Abstract
Retroviral replication requires the integration of reverse-transcribed viral cDNA into a cell chromosome. A key barrier to forming the integrated provirus is the nuclear envelope, and numerous regions in human immunodeficiency virus type 1 (HIV-1) have been shown to aid the nuclear localization of viral preintegration complexes (PICs) in infected cells. One region in integrase (IN), composed of Val-165 and Arg-166, was reportedly essential for HIV-1 replication and nuclear localization in all cell types. In this study we confirmed that HIV-1V165A and HIV-1R166A were replication defective and that less mutant viral cDNA localized to infected cell nuclei. However, we present three lines of evidence that argue against a specific role for Val-165 and Arg-166 in PIC nuclear import. First, results of transient transfections revealed that V165A FLAG-tagged IN and green fluorescent protein-IN fusions carrying either V165A or R166A predominantly localized to cell nuclei. Second, two different strains of previously described class II IN mutant viruses displayed similar nuclear entry profiles to those observed for HIV-1V165A and HIV-1R166A, suggesting that defective nuclear import may be a common phenotype of replication-defective IN mutant viruses. Third, V165A and R166A mutants were defective for in vitro integration activity, when assayed both as PICs isolated from infected T-cells and as recombinant IN proteins purified from Escherichia coli. Based on these results, we conclude that HIV-1V165A and HIV-1R166A are pleiotropic mutants primarily defective for IN catalysis and that Val-165 and Arg-166 do not play a specific role in the nuclear localization of HIV-1 PICs in infected cells.
Retroviral replication requires the accomplishment of certain key steps in the early phase of the viral life cycle. Soon after entering a cell, the viral enzyme reverse transcriptase (RT) copies genomic RNA into linear double-stranded cDNA. The viral enzyme integrase (IN) then inserts this DNA into a host cell chromosome. In vivo, reverse transcription and integration take place in the context of large nucleoprotein complexes that are called reverse transcription complexes (RTCs) and preintegration complexes (PICs), respectively (reviewed in reference 29).
Two different IN activities are required for integration. During an initial 3′ processing reaction, IN cleaves each cDNA end adjacent to the phylogenetically conserved sequence CA. For both Moloney murine leukemia virus (MoMLV) (7, 28, 45) and human immunodeficiency virus type 1 (HIV-1) (12, 13, 39), 3′ processing can occur in the cell cytoplasm. Following nuclear entry, IN transfers the processed 3′ ends to the 5′ phosphates of a double-stranded staggered cut in chromosomal DNA (7, 28). The final step of the integration process, which involves repairing the gaps between the unjoined viral 5′ ends and the chromosome, can be completed by host cell enzymes (4, 54).
The double lipid bilayer that envelops animal cell nuclei presents a formidable barrier for retroviral PICs. The nuclear envelope contains numerous nuclear pore complexes (NPCs) that permit the passive diffusion of relatively small macromolecules with diameters up to about 9 nm, which corresponds roughly to a 40- to 60-kDa globular protein (reviewed in reference 37). The relatively large size of retroviral PICs, estimated to be roughly the size of a ribosome for HIV-1 (25), precludes their passive transport through intact NPCs (reviewed in reference 27).
Different retroviruses have apparently evolved different strategies to access the cell nucleus. HIV-1, for example, can be transported by an energy-dependent process in nondividing cells (9), suggesting that HIV-1 PICs contain specific nuclear localization sequences (NLSs) that govern their transport through intact NPCs (reviewed in references 27 and 52). Productive infection by MoMLV, in contrast, requires cells to pass through the M phase of the cell cycle (36, 44). Since animal cell nuclei disassemble during mitosis, MoMLV apparently reaches cell chromosomes in the absence of active nuclear transport by waiting for the dissolution of nuclear membranes.
Recently, replication-defective MoMLV (55) and HIV-1 (3, 57) mutants were described as blocked at the nuclear import step in rapidly dividing cells. This suggested that retroviruses might interact with specific host cell factors to gain nuclear entry even when cells disassemble their nuclear envelope. To address the mechanism of HIV-1 nuclear localization under these conditions, we characterized viral replication and nuclear import phenotypes of mutants defective for the central DNA flap (57) and Val-165 and Arg-166 in IN, also known as the IN NLS (3). In a related study (A. Limón, N. Nakajima, R. Lu, H. Z. Ghory, and A. Engelman, submitted for publication), we determined that flap-minus mutants displayed the wild-type (WT) levels of replication and nuclear localization under a variety of infection conditions, calling into question the importance of the DNA flap in HIV-1 replication and nuclear import. In this study, we confirmed that IN NLS mutant viruses were replication defective. However, in contrast to the previous claim, we found the mutants to be defective primarily for IN 3′ processing and DNA strand transfer activities. Although these mutant PICs entered dividing cell nuclei less efficiently than the WT PICs did, a similar import defect was observed for previously described replication-defective class II IN mutant viruses. Since we also determined that mutant fusion proteins efficiently localized to cell nuclei in transient-transfection assays, we conclude that Val-165 and Arg-166 in IN do not play a specific role in the nuclear localization of HIV-1 PICs.
MATERIALS AND METHODS
Plasmids.
IN sequences amplified from codon-optimized HIV-1NL4-3 Gag-Pol (pHDM.Hgpm2; a kind gift of R. C. Mulligan and J. Gray, Harvard Medical School) were inserted into pECMV-FLAG (a kind gift of G. Adelmant, Dana-Farber Cancer Institute). The V165A change was introduced using the QuikChange Site-Directed Mutagenesis Kit as recommended by the manufacturer (Stratagene, La Jolla, Calif.). The pFLAG-IN and pFLAG-IN/V165A expression vectors described by Bouyac-Bertoia et al. (3) were a kind gift of M. H. Malim, King's College London.
Plasmids pCMX-SAH/Y145F and pGFP-IN (49), expressing green fluorescent protein (GFP) and a fusion between GFP and HIV-1NL4-3 IN, respectively, were kindly provided by Ruth Yu and Takao Masuda, Tokyo Medical and Dental University. V165A and R166A changes were introduced into pGFP-IN by QuikChange Mutagenesis.
R166A and R166E changes were incorporated into pNL4-3 (1) by overlapping PCR as described previously (23). Plasmid pUCWTpol was made by inserting the 1.8-kb AgeI-PflMI fragment from pNL4-3 into AgeI-PflMI-digested pUC(A/P) (40). The V165A change was introduced into pUCWTpol by QuikChange mutagenesis, and the resulting 1.8-kb mutant fragment was swapped for the WT fragment in pNL43/XmaI (5). Derivatives of pNL4-3 carrying D64N/D116N, 1-212 (40), and F185K (32) were described previously. The F185K change was incorporated into pHXBH10-vpu+ (30), which encoded a Vpu-plus version of Vpr− Nef− HIV-1HXBc2 (26), by overlapping PCR.
