Abstract
Systemic spread of viruses in plants involves local movement from cell to cell and long-distance transport through the vascular system. The cell-to-cell movement of the Beet yellows virus (BYV) is mediated by a movement protein that is an Hsp70 homolog (Hsp70h). This protein is required for the assembly of movement-competent virions that incorporate Hsp70h. By using the yeast two-hybrid system, in vitro coimmunoprecipitation, and in planta coexpression approaches, we show here that the Hsp70h interacts with a 20-kDa BYV protein (p20). We further demonstrate that p20 is associated with the virions presumably via binding to Hsp70h. Genetic and immunochemical analyses indicate that p20 is dispensable for assembly and cell-to-cell movement of BYV but is required for the long-distance transport of virus through the phloem. These results reveal a novel activity for the Hsp70h that provides a molecular link between the local and systemic spread of a plant virus by docking a long-distance transport factor to virions.
The systemic invasion of plants by viruses was conceptualized and later visualized as a two-phase process that involves relatively slow movement of virus from cell to cell and much faster long-distance transport through the vascular system (8, 12, 20, 28). The cell-to-cell movement of viruses proceeds via the plasmodesmata (30, 34) and is potentiated by virus-coded movement proteins (MPs) (26). Despite large variation in their functional profiles, several common activities were revealed in diverse MP species. Among these are the plasmodesmatal targeting of the MPs, their ability to bind nucleic acids, and interaction of the MPs with the cytoskeleton and with the endoplasmic reticulum (26). At least one of the MPs was recently implicated in the suppression of RNA silencing (52), which is a host defense response targeting viral RNAs for degradation (9, 51). Although the comprehensive theory of the viral cell-to-cell movement has yet to emerge, it will certainly include MP-driven modifications of the plasmodesmata, cytoskeleton, and other compartments of the plant cell.
Interestingly, several groups of the rod-shaped plant viruses move from cell to cell in a capsid protein (CP)-independent manner (reviewed in reference 7). In contrast, filamentous viruses generally require CP for cell-to-cell movement (10, 15). Among spherical viruses, some do not require CP for movement, whereas others do (7). Variation in CP requirements irrespective of the virion morphology presumes different mechanisms of cell-to-cell movement or a different distribution of functional assignments among individual virus proteins.
Long-distance transport of the plant viruses proceeds in the phloem. The entry into and exit from the phloem involves extensive, tissue-specific interactions between virus-coded proteins and plant proteins restricting viral transport (11). Due to important yet poorly understood specialization of the plasmodesmata interconnecting the cells within the phloem, viral loading into and unloading from the phloem has more stringent requirements than cell-to-cell movement in mesophyll (25, 36, 45). This tendency is illustrated by the large number of viruses that require virion formation for long-distance, but not cell-to-cell, transport (7, 8). In fact, only a few viruses are capable of spreading systemically in a nonvirion form (29, 41, 44, 47). These viruses, however, code for long-distance transport factors (LTFs) that are specifically required for efficient systemic spread (13, 14, 44, 48). Strikingly, several LTFs are suppressors of RNA silencing. Correlation between silencing suppression and transport activities of LTFs suggests that the ability of some viruses to spread systemically is linked to overcoming systemic RNA silencing (9, 21, 24, 51). As a general picture of viral long-distance transport emerges, the following two questions need to be addressed. What is the mechanistic connection between cell-to-cell movement and long-distance transport processes that are mediated by the MPs and LTFs, respectively? For the viruses whose transport requires both virion formation and LTF function, does the LTF bind virions? In this work, we address these questions with Beet yellows virus (BYV) as a model. According to present taxonomy, BYV belongs to the genus Closterovirus within the large family Closteroviridae (23).
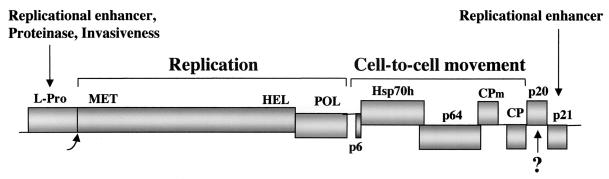
Closteroviruses possess complex, positive-strand RNA genomes (Fig. 1) that are encapsidated into exceptionally long filamentous virions (23). These virions possess two distinct parts, a virion body that is assembled by major CP and a tail assembled by a minor capsid protein (CPm) (1). Genetic analyses revealed that at least five proteins are required for BYV cell-to-cell movement (Fig. 1) (2, 40). These proteins include CP, CPm, and three MPs, namely, the 6-kDa protein (p6), the Hsp70homolog (Hsp70h), and the 64-kDa protein (p64). Intriguingly, the two latter proteins were found in tight association with the virions and were implicated in virion formation (32, 46, 49). Further analyses revealed strong correlation between BYV cell-to-cell movement and virion assembly and showed that Hsp70h is required for tail formation (3). Accordingly, the tail was conceptualized as an Hsp70h-powered movement device attached to the virion body (3).
FIG. 1.
Genome map of BYV. L-Pro, leader proteinase (the bent arrow shows the site of autocatalytic processing); MET, HEL, and POL, the methyltransferase, RNA helicase, and RNA polymerase domains of the BYV replicase, respectively.
It is well established that the molecular chaperones of the Hsp70 family function via ATP-regulated interaction with their protein targets (6, 37). All Hsp70s, including closteroviral Hsp70h, possess two major domains: a conserved, N-terminal, ATPase domain, and a less-conserved, C-terminal domain (5, 19, 54). By analogy to the cellular Hsp70s, we hypothesized that the closteroviral Hsp70h acts via protein-protein interactions. In this work we describe the interaction between Hsp70h and a 20-kDa BYV protein (p20) and show that Hsp70h provides a site for docking p20 to virions. Moreover, we demonstrate that p20 is a BYV LTF. Taken together these data provide a first example of an LTF that interacts with the MP and virions, thus linking the processes of cell-to-cell movement and long-distance transport.
MATERIALS AND METHODS
Yeast two-hybrid screens.
