV (32) and M-PMV MA (10) and of HIV CA (18, 21, 33). Chimeras C and D essentially are fusions of the N-terminal half of M-PMV Gag, including the p12 domain, with the C-terminal half of HIV Gag, but they were designed to maintain the N-terminal cleavage site before CA in M-PMV and HIV, respectively. The deletion of some C-terminal sequences of p12 were thought unlikely to affect this domain's ability to modulate the efficiency of assembly, since the entire C-terminal third of this domain, amino acids 54 to 83, may be deleted without any apparent effect upon M-PMV Gag assembly both in vivo (41) and in vitro (38). It should be pointed out that chimeras A to D each contain two proline-rich late-budding domains, PPPY in pp24 (46) and PTAP in p6 (20). It is unlikely that this duplication would affect assembly, since similar duplications constructed with RSV and HIV sequences efficiently released particles from cells (34).
Chimeras 1 and 2 (Fig. 1) replace M-PMV MA sequences with those of HIV MA. Chimera 1 joins a truncated HIV MA to the region containing the M-PMV MA-pp24 protease cleavage site. Replacement of the C terminus of HIV MA with M-PMV sequences was thought unlikely to have an effect upon assembly, since there has been no description of mutations within this region alone which affect assembly (reviewed in reference 28). Chimera 2 joins the region containing the HIV MA-CA cleavage site to the region containing the M-PMV pp24-p24 cleavage site, thus deleting the majority of pp24.
Construction of p12 insertion chimeras.
During the construction and analysis of the above panel of chimeric gag genes, we firmly established that the region within M-PMV Gag responsible for its efficient assembly lies within the p12 domain. This property of p12 is particularly evident in the in vitro assay, where its function is required for assembly (38, 41). The critical region within p12 has been thus termed the internal scaffold domain (ISD) since it appears to function in a manner analogous to the scaffolding proteins of phage and other viruses (38).
Thus, we designed a second panel of chimeric proteins that consist of either the entire, or the major portion of, HIV Gag with the M-PMV p12 domain added or inserted. Chimeras 3 through 7 (Fig. 1) were constructed to place the entire p12 domain, and thus also the ISD, at either the C terminus of the molecule (chimeras 3, 3a, and 4) or the N terminus of the molecule (chimeras 5 and 6), or to insert it in a position in HIV Gag analogous to its position in M-PMV Gag between MA and CA (chimera 7). Chimeras 4, 5, and 7 contain the complete sequence of HIV Gag. The remaining chimeras contain either N- or C-terminal truncations of the HIV Gag sequence (Fig. 1). For each of the chimeras 3 through 7, the points of fusion are to the N- and C-terminal residues corresponding to the cleavage sites of the respective viral protease. In addition, several Gag deletion species, Δp6, Δp1Δp6, and (−)MA, were created to serve as controls for chimeras 3, 3a, and (−)MA, respectively (Fig. 1). These were constructed to test the formal possibility that the results we observed could be due to the deletion of HIV sequences within these chimeras rather than to the addition of p12.
Assembly of ISD-enabled HIV Gag species proceeds with delayed kinetics.
Analysis of the above chimeras was performed in the in vitro synthesis and assembly system with a 2-h incubation followed by gradient sedimentation, essentially as described previously for M-PMV Gag (38, 39). Under these conditions, only chimeras 1 and 2, in addition to M-PMV Gag, produced particulate material that sedimented into sucrose gradients similar to that produced by M-PMV Gag (data not shown). All the remaining chimeras, as well as HIV, which had previously been analyzed in this system (39), failed to produce sedimenting material indicative of immature capsid assembly, with the majority of the Gag protein remaining at or near the top of the gradient.
While chimeras 1 and 2 could assemble in our assay, both of these consisted largely of M-PMV sequences with essentially only the matrix domain of HIV substituted. Although HIV MA has been shown to assist in the interaction of HIV Gag with itself in the yeast two-hybrid system, the region most responsible for this association lies within sequences in NC and the C-terminal region of CA (4, 48). Moreover, additional experiments have shown that HIV MA is not needed for virus assembly or even infectivity (36).