D64N/D116N, 1-212, F185K, R166A, and R166E AgeI-PflMI fragments were shuttled from pNL4-3 into pNL43/XmaI. Envelope (Env)-deleted (Env−) pNLXΔenv plasmids for real-time quantitative (RQ)-PCR assays were constructed by digestion with NheI and StuI, filling in with Klenow, and religating. R166A and V165A changes were introduced into pHXBH10ΔenvCAT-vpu+, which encoded Env− HIV-1HXBc2 (43), by overlapping PCR. Whereas the 1.8-kb PflMI fragment from pHXBH10-vpu+/F185K was swapped for the corresponding fragment in pHXBH10ΔenvCAT-vpu+, the Env− D116A single-round vector was previously described (43).
Plasmid pUC19.2LTR was built by amplifying two long terminal repeat (LTR)-containing circles from HIV-1NL4-3-infected cell Hirt supernatant by using EcoRI-tagged 5′-ACTGACGAATTCGAGCTTGCTACAAGG-3′ and BamHI-tagged 5′-CATGCAGGATCCCAGGGTGTAACAAGCTGG-3′, digestion and ligating to EcoRI-BamHI-cut pUC19.
Bacterial expression vector pINSD.His encoded HIV-1NL4-3 IN fused to a 21-residue amino-terminal tag with six His residues (18). Whereas pINSD.His/F185K was previously described (24), V165A and R166A changes were introduced by QuikChange mutagenesis. DNA sequencing was used to verify the regions of all plasmids built by PCR.
The HIV-1HXBc2 Env expression vector pSVIII-env was previously described (31). An HIV-1NL4-3 derivative was made by swapping the 2.1-kb KpnI-BamHI env fragment from pNL43/XmaI for the corresponding pSVIII-env fragment. This resulted in a chimeric Env where amino acids 42 to 750, comprising most of gp120 and gp41, were HIV-1NL4-3 derived.
Nuclear localization of IN in transient transfections.
One day prior to transfection, HeLa cells were seeded at 10,000 cells per well in Nunc Lab-Tek II chamber slides (Nalge Nunc International, Rochester, N.Y.). The cells were transfected with 0.5 μg of plasmid using the FuGENE 6 Transfection Reagent (Roche Molecular Biochemicals, Indianapolis, Ind.).
To detect FLAG-tagged IN by immunofluorescence, cells washed with phosphate-buffered saline (PBS) (pH 7.4) 2 days posttransfection were fixed for 10 min in 3.7% formaldehyde in PBS, permeabilized for 10 min in 0.2% Triton X-100, and blocked for 30 min in 5% bovine serum albumin (BSA) in PBS. Fixed cells were incubated for 1 h at ambient temperature with anti-FLAG M2 antibody (Sigma-Aldrich, St. Louis, Mo.) diluted 1:500 in 5% BSA. After three 5-min washes with PBS, the cells were incubated for 1 h with fluorescein isothiocyanate-conjugated donkey anti-mouse antibody (Jackson ImmunoResearch Laboratories, Inc., Bar Harbor, Maine) diluted 1:250 in 5% BSA. After three 5-min washes with PBS, the cells were mounted with Vectashield mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories Inc., Burlingame, Calif.) and covered with a coverslip. Images were acquired and processed on a DeltaVision platform (Applied Precision Inc., Issaquah, Wash.). Data collected in 1-μm sections were subjected to five cycles of iterative deconvolution. A central plane from each sample is presented. To detect GFP, cells were fixed for 10 min in 1.5% formaldehyde 1 day posttransfection.
Spreading-infection assays.
Viral stocks were prepared in 293T cells by transfection with calcium phosphate. Viruses filtered through 0.45-μm-pore-size filters were subjected to titer determination using a 32P-labeled RT assay as described previously (23, 40). Jurkat T-cells (2 × 106) infected for 18 h with 10 7 RT cpm of WT and mutant virus were washed and seeded in 5 ml of RPMI 1640 medium containing 10% fetal calf serum. Each infection was performed at least twice.
RQ-PCR assays.
Single-round viruses were prepared by cotransfecting pNLXΔenv or pHXBH10ΔenvCAT-vpu+ with the appropriate Env expression vector by using FuGENE 6. Filtered supernatants were treated with DNase I as described previously (14).
Jurkat cells (6 × 106) were infected with 6 × 106 RT cpm of pseudotyped HIV-1NL4-3 or 3 × 107 RT cpm of pseudotyped HIV-1HXB2 by spinoculation (41) at 480 × g for 2 h. DNA was extracted from infected cells using the DNeasy tissue kit as recommended by the manufacturer (Qiagen, Valencia, Calif.). Infections were performed in duplicate, and each infection was analyzed in duplicate by RQ-PCR.
Two different HIV-1 amplicons, representing products of late reverse transcription (LRT) and 2-LTR circles, were quantified essentially as previously described (11, 16, 33, 35). Primers, probes, and PCR conditions were as described previously (11) except that the minus-strand primer for amplifying HIV-1 NL4-3 LRT products was 5′-CCTGCGTCGAGAGATCTCCTCTGG-3′ (nucleotides 695 to 672). Whereas half-genome plasmids p83-2/XmaI and pSVC21/5′-LTR (5) were used to generate standard curves for HIV-1NL4-3 and HIV-1 HXBc2 LRT products, respectively, pUC19.2LTR was used for both strains.
Human endogenous retrovirus-3 (ERV-3) was amplified essentially as previously described (56) to quantify total cellular DNA. Levels of HIV-1 LRT and 2-LTR circles normalized to ERV-3 are presented in arbitrary units (AU). The 1.7-kb env fragment from ERV-3, a generous gift from D. Waters, Frederick Cancer Research and Development Center, was used to generate the standard curve.
Preparation of HIV-1 PICs and in vitro PIC activity.
PICs prepared from cytoplasmic extracts of C8166 T cells (2 × 107) at 7 h postinfection were reacted with linearized φX174 target DNA as previously described (13, 14). Mixtures were deproteinized, fractionated through agarose, and analyzed by Southern blotting as previously described (14). Integration activity, calculated as the percentage of linear HIV-1 converted into the 15.1-kb integration product, was quantified by PhorphorImager analysis using ImageQuant version 1.11 (Molecular Dynamics, Sunnyvale, Calif.).
Purification of recombinant IN from Escherichia coli.
E. coli BL21(DE3) (46) or BL21(DE3)pLysS (Novagen Inc., Madison, Wisc.) strains grown at 37°C in 500 ml of Terrific broth containing 100 μg of ampicillin per ml were induced for IN expression as previously described (18). Cells harvested by centrifugation were resuspended in 35 ml of 25 mM HEPES (pH 7.6)-0.1 mM EDTA at 4°C, frozen in liquid N2, and stored at −80°C. The remainder of the preparation was done at 4°C.