The β-glucuronidase (GUS) open reading frame (ORF) and BYV ORFs encoding L-Pro (leader proteinase), p6, Hsp70h, p64, CPm, CP, p20, and p21 (Fig. 1) were PCR amplified by using pBYV-4 as a template (40), with the concomitant addition of the NcoI and BamHI restriction endonuclease sites to the termini of the resulting products. These products were digested with NcoI and BamHI and were ligated into appropriately digested plasmids pACT-2 and pAS-2 (Clontech). Yeast strain Y190 was used throughout the study. All protocols for yeast transformation and DNA isolation were as described previously (17). Yeast two-hybrid analyses were conducted with protocols described by Finley and Brent (18). Quantification of the β-galactosidase (β-Gal) activity in units was performed by using the following formula: 1,000 × [OD420 − (1.75 × OD550)]/(T × V × OD600), where optical density at 420 nm (OD420) and OD550 are the absorbances of the reaction mixture, OD600 is the cell density of the culture, T is the reaction time in minutes, and V is the volume in milliliters.
In vitro translation and immunochemical analyses.
Preparation and translation of the capped mRNAs as well as immunochemical analyses were conducted as described previously (32). Histidine-tagged p20 was expressed in plants by using a Tobacco etch virus-derived gene expression vector and was isolated as described previously (16). Each immunoprecipitation reaction mixture, except for the negative control, contained 0.5 μg of p20 and monoclonal anti-histidine tag antibody (Amersham-Pharmacia Biotech) in 1:500 dilution. Detection of p20 in virions was conducted with polyclonal antiserum against synthetic, C-terminal oligopeptide of p20 (CELDKSGGELEILTFSKNEVFL) that was generated at Genemed Synthesis, Inc. (San Francisco, Calif.). Alternatively, anti-p20 antiserum to full-size, recombinant p20 kindly provided by R. Creamer (New Mexico State University) was used. The anti-Hsp70h (32) as well as anti-CP and anti-CPm sera (3) were described previously.
Agroinfection of plants and transient protein expression.
The previously described plasmid pBYV-GFP (40) was used as the source of virus sequences. A fragment of BYV cDNA (nucleotides 1 to 2548) was PCR amplified with concomitant addition of XbaI sites at either end of the product and was cloned into an XbaI site downstream from the Cauliflower mosaic virus 35S RNA polymerase promoter cassette. This cassette was previously generated by insertion of the 35S promoter between NotI and XbaI sites in a pBlueScript SK(II) plasmid (Stratagene) and contained nucleotides −417 to −1 relative to the transcription initiation site. A primer (5′-CATTTCATTTGGAGAGCAGTTTTTAACCATCCTTC) containing the 3′ end of the 35S promoter and the 5′ end of the BYV cDNA (underlined) was used for site-directed mutagenesis to remove nonvirus sequences as described previously (50). The DNA fragment containing the 35S promoter and 5′ region of the BYV cDNA was PCR amplified by using the primers 5′-TAGAGCTCAACATGGTGGAGCAC and 5′-CATCTAGAAGTTCACCCGGAG and was cloned into minibinary vector pCB301 digested with SacI and XbaI (53) to yield a pCB-35S-5BYV-NOS plasmid. A primer complementary to the 3′ end of the BYV sequence followed by a self-cleaving ribozyme (27) was synthesized (5′-TACCCGGGCCGTTTCGTCCTCACGGACTCATCAGAAGACATGTGAATCATGTCTTGACGGCCCTTATTTTTTCTTC) and used in combination with the upstream primer, 5′-ATTGGCAAACGCGGGATCCCGT, to amplify the 3′ region of the BYV-green fluorescent protein (GFP) cDNA and to add ribozyme sequence. This PCR product was digested with BamHI and XmaI and was cloned into pCB-35S-5BYV-NOS plasmid to produce pCB-35S-5′3′BYV-Rib-NOS. Finally, a XbaI-BamHI fragment of the BYV-GFP cDNA was transferred into pCB-35S-5′3′BYV-Rib-NOS to yield p35S-BYV-GFP.
The ORFs encoding BYV p20, Hsp70h, the 42-kDa N-terminal domain (N42), and the 23-kDa C-terminal domain (C23) were cloned into minibinary vector for Agrobacterium-mediated, transient expression in a free form or as a fusion with GFP as described previously (38). The resulting plasmids were mobilized into Agrobacterium tumefaciens strain EHA 105 by electroporation. Young Nicotiana benthamiana (6- to 8-leaf stage) plants were inoculated by using a 3-ml plastic syringe to infiltrate the leaf tissue. The same protocol and A. tumefaciens strain C58 were used for plant inoculation with the p35S-BYV-GFP plasmid.
Generation and analyses of the mutant BYV variants.
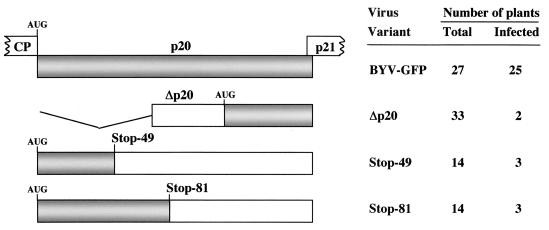
Mutant BYV variant Δp20 was described previously (2), whereas mutations Stop-49 and Stop-81 were generated by using the primers 5′-CTCCACGATCTCTAGTTGAATGTTAAC and 5′-GGTCTCTTACACTTGAGTGAGTTCTGG, respectively (the premature stop codons are underlined). Each of the Stop mutations was introduced into pBYV-GFP and was assayed for cell-to-cell movement by using inoculation of Claytonia perfoliata leaves as described previously (40). In addition, each of the four mutations was introduced into p35S-BYV-GFP via transferring BamHI-BstEII fragments from the corresponding pBYV-GFP variant. Cell-to-cell movement in C. perfoliata and long-distance transport in N. benthamiana were monitored with epifluorescent microscopy (40), whereas subcellular localization of the GFP-tagged proteins in epidermal cells of N. benthamiana was imaged by using confocal laser scanning microscopy as described previously (38). Isolation of the virions, sucrose density gradient centrifugation, and characterization of the virions' protein composition were as described previously (32).
RESULTS
Yeast two-hybrid screens reveal that p20 interacts with Hsp70h and itself.
In order to identify BYV proteins that interact with Hsp70h, we employed a yeast two-hybrid system. Each of the BYV genes encoding Hsp70h, L-Pro, p6, p64, CPm, CP, p20, and p21 (Fig. 1) was expressed in yeast cells as a fusion product with either a GAL4 activation domain (pACT series of plasmids) or a GAL4 DNA-binding domain (pAS series). In addition, we generated analogous plasmids expressing fusions of each of the Hsp70h principal domains, N42 and C23. Protein-protein interactions were assayed by GAL4-mediated activation of the two reporter genes, HIS3 and lacZ. The yeast strains that grew efficiently on His-deficient medium were further tested in β-Gal filter assays. The examined combinations of fusion proteins are shown in Table 1.