Although we have never observed the production of HIV immature capsids in an in vitro translation-assembly system, others have reported such structures in both reticulocyte lysate and wheat germ extract systems (31, 40, 42). We therefore investigated whether modifications to the assay might facilitate HIV Gag assembly. In one series of experiments, the temperature of incubation was varied between 25 and 37°C. In another, additional divalent cations (Mg2+) were added to the reaction. Neither of these modifications to the assay resulted in more extensive chimeric Gag assembly (data not shown). We then attempted a third series of experiments where we simply allowed the incubation to extend over a period of 24 h at the standard temperature of 30°C. Previous analysis of M-PMV and HIV Gag had shown that both proteins are stable over this period (M. Sakalian, unpublished data).
Analysis of each chimera in a 24-h time course revealed that several chimeric Gag proteins could produce sedimenting material indicative of assembled Gag if the reactions were allowed to incubate for an extended period of at least 9 h (Fig. 2 and 3; Table 1). HIV Gag and other chimeric Gag molecules that failed to produce this material by 9 h remained negative even after further incubation (Fig. 2). Chimeras 1 to 3, 3a, 4, and 7 all yielded evidence of assembly in this assay. The remainder, chimeras A to D, 5, and 6, and also HIV, failed to produce a peak of material in fractions 8 through 11 (Fig. 3), similar to that observed for M-PMV Gag, although in some a small amount of the translated Gag sedimented into the gradient or to the bottom of the tube, especially by 24 h (Fig. 2). None of the control deleted forms of HIV-1 Gag, Δp6, Δp1Δp6, and (−)MA were able to assemble in this in vitro assay. Thus, the addition of the M-PMV ISD to HIV Gag appeared to enable the assembly of several chimeric molecules in the in vitro system, but with delayed kinetics compared to M-PMV.
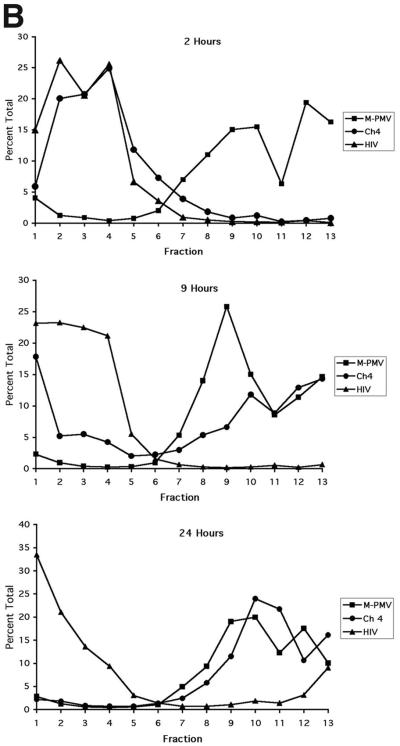
FIG. 2.
Example of time course of assembly by sucrose gradient analysis of chimera 4 versus M-PMV and HIV. (A) Aliquots of gradient fractions were electrophoresed on an SDS-10% polyacrylamide gel. Columns of panels are labeled at the top with the gag gene used to program the translation reaction. Rows of panels are of reactions allowed to incubate for 2, 9, and 24 h, as indicated at the left. Lane numbers indicate gradient fractions, beginning with the top fraction at no. 1. Lanes L contain an equivalent aliquot of the translation reactions before gradient fractionation. Lanes P contain material remaining as a pellet in the tube after removal of the gradient. Lanes 8 to 11 contain material indicative of immature capsid structures. The numbers to the left indicate the approximate positions of prestained molecular size standards in kilodaltons. Two major Gag species are visible for M-PMV Gag, Pr78gag and a slightly larger, truncated Gag-Pro protein that results from a frameshift. For both chimera 4 and HIV, only one major Gag species, Pr65.3 and Pr55, respectively, is produced due to the presence in these clones of a mutation to remove the frameshift. For all species, numerous smaller visible bands are presumed to be derived from internal initiations and/or abortive translations. (B) Quantitative representation of gel data. Radioactivity of Gag protein for each gradient fraction was quantified by phosphorimager analysis and is represented as a percentage of the total in lanes 1 to 12, plus P. Fraction 13 on the graphs corresponds to lane P on the gels.
FIG. 3.