NaCl, β-mercaptoethanol (β-ME), imidazole, and lysozyme were added to thawed cells to 150 mM, 2 mM, 5 mM, and 0.2 mg/ml, respectively. After 30 min, the cells were sonicated essentially as previously described (18). The lysate was centrifuged at 39,800 × g for 45 min, the supernatant (fraction I) was saved, and the pellet was resuspended by homogenization in 35 ml of buffer A (20 mM Tris-HCl [pH 8.0], 2 M NaCl, 2 mM β-ME, 0.1 mM EDTA, 5 mM imidazole). After 30 min, the suspension was recentrifuged, the fraction II supernatant was saved, and the pellet was homogenized in 35 ml of buffer B (20 mM Tris-HCl [pH 8.0], 0.5 M NaCl, 6 M guanidine-HCl, 2 mM β-ME, 0.1 mM EDTA, 5 mM imidazole). After 30 min, the suspension was centrifuged and the fraction III supernatant was saved. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis revealed that the vast majority of V165A and R166A IN partitioned to fraction III. Because of this, IN was purified by nickel affinity chromatography under denaturing conditions essentially as previously described (18, 24). No differences in WT or mutant activities have been observed for proteins purified under native versus denaturing conditions (reference 24 and references therein). Purified WT and mutant IN were refolded as follows.
EDTA was added to 5 mM to pooled column fractions, and dialysis was performed against 6 M guanidine-HCl-20 mM HEPES (pH 7.6)-2 mM EDTA-2 mM β-ME. The protein concentration was adjusted to 1 mg/ml in dialysis buffer, an equal volume of 1 M NaCl-20 mM HEPES (pH 7.6)-2 mM EDTA-2 mM β-ME was added, and dialysis was performed against 2 M urea-20 mM HEPES (pH 7.6)-0.5 M NaCl-2 mM EDTA-10 mM β-ME-10 mM [(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) followed by dialysis against 20 mM HEPES (pH 7.6)-1 M NaCl-1 mM EDTA-1 mM dithiothreitol (DTT)-10 mM CHAPS-10% (wt/vol) glycerol followed by a third dialysis against buffer C (20 mM HEPES [pH 7.6], 0.5 M NaCl, 1 mM EDTA, 1 mM DTT, 10 mM CHAPS, 10% glycerol). Following centrifugation at 19,000 × g for 10 min, the amino-terminal hexahistidine tag was removed by digesting with 30 National Institutes of Health units of human thrombin (Sigma-Aldrich) per mg of IN for 40 min at room temperature, followed by an additional 30 U for 40 min. Thrombin was removed by passage over a benzamidine-Sepharose 4 Fast Flow (Amersham Pharmacia Biotech., Piscataway, N.J.) column equilibrated in buffer C, and the flowthrough was dialyzed against buffer D (20 mM HEPES [pH 7.6], 0.5 M NaCl, 1 mM EDTA, 5 mM DTT, 10 mM CHAPS, 10% glycerol). After centrifugation at 19,000 × g for 10 min, soluble IN was frozen in liquid N2 and stored at −80°C. Densitometric scans of sodium dodecyl sulfate-polyacrylamide gels stained with Coomassie blue revealed that each IN preparation was greater than 95% pure.
IN assays.
The DNA substrate for assaying 3′ processing and DNA strand transfer activities corresponded to the terminal 21 bp of the U5 end of HIV-1. A second substrate, the branched Y-mer oligonucleotide, was used to measure disintegration activity. Both substrates were prepared essentially as previously described (18, 22).
IN reaction mixtures (16 μl) contained 25 mM morpholinepropanesulfonic acid (MOPS) (pH 7.2), 0.1 mg of BSA per ml, 10 mM β-ME, 10% glycerol, 25 nM labeled DNA substrate, 25 mM NaCl, 7.5 mM MnCl2, and either 0.25 or 0.5 μM IN. The products of reactions terminated after 60 min at 37°C were electrophoresed through denaturing polyacrylamide sequencing gels as previously described (18, 22). The results were visualized by autoradiography and quantitated by PhosphorImager analysis. Whereas 3′ processing activity was calculated as the percent conversion of the 21-base strand into the 19-base cleavage product, disintegration activity was calculated as the percent conversion of the labeled 15-base strand into the 30-base product (18, 22, 24).
RESULTS
V165A and R166A IN proteins display the WT pattern of nuclear localization in transiently transfected cells.
Results of recent experiments showed that a 13-residue peptide composed of amino acids 161 to 173 in HIV-1 IN behaved as a transferable NLS (3). When fused to the carboxyl terminus of pyruvate kinase, the peptide directed the nuclear accumulation of the otherwise strictly cytoplasmic pyruvate kinase protein in transiently transfected cells. Results of alanine scanning identified Val-165 and Arg-166 as the critical NLS residues within the 13-mer peptide (3). Because these results indicated that Val-165 and Arg-166 directed the nuclear localization of IN, we constructed codon-optimized FLAG-tagged WT and V165A IN expression vectors as a means of investigating host cell factors involved in HIV-1 IN nuclear transport.
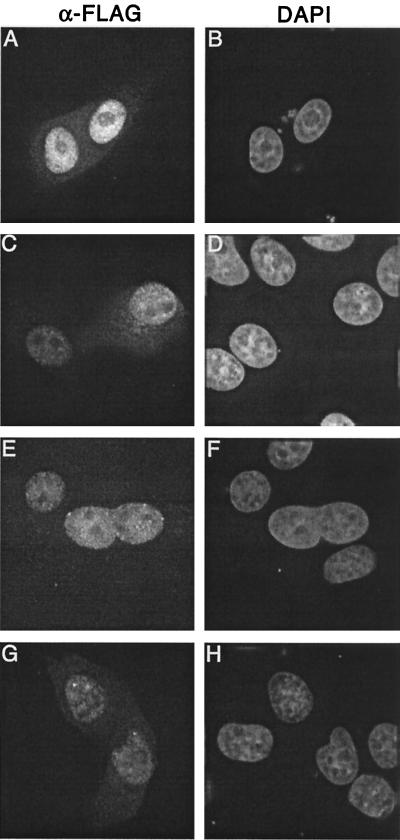
As expected (3, 42), cells transfected with our WT expression vector showed prominent nuclear staining for IN (Fig. 1A and B). To our surprise, however, codon-optimized V165A IN was also predominantly nuclear (Fig. 1C and D). This was unexpected because a similar FLAG-tagged V165A construct was precluded from nuclear entry (3). We therefore considered that the higher protein expression levels predicted for our codon-optimized constructs might have affected the outcome of V165A IN nuclear localization. Because of this, we next tested the previously described WT and V165A FLAG-tagged constructs. As described in previous reports (3, 42), the WT IN protein localized exclusively to the nuclei of transfected cells (Fig. 1E and F). In contrast to the report by Bouyac-Bertoia et al. (3), FLAG-tagged V165A was also predominantly nuclear (Fig. 1G and H). Western blot analysis indicated that full-length FLAG-tagged IN was expressed in all transfections (data not shown). Based on these results, we conclude that Val-165 is not a critical determinant of FLAG-tagged HIV-1 IN nuclear localization in transiently transfected cells.
FIG. 1.
FLAG-tagged WT and V165A IN proteins localize to cell nuclei. (A and B) HeLa cells transfected with the codon-optimized WT expression vector. (C and D) Cells transfected with the codon-optimized V165A vector. (E and F) Cells transfected with the previously described (3, 42) FLAG-tagged WT IN expression vector. (G and H) Cells transfected with the previously described V165A vector (3). Fixed cells were mounted in DAPI-containing medium and visualized by indirect immunofluorescence microscopy. Whereas DAPI stained the nuclear DNA of all cells, the anti-FLAG antibody detected IN only in cells that were transfected.