TABLE 1.
Yeast two-hybrid screensa
| ACT fusion | DB fusion | No. of β-Gal-positive colonies |
|---|---|---|
| Hsp70h | GUS | 0 |
| Hsp70h | 8 × ORFsb | 9 |
| N42 | 8 × ORFs | 11 |
| C23 | 8 × ORFs | 9 |
| Hsp70h | p20 | 17 |
| N42 | p20 | 38 |
| C23 | p20 | 33 |
| GUS | p20 | 0 |
| p20 | N42 | 22 |
| p20 | C23 | 24 |
| p20 | p20 | 117 |
Data from one representative experiment out of at least three experiments are shown.
8× ORFs, a mixture of eight variants that harbor BYV ORFs encoding L-Pro, p6, Hsp70h, p64, CPm, CP, p20, and p21.
In the first series of experiments, yeast strains possessing pACT-Hsp70h, pACT-N42, or pACT-C23 were transformed with the mixture of eight pAS variants harboring the BYV genes listed above. Each of these experiments yielded β-Gal-positive colonies (Table 1). In contrast, no positive colonies were recovered when the pACT-Hsp70h-containing strain was transformed with pAS-GUS, a plasmid that expressed bacterial GUS fusion as a negative control. It was found that all of the analyzed β-Gal-positive clones contained identical pAS plasmids that harbored a p20 coding region (data not shown). These results strongly suggested that full-size Hsp70h, as well as each of its domains, is capable of interacting with p20.
To reconfirm these observations, we transformed yeast strains possessing pACT-Hsp70h, pACT-N42, or pACT-C23 with pAS-p20. As expected, we recovered a number of colonies that grew on the medium lacking His and were also β-Gal positive (Table 1). To further determine if the interaction between Hsp70h and p20 is specific, we tested combination of pACT-GUS with pAS-p20. The fact that no colonies were found (Table 1) confirmed the specificity of Hsp70h-p20 interaction. We have also tested combinations of pACT-p20 with pAS-N42 and pAS-C23 and recovered β-Gal-positive colonies in each experiment (Table 1). It should be noted, however, that the intensity of blue staining was lower than that of the reciprocal combinations. Apparently, the level of interaction between p20 and Hsp70h domains in yeast may be affected by the structure of the fusion products.
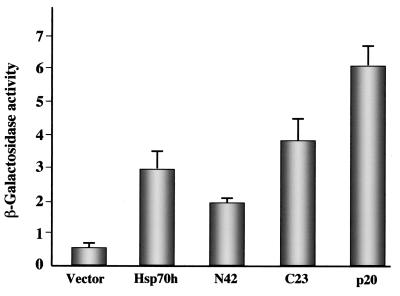
To examine the ability of p20 to interact with itself, we tested the combination of pACT-p20 and pAS-p20. Since the colonies were of intense blue color (Table 1, final row), we concluded that p20 is capable of efficient di- or multimerization. The observed interactions between p20 and Hsp70h, N42, and C23 as well as p20 self-interaction were also quantified by using in vitro β-Gal assays. As seen in Fig. 2, the results of these assays are in full agreement with the qualitative observations summarized in Table 1.
FIG. 2.
β-Gal assay for yeast two-hybrid interactions with BYV p20. Interaction in yeast was tested between p20 and Hsp70h, N42, C23, p20 (self-interaction), or empty vector (negative control). The β-Gal activity in units was calculated as described in Materials and Methods. The data represent means and standard deviations of four experiments.
In vitro interactions between p20 and Hsp70h domains.
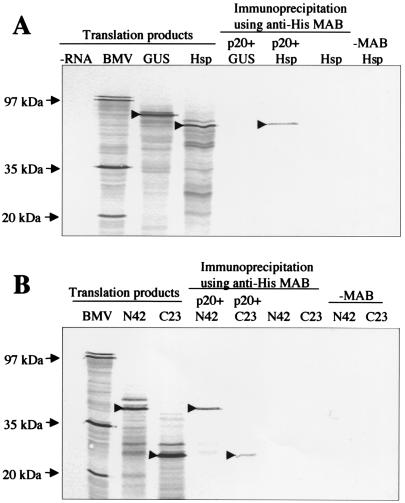
Since the yeast two-hybrid system is prone to artifacts, we employed an alternative experimental system to confirm interactions between p20 and Hsp70h domains. To this end, we expressed His-tagged p20 in plants by using a Tobacco etch virus gene vector (16). Purified, His-tagged p20 was incubated with 35S-labeled Hsp70h and was immunoprecipitated by His-tag-specific monoclonal antibody. Presence of labeled Hsp70h in the pelleted material was indicative of its coimmunoprecipitation with His-tagged p20 (Fig. 3A). In a control experiment, 35S-labeled GUS was not coimmunoprecipitated with p20 by the same antibody. Likewise, no Hsp70h was detected in the pellet when p20 or antibody was absent from the incubation mixture (Fig. 3A). These data indicated that p20 and Hsp70h can physically interact in vitro.
FIG. 3.
Coimmunoprecipitation of the 35S-labeled Hsp70h (A) and N42 and C23 (B) with unlabeled, histidine-tagged p20. For lanes under the heading translation products, the isotope-labeled products of in vitro translation reactions programmed with no RNA (negative control; −RNA lane), Brome mosaic virus RNA (positive control; BMV lane), GUS mRNA (GUS lane), Hsp70h mRNA (Hsp lane), N42 mRNA (N42 lane), or C23 mRNA (C23 lane) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, blotted onto a nitrocellulose membrane, and subjected to autoradiography. For lanes under the heading immunoprecipitation using anti-His MAB, the indicated labeled translation products were mixed with histidine-tagged, unlabeled p20 and were immunoprecipitated by using monoclonal, histidine tag-specific antibody and protein A-Sepharose. Negative controls in which p20 or monoclonal antibody were omitted are also shown. The arrows at the left mark the positions of corresponding BMV proteins, whereas the arrowheads within the autoradiograms mark the major translation products of the mRNAs that encode GUS, Hsp70h, N42, and C23 before or after immunoprecipitation.