Comparison of assembly by sucrose analysis of wild-type and chimeric Gag proteins. The experiment was performed and the figure is labeled as for Fig. 2, except that all panels show analysis of 9-h incubations only. The calculated molecular size for each Gag species, as determined from the amino acid composition, is indicated at the right of each panel. For HIV and chimeras A, B, C, and D, Gag species of greater apparent size than the indicated band result from frameshift into a truncated pro sequence. For chimera 1 and chimera 2, multiple Gag bands are visible due to frameshift products derived from both Gag and an internal initiation product. For chimeras 3 through 7, there are no frameshift products due to the presence of a mutation in the ribosome “shift” sequence. For all species, smaller bands are the presumed products of internal initiations and/or abortive translations.
TABLE 1.
Assembly competence of Gag species
| Gag protein | Gradient analysisa | EM analysisb |
|---|---|---|
| HIV | − | − |
| M-PMV | + | + |
| Ch A | − | − |
| Ch B | − | − |
| Ch C | − | NDc |
| Ch D | − | ND |
| Ch 1 | + | ND |
| Ch 2 | + | ND |
| Ch 3 | + | + |
| Ch 3a | + | + |
| Ch 4 | + | + |
| Ch 5 | − | ND |
| Ch 6 | − | ND |
| Ch 7 | + | ND |
| Δp1Δp6 | − | ND |
| Δp6 | − | − |
| −MA | − | ND |
Production of sedimentable material indicative of immature assembly structures by 9 h.
Observation of immature assembly structures in thin sections; EM, electron microscopy.
ND, not determined.
Chimeras produce structures similar to assembling immature capsids.
Some of the chimeric Gag proteins produced material suggestive of assembled immature capsid structures in sucrose gradients, and so we performed electron microscopy to provide direct visual proof that such structures were made. Synthesis and assembly reactions for several chimeras were centrifuged at high speed and the resulting pellets processed for thin section electron microscopy. In addition, for each occasion when chimera samples were prepared, control samples of HIV and M-PMV were prepared in parallel. For all samples, the assembly reactions were allowed to incubate for 24 h before processing in order to allow the maximum time for potential structures to assemble. For chimeras 3, 3a, and 4, we found crescent-shaped partial immature capsid-like structures reminiscent of those that can be observed assembling under the plasma membrane of HIV-infected cells (Fig. 4). For the remainder of the chimeric Gag molecules analyzed, we were unable to observe similar structures (Table 1). This was also true for wild-type HIV-1 Gag translation products analyzed in parallel. In contrast, assembled immature capsids were consistently observed for M-PMV (Fig. 4A, D, and G).
FIG. 4.
Thin-section electron microscopy analysis of in vitro-assembled structures. M-PMV, chimera 4, chimera 3a, and chimera 3 gag genes were expressed in vitro and allowed to assemble for a total of 24 h, then the reaction mixtures were diluted and centrifuged at high speed, and the resulting pellets were fixed and processed for microscopy. (A, D, and G) Pelleted material from lysates expressing M-PMV; (B and C) chimera 4; (E and F) chimera 3a; (H and I) chimera 3. Each row of panels is of samples prepared in parallel. Arrows indicate immature retrovirus particle-like assembled structures. Bars = 100 nm.
In contrast to the apparently completed spheres produced by M-PMV Gag, the assembly-competent chimeric proteins produced only partial structures. In addition, the curvature of these crescents suggests a potential particle of approximately 150 nm in diameter rather than the 100-nm structures produced by M-PMV Gag. These differences may reflect altered assembly morphology due to the presence of p12 in these chimeric precursors. However, the potential spherical size of the chimeric structures is consistent with the diameters of HIV-1 immature particles determined from cryo-electron microscopy, where the modal width of these heterogeneous and imperfectly spherical particles was found to be between 140 and 160 nm with numerous larger-sized particles evident (17).
HIV CA domain function is required for chimera assembly.
The ability of chimeric Gag proteins to assemble after insertion of M-PMV p12 raised the possibility that this assembly phenotype was mediated primarily by the ISD. We therefore sought to determine whether there was a continued requirement for HIV Gag domain function within a chimera by incorporating a mutation, previously shown to render HIV assembly defective, into the CA sequence of chimera 4. The C-terminal domain of HIV CA contains a hydrophobic dimerization domain critical to the function of CA. Previous work by Gamble et al. (18) had shown that a mutation, M185A, within this hydrophobic sequence can block the dimerization of the CA protein in vitro and can significantly reduce the production of virus from cells. Furthermore, the mutation rendered such virus completely noninfectious. Thus, the dimer interface of CA must function in several stages of virus replication, including the assembly of immature virus particles. The M185A mutation, kindly provided Wesley Sundquist, was incorporated into chimera 4, and the mutant construct was tested for assembly in vitro. As shown in Fig. 5, this mutation effectively blocked the production of immature assembly products. Thus, despite the presence of p12 and the ISD in this Gag species, a functioning assembly domain within the HIV sequences was required for assembly of the chimeric Gag protein.