Since the approximately 34-kDa monomeric mass of FLAG-tagged IN is below the predicted 40- to 60-kDa exclusion limit of intact NPCs, we next tested the effects of V165A and R166A on the cellular distribution of an HIV-1 IN fusion protein whose predicted monomeric mass was closer to this limit. For this, we analyzed a previously characterized GFP-IN fusion (49). In contrast to the dispersed cytoplasmic-nuclear localization of the approximate 27-kDa parental GFP, the 60-kDa fusion protein efficiently localized to the nuclei of transiently transfected cells (49).
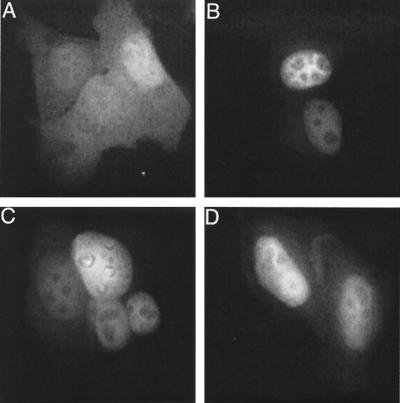
In agreement with the previous report, GFP-IN was predominantly nuclear (Fig. 2B) under conditions where parental GFP was dispersed throughout the cell cytoplasm and nucleus (Fig. 2A). GFP-IN/V165A and GFP-IN/R166A also localized predominantly to cell nuclei (Fig. 2C and D), indicating that neither Val-165 nor Arg-166 played a critical role in the nuclear localization of GFP-IN under these assay conditions.
FIG. 2.
Nuclear localization of GFP-IN fusion proteins. (A) HeLa cells were transfected with the pCMX-SAH/Y145F GFP expression vector. (B through D) Cells were transfected with pGFP-IN, pGFP-IN/V165A, and pGFP-IN/R166A expression vectors, respectively. Fixed cells were examined by fluorescence microscopy at 24 h posttransfection.
HIV-1V165A, HIV-1R166A, and HIV-1R166E fail to replicate in Jurkat T-cells.
The previous report indicated that Val-165 and Arg-166 played an essential role in PIC nuclear import (3). Our transient-transfection results, however, began to cast doubt on this interpretation. Because of this, we next examined Val-165 and Arg-166 mutants in HIV-1-infected cells. V165A, R166A, and R166E changes were introduced into HIV-1NL4-3, and WT and IN mutant viruses were produced by transiently transfecting 293T cells. Because various changes in IN have previously been shown to disrupt the proper assembly and release of HIV-1 virions (2, 8, 23, 34, 48), the previously described release-defective F185K class II IN mutant (24, 32) was included in this experiment. Replication-defective HIV-1 IN mutant viruses can be divided into two classes based on the specificity of the viral replication block (21). Class II mutants are pleiotropic, displaying defects in particle assembly and/or reverse transcription in addition to integration. In contrast, class I mutants, typified by changes in the IN active site, are specifically blocked at the integration step (reviewed in reference 21).
Cells transfected with HIV-1F185K yielded 40 to 60% of the levels of WT HIV-1NL4-3 particles in repeated experiments (data not shown). In contrast, HIV-1V165A, HIV-1R166A, and HIV-1R166E were released at 90 to 120% of the WT level in repeated experiments. Thus, as previously noted by Bouyac-Bertoia et al. (3), changes in Val-165 and Arg-166 do not appear to perturb HIV-1 assembly and release.
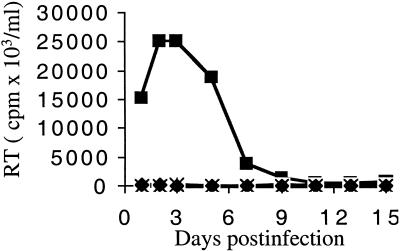
Jurkat cells were infected with 107 RT cpm of WT and mutant viruses, which equated to a multiplicity of infection of approximately 0.4 (40). HIV-1NL4-3 reached its peak replication 2 to 3 days postinfection under these conditions (Fig. 3). In contrast, HIV-1F185K, HIV-1V165A, HIV-1R166A, and HIV-1R166E showed no signs of replication over 2 months of observation (Fig. 3 and data not shown). These results are consistent with previous reports that HIV-1V165A (3) and HIV-1R166A (3, 53) were replication defective.
FIG. 3.
HIV-1V165A, HIV-1R166A, and HIV-1R166E are replication- defective in Jurkat T cells. Supernatants of cells infected with WT HIV-1NL4-3 (▪), HIV-1V165A (□), HIV-1R166A (•), HIV-1R166E (○), HIV-1F185K (⧫), or supernatant from mock-transfected cells (×) were assayed for RT activity at the indicated times.
HIV-1V165A, HIV-1R166A, and HIV-1R166E support levels of nuclear entry similar to previously described class II IN mutant viruses.
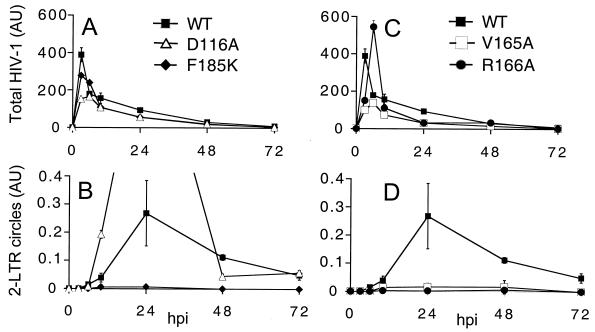
We next characterized the step in the HIV-1 life cycle at which the HIV-1V165A, HIV-1R166A, and HIV-1R166E mutants were blocked. Because each virus was released from transfected cells at the WT level, we focused our attention on the early steps of HIV-1 replication. Soon after entry, RT copies HIV-1 RNA into linear duplex cDNA containing an LTR at each end, and this linear cDNA is the substrate for IN-mediated DNA recombination (reviewed in reference 17). Not all linear molecules, however, become integrated. For example, a fraction of retroviral cDNA is circularized by host recombination enzymes after nuclear entry. One circular DNA product in particular, containing a tandem repeat of the LTR, has been extensively used as a marker for nuclear localization because its LTR-LTR junction affords a convenient template for PCR (17). Here we monitored HIV-1 cDNA synthesis and nuclear import by quantifying levels of total HIV-1 cDNA and 2-LTR circles in acute infections using RQ-PCR. The fraction of total viral DNA present as 2-LTR circles was then used as an indicator of PIC nuclear import. Although this analysis did not detect the total level of nuclear HIV-1 DNA, it nonetheless afforded a quantitative comparison of WT and mutant nuclear entry. HIV-1 vectors were used in RQ-PCR assays to restrict cDNA synthesis and nuclear import to single infectious cycles.