In a similar series of experiments, we found that recombinant p20 is capable of binding each of the two Hsp70h domains (Fig. 3B). Thus, the in vitro immunochemical analysis confirmed the results of yeast two-hybrid screening.
Hsp70h changes subcellular localization of p20.
It was demonstrated previously that Hsp70h is associated with the plasmodesmata and virions in the BYV-infected cells (31, 32). We were interested to determine if p20 and Hsp70h possess autonomous signals that would target them to a specific cellular compartment and if coexpression of these proteins would result in their relocalization. To address these questions, we employed Agrobacterium-mediated, transient expression of p20 and Hsp70h fused with GFP. The GUS-GFP fusion was used as a control because neither GFP nor GUS possesses specific targeting signals (38).
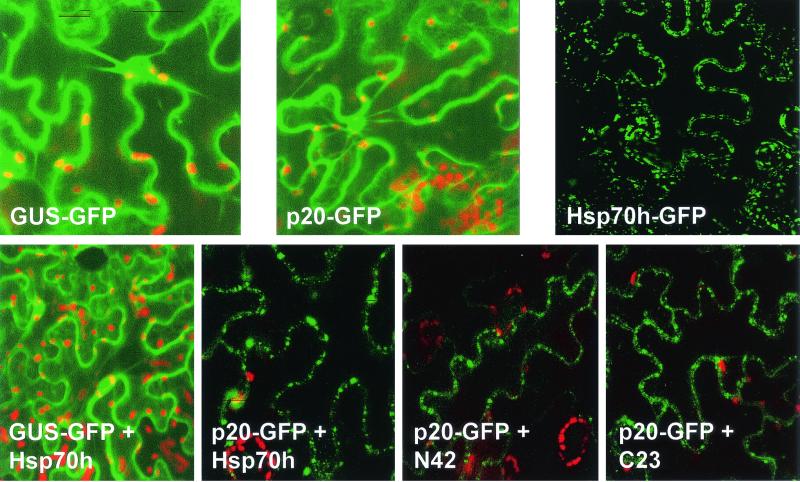
The epifluorescent microscopy analyses revealed that both GUS-GFP and p20-GFP are uniformly distributed throughout the cortical cytoplasm, transvacuolar cytoplasmic strands, and perinuclear regions (Fig. 4). In contrast, Hsp70h-GFP formed punctate bodies that were aligned with the cell wall. These results indicated that p20 does not possess an autonomous targeting signal, whereas Hsp70h is targeted to the cell periphery.
FIG. 4.
In planta colocalization of p20 and Hsp70h, N42, or C23. The green color corresponds to the GFP fluorescence; the red spots represent the autofluorescent chloroplasts. Upper row, transient expression of the GUS-GFP, p20-GFP, and Hsp70h-GFP fusion products. GFP-GUS was used as a control product that is distributed uniformly throughout the cortical cytoplasm and cytoplasmic strands. Lower row, coexpression of GUS-GFP or p20-GFP fusion products with free Hsp70h, N42, or C23.
In the next series of experiments we coexpressed GFP fusions with individual nonfused proteins. As expected, coexpression of GUS-GFP and free Hsp70h did not result in relocalization of the fluorescent product due to a lack of interaction between Hsp70h and GUS or GFP. However, p20-GFP did localize to the punctate bodies upon coexpression with nonfused Hsp70h (Fig. 4). The same pattern of p20-GFP localization was observed in the presence of N42-GFP and C23-GFP fusions. These data indicate that Hsp70h and each of its principal domains possess a signal for targeting to cell periphery. Moreover, coexpression of Hsp70h, N42, or C23 with p20-GFP results in relocalization of the latter product, suggesting that p20 can interact with Hsp70h or its domains in live plant cells.
p20 is a long-distance transport factor.
What is the primary function of p20 in the BYV life cycle? Previous work demonstrated that p20 is dispensable for BYV replication (39) and is not essential for virus cell-to-cell movement, although it might have an accessory role in the latter process (2). The interaction of p20 with Hsp70h, which is one of the three BYV MPs (40), prompted us to revisit the role of p20 in cell-to-cell movement.
A previously characterized p20 mutant, Δp20, was potentially capable of expressing C-terminal fragments of p20 (Fig. 5). It could not be excluded that this fragment was partially functional in the cell-to-cell movement of the corresponding mutant variant (2). Deletion of the RNA region encoding this fragment was impractical because it would inactivate a subgenomic promoter that governs expression of a replication-associated BYV protein, p21 (39). Consequently, we designed two additional point mutations by introducing premature stop codons in place of the 49th and 81st p20 codons. These mutants could direct the translation of only short, N-terminal p20 peptides (Fig. 5). Each mutation was introduced into GFP-tagged BYV, and the resulting mutant BYV-GFP variants were inoculated into the local lesion host plants (40). Examination of these mutants demonstrated that each of them was capable of cell-to-cell movement. The Stop-49 and Stop-81 mutants produced multicellular infection foci with mean diameters of 4.0 ± 2.3 and 2.1 ± 1.1 cells, respectively. The infection foci formed by the parental BYV-GFP variant had mean diameters of 5.1 ± 2.8 cells. These data confirmed our previous conclusion that p20 is not essential for the BYV cell-to-cell movement, although its inactivation results in a less efficient local spread.
FIG. 5.
Mutations introduced into the p20 ORF and systemic infectivity of the corresponding BYV-GFP variants. Gray rectangles represent the wild-type p20 ORF (upper diagram) or translatable parts of this ORF in the mutants. The parts of flanking CP and p21 ORFs are shown. Empty rectangles correspond to untranslatable parts in the mutant ORFs, whereas the broken lines mark the region that is deleted in the Δp20 variant. AUG, the 5′-most translation start sites in p20 ORFs. Stop-49 and Stop-81, mutants in which premature stop codons were introduced in place of the 49th or 81st codons in the p21 ORF. The plants were considered systemically infected if at least one green fluorescent area was detected in any of the noninoculated leaves.
We were next interested to test a possible role of p20 in the long-distance transport of BYV. Since the mechanical inoculation of the BYV systemic hosts is problematic (43), we developed a more efficient agroinfection procedure. BYV-GFP cDNA was cloned into a minibinary vector under control of the Cauliflower mosaic virus 35S RNA polymerase promoter and the Nos-terminator. The ribozyme cDNA was inserted downstream from the 3′ end of BYV cDNA to facilitate formation of an authentic 3′ terminus of viral RNA. The suspension of Agrobacterium cells transformed with the resulting plasmid was infiltrated into leaves of N. benthamiana. Six weeks postinfiltration, 93% of the agroinoculated plants were systemically infected by BYV-GFP, which was determined by symptom appearance and GFP accumulation in the noninoculated leaves (Fig. 5 and 6).