FIG. 5.
Comparison of assembly by sucrose analysis of chimera 4 and of chimera 4 containing the M185A substitution in the HIV-derived CA domain. (A) The experiment was performed and the figure is labeled as for Fig. 2A, except that the incubation time for both panels is 16 h. (B) Quantitative representation of gel data, as in Fig. 2B.
Chimeric Gag species and the in vitro assay can be used to test inhibitors of assembly.
The assembly of chimeric Gag proteins and the demonstration that HIV sequence function is necessary for this assembly suggest the use of this system to test in vitro for inhibitors of HIV Gag. Previously, we have shown the inhibition of M-PMV Gag assembly by anti-Gag antibodies (39). While this demonstrated the utility of the in vitro assay for such studies, the ability of a large molecule, such as an antibody, to inhibit the function of its ligand is not surprising and is probably achieved by simple steric hindrance.
To demonstrate the utility of the system for the analysis of potential small-molecule inhibitors, we examined the effect of bis-ANS on the assembly of chimera 4. The compound bis-ANS is a small, hydrophobic, polyaromatic compound that has previously been shown to block the assembly of P22 bacteriophage and hepatitis B virus capsids in vitro (44, 51). Using our established protocol for inhibition studies (see Materials and Methods and reference 39), bis-ANS was added at a final concentration of 1 mM and the reaction assayed by sucrose gradient analysis (Fig. 6). For both chimera 4 and M-PMV Gag, the result was the same. The addition of bis-ANS blocks the formation of immature assembly products.
FIG. 6.
Inhibition of chimera 4 and M-PMV immature capsid structure assembly in vitro by bis-ANS. Translation reactions were treated with cycloheximide either alone (left panels) or in combination with bis-ANS (right panels) after 30 min of incubation. Reactions were further incubated for 23.5 h. Assembled structures were analyzed by sucrose gradient fractionation and the figure is labeled as for Fig. 2.
DISCUSSION
The experiments described here demonstrate the ability of the M-PMV ISD to enable HIV Gag assembly in a reticulocyte lysate protein synthesis system. The structures produced are, by gradient analysis, comparable to those produced by M-PMV, and by thin-section electron microscopy they are morphologically similar to the immature assembly structures seen associated with the plasma membrane in HIV-infected cells. The system, where all one needs is plasmid DNA and a commercially available translation system, might constitute a convenient assay for potential assembly inhibitors. Furthermore, the in vitro translation assembly assay, unlike those that have been devised from purified proteins, allows the analysis of the complete HIV Gag sequence, albeit in the context of a chimeric molecule (chimeras 4 and 7).
At the outset of this study, we considered the ability of the RSV/HIV chimera, RHB (2), to function independently of a rescuing wild-type Gag molecule as evidence that gross fusions of M-PMV and HIV sequences would provide the most productive approach to chimera construction, provided that fusions within known Gag conformational domains were avoided. However, despite our careful analysis of structural data, the failure of chimeras A to D to assemble may indicate that the structures of the individual domains do not accurately reflect those within the whole Gag precursor. Indeed, recent evidence suggests that there is a direct interaction between the N- and C-terminal domains of CA that would have been impossible to predict from the available structures of the separate domains (30). In RSV CA, second-site suppressor mutations were found in the N-terminal domain of CA that rescued mutants with lethal substitutions in the MHR of the C-terminal domain (3). More recently, it was shown in a kinetic analysis of HIV CA assembly that the N-terminal domain can relieve inhibition of full-length CA assembly by the C-terminal domain, thus providing direct biochemical evidence for an interaction between the domains within the matured core (30).
