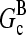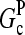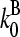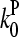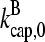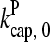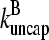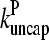Abstract
The dependences of the steady-state critical concentration and average filament length of actin solutions, on the filament branching and capping rates, are calculated using a rate methodology based on the total number of actin filaments. The methodology generalizes calculations of the “treadmilling” actin concentration at which an average filament has net zero growth rate. The predictions of the rate methodology are validated by comparison with stochastic-growth simulations that track the positions of all filament subunits over time. For side branching, the critical concentration drops proportionally to the square root of the branching rate; for end branching the drop is linear. The polymerization response to branching has a maximum as a function of the capping-protein concentration. The average filament length drops with increasing branching, because the critical concentration drops. Even small rates of filament uncapping have a large impact on the average filament length in vitro. The potential significance of these phenomena for cell behavior is evaluated.
INTRODUCTION
The motility of cells, the formation of protrusions such as filopodia and lamellipodia, and the motions of intracellular pathogens, are strongly influenced by extracellular or intracellular factors that stimulate actin polymerization (1,2).
One channel by which actin polymerization can be stimulated is the activation of Arp2/3 complex, a seven-subunit complex of actin-related proteins that can bind to an existing filament and initiate a new branch at the binding site. The newly generated filaments have “barbed” and “pointed” ends, with rapid growth taking place at the barbed ends. The pointed ends are attached to Arp2/3 complex. Arp2/3 complex is constitutively inactive, but can be activated by several intracellular proteins. The activation path can be direct, as in the case of the ActA bacterial surface protein, or proceed via a signaling cascade ending in interactions between Arp2/3 complex and proteins such as those of the Wasp/Scar family (2,3). Filament growth is limited, and an adequate supply of free monomers maintained, by the presence of capping proteins that block the filaments' barbed ends from assembly (4). Capping, however, can be suppressed by the presence of membrane-bound phosphoinositides such as PIP2, which thus act as polymerization stimulants. Arp2/3 complex also caps pointed ends (5). At present there is no quantitative understanding of the extent of polymerization or changes in filament length caused by Arp2/3-complex-induced branching. Although there have been numerical modeling studies of actin polymerization in vitro in the presence of Arp2/3 complex and capping protein (6,7), there is no straightforward mathematical formula that gives the extent of polymerization or the filament lengths in terms of the relevant protein concentrations, either in vitro or in vivo.
This article takes a first step toward a quantitative understanding of the polymerization response to branching by calculating the critical concentration and average filament length in a simple model of actin polymerizing in vitro. The analysis treats steady-state properties, as might be obtained by allowing a polymerization experiment to run for a long time. Understanding the steady-state properties is a prerequisite for understanding the dynamics, and some of the phenomena thus elucidated will also be present in the dynamic behavior of cells. The model includes polymerization/depolymerization, branching/debranching, and capping/uncapping effects. It is based on a simple rate equation expressing the constancy of the number of filaments in steady state. Within this framework, balancing filament “birth” rates from branching and “death” rates from debranching and depolymerization fixes the critical concentration, which in turn determines the average filament length. The filament length enters the calculation self-consistently because it affects the filament birth and death rates.
Using this model, we develop formulas for the critical concentration and average filament length in terms of the relevant rate parameters. The formulas are backed up by stochastic growth simulations using rate parameters obtained from recent fits to kinetic data. This work has three main goals. First, to obtain a general understanding of branching polymerization that may be transferable to cellular processes, and may be used to make predictions that can be experimentally tested. Second, to establish relationships between the extent of polymerization and filament lengths on one hand, and rate parameters on the other hand, which can be used in combination with in vitro experiments to measure or constrain the rate parameters. Third, to develop key inputs for mathematical modeling of whole-cell behavior based on spatially varying concentrations of actin and related proteins, for example, as applied recently to keratocytes (8); if such modeling studies incorporate the mechanical properties of the actin network, the filament length is also an important input.
The organization of the remainder of the article is as follows. The next section defines the model. Subsequently, we derive the steady-state relation for the filament number, calculate the average filament length in terms of the free-actin concentration, and combine these results to obtain an analytic expression for the critical concentration. We then validate the analytic theory by comparing its predictions with simulation results. Next we discuss the limits of the model used. Finally, we conclude the article with a discussion of the potential significance of the results for actin polymerization in vitro and in vivo.