FIG. 6.
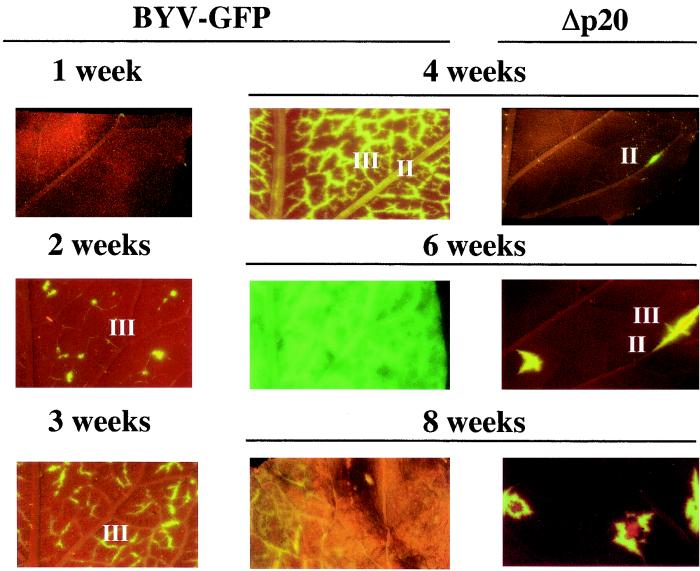
Systemic transport and unloading of the BYV-GFP variants into the upper, noninoculated leaves of N. benthamiana. II and III, class II and III leaf veins. Images under the BYV-GFP heading show the progression of the virus-infected, green fluorescent areas in the leaves of plants inoculated with parental BYV-GFP from 1 to 8 w.p.i. Note that the infected cells appear first at 2 w.p.i. in the class III veins and spread throughout the leaf by 6 w.p.i. At 8 w.p.i., the leaves become necrotic and show only the residual fluorescence. The images under the heading Δp20 show that occasional virus unloading from class II veins is observed at 4 w.p.i., whereas limited secondary spread into class III veins is detected only by 6 w.p.i.
The high efficiency of agroinfection allowed us to examine the long-distance transport phenotypes of the p20 mutants. It was found that each mutant was capable of only sporadic systemic transport (Fig. 5) that was detected by the occurrence of small, isolated green fluorescent areas. In total, only 8 out of 61 plants inoculated with Δp20, Stop-49, and Stop-81 mutants showed such fluorescent areas (Fig. 5 and 6).
It was demonstrated recently that GFP expressed in the phloem companion cells of healthy transgenic plants is capable of trafficking through the phloem (35). To test whether the GFP accumulation observed in our experiments was due to BYV-GFP transport and amplification rather than to virus-independent trafficking of GFP, we isolated RNAs from the fluorescent tissues and conducted Northern hybridization and reverse transcription-PCR analyses. These analyses, followed by nucleotide sequencing, confirmed that the viral RNA harboring original mutations was present in the green fluorescent areas of the infected plants (data not shown). Thus, gross debilitation of systemic spread due to mutations in p20 allowed us to conclude that p20 plays a critical role in long-distance transport and represents a BYV LTF.
Analysis of virus unloading.
Previous work with GFP-tagged Potato virus X (PVX) established that long-distance transport results in virus unloading into class III veins of the sink leaves, with subsequent cell-to-cell movement into mesophyll cells and minor veins (42). To determine if BYV follows this pathway, we conducted temporal and spatial analyses of the infection by BYV-GFP and its mutant variants. Figure 6 shows that BYV-GFP in noninoculated leaves appears at 2 weeks postinoculation (w.p.i), while PVX-GFP in noninoculated leaves appears at ∼1 w.p.i. (42). Despite this slow spread, the pattern of BYV-GFP unloading matches that found in PVX-GFP: GFP accumulation is detected first in association with class III veins. At 6 w.p.i., virus egress into leaf mesophyll cells and subsequent cell-to-cell movement results in a global colonization of the leaf tissues.
The timing of virus unloading for Δp20 mutant was dramatically different: the first fluorescent cells did not appear till 4 w.p.i. Furthermore, only sporadic unloading that involved preferentially class II veins was detected (Fig. 6). The subsequent local spread of virus from the sites of primary unloading indicated that mutant virus was capable of cell-to-cell movement. Hence, the timing, extent, and pattern of virus unloading were drastically affected in the Δp20 mutant. These results reconfirmed that functional p20 is required for efficient passage of BYV through and/or unloading from the phloem of infected plants.
p20 is associated with the virions.
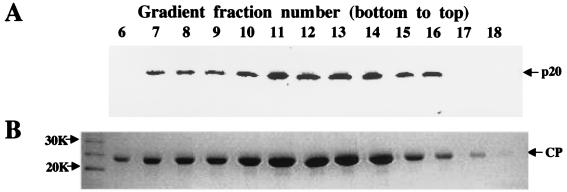
Taking into account the ability of p20 to bind a virion-associated Hsp70h, we hypothesized that p20 functions in long-distance transport via attachment to the virions. To address this hypothesis, we used a p20-specific antiserum for the analysis of BYV virions separated in a sucrose gradient. As seen in Fig. 7A, p20 was confidently detected in the virion preparation. Moreover, p20 and CP peaked in the same fractions of the gradient (Fig. 7A and B) that were previously shown to contain intact, full-length BYV virions (32). These results showed that p20 is physically associated with the BYV virions.
FIG. 7.
Comigration of BYV p20 and virions in sucrose density gradient. (A) Immunoblot analysis of the gradient fractions (numbered from the bottom up) using antiserum to the synthetic, C-terminal p20 peptide. (B) The same gradient fractions separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and stained with Coomassie brilliant blue. The position of the BYV CP is marked by an arrow; the numbers at the left indicate molecular sizes of the protein markers.