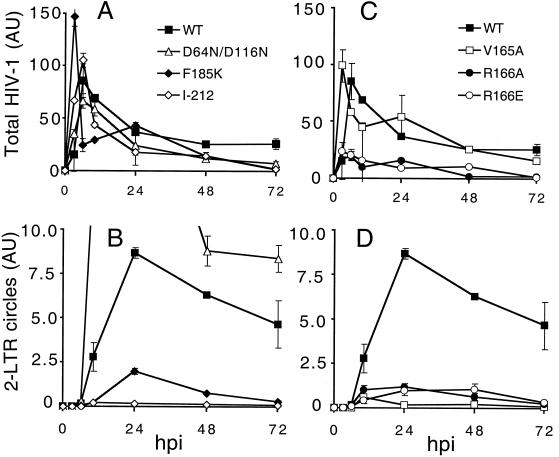
Examples of each of the two previously described classes of HIV-1 IN mutant viruses were included as controls. WT HIV-1NL4-3, class I mutant HIV-1D64N/D116N (40), and class II mutants HIV-11-212 (40) and HIV-1F185K (32) synthesized similar levels of total HIV-1 DNA in acutely infected Jurkat cells (Fig. 4A). As previously reported (23, 34, 53), the class I IN active-site mutant formed more 2-LTR circles than did the WT virus (Fig. 4A and B). We note that this is not interpreted as increased nuclear import of class I IN mutant PICs. Instead, cells infected with retroviral mutants specifically blocked at the integration step yield transient increases in the level of unintegrated viral DNA (reviewed in reference 21). In contrast to the results with HIV-1D64N/D116N, the HIV-1F185K and HIV-11-212 class II mutants formed fewer 2-LTR circles than did the WT (Fig. 4A and B). Thus, these pleiotropic class II IN mutant viruses were apparently defective for cDNA nuclear localization. This was not interpreted as evidence that IN plays a direct role in PIC nuclear import. Instead, defective nuclear import is apparently another phenotype associated with pleiotropic HIV-1 IN mutant viruses.
FIG. 4.
HIV-1V165A, HIV-1R166A, HIV-1R166E, and pleiotropic class II IN mutants form similar levels of 2-LTR circles in acute infections. (A) DNA prepared at the indicated times from cells infected with WT HIV-1NL4-3, HIV-1D64N/D116N, HIV-1F185K, or HIV-11-212 was assayed for total HIV-1 content by RQ-PCR. (B) DNA prepared as described in panel A was assayed for 2-LTR circles by RQ-PCR. (C) DNA prepared from cells infected with WT HIV-1NL4-3, HIV-1V165A, HIV-1R166A, or HIV-1R166E was assayed for total HIV-1 cDNA by RQ-PCR. (D) DNA prepared as described in panel C was assayed for 2-LTR circles. Panels B and D are presented with identical y-axis ranges for ease of comparing class II and HIV-1V165A, HIV-1R166A, and HIV-1R166E 2-LTR circle levels. Error bars represent variation between duplicate RQ-PCR assays. The class I HIV-1D64N/D116N mutant in panel B formed approximately 21 AU of 2-LTR circles at 24 h. AU, arbitrary units; hpi, hours postinfection.
HIV-1V165A, HIV-1R166A, and HIV-1R166E resembled the class II mutants in their cDNA synthesis and 2-LTR circle formation profiles. Whereas HIV-1V165A supported the WT level of cDNA synthesis, HIV-1R166A and HIV-1R166E synthesis was reduced approximately fourfold from WT levels in repeated experiments (Fig. 4C and data not shown). The nuclear import profile of HIV-1V165A closely resembled those of the class II mutants (Fig. 4), implying that HIV-1V165A might be a pleiotropic viral mutant. Levels of 2-LTR circles formed by HIV-1R166A and HIV-1R166E also resembled those in the class II mutants (Fig. 4B and D). However, since these viruses synthesized less total cDNA than did HIV-1NL4-3 and HIV-1V165A, the fraction of total HIV-1R166A and HIV-1R166E converted to 2-LTR circles was similar to that in the WT in repeated experiments (Fig. 4C, and D and data not shown).
Although the T-cell-tropic strain HIV-1HXBc2 is 97.6% identical to HIV-1NL4-3 (38), we recently determined that HIV-1HXBc2 yielded only about 10% of the level of HIV-1NL4-3 LRT RQ-PCR products under identical infection conditions (Limón et al., submitted). Because of this, we next determined WT and mutant nuclear import activities under these somewhat more limited conditions of cDNA synthesis. HIV-1HXBc2 infections were initiated with fivefold more virus inoculum to circumvent this relatively low cDNA yield.
The HIV-1D116A class I and HIV-1F185K class II mutant control viruses synthesized levels of total cDNA that were similar to those in WT HIV-1HXBc2 in repeated experiments (Fig. 5A and data not shown). As expected from the results presented in Fig. 4, HIV-1F185K formed fewer 2-LTR circles under conditions where the class I mutant formed more 2-LTR circles than the WT (Fig. 5B).
FIG. 5.
The nuclear localization of HIV-1HXBc2 V165A and R166A mutants resembles the pleiotropic class II F185K mutant. (A) DNA prepared at the indicated times from Jurkat cells infected with WT HIV-1HXBc2, HIV-1D116A, or HIV-1F185K was assayed for total HIV-1 by RQ-PCR. (B) The DNA prepared in panel A was assayed for 2-LTR circles. (C) DNA prepared from cells infected with WT HIV-1HXBc2, HIV-1V165A, or HIV-1R166A was assayed for total HIV-1. (D) DNA prepared in panel C was assayed for 2-LTR circles. HIV-1D116A formed about 1.0 AU of 2-LTR circles at 24 h (panel B). Other labeling is as in Fig. 4.
WT HIV-1HXBc2, HIV-1V165A, and HIV-1R166A supported similar levels of reverse transcription in repeated experiments (Fig. 5A and data not shown). And, as observed for the HIV-1NL4-3 strains, HIV-1V165A and HIV-1R166A formed levels of 2-LTR circles that were similar to the levels formed by the HIV-1F185K control (Fig. 5B and D).
V165A and R166A mutant PICs do not support detectable levels of in vitro integration activity.
The conclusion that HIV-1V165A and HIV-1R166A were specifically defective for nuclear entry was based in part on inferred WT catalytic activities of V165A and R166A IN (3). For example, the in vivo integration defect of a catalytically inactive HIV-1D64A class I mutant virus was rescued by incorporating V165A IN in trans. This result led to the conclusion that V165A IN was catalytically active in vivo. Due to our results that different strains of HIV-1V165A and HIV-1R166A displayed nuclear localization phenotypes similar to class II IN mutant viruses, we next examined the catalytic activities of V165A and R166A IN.
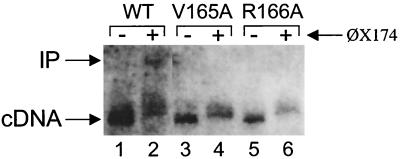
Retroviral PICs isolated from infected cells support endogenous cDNA integration in vitro (6, 7, 20, 25, 28). HIV-1NL4-3, HIV-1V165A, and HIV-1R166A PICs were assayed for in vitro integration activity as previously described (13, 14). About 38% of the WT cDNA was converted into the integration product (Fig. 6, lanes 1 and 2). In contrast, neither HIV-1V165A nor HIV-1R166A supported a detectable level of in vitro PIC activity in repeated experiments (Fig. 6 and data not shown).
FIG. 6.