While chimeras 1 and 2 could assemble in the in vitro assembly assay, both of these consist largely of M-PMV sequences with only the MA domain of HIV substituted. Although HIV MA has been shown to assist in the interaction of HIV Gag with itself in the yeast two-hybrid system, the region most responsible for this association lies within sequences in NC and the C-terminal region of CA (4, 48). Indeed, with the exception of the short N-terminal membrane interaction region, including the myristate moiety, HIV MA is dispensable for particle production and even infectivity (36). Thus, these chimeras appear to posses little utility for the analysis of Gag assembly and its inhibition.
We next turned to the construction of a more-defined set of chimeras in which only the M-PMV p12 domain is fused to HIV Gag sequences. It is interesting to note which of these chimeric species could assemble with respect to the placement of the ISD within them. Chimeras 3, 3a, 4, and 7, where the N terminus of p12 is fused to other sequences, could assemble, while chimeras 5 and 6, where the N terminus is free, could not. Indeed, the only difference between chimeras 4 and 5 is the position of the p12 domain; both contain the complete HIV Gag plus p12. The functional element within p12, the ISD, has been localized to the N-terminal third of the p12 domain (38, 41). Perhaps this region must be anchored in order to perform its function. Alternatively, the position of the ISD within the Gag protein itself might be critical. However, the ISD apparently functions when fused to the C terminus of Gag, a position it does not occupy in M-PMV Gag. Thus, it is more likely that the ISD is conformationally or functionally dependent upon an N-terminal anchor. Studies to determine the precise structure and, thus, the mechanism by which the ISD functions are ongoing.
The ability of other investigators to observe nonchimeric HIV Gag assembly in a similar cell-free translation system is not consistent with the results presented here. In contrast to reports where particles have been found in a reticulocyte lysate (42), we have never been able to observe HIV Gag assembly in the system described here, either by gradient analysis or by electron microscopy (present report and reference 39). What might account for the discrepancy? For the reticulocyte lysate system, we have observed occasional batch-dependent differences in the efficiency of M-PMV Gag assembly, and in at least one instance a production lot of lysate completely failed to support assembly of synthesized Gag. Given this potential variability, it is possible that a small amount of HIV Gag assembly may occur in reticulocyte lysates under optimal conditions that we have not been able to achieve. A second, and perhaps related, possibility is that assembly is concentration dependent. Under ideal conditions it is possible that sufficient protein is synthesized to achieve assembly. This idea is supported by the efficient assembly of a relatively high concentration of recombinant HIV Gag into morphologically authentic immature particles when added to reticulocyte lysate (5). Particle assembly has also been described in the wheat germ translation system, where a chaperone critical for assembly was identified (50). However, HIV Gag assembly in this system requires the addition of cellular membranes and myristic acid, neither of which is required for M-PMV assembly or for assembly of the chimeras described here.
Finally, as discussed previously, we have postulated that the ISD may function to increase the effective concentration of Gag to make intracytoplasmic assembly more efficient. In contrast, viruses that assemble on the plasma membrane may, through that interaction, achieve a similarly effective concentration (38). Both M-PMV Gag, with the ISD deleted, and HIV or RSV Gag, in the absence of membrane, can efficiently assemble if present in sufficient concentration (5-7, 26, 38, 41, 47). The ability of the ISD to facilitate HIV Gag assembly in a system where transport to the plasma membrane is absent suggests that the ISD and the plasma membrane provide similar and interchangeable concentrating functions in retrovirus assembly. Whether plasma membrane interaction can substitute for the ISD in the context of M-PMV Gag is currently under investigation.
Acknowledgments
We are grateful to Leigh Millican at the UAB Electron Microscopy Core Facility for her expertise with electron microscopy.
This work was supported by National Institutes of Health grants CA-27834 to E.H. and AI-43230 to M.S.