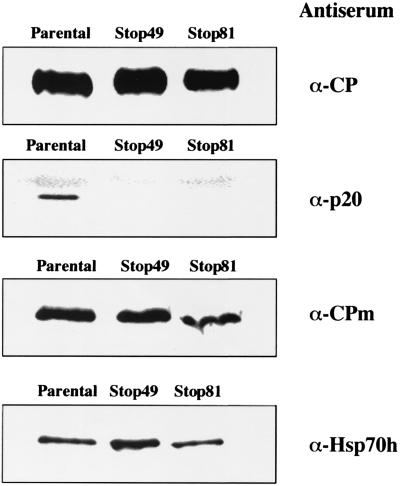
It was demonstrated that the assembly of the tailed BYV virions is a prerequisite for virus cell-to-cell movement (2, 3). Although CP is apparently the only protein required for virion body assembly, tail formation requires CPm and Hsp70h (3). Presence of the p20 in virions prompted us to ask if this protein is also required for tail assembly. To test for the formation of virion bodies and tails, the virions were isolated from green fluorescent tissues infected by parental BYV-GFP as well as by mutant variants Stop-49 and Stop-81. It was found that each of the preparations of mutant virions contained CP, CPm, and Hsp70h in amounts similar to those present in wild-type virions (Fig. 8). In contrast, either antiserum to the synthetic, C-terminal p20 peptide (Fig. 8) or antiserum to the full-size, recombinant p20 (data not shown) detected p20 in parental but not mutant virions. Taken together, these results indicate that inactivation of p20 does not affect assembly of the tailed BYV virions, and they suggest that the association of p20 with the virions is required to facilitate their long-distance transport.
FIG. 8.
Protein composition of the virions isolated from plants infected with parental and mutant BYV-GFP variants. Immunoblot analyses of the virions were done by using the specific antisera indicated at the right of each panel.
DISCUSSION
Phloem is among the most complex and least experimentally amenable tissues of vascular plants. Viruses, a great majority of which rely on the phloem in their long-distance transport, provide powerful tools to probe this tissue. Viral loading into, passage through, and unloading from the phloem proceed via utilization and modification of normal phloem functions. With a few exceptions, viruses traffic in the phloem in a form of virions. Other recognized key players in viral long-distance transport are MPs and LTFs. The interplay between the virions, MPs, and LTFs largely defines the virus-specific part of the long-distance transport machinery. However, little is known about how virions, MPs, and LTFs work together to ensure successful virus spread through the phloem.
In a previous work we developed the concept of a BYV virion as a morphologically and functionally bipartite entity (3). One principal part of the virion, the body made up of CP and RNA, carries the function of genome protection. The other part, the tail assembled by CPm, represents a specialized device for cell-to-cell movement. We have also demonstrated that one of the BYV MPs, Hsp70h, is an integral part of the virion (32) that is required for tail assembly (3). It was further hypothesized that the virion translocation through the plasmodesma occurs in a directional, tail-first manner and is powered by ATPase activity of Hsp70h. In this work we show that in addition to its central role in BYV cell-to-cell movement, Hsp70h provides a docking site for p20, an accessory virion protein that is critical for long-distance transport of the virus.
Initially, p20 was identified as the BYV protein that interacts with Hsp70h in yeast, in vitro, and in plant cells. Further experiments revealed that each of the principal domains of the Hsp70h was capable of binding p20. This observation suggests that each Hsp70h molecule can bind at least two p20 molecules. Alternatively, the interface between Hsp70h and p20 in a 1:1 complex may overlap each of the Hsp70h domains. Additional p20 molecules could be included in a complex via p20 self-interaction.
To address the significance of Hsp70h-p20 interaction, we investigated the p20 function in the BYV life cycle and revealed its primary role in the long-distance transport of BYV. Indeed, mutation of the p20 gene resulted in profound defects in a phloem-associated virus spread. First, the mutants were unable to spread systemically in a majority of inoculated plants. Second, in those few plants where systemic spread was detected, it was delayed by at least two weeks. Third, the extent and pattern of virus unloading were affected by mutations. Instead of the massive unloading from class III veins seen with parental BYV-GFP (Fig. 6) as well as with other viruses (42), mutant BYV-GFP variants exhibited patchy unloading from class III and/or class II veins. Taken together, these results established p20 as a BYV LTF.
Further analyses revealed that although p20 is not essential for virion assembly or cell-to-cell movement, it is attached to the virions, presumably via interaction with the BYV MP, Hsp70h. This association between virion, MP, and LTF suggests that the processes of the local and systemic translocation of BYV are mechanistically linked to each other via physical interaction between virion-associated MP and LTF. Furthermore, it prompts a novel paradigm of a BYV virion as a dynamic entity that mediates virus transport via recruitment of the MP and LTF.
It should be emphasized that the transport mechanisms may not be uniform among the viruses in the Closteroviridae family, as BYV is the only known closterovirus that is capable of exiting the phloem. On the other hand, interaction between the virions and MP was recently described for a Potexvirus, suggesting that recruitment of the transport proteins by virions could be a widespread phenomenon (4).
What is a mechanism of p20 function in long-distance transport of BYV? One possibility is that p20 facilitates either entry of virions into or exit from the phloem, e.g., via modification of the plasmodesmatal function of the Hsp70h. This modification may be required to account for the specialized nature of plasmodesmata that interconnect cells within phloem. Another possibility is stabilization of the virions inside the phloem. In particular, p20 may prevent the disassembly of the virions and damage of RNA due to alkaline pH of the phloem sap (33). Alternatively, p20 could protect virions from inactivation by plant defense proteins that are present in the phloem (11).
The third mechanistic possibility is involvement of p20 with suppression of plant RNA silencing response, which is a likely mechanism of action for several LTFs (1, 24). Our attempt to reveal such suppressor activity of p20 with a model system in which RNA silencing is induced by transient expression of the double-stranded RNA (22) was unsuccessful (J. Reed, K. Kasschau, J. Carrington, and V. V. Dolja, unpublished results). However, it cannot be excluded that p20 interferes with something other than the double-stranded-RNA-induced facet of the RNA silencing pathway.
Phylogenetically, p20 represents a novel class of viral LTFs. A database search revealed that p20 shows only marginal similarity to other ∼20-kDa proteins encoded by related members of the Closterovirus genus (V. V. Dolja, unpublished data). In general, none of the presently characterized LTFs exhibits sequence similarity to any other proteins except for their orthologs within the same virus taxon. Because of that, LTFs appear to be a relatively recent evolutionary invention that may provide valuable mechanistic clues as to virus-host interactions within the phloem.