HIV-1V165A and HIV-1R166A do not support detectable levels of in vitro PIC activity. PICs were incubated in the absence or presence of φX174 target DNA as indicated. The lower mobility of the cDNA substrate in lanes 2, 4, and 6 was due to the large excess of target DNA (1.5 μg) present in the integration reaction mixtures. cDNA, 9.7-kb linear HIV-1 substrate; IP, 15.1-kb integration product.
V165A and R166A mutants are defective for in vitro 3′ processing and DNA strand transfer activities.
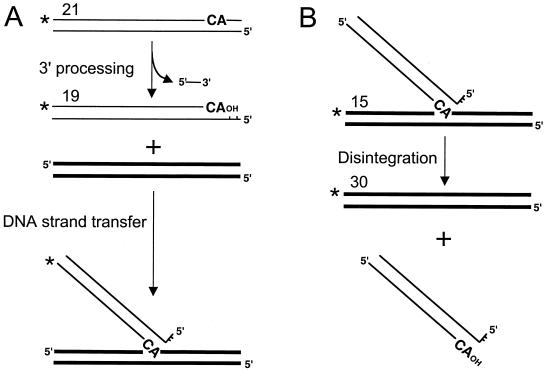
Two IN activities, 3′ processing and DNA strand transfer, are required for integration. During 3′ processing, IN removes a dinucleotide from each 3′ end of blunt-ended HIV-1. This reaction can be modeled using oligonucleotide substrates that mimic the ends of endogenous cDNA (reviewed in reference 18). For example, if the DNA substrate is labeled at the 5′ end of the strand that is to be cleaved by IN, 3′ processing yields a labeled product 2 bases shorter than the starting substrate (Fig. 7A). IN can also integrate the processed 3′ end into a second oligonucleotide in the reaction mixture. Since virtually any DNA sequence can support integration, DNA strand transfer yields a population of products that migrate more slowly than does substrate DNA (18).
FIG. 7.
Schematic of in vitro 3′ processing, DNA strand transfer, and disintegration reactions. (A) During 3′ processing, IN cleaves a dinucleotide from the 3′ end of HIV-1 DNA, yielding a product 2 bases shorter than the substrate. During DNA strand transfer, IN joins the processed 3′ end to a 5′ phosphate in target DNA, yielding a population of products longer that the processed DNA strand. (B) Disintegration is essentially a reversal of the DNA strand transfer reaction. Here, IN resolves a branched Y-mer oligonucleotide into separate viral and target DNA duplexes. Thin lines, the U5 end of HIV-1 cDNA; bold lines, target DNA. Numbers refer to lengths of radiolabeled substrate and product DNA strands. ∗, labeled 5′ ends.
In addition to the physiologically relevant 3′ processing and DNA strand transfer reactions, IN catalyzes a third in vitro activity termed disintegration (15). In this reaction, IN resolves a substrate that mimics the product of DNA strand transfer into its separate viral and target DNA components (Fig. 7B). Many purified mutant INs, such as amino- and/or carboxyl-terminal deletion proteins, display efficient disintegration activity yet fail to support detectable levels of in vitro 3′ processing and DNA strand transfer activities (10, 51). In contrast, single-amino-acid substitutions in the IN active site abolish all three enzyme activities (22, 50).
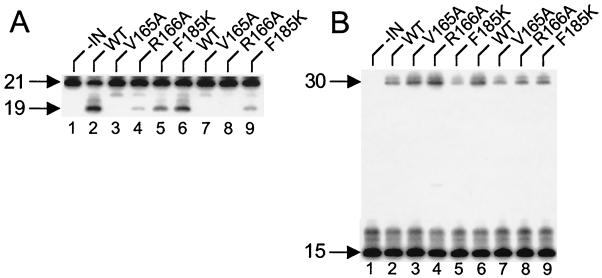
WT, V165A, R166A, and F185K proteins purified from E. coli were assayed for in vitro 3′ processing, DNA strand transfer, and disintegration activities at 0.25 and 0.5 μM IN. At 0.5 μM IN, the 3′ processing activity of V165A was reduced 30- to 40-fold compared to WT in repeated experiments (Fig. 8A, lanes 2 and 3). R166A, somewhat more active than V165A, displayed about 9% of WT activity (lane 4). At 0.25 μM IN, V165A and R166A displayed only about 1 and 3% of WT activity, respectively (lanes 6 to 8). In contrast, the activity of the F185K mutant was reduced only three to fourfold from the WT activity at both concentrations of IN (lanes 5 and 9). Long autoradiographic exposures revealed that WT and mutant DNA strand transfer activities mirrored 3′ processing activities (data not shown).
FIG. 8.
V165A and R166A IN proteins are defective for 3′ processing and DNA strand transfer activities. (A) 3′ processing activity. IN was omitted from the reaction in lane 1. Lanes 2 to 5, 0.5 μM indicated IN protein; lanes 6 to 9, 0.25 μM IN; 21, migration position of the 21-base labeled strand in substrate DNA; 19, migration position of the cleaved 19-base product (Fig. 7A). (B) Disintegration activity. 15, migration position of the 15-base substrate DNA strand; 30, migration position of the 30-base product (Fig. 7B). Other labeling is as in panel A.
In agreement with the previous report (3), V165A and R166A displayed close to the WT level of IN disintegration activity (Fig. 8B). Thus, recombinant V165A and R166A proteins are not dead for IN catalysis. However, our results highlight that these mutants were selectively defective for the 3′ processing and DNA strand transfer activities required for HIV-1 integration in vivo.
DISCUSSION
Productive retroviral replication requires the integration of the cDNA reverse transcript into a cell chromosome. In vivo, integration is mediated by PICs whose large size precludes their passive transport through NPCs in intact nuclear membranes. Whereas simple oncoretroviruses like MoMLV appear to circumvent the nuclear membrane by waiting for its dissolution during the M phase of the cell cycle, lentiviruses like HIV-1 can be actively transported in nondividing cells under conditions where the nuclear membrane remains intact. Thus, in contrast to simple retroviruses, lentiviruses appear to have evolved NLSs to guide their nuclear localization in nondividing cells (reviewed in reference 27, 29, and 52).
Mutants of MoMLV (55) and HIV-1 (3, 57) were recently reported to be defective for nuclear localization in rapidly dividing cells, suggesting that retroviruses might interact with specific host cell factors to gain nuclear access even when membranes are actively disassembled. One region of HIV-1 implicated in this novel class of nuclear import was Val-165 and Arg-166 in IN (3). To investigate the role of this NLS in HIV-1 replication and nuclear localization, we analyzed WT and mutant IN activities under a variety of assay conditions.
Defective HIV-1V165A and HIV-1R166A replication and nuclear import.
Although our results confirmed earlier reports that HIV-1V165A (3) and HIV-1R166A (3, 53) were replication defective and transported less of their cDNA to cell nuclei than did WT controls, our results discount a specific role for Val-165 and Arg-166 in PIC nuclear localization for a variety of reasons. First, FLAG-tagged V165A IN (Fig. 1) and GFP-IN/V165A and GFP-IN/R166A (Fig. 2) fusion proteins localized predominantly to cell nuclei. Second, HIV-1V165A and HIV-1R166A displayed reverse transcription and 2-LTR circle formation profiles similar to those of previously described replication-defective class II IN mutant viruses (Fig. 4 and 5). Although the class II IN mutants analyzed here were defective for particle assembly and release (32, 40), a large number of other replication-defective IN mutant viruses that assembled normally also yielded low levels of 2-LTR circles in acute infections (24, 49, 53). Based on this, we speculate that defective nuclear import may be a phenotype in common to a variety of replication-defective IN mutant viruses.