REFERENCES
- 1.Accola, M. A., B. Strack, and H. G. Göttlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, R. P., T. D. Nelle, and J. W. Wills. 1993. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 67:6487-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowzard, J. B., J. W. Wills, and R. C. Craven. 2001. Second-site suppressors of Rous sarcoma virus CA mutations: evidence for interdomain interactions. J. Virol. 75:6850-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, S., R. J. Fisher, E. M. Towler, S. Fox, H. J. Issaq, T. Wolfe, L. R. Phillips, and A. Rein. 2001. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc. Natl. Acad. Sci. USA 98:10875-10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, S., and A. Rein. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, S., and V. M. Vogt. 1997. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J. Virol. 71:4425-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, S., and V. M. Vogt. 1995. Self assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, B. K., I. Rousso, S. Shim, and P. S. Kim. 2001. Efficient assembly of an HIV-1/MLV Gag-chimeric virus in murine cells. Proc. Natl. Acad. Sci. USA 98:15239-15244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conte, M. R., M. Klikova, E. Hunter, T. Ruml, and S. Matthews. 1997. The three-dimensional solution structure of the matrix protein from the type D retrovirus, the Mason-Pfizer monkey virus. EMBO J. 16:5819-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deminie, C. A., and M. Emerman. 1994. Functional exchange of an oncoretrovirus and a lentivirus matrix protein. J. Virol. 68:4442-4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deminie, C. A., and M. Emerman. 1993. Incorporation of human immunodeficiency virus type 1 Gag proteins into murine leukemia virus virions. J. Virol. 67:6499-6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupraz, P., and P.-F. Spahr. 1992. Specificity of Rous sarcoma virus nucleocapsid protein in genomic RNA packaging. J. Virol. 66:4662-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehrlich, L. S., B. E. Agresta, and C. A. Carter. 1992. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 66:4874-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehrlich, L. S., T. Liu, S. Scarlata, B. Chu, and C. A. Carter. 2001. HIV-1 capsid protein forms spherical (immature-like) and tubular (mature-like) particles in vitro: structure switching by pH-induced conformational changes. Biophys. J. 81:586-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Erickson-Viitanen, S., J. Manfredi, P. Viitanen, D. E. Tribe, R. Tritch, C. A. Hutchison III, D. D. Loeb, and R. Swanstrom. 1989. Cleavage of HIV-1 gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res. Hum. Retrovir. 5:577-591. [DOI] [PubMed] [Google Scholar]
- 17.Fuller, S. D., T. Wilk, B. E. Gowen, H.-G. Kräusslich, and V. M. Vogt. 1997. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr. Biol. 7:729-738. [DOI] [PubMed] [Google Scholar]
- 18.Gamble, T. R., S. Yoo, F. F. Vajdos, U. K. von Schwedler, D. K. Worthylake, H. Wang, J. P. McCutcheon, W. I. Sundquist, and C. P. Hill. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 19.Ganser, B. K., S. Li, V. Y. Klishko, J. T. Finch, and W. I. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80-83. [DOI] [PubMed] [Google Scholar]
- 20.Garnier, L., J. W. Wills, M. F. Verderame, and M. Sudol. 1996. WW domains and retrovirus budding. Nature 381:744-745. [DOI] [PubMed] [Google Scholar]
- 21.Gitti, R. K., B. M. Lee, J. Walker, M. F. Summers, S. Yoo, and W. I. Sundquist. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273:231-235. [DOI] [PubMed] [Google Scholar]
- 22.Gross, I., H. Hohenberg, C. Huckhagel, and H.-G. Kräusslich. 1998. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J. Virol. 72:4798-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gross, I., H. Hohenberg, T. Wilk, K. Wiegers, M. Grättinger, B. Müller, S. Fuller, and H.-G. Kräusslich. 2000. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 19:103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter, E., J. Casey, B. Hahn, M. Hayami, B. Korber, R. Kurth, J. Neil, A. Rethwilm, P. Sonigo, and J. Stoye. 1999. Retroviridae, p. 369-387. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. Carstens, M. K. Estes, S. Lemon, J. Maniloff, M. A. Mayo, D. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy. Seventh Report of the International Committee on Taxonomy of Viruses. Academic Press, London, England.