Acknowledgments
We are grateful to James C. Carrington, Kenneth B. Johnson, and Michael E. Taliansky for useful discussions and critical reading of the manuscript. We thank Rebecca Creamer for the anti-p20 serum and Jin Hong and Xiao-Hua Li for their help with the yeast two-hybrid screening.
This work was supported by grants from the National Institutes of Health (R1GM53190B), from the U.S. Department of Agriculture (CSREES 2001-35319-10875), and from the Large Scale Biology Corporation (2000-13) to V.V.D.
REFERENCES
- 1.Agranovsky, A. A., D. E. Lesemann, E. Maiss, R. Hull, and J. G. Atabekov. 1995. “Rattlesnake” structure of a filamentous plant RNA virus built of two capsid proteins. Proc. Natl. Acad. Sci. USA 92:2470-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alzhanova, D. V., Y. Hagiwara, V. V. Peremyslov, and V. V. Dolja. 2000. Genetic analysis of the cell-to-cell movement of beet yellows closterovirus. Virology 267:192-200. [DOI] [PubMed] [Google Scholar]
- 3.Alzhanova, D. V., A. J. Napuli, R. Creamer, and V. V. Dolja. 2001. Cell-to-cell movement and assembly of a plant closterovirus: roles for the capsid proteins and Hsp70homolog. EMBO J. 20:6997-7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atabekov, J. G., N. P. Rodionova, O. V. Karpova, S. V. Kozlovsky, and V. Y. Poljakov. 2000. The movement protein-triggered in situ conversion of potato virus X virion RNA from a nontranslatable into a translatable form. Virology 271:259-263. [DOI] [PubMed] [Google Scholar]
- 5.Boorstein, W. R., T. Ziegelhoffer, and E. A. Craig. 1994. Molecular evolution of the Hsp70 multigene family. J. Mol. Evol. 38:1-17. [DOI] [PubMed] [Google Scholar]
- 6.Bukau, B., and A. L. Horwich. 1998. The HSP70 and HSP60 chaperone machines. Cell 92:351-366. [DOI] [PubMed] [Google Scholar]
- 7.Callaway, A., D. Giesman-Cookmeyer, E. T. Gillock, T. L. Sit, and S. A. Lommel. 2001. The multifunctional capsid proteins of plant RNA viruses. Annu. Rev. Phytopathol. 39:419-460. [DOI] [PubMed] [Google Scholar]
- 8.Carrington, J. C., K. D. Kasschau, S. K. Mahajan, and M. C. Schaad. 1996. Cell-to-cell and long-distance transport of viruses in plants. Plant Cell 8:1669-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrington, J. C., K. D. Kasschau, and L. K. Johansen. 2001. Activation and suppression of RNA silencing by plant viruses. Virology 281:1-5. [DOI] [PubMed] [Google Scholar]
- 10.Chapman, S., G. Hills, J. Watts, and D. Baulcombe. 1992. Mutational analysis of the coat protein gene of potato virus X: effects on virion morphology and viral pathogenicity. Virology 191:223-230. [DOI] [PubMed] [Google Scholar]
- 11.Chisholm, S. T., M. A. Parra, R. J. Anderberg, and J. C. Carrington. 2001. Arabidopsis RTM1 and RTM2 genes function in phloem to restrict long-distance movement of tobacco etch virus. Plant Physiol. 127:1667-1675. [PMC free article] [PubMed] [Google Scholar]
- 12.Citovsky, V., and P. Zambryski. 2000. Systemic transport of RNA in plants. Trends Plant Sci. 5:52-54. [DOI] [PubMed] [Google Scholar]
- 13.Cronin, S., J. Verchot, R. Haldeman-Cahill, M. Shaad, and J. C. Carrington. 1995. Long-distance movement factor: a transport function of the potyvirus helper component proteinase. Plant Cell 7:549-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding, S.-W., W. X. Li, and R. H. Symons. 1995. A novel naturally occurring hybrid gene encoded by a plant RNA virus facilitates long distance virus movement. EMBO J. 14:5762-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dolja, V. V., R. Haldeman, N. Robertson, W. G. Dougherty, and J. C. Carrington. 1994. Distinct functions of the capsid protein in assembly and cell-to-cell movement of tobacco etch potyvirus in plants. EMBO J. 13:1482-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dolja, V. V., V. V. Peremyslov, K. E. Keller, R. R. Martin, and J. Hong. 1998. Isolation and stability of histidine-tagged proteins produced in plants via potyvirus gene vectors. Virology 252:269-274. [DOI] [PubMed] [Google Scholar]
- 17.Dunn, B., and R. C. Wobbe. 1993. Saccharomyces cerevisiae, p. 13.0.1-13.13.9. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology, vol. 2. John Wiley & Sons, Inc., New York, N.Y.