Defective V165A and R166A IN catalytic activity.
A third line of reasoning to argue against a specific role for Val-165 and Arg-166 in PIC nuclear import was based on results of in vitro integration assays. The conclusion that HIV-1V165A and HIV-1R166A were primarily defective for nuclear import was in large part based on the inferred WT activities of V165A and R166A IN. This came from two lines of investigation. First, the in vivo integration defect of IN active-site mutant HIV-1D64A was rescued by incorporating a Vpr-V165A IN fusion protein in trans into phenotypically mixed virions. Second, recombinant V165A and R166A IN proteins displayed the WT level of disintegration activity (3). Although our results confirmed that recombinant V165A and R166A IN each displayed the WT level of disintegration activity (Fig. 8B), we determined that these proteins were defective for the 3′ processing and DNA strand transfer activities required for integration in vivo (Fig. 6 and 8A).
To investigate the reported complementation between Vpr-V165A and D64A IN, purified V165A and active-site mutant D116N (22) were mixed prior to their addition to in vitro integration reaction mixtures. At 0.25 μM total IN protein, this mixture supported approximately 6% of the WT 3′ processing activity, which was approximately fivefold more activity than that observed with V165A alone (data not shown). Although this level of IN activity was apparently well below the observed level of in vivo complementation (3), our results are consistent with the interpretation that a dead active-site IN mutant can stimulate the activity of a highly defective core domain mutant protein in trans. We previously observed a similar low level of in vitro complementation by premixing D116N with the highly defective P109S IN mutant (47).
Taken together, our results discount a specific role for Val-165 and Arg-166 in HIV-1 PIC nuclear localization and instead suggest that impaired IN catalytic activity was central to the replication defects of HIV-1V165A and HIV-1R166A.
Nuclear localization of HIV-1 PICs.
Since HIV-1 productively infects nondividing target cells, there is reason to believe that NLSs function to import PICs through NPCs; matrix, Vpr, and IN mutants specifically blocked at nuclear import in nondividing cells have been identified. However, the precise contributions of each of these proteins to PIC nuclear import in nondividing cells remain unclear (3, 43; reviewed in references 19, 27, and 29).
Since mutations in MoMLV p12 Gag (55) and HIV-1 (3, 57) reportedly blocked replication at the nuclear import step in dividing cells, there was precedence that host cell factors might in large part regulate retroviral PIC nuclear localization independent of cell division (19). However, our analyses of the HIV-1 DNA flap and Val-165/Arg-166 in IN argue against this perspective. Since we were unable to ascribe specific nuclear localization functions to either the DNA flap or Val-165/Arg-166, there is presently no irrefutable evidence for a specific block(s) in HIV-1 nuclear import in dividing cells. Since there is no obvious counterpart of MoMLV p12 Gag in HIV-1, it is currently unclear whether HIV-1 mutagenesis will reveal an import-defective replication block in dividing cell types.
Our results indicate that pleiotropic IN mutants import viral cDNA less efficiently than does WT HIV-1 in dividing cell types. This could be interpreted as evidence that IN plays an important role in nuclear import, either through an as yet unidentified although perhaps easily disturbed NLS or indirectly by altering the structure and/or stability of the incoming RTC or PIC. The pleiotropic nature of many IN mutations has complicated the analysis of its precise role in PIC nuclear import. Continued analyses of the reverse transcription, nuclear localization, integration, and particle assembly steps in the HIV-1 life cycle should help define viral sequences that govern PIC nuclear localization in dividing as well as nondividing cells.
Acknowledgments
We thank G. Adelmant, H. Göttlinger, J. Gray, M. H. Malim, T. Masuda, R. C. Mulligan, D. Waters, and R. Yu for providing plasmid DNAs; A. Hitchcock for assisting with the DeltaVision microscope; and M. H. Malim for sharing results prior to publication.
This work was supported by NIH grants AI39394 (A.E.), AI45313 (A.E.), GM36373 (P.A.S.), AI28691 (Center for AIDS Research), and a Predoctoral Fellowship from the Howard Hughes Medical Institute (E.D.).
REFERENCES
- 1.Adachi, A., H. E. Gendelman, S. Koenig, T. Folks, R. Willey, A. Rabson, and M. A. Martin. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and non-human cells transfected with an infectious molecular clone. J. Virol. 59:284-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansari-Lari, M. L., L. A. Donehower, and R. A. Gibbs. 1995. Analysis of human immunodeficiency virus type 1 integrase mutants. Virology 211:332-335. [DOI] [PubMed] [Google Scholar]
- 3.Bouyac-Bertoia M., J. D. Dvorin, R. A. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 4.Brin, E., J. Yi, A. M. Skalka, and J. Leis. 2000. Modeling the late steps in HIV-1 retroviral integrase-catalyzed DNA integration. J. Biol. Chem. 275:39287-39295. [DOI] [PubMed] [Google Scholar]
- 5.Brown, H. E. V., H. Chen, and A. Engelman. 1999. Structure-based mutagenesis of the human immunodeficiency virus type 1 DNA attachment site: effects on integration and cDNA synthesis. J. Virol. 73:9011-9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, P. O., B. Bowerman, H. E. Varmus, and J. M. Bishop. 1987. Correct integration of retroviral DNA in vitro. Cell 49:347-356. [DOI] [PubMed] [Google Scholar]
- 7.Brown, P. O., B. Bowerman, H. E. Varmus, and J. M. Bishop. 1989. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc. Natl. Acad. Sci. USA 86:2525-2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukovsky, A., and H. Göttlinger. 1996. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J. Virol. 70:6820-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukrinsky, M. I., N. Sharova, M. P. Dempsey, T. L. Stanwick, A. G. Bukrinskaya, S. Haggerty, and M. Stevenson. 1992. Active nuclear import of human immunodeficiency virus preintegration complexes. Proc. Natl. Acad. Sci. USA 89:6580-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bushman, F. D., A. Engelman, I. Palmer, P. Wingfield, and R. Craigie. 1993. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc. Natl. Acad. Sci. USA 90:3428-3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butler S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7:631-634. [DOI] [PubMed] [Google Scholar]
- 12.Chen, H., and A. Engelman. 2000. Characterization of a replication-defective human immunodeficiency virus type 1 att site mutant that is blocked after the 3′ processing step of retroviral integration. J. Virol. 74:8188-8193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, H., and A. Engelman. 2001. Asymmetric processing of human immunodeficiency virus type 1 cDNA in vivo: implications for functional end coupling during the chemical steps of DNA transposition. Mol. Cell. Biol. 21:6758-6767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, H., S.-Q. Wei, and A. Engelman. 1999. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type 1 intasome. J. Biol. Chem. 274:17358-17364. [DOI] [PubMed] [Google Scholar]
- 15.Chow, S. A., K. A. Vincent, V. Ellison, and P. O. Brown. 1992. Reversal of integration and DNA splicing mediated by integrase of human immunodeficiency virus. Science 255:723-726. [DOI] [PubMed] [Google Scholar]