MODEL
Our model describes the structure of actin in solution with activated Arp2/3 complex and capping protein, having concentrations [Arp2/3] and [CP], respectively. A simplified solution of this model has previously been used to study the dynamics of actin filament cluster sizes (9). The processes included in the model are filament polymerization/depolymerization, capping/uncapping, and branching/debranching. Polymerization is described by a net barbed end polymerization rate 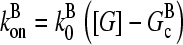 and a net pointed-end depolymerization rate
and a net pointed-end depolymerization rate 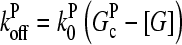 (measured in subunits per second), where
(measured in subunits per second), where 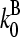 and
and 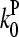 are concentration-independent rate parameters, and [G] is the free-monomer concentration.
are concentration-independent rate parameters, and [G] is the free-monomer concentration. 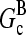 is the barbed-end critical concentration, at which the barbed-end on- and off-rates cancel;
is the barbed-end critical concentration, at which the barbed-end on- and off-rates cancel; 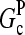 is defined similarly for the pointed end. Both
is defined similarly for the pointed end. Both 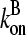 and
and 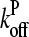 are positive, because
are positive, because 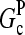 is known to be larger than
is known to be larger than 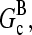 and [G] is between
and [G] is between 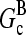 and
and 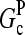 in steady state. Capping is described by barbed- and pointed-end capping and uncapping rates
in steady state. Capping is described by barbed- and pointed-end capping and uncapping rates 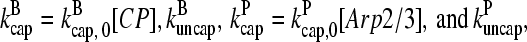 where
where 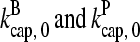 are rate parameters. The effects of capping are conveniently summarized by the parameters
are rate parameters. The effects of capping are conveniently summarized by the parameters 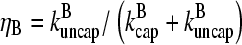 and
and 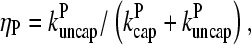 which give the equilibrium probabilities for the barbed and pointed ends of a filament, respectively, to be uncapped; the net steady-state growth and depolymerization rates are then
which give the equilibrium probabilities for the barbed and pointed ends of a filament, respectively, to be uncapped; the net steady-state growth and depolymerization rates are then 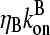 and
and 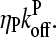
Branching in vitro occurs mainly along filament sides, as shown by several studies (7,10–12). Therefore, our calculations focus mainly on side branching, which is described by a branching rate per filament subunit:
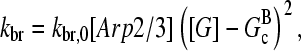 |
(1) |
where kbr, 0 is a rate parameter and the power of two is taken from a recent kinetic analysis (7). Because branching in cells is often restricted to regions very near the plasma membrane, we also include calculations for end branching, in which kbr is the branching rate per filament; it is taken to have the same functional form as the side branching rate. Debranching, either spontaneous or induced by actin-severing proteins such as ADF/cofilin, is described by the average time τdis required for a branch to dissociate. Initiation of branching polymerization requires the “de novo” nucleation of at least one seed filament that does not grow from a preexisting filament. However, in steady state such nucleation effects are small in comparison with branching, because their rates are very low. Therefore, we ignore de novo filament nucleation processes. Severing, also ignored, is potentially more important; this is discussed below under “Critique of model”.
Analytic theory of critical concentration and filament lengths
The extent of polymerization in steady state is determined by the critical concentration Gc, which is the maximum concentration of free actin that can remain unpolymerized indefinitely. In the absence of rapid filament nucleation, Gc is nearly equal to the “treadmilling” concentration Gtr at which polymerization of barbed ends is precisely balanced by depolymerization of pointed ends, both in their equilibrium capping states. Balancing these rates at [G] = Gtr, one obtains
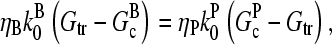 |
(2) |
so that
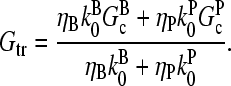 |
(3) |
When [G] = Gtr, the change in a filament's length over time is parallel to that of an unbiased random walker moving in one dimension with an absorbing boundary. The random walker is unbiased because forward (increasing length) steps are as likely as backward (decreasing length) steps, and the absorbing boundary corresponds to filaments disappearing when they reach a very short length corresponding to the critical nucleus size. The average displacement for such a walker, before being absorbed, is infinite (13). Therefore, in the absence of severing, the steady-state average filament length will be infinite when [G] = Gtr.
However, in the presence of rapid filament nucleation caused by branching, the time-averaged capping states of the filament ends will differ from the equilibrium values. When a filament is “born”, for example, it is capped at the pointed end and uncapped at the barbed end. If the time for it to reach the equilibrium capping state is a sizeable fraction of the filament lifetime, the critical concentration will differ noticeably from Gtr. We account for this effect by balancing filament creation and destruction rather than polymerization and depolymerization of single filaments. We envisage