Abstract
In vitro-assembled core-like particles produced from alphavirus capsid protein and nucleic acid were studied by cryoelectron microscopy. These particles were found to have a diameter of 420 Å with 240 copies of the capsid protein arranged in a T=4 icosahedral surface lattice, similar to the nucleocapsid core in mature virions. However, when the particles were subjected to gentle purification procedures, they were damaged, preventing generation of reliable structural information. Similarly, purified nucleocapsid cores isolated from virus-infected cells or from mature virus particles were also of poor quality. This suggested that in the absence of membrane and glycoproteins, nucleocapsid core particles are fragile, lacking accurate icosahedral symmetry.
Alphavirus virions consist of a nucleocapsid core, a host-derived phospholipid membrane, and an outer glycoprotein layer with 80 trimeric spikes (24). The nucleocapsid core in the mature alphavirus particle has a diameter of ∼420 Å and consists of 240 copies of the capsid protein surrounding an 11.7-kb viral RNA genome. Each spike on the virus surface is a trimer of E1-E2 glycoprotein heterodimers. Cryoelectron microscopy (cryoEM) and three-dimensional image reconstructions of numerous alphaviruses (3, 7, 14, 16, 17, 26, 27) have shown that the capsid protein subunits in the nucleocapsid core and the external glycoprotein spikes have matching T=4 icosahedral symmetry.
The assembly of alphaviruses is a multistep process involving nucleocapsid core formation followed by glycoprotein attachment and virus budding (24). Upon translation from a 26S subgenomic mRNA, the capsid protein is autocatalytically cleaved from the polyprotein, the viral genome RNA is encapsidated, and nucleocapsid cores are assembled in the cytoplasm. Simultaneously, the glycoproteins are translated and processed through the endoplasmic reticulum and Golgi and, subsequently, they are transported to the plasma membrane. The cytoplasmic nucleocapsid cores interact with the transmembrane E1-E2 glycoproteins at the plasma membrane, allowing the mature virions to bud from the cell. It has been suggested that this interaction is in part accomplished by 2 of the 33 cytoplasmic residues of E2 that bind into a hydrophobic pocket in the capsid protein (11, 21).
Forsell et al. (7) have suggested that membrane glycoproteins have a direct role in organizing the structure of the mature alphavirus particle. They suggest that both spike-spike and spike-capsid protein interactions are responsible for proper virus symmetry. More recently, Pletnev et al. (19) and Lescar et al. (12) independently suggested that it is the E1 glycoprotein that forms an icosahedral scaffold on the virus surface during assembly. Pletnev et al. (19) further suggested that cytoplasmic nucleocapsid cores lack accurate icosahedral symmetry and require association with the glycoproteins at the plasma membrane to gain a well-defined icosahedral structure.
The nucleocapsid core can be divided into three sections defined by their protein and RNA content. The protein layer is defined by 30 hexamers and 12 pentamers around the periphery of the nucleocapsid core between radii of 180 and 210 Å. The carboxy-terminal domain of the Sindbis virus capsid protein (residues 114 to 264) (4) has been fitted into this layer of the Sindbis virus cryoEM density, and the resulting outer surface charge distribution is largely positive, which is important for close contacts with the acidic phospholipid membrane (27). The base of the nucleocapsid core, between radii of 150 and 180 Å, is a mixture of the positively charged amino-terminal domain of the capsid protein and parts of the RNA genome and contributes to the stabilization of the nucleocapsid core (7, 18) during assembly in the cytoplasm. The cryoEM density of this protein-RNA mixed region is about 80% in height of the density of the outer protein layer, suggesting a slightly less-well-ordered structure. Nevertheless, alphavirus virions also maintain T=4 icosahedral symmetry in this region (27). Internal to a radius of 150 Å is the core that consists mainly of genomic RNA.
Nucleocapsid core-like particles (CLPs) can be assembled in vitro by using purified capsid protein with single-stranded nucleic acid (22, 25) or in vivo using a baculovirus system expressing capsid protein (data not shown). CLPs from either system are the same size, as determined by negative stain and cryoEM, and have similar sedimentation properties as cytoplasmic nucleocapsid cores isolated from virus-infected cells (reference 22 and data not shown). However, until now, there has been no three-dimensional structural evidence that has shown that CLPs resemble either cytoplasmic nucleocapsid cores or the nucleocapsid core found in mature virus particles.
Here, we present the cryoEM reconstructions of in vitro-assembled CLPs from Ross River virus (RRV) and western equine encephalitis virus (WEEV) capsid proteins expressed in Escherichia coli and assembled with a synthetic 48-nucleotide oligomer (22, 25). Only after numerous trials and failed attempts at determining the structure of purified CLPs and cytoplasmic nucleocapsid cores was it concluded that extensive centrifugation steps, such as those required for sedimentation through density gradients, were detrimental and consequently were eliminated from the procedure. The cryoEM reconstructions of nonpurified CLPs, although only at 30 Å resolution, show that they contain the same T=4 quasi-symmetric arrangement of hexamers and pentamers as nucleocapsid cores found in mature virions. Inability to improve the resolution of the reconstruction suggested that the CLPs vary slightly in structure among themselves and may deviate from exact icosahedral symmetry, consistent with the suggestions of Pletnev et al. (19).
Capsid protein expression and purification and CLP assembly.
RRV capsid protein (amino acid residues 1 to 270) from the T48 strain (10) was cloned into pET29b (Novagen, Madison, Wis.) as previously described (22) and expressed in BL21(DE3)RIL cells. Cells were grown in Luria broth at 37°C until an optical density at 600 nm (OD600) of 0.5 was reached. At that time, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to obtain a final concentration of 1 mM. Cells were allowed to grow for an additional 6 h at 37°C before being harvested by centrifugation at 4,000 × g at 4°C for 10 min. Cells were resuspended in 20 mM HEPES (pH 7.5), 0.25 M NaCl, and 5 mM EDTA (30 ml of buffer/liter of cells) and lysed after two passages through a cold French Press cell. The lysate was clarified by centrifugation at 23,000 × g at 4°C for 30 min, and then the supernatant was loaded onto a 6-ml Resource S column (Amersham Biosciences, Piscataway, N.J.) that was preequilibrated with lysis buffer. A step gradient was applied, and RRV capsid protein was eluted in two peaks at 0.66 M and 0.94 M NaCl. The protein that eluted without nucleic acid at 0.66 M NaCl was concentrated and exchanged into assembly buffer (25 mM HEPES [pH 7.4], 100 mM potassium acetate, 1.7 mM magnesium acetate) and used for in vitro assembly reactions.
WEEV capsid protein (amino acid residues 1 to 259) from strain BFS1703 (9) was cloned into pSBET (20) and expressed in BL21(DE3) cells. Cells were grown in Luria broth at 37°C until an OD600 of 0.4 was reached, at which time IPTG was added to obtain a final concentration of 1 mM; cells continued to grow for an additional 10 h at 37°C. The cells were pelleted at 4,000 × g at 4°C for 10 min and resuspended in buffer A (25 mM Tris-Cl [pH 7.4], 5 mM EDTA, 5% glycerol [vol/vol], 5 mM dithiothreitol) and 50 mM NaCl (10 ml of buffer/liter of cells). Cells were lysed using a cold French press, and the soluble fraction was obtained after centrifugation at 23,000 × g at 4°C for 30 min. The clarified cell lysate was loaded onto a 20-ml SP Sepharose Fast Flow column (Amersham Biosciences) equilibrated with buffer A. A linear gradient was applied, and the WEEV capsid protein was eluted with 0.6 M NaCl in buffer A. The capsid protein was concentrated with a Centriprep-10 concentrator (Millipore, Bedford, Mass.) and exchanged into buffer A and 0.12 M NaCl. The concentrated protein was applied to a Superdex 75 10/30 column (Amersham Biosciences), and the fractions containing WEEV capsid protein were concentrated and exchanged into assembly buffer.
In contrast to Sindbis virus capsid protein (22), full-length RRV and WEEV capsid proteins could be expressed and purified. In vitro assembly of WEEV CLPs was essentially as described for Sindbis virus and RRV CLPs (22).
Purification of cytoplasmic nucleocapsid cores.
Preassembled nucleocapsid cores were isolated from virus-infected cells. Cells were lysed with detergent (6, 18) to release the nucleocapsid cores from internal membranes (8, 13), and sedimentation centrifugation was used to further purify them from the cell lysate. To eliminate the possibility that nucleocapsid cores were sensitive to the detergent present during their isolation, nucleocapsid cores from the mutant E2-L402Y Sindbis virus, which accumulated in the cytoplasm independent of membrane association (15) and hence did not require detergent for their isolation, were also purified and used for cryoEM studies. Furthermore, some of the purified cytoplasmic nucleocapsid cores were cross-linked with dimethyl suberimidate (23) to enhance their stability.
CryoEM of purified cytoplasmic nucleocapsid cores and CLPs.
CryoEM reconstructions (2) were not successful when the cytoplasmic nucleocapsid cores or CLPs were purified by sedimentation centrifugation (A. E. Hamburger, S. Lee, S. Mukhopadhyay, T. L. Tellinghuisen, and W. Zhang, unpublished data). The failed image reconstruction attempts included efforts using both the common lines method (5) and the model-based approach of the polar Fourier transform method (1). The nucleocapsid core from the previously published whole-virus cryoEM structure of RRV (3) and the outer protein layer of the RRV nucleocapsid core were used as starting models. However, in these procedures, the particle orientation and centers varied greatly from cycle to cycle and the correlation coefficients approached zero with further iterations, resulting in a smooth ball of density.
It was possible that purified Sindbis virus capsid protein CLPs did not produce reliable reconstructions because an N-terminal truncated Sindbis virus capsid protein (amino acid residues 19 to 264) (22) was used and, as a result, the CLPs were less stable. However, purified CLPs using full-length RRV capsid protein also did not yield satisfactory structural results. Furthermore, cytoplasmic nucleocapsid cores that were cross-linked (23) did not improve the reconstruction, and it was concluded that perhaps the centrifugation step had already damaged the cytoplasmic nucleocapsid cores. In contrast, several high-resolution cryoEM structures have been obtained from mature alphavirus particles that were purified by different methods, including sedimentation centrifugation (14, 27; P. R. Chipman, personal communication). These results suggested that cytoplasmic nucleocapsid cores and CLPs, unlike mature virus particles, are fragile and could be damaged during centrifugation, although the damage could only be detected by an inability to produce good three-dimensional reconstructions.
CryoEM of nonpurified CLPs.
A successful reconstruction of in vitro-assembled RRV and WEEV CLPs was achieved when the particles were not purified by sedimentation gradient centrifugation after being assembled (Table 1). In contrast to the results with purified CLPs, the reconstruction of the nonpurified CLPs showed T=4 quasi-symmetry, consistent with cryoEM reconstructions of the whole virus (Fig. 1 ). The 30 Å resolution limit and the need to reject 70% of all boxed particles for both CLP reconstructions suggests that CLPs, and most likely cytoplasmic nucleocapsid cores, are fragile and that their structure can be easily damaged during purification (see above). In comparison, reconstructions of mature alphaviruses have at least 20 Å resolution and can readily be extended to 10 Å resolution by increasing the number of particles (14, 27). Presumably, the nucleocapsid core becomes a rigid icosahedral structure only after interactions with the phospholipid membrane and the glycoproteins (7, 19). It was not possible to obtain a successful reconstruction of the cytoplasmic nucleocapsid cores because centrifugation was necessary to produce a sufficiently clean preparation. Biochemical analyses (Table 2) (22) have demonstrated that the in vitro-assembled CLPs have similar properties to cytoplasmic nucleocapsid cores. Thus, it is reasonable to postulate that the cytoplasmic nucleocapsid cores have a similar structure to the CLPs and to the nucleocapsid core of the mature virus.
TABLE 1.
CryoEM and three-dimensional image reconstruction dataa
| Sample | No. of particles in map | No. of particles boxed | Resolution (Å) | Correl. coeff.b | Dose (e−/Å2) | Defocus range (μm) | Starting model |
|---|---|---|---|---|---|---|---|
| Mature RRV | 961 | 1,652 | 20 | 0.50 | 9 | 1.12-3.64 | Sindbis virus |
| 30 | 0.93 | ||||||
| RRV CLP | 671 | 2,339 | 30 | 0.48 | 26 | 1.70-3.84 | RRV corec |
| WEEV CLP | 581 | 2,092 | 28 | 0.52 | 24 | 1.21-3.81 | RRV corec |
| 30 | 0.85 |
A Phillips CM300 FEG transmission electron microscope was used to record images on Kodak SO-163 film under low-dose conditions. All micrographs were digitized using a Zeiss-SCAI microdensitometer at 14-μm intervals. CryoEM reconstructions were performed as previously described (2).
Correlation coefficient is defined as CMP CC in Baker and Cheng (1).
RRV core refers to the nucleocapsid core from the cryoEM reconstruction of the mature RRV particle described above.
FIG. 1.
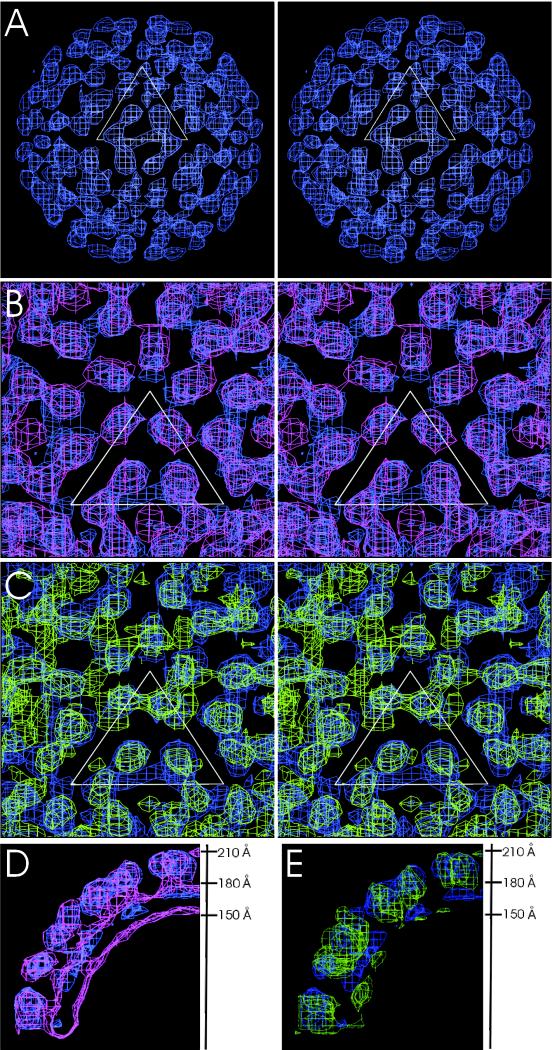
CryoEM density maps of RRV CLPs, WEEV CLPs, and the nucleocapsid core excised from the reconstruction of mature RRV. (A to C) Stereo diagrams, where only the top hemisphere of the map is shown. All maps are at 30 Å resolution and contoured at 2.5 σ. The white triangle represents one icosahedral asymmetric unit at a radius of 170 Å. The top vertex of the triangle is at the fivefold-symmetry axis. (A) The RRV CLP structure with density shown between radii of 150 and 210 Å. (B) The densities of both RRV CLP (blue; radii of 150 to 210 Å) and WEEV CLP (pink; radii of 180 to 210 Å). Note the quasi-symmetry present in both CLP reconstructions. (C) Superimposed densities of RRV CLP (blue; radii of 150 to 210 Å) and the nucleocapsid core from mature RRV (green; radii of 150 to 210 Å). (D) Central cross section along twofold axis of RRV CLP (blue) and WEEV CLP (pink), both from radii of 150 to 210 Å. (E) Same orientation as shown in panel D, with RRV CLP (blue) and the nucleocapsid core from the mature RRV particle (green).
TABLE 2.
Biochemical characterization of in vitro-assembled CLPs and in vivo-assembled nucleocapsid cores
| Property | Cytoplasmic nucleocapsid cores or CLPs | Assay methoda,b |
|---|---|---|
| Ionic strength | Unstable in assembly buffer plus 0.5 M NaCl | Agarose gel, EM |
| pH | Stable between pH 5.2 and 8.2 | Agarose gel, EM, gra- dient sedimentation |
| Temperature | Unstable above 55°C | Agarose gel |
| Nuclease sensitivity | Yes | Agarose gel, EM |
| Particle size | ≈410 ± 10 Å | CryoEM |
All assays were performed as described by Tellinghuisen et al. (22).
EM refers to negative-stain EM.
The most obvious difference between the structures of the RRV CLPs and the RRV nucleocapsid core of the mature virus is that the orientation of the pentamers is rotated by about 10° counterclockwise with respect to the icosahedral axial system when viewed from the outside (Fig. 1). In addition, in the RRV CLPs the density around the fivefold axes projects outwards, away from the center of the particle. In contrast, the orientation of the monomers in the hexamers between the RRV CLPs and the RRV nucleocapsid core appears to be the same (Fig. 1). Neither CLPs nor cytoplasmic nucleocapsid cores have yet been in contact with membrane proteins; thus, these particles probably represent an earlier stage of assembly. Hence, the difference at the fivefold axes between CLPs compared to the mature nucleocapsid core might be the result of different stages in the virus assembly process.
In contrast to the mature virus, the density in the base of the RRV CLPs has no T=4 quasi-symmetry, and the maximum density in this region is only about 35% in height of the density in the protein layer. This difference in the base of the CLP compared to the nucleocapsid core of the mature virion suggests specific protein-protein and protein-RNA interactions in the mature particle that are weaker in the CLPs, possibly because a DNA oligonucleotide rather than viral RNA was used in the assembly reactions. Furthermore, in the protein layer of the RRV CLPs, the volume of density representing each capsid protein monomer is less than its corresponding volume in the RRV nucleocapsid core of the mature virus.
In both the RRV and WEEV CLP reconstructions, some additional density appears in the middle of each hexamer (but not pentamer) at a radial distance between 195 and 225 Å. Although this extra density might represent protein or noise, the most probable interpretation is that it represents a DNA oligonucleotide that binds to the external surface of the positively charged hexameric arrangement of capsid proteins (27).
Paredes et al. (16) suggested that there is a difference in the nucleocapsid core of Old World (RRV, Sindbis virus) and New World (WEEV, Venezuelan equine encephalomyelitis virus) alphaviruses. Relative to Sindbis virus, the orientations of the hexamers and pentamers in Venezuelan equine encephalomyelitis virus were rotated by approximately 11° and 4° clockwise, respectively (16). In contrast to what has been observed in the mature virus particles, no significant difference in the orientation of the hexamers and pentamers was seen between the RRV CLPs and the WEEV CLPs (Fig. 1). This suggests that CLPs and cytoplasmic nucleocapsid cores are similar in their organization, regardless of the alphavirus. Only after their interaction with the glycoproteins and lipid membrane will they attain a conformation that may be specific to Old and New World viruses.
Implications for particle assembly and structure.
In vitro-assembled CLPs are a reliable representation of the nucleocapsid cores found in the cytoplasm of infected cells. Biochemical analyses showed that CLPs and cytoplasmic nucleocapsid cores were similar in size and stability, and cryoEM reconstructions showed that CLPs closely resemble the nucleocapsid core found in the mature virus. Thus, in vitro-assembled CLPs are good models for studying alphavirus nucleocapsid core assembly, permitting the isolation and characterization of assembly intermediates as well as of fully assembled CLPs.
Acknowledgments
We thank Cheryl Towell and Sharon Wilder for help in preparation of the manuscript, and Rob Ashmore and Chuan Xiao for assistance with RobEM and other programs (see http://bilbo.bio.purdue.edu/∼viruswww/Rossmann_home/softwares.shtml).
The work was supported by an NIH Program project grant to R.J.K. and M.G.R. and others (AI45976), an NIH grant to R.J.K. (GM56279), a Purdue University reinvestment grant, and a grant from the Keck Foundation.
REFERENCES
- 1.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120-130. [DOI] [PubMed] [Google Scholar]
- 2.Baker, T. S., N. H. Olson, and S. D. Fuller. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 63:862-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng, R. H., R. J. Kuhn, N. H. Olson, M. G. Rossmann, H. K. Choi, T. J. Smith, and T. S. Baker. 1995. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell 80:621-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi, H. K., L. Tong, W. Minor, P. Dumas, U. Boege, M. G. Rossmann, and G. Wengler. 1991. Structure of Sindbis virus core protein reveals a chymotrypsin-like serine proteinase and the organization of the virion. Nature (London) 354:37-43. [DOI] [PubMed] [Google Scholar]
- 5.Crowther, R. A. 1971. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Phil. Trans. R. Soc. Lond. B 261:221-230. [DOI] [PubMed] [Google Scholar]
- 6.Forsell, K., G. Griffiths, and H. Garoff. 1996. Preformed cytoplasmic nucleocapsids are not necessary for alphavirus budding. EMBO J. 15:6495-6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forsell, K., L. Xing, T. Kozlovska, R. H. Cheng, and H. Garoff. 2000. Membrane proteins organize a symmetrical virus. EMBO J. 19:5081-5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 107:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahn, C. S., S. Lustig, E. G. Strauss, and J. H. Strauss. 1988. Western equine encephalitis virus is a recombinant virus. Proc. Natl. Acad. Sci. USA 85:5997-6001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuhn, R. J., H. G. Niesters, Z. Hong, and J. H. Strauss. 1991. Infectious RNA transcripts from Ross River virus cDNA clones and the construction and characterization of defined chimeras with Sindbis virus. Virology 182:430-441. [DOI] [PubMed] [Google Scholar]
- 11.Lee, S., K. E. Owen, H. K. Choi, H. Lee, G. Lu, G. Wengler, D. T. Brown, M. G. Rossmann, and R. J. Kuhn. 1996. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure 4:531-541. [DOI] [PubMed] [Google Scholar]
- 12.Lescar, J., A. Roussel, M. W. Wein, J. Navaza, S. D. Fuller, G. Wengler, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 13.Lopez, S., J. S. Yao, R. J. Kuhn, E. G. Strauss, and J. H. Strauss. 1994. Nucleocapsid-glycoprotein interactions required for assembly of alphaviruses. J. Virol. 68:1316-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mancini, E. J., M. Clarke, B. Gowen, T. Rutten, and S. D. Fuller. 2000. Cryo-electron microscopy reveals the functional anatomy of an enveloped virus, Semliki Forest virus. Mol. Cell 5:255-266. [DOI] [PubMed] [Google Scholar]
- 15.Owen, K. E., and R. J. Kuhn. 1997. Alphavirus budding is dependent on the interaction between the nucleocapsid and hydrophobic amino acids on the cytoplasmic domain of the E2 envelope glycoprotein. Virology 230:187-196. [DOI] [PubMed] [Google Scholar]
- 16.Paredes, A., K. Alwell-Warda, S. C. Weaver, W. Chiu, and S. J. Watowich. 2001. Venezuelan equine encephalomyelitis virus structure and its divergence from Old World alphaviruses. J. Virol. 75:9532-9537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paredes, A. M., D. T. Brown, R. Rothnagel, W. Chiu, R. J. Schoepp, R. E. Johnston, and B. V. V. Prasad. 1993. Three-dimensional structure of a membrane-containing virus. Proc. Natl. Acad. Sci. USA 90:9095-9099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perera, R., K. E. Owen, T. L. Tellinghuisen, A. E. Gorbalenya, and R. J. Kuhn. 2001. Alphavirus nucleocapsid protein contains a putative coiled-coil alpha-helix important for core assembly. J. Virol. 75:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pletnev, S. V., W. Zhang, S. Mukhopadhyay, B. R. Fisher, R. Hernandez, D. T. Brown, T. S. Baker, M. G. Rossmann, and R. J. Kuhn. 2001. Locations of carbohydrate sites on Sindbis virus glycoproteins show that E1 forms an icosahedral scaffold. Cell 105:127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schenk, P. M., S. Baumann, R. Mattes, and H. H. Steinbiss. 1995. Improved high-level expression system for eukaryotic genes in Escherichia coli using T7 RNA polymerase and rare ArgtRNAs. BioTechniques 19:196-200. [PubMed] [Google Scholar]
- 21.Skoging, U., M. Vihinen, L. Nilsson, and P. Liljeström. 1996. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure 4:519-529. [DOI] [PubMed] [Google Scholar]
- 22.Tellinghuisen, T. L., A. E. Hamburger, B. R. Fisher, R. Ostendorp, and R. J. Kuhn. 1999. In vitro assembly of alphavirus cores by using nucleocapsid protein expressed in Escherichia coli. J. Virol. 73:5309-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tellinghuisen, T. L., and R. J. Kuhn. 2000. Nucleic acid-dependent cross-linking of the nucleocapsid protein of Sindbis virus. J. Virol. 74:4302-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tellinghuisen, T. L., R. Perera, and R. J. Kuhn. 2001. Genetic and biochemical studies on the assembly of an enveloped virus. Genet. Eng. 23:83-112. [DOI] [PubMed] [Google Scholar]
- 25.Wengler, G., U. Boege, G. Wengler, H. Bischoff, and K. Wahn. 1982. The core protein of the alphavirus Sindbis virus assembles into core-like nucleoproteins with the viral genome RNA and with other single-stranded nucleic acids in vitro. Virology 118:401-410. [DOI] [PubMed] [Google Scholar]
- 26.Zhang, W., B. R. Fisher, N. H. Olson, J. H. Strauss, R. J. Kuhn, and T. S. Baker. 2002. Aura virus structure suggests that the T=4 organization is a fundamental property of viral structural proteins. J. Virol. 76:7239-7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang, W., S. Mukhopadhyay, S.V. Pletnev, T. S. Baker, R. J. Kuhn, and M. G. Rossmann. Placement of the structural proteins in Sindbis virus. J. Virol., in press. [DOI] [PMC free article] [PubMed]