- 16.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Rescue of multiple viral functions by a second-site suppressor of a human immunodeficiency virus type 1 nucleocapsid mutation. J. Virol 74:4273-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coffin, J. M. 1996. Retroviridae: the viruses and their replication, p. 1767-1848. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Field's virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 18.Craigie, R., A. B. Hickman, and A. Engelman. 1995. Integrase, p. 53-71. In J. Karn (ed.), HIV volume II: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 19.Cullen, B. R. 2001. Journey to the center of the cell. Cell 105:697-700. [DOI] [PubMed] [Google Scholar]
- 20.Ellison, V., H. Abrams, T. Roe, J. Lifson, and P. O. Brown. 1990. Human immunodeficiency virus integration in a cell-free system. J. Virol. 64:2711-2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Engelman, A. 1999. In vivo analysis of retroviral integrase structure and function. Adv. Virus Res. 52:411-416. [DOI] [PubMed] [Google Scholar]
- 22.Engelman, A., and R. Craigie. 1992. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol. 66:6361-6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelman, A., G. Englund, J. M. Orenstein, M. A. Martin, and R. Craigie. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engelman, A., Y. Liu, H. Chen, M. Farzan, and F. Dyda. 1997. Structure-based mutagenesis of the catalytic domain of human immunodeficiency virus type 1 integrase. J. Virol. 71:3507-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farnet, C. M., and W. A. Haseltine. 1990. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc. Natl. Acad. Sci. USA 87:4164-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher, A. G., E. Collati, L. Ratner, R. C. Gallo, and F. Wong-Staal. 1985. A molecular clone of HTLV-III with biological activity. Nature 316:262-265. [DOI] [PubMed] [Google Scholar]
- 27.Fouchier, R. A., and M. H. Malim. 1999. Nuclear import of human immunodeficiency virus type-1 preintegration complexes. Adv. Virus Res. 52:275-299. [DOI] [PubMed] [Google Scholar]
- 28.Fujiwara, T., and K. Mizuuchi. 1988. Retroviral DNA integration: structure of an integration intermediate. Cell 54:497-504. [DOI] [PubMed] [Google Scholar]
- 29.Goff, S. P. 2001. Intracellular trafficking of retroviral genomes during the early phase of infection: viral exploitation of cellular pathways. J. Gene Med. 3:517-528. [DOI] [PubMed] [Google Scholar]
- 30.Göttlinger H. G., T. Dorfman, E. A. Cohen, and H. A. Haseltine. 1993. Vpu protein of human immunodeficiency virus type 1 enhances the release of capsids produced by gag gene constructs of widely divergent retroviruses. Proc. Natl. Acad. Sci. USA 90:7381-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Helseth, E., M. Kowalski, D. Gabuzda, U. Olshevsky, W. Haseltine, and J. Sodroski. 1990. Rapid complementation assays measuring replicative potential of human immunodeficiency virus type 1 envelope glycoprotein mutants. J. Virol. 64:2416-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenkins, T. M., A. Engelman, R. Ghirlando, and R. Craigie. 1996. A soluble active mutant of HIV-1 integrase: involvement of both the core and C-terminal domains in multimerization. J. Biol. Chem. 271:7712-7718. [DOI] [PubMed] [Google Scholar]
- 33.Julias J. G., A. L. Ferris, P. L. Boyer, and S. H. Hughes. 2001. Replication of phenotypically mixed human immunodeficiency virus type 1 virions containing catalytically active and catalytically inactive reverse transcriptase. J. Virol. 75:6537-6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leavitt, A. D., G. Robles, N. Alesandro, and H. E. Varmus. 1996. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate DNA efficiently during infection. J. Virol. 70:721-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lewin, S. R., M. Vesanen, L. Kostrikis, A. Hurley, M. Duran, L. Zhang, D. D. Ho, and M. Markowitz. 1999. Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. J. Virol. 73:6099-6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lewis, P. F., and M. Emerman. 1994. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J. Virol. 68:510-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mattaj, I. W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265-306. [DOI] [PubMed] [Google Scholar]
- 38.Meyers, G., S. Wain-Hobson, B., Korber, R. F. Smith, and G. N. Pavlakis. 1993. Human retroviruses and AIDS. A compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.M.
- 39.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency type 1 preintegration complexes: studies of organization and function. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakajima, N., R. Lu, and A. Engelman. 2001. Human immunodeficiency virus type 1 replication in the absence of integrase-mediated DNA recombination: definition of permissive and nonpermissive T-cell lines. J. Virol. 75:7944-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petit, C., O. Schwartz, and F. Mammano. 1999. Oligomerization within virions and subcellular localization of human immunodeficiency virus type 1 integrase. J. Virol. 73:5079-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Göttlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roe, T., T. C. Reynolds, G. Yu, and P. O. Brown. 1993. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 12:2099-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roth, M. J., P. L. Schwartzberg, and S. P. Goff. 1989. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell 58:47-54. [DOI] [PubMed] [Google Scholar]
- 46.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 47.Taddeo, B., F. Carlini, P. Verani, and A. Engelman. 1996. Reversion of a human immunodeficiency virus type 1 integrase mutant at a second site restores enzyme function and virus infectivity. J. Virol. 70:8277-8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taddeo, B., W. A. Haseltine, and C. M. Farnet. 1994. Integrase mutants of human immunodeficiency virus type 1 with a specific defect in integration. J. Virol. 68:8401-8405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsurutani, N., M. Kubo, Y. Maeda, T. Ohashi, N. Yamamoto, M. Kannagi, and T. Masuda. 2000. Identification of critical amino acid residues in human immunodeficiency virus type 1 IN required for efficient proviral DNA formation at steps prior to integration in dividing and nondividing cells. J. Virol. 74:4795-4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Gent, D. C., A. A. M. Oude Groeneger, and R. H. A. Plasterk. 1992. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc. Natl. Acad. Sci. USA 89:9598-9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vink, C., A. A. M. Oude Groeneger, and R. H. A. Plasterk. 1993. Identification of the catalytic and DNA-binding region of human immunodeficiency virus type 1 integrase protein. Nucleic Acids Res. 21:1419-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whittaker, G. R., M. Kann, and A. Helenius. 2000. Viral entry into the nucleus. Annu. Rev. Cell Dev. Biol. 16:627-651. [DOI] [PubMed] [Google Scholar]
- 53.Wiskerchen, M., and M. A. Muesing. 1995. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct gene expression from unintegrated viral DNA templates, and sustain propagation in primary cells. J. Virol. 69:376-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yoder, K. E., and F. D. Bushman. 2000. Repair of gaps in retroviral DNA integration intermediates. J. Virol. 74:11191-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the Moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan, C. C., W. Miley, and D. Waters. 2001. A quantification of human cells using an ERV-3 real time PCR assay. J. Virol. Methods 91:109-117. [DOI] [PubMed] [Google Scholar]
- 57.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]