- 25.Jacks, T., M. D. Power, F. R. Masiarz, P. A. Luciw, P. J. Barr, and H. E. Varmus. 1988. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331:280-283. [DOI] [PubMed] [Google Scholar]
- 26.Joshi, S. M., and V. M. Vogt. 2000. Role of the Rous sarcoma virus p10 domain in shape determination of Gag virus-like particles assembled in vitro and within Escherichia coli. J. Virol. 74:10260-10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klikova, M., S. S. Rhee, E. Hunter, and T. Ruml. 1995. Efficient in vivo and in vitro assembly of retroviral capsids from Gag precursor proteins expressed in bacteria. J. Virol. 69:1093-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kräusslich, H.-G., and R. Welker. 1996. Intracellular transport of retroviral capsid components. Curr. Top. Microbiol. Immunol. 214:25-64. [DOI] [PubMed] [Google Scholar]
- 29.Krishna, N. K., and J. W. Wills. 2001. Insertion of capsid proteins from nonenveloped viruses into the retroviral budding pathway. J. Virol. 75:6527-6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lanman, J., J. Sexton, M. Sakalian, and P. E. J. Prevelige. 2002. Kinetic analysis of the role of intersubunit interactions in human immunodeficiency virus type 1 capsid protein assembly in vitro. J. Virol. 76:6900-6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lingappa, J. R., R. L. Hill, M. L. Wong, and R. S. Hegde. 1997. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J. Cell Biol. 136:567-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matthews, S., P. Barlow, J. Boyd, G. Barton, R. Russell, H. Mills, M. Cunningham, N. Meyers, N. Burns, N. Clark, S. Kingsman, A. Kingsman, and I. Campbell. 1994. Structural similarity between the p17 matrix protein of HIV-1 and interferon-γ. Nature 370:666-668. [DOI] [PubMed] [Google Scholar]
- 33.Momany, C., L. C. Korvari, A. J. Prongay, W. Keller, R. K. Gitti, B. M. Lee, A. E. Gorbalenya, L. Tong, J. McClure, L. S. Ehrlich, M. F. Summers, C. Carter, and M. Rossmann. 1996. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 3:763-770. [DOI] [PubMed] [Google Scholar]
- 34.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reed, M., R. Mariani, L. Sheppard, K. Pekrun, N. R. Landau, and N.-W. Soong. 2002. Chimeric human immunodeficiency virus type 1 containing murine leukemia virus matrix assembles in murine cells. J. Virol. 76:436-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Göttlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakalian, M., and E. Hunter. 1998. Molecular events in the assembly of retrovirus particles. Adv. Exp. Med. Biol. 440:329-339. [DOI] [PubMed]
- 38.Sakalian, M., and E. Hunter. 1999. Separate assembly and transport domains within the Gag precursor of Mason-Pfizer monkey virus. J. Virol. 73:8073-8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakalian, M., S. D. Parker, R. A. Weldon, Jr., and E. Hunter. 1996. Synthesis and assembly of retrovirus Gag precursors into immature capsids in vitro. J. Virol. 70:3706-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh, A. R., R. L. Hill, and J. R. Lingappa. 2001. Effect of mutations in Gag on assembly of immature human immunodeficiency virus type 1 capsids in a cell-free system. Virology 279:257-270. [DOI] [PubMed] [Google Scholar]
- 41.Sommerfelt, M. A., S. S. Rhee, and E. Hunter. 1992. Importance of p12 protein in Mason-Pfizer monkey virus assembly and infectivity. J. Virol. 66:7005-7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spearman, P., and L. Ratner. 1996. Human immunodeficiency virus type 1 capsid formation in reticulocyte lysates. J. Virol. 70:8187-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Press, Cold Spring Harbor, N.Y. [PubMed]
- 44.Teschke, C. M., J. King, and P. E. J. Prevelige. 1993. Inhibition of viral capsid assembly by 1,1′-bi(4-anilinonaphthalene)-5-sulfonic acid. Biochemistry 32:10658-10665. [DOI] [PubMed] [Google Scholar]
- 45.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yasuda, J., and E. Hunter. 1998. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J. Virol. 72:4095-4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu, F., S. M. Joshi, Y. M. Ma, R. L. Kingston, M. N. Simon, and V. M. Vogt. 2001. Characterization of Rous sarcoma virus Gag particles assembled in vitro. J. Virol. 75:2753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zábranský, A., E. Hunter, and M. Sakalian. 2002. Identification of a minimal HIV-1 Gag domain sufficient for self-association. Virology 294:141-150. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72:1782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zimmerman, C., K. C. Klein, P. K. Kiser, A. R. Singh, B. L. Firestein, S. C. Riba, and J. R. Lingappa. 2002. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature 415:88-92. [DOI] [PubMed] [Google Scholar]
- 51.Zlotnik, A., P. Ceres, S. Singh, and J. M. Johnson. 2002. A small molecule inhibits and misdirects assembly of hepatitis B virus capsids. J. Virol. 76:4848-4854. [DOI] [PMC free article] [PubMed] [Google Scholar]