- 18.Finley, R. L., and R. Brent. 1996. Interaction trap cloning with yeast, p. 163-203. In D. Glover and D. B. Hames (ed.), DNA cloning-expression systems: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 19.Flaherty, K. M., C. DeLuca-Flaherty, and D. B. McKay. 1990. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 346:623-628. [DOI] [PubMed] [Google Scholar]
- 20.Gilbertson, R. L., and W. J. Lucas. 1996. How do viruses traffic on the ′vascular highway'? Trends Plant Sci. 1:260-267. [Google Scholar]
- 21.Guo, H. S., and S. W. Ding. 2002. A viral protein inhibits the long range signaling activity of the gene silencing signal. EMBO J. 21:398-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansen, L. K., and J. C. Carrington. 2001. Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 126:930-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karasev, A. V. 2000. Genetic diversity and evolution of closteroviruses. Annu. Rev. Phytopathol. 38:293-324. [DOI] [PubMed] [Google Scholar]
- 24.Kasschau, K. D., and J. C. Carrington. 2001. Long-distance movement and replication maintenance functions correlate with silencing suppression activity of potyviral HC-Pro. Virology 286:71-81. [DOI] [PubMed] [Google Scholar]
- 25.Knoblauch, M., and A. J. E. van Bel. 1998. Sieve tubes in action. Plant Cell 10:35-50. [Google Scholar]
- 26.Lazarowitz, S. G., and R. N. Beachy. 1999. Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell 11:535-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leiser, R.-M., V. Ziegler-Graff, A. Reutenauer, E. Herrbach, O. Lemaire, H. Guilley, K. Richards, and G. Jonard. 1992. Agroinfection as an alternative to insects for infecting plants with beet western yellows luteovirus. Proc. Natl. Acad. Sci. USA 89:9136-9140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maule, A. J. 1994. Plant-virus movement: de novo process or redeployed machinery? Trends Microbiol. 2:305-306. [DOI] [PubMed] [Google Scholar]
- 29.McGeachy, K. D., and H. Barker. 2000. Potato mop-top virus RNA can move long distance in the absence of coat protein: evidence from resistant, transgenic plants. Mol. Plant-Microbe Interact. 13:125-128. [DOI] [PubMed] [Google Scholar]
- 30.McLean, B. G., F. D. Hempel, and P. C. Zambryski. 1997. Plant intercellular communication via plasmodesmata. Plant Cell 9:1043-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Medina, V., V. V. Peremyslov, Y. Hagiwara, and V. V. Dolja. 1999. Subcellular localization of the Hsp70-homolog encoded by beet yellows closterovirus. Virology 260:173-181. [DOI] [PubMed] [Google Scholar]
- 32.Napuli, A. J., B. W. Falk, and V. V. Dolja. 2000. Interaction between Hsp70homolog and filamentous virions of the beet yellows virus. Virology 274:232-239. [DOI] [PubMed] [Google Scholar]
- 33.Nelson, R. S., and A. J. E. van Bel. 1998. The mystery of virus trafficking into, through, and out of vascular tissue. Prog. Bot. 59:476-533. [Google Scholar]
- 34.Oparka, K. J., and A. G. Roberts. 2001. Plasmodesmata. A not so open-and-shut case. Plant Physiol. 125:123-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oparka, K. J., A. G. Roberts, P. Boevnik, S. Santa Cruz, I. Roberts, K. S. Pradel, A. Imlau, G. Kotlizky, N. Sauer, and B. Epel. 1999. Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell 97:743-754. [DOI] [PubMed] [Google Scholar]
- 36.Oparka, K. J., and R. Turgeon. 1999. Sieve elements and companion cells-traffic control centers of the phloem. Plant Cell 11:739-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pelham, H. R. B. 1986. Speculations on the functions of the major heat shock and glucose-regulated proteins. Cell 46:959-961. [DOI] [PubMed] [Google Scholar]
- 38.Peng, C.-W., V. V. Peremyslov, A. R. Mushegian, W. O. Dawson, and V. V. Dolja. 2001. Functional specialization and evolution of the leader proteinases in the family Closteroviridae. J. Virol. 75:12153-12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peremyslov, V. V., Y. Hagiwara, and V. V. Dolja. 1998. Genes required for replication of the 15.5-kilobase RNA genome of a plant closterovirus. J. Virol. 72:5870-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peremyslov, V. V., Y. Hagiwara, and V. V. Dolja. 1999. HSP70homolog functions in cell-to-cell movement of a plant virus. Proc. Natl. Acad. Sci. USA 96:14771-14776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petty, I. T. D., and A. O. Jackson. 1990. Mutational analysis of barley stripe mosaic virus RNA β. Virology 179:712-718. [DOI] [PubMed] [Google Scholar]
- 42.Roberts, A. G., S. Santa Cruz, I. M. Roberts, D. A. M. Prior, R. Turgeon, and K. J. Oparka. 1997. Phloem unloading in sink leaves of Nicotiana bentamiana: comparison of a fluorescent solute with a fluorescent virus. Plant Cell 9:1381-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russel, G. E. 1963. Isolation and individual strains of beet yellows virus. Nature 197:623-624. [Google Scholar]
- 44.Ryabov, E. V., D. J. Robinson, and M. E. Taliansky. 1999. A plant virus-encoded protein facilitates long-distance movement of heterologous viral RNA. Proc. Natl. Acad. Sci. USA 96:1212-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Santa Cruz, S. 1999. Perspective: phloem transport of viruses and macromolecules-what goes in must come out. Trends Microbiol. 7:237-241. [DOI] [PubMed] [Google Scholar]
- 46.Satyanarayana, T., S. Gowda, M. Mawassi, M. R. Albiach-Marti, M. A. Ayllon, C. Robertson, S. M. Garnsey, and W. O. Dawson. 2000. Closterovirus encoded Hsp70homolog and p61 in addition to both coat proteins function in efficient virion assembly. Virology 278:253-265. [DOI] [PubMed] [Google Scholar]
- 47.Scholthof, H. B., T. J. Morris, and A. O. Jackson. 1993. The capsid protein gene of tomato bushy stunt virus is dispensable for systemic movement and can be replaced for localized expression of foreign genes. Mol. Plant-Microbe Interact. 6:309-322. [Google Scholar]
- 48.Scholthof, H. B., K. B. G. Scholthof, M. Kikkert, and A. O. Jackson. 1995. Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology 213:425-438. [DOI] [PubMed] [Google Scholar]
- 49.Tian, T., L. Rubio, H. H. Yeh, B. Crawford, and B. W. Falk. 1999. Lettuce infectious yellows virus: in vitro acquisition analysis using partially purified virions and the whitefly, Bemisia tabaci. J. Gen. Virol. 80:1111-1117. [DOI] [PubMed] [Google Scholar]
- 50.Turpen, T. H., A. M. Turpen, N. Weinzettl, M. H. Kumagi, and W. O. Dawson. 1993. Transfection of whole plants from wounds inoculated with Agrobacterium tumefaciens containing cDNA of tobacco mosaic virus. J. Virol. Methods 42:227-240. [DOI] [PubMed] [Google Scholar]
- 51.Vance, V., and H. Vaucheret. 2001. RNA silencing in plants-defense and counterdefense. Science 292:2277-2280. [DOI] [PubMed] [Google Scholar]
- 52.Voinnet, O., C. Lederer, and D. C. Baulcombe. 2000. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell 103:157-167. [DOI] [PubMed] [Google Scholar]
- 53.Xiang, C., P. Han, I. Lutziger, K. Wang, and D. J. Oliver. 1999. A mini binary vector series for plant transformation. Plant. Mol. Biol. 40:711-717. [DOI] [PubMed] [Google Scholar]
- 54.Zhu, X., X. Zhao, W. F. Burkholder, A. Gragerov, C. M. Ogata, M. E. Gottesman, and W. A. Hendrikson. 1996. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272:1606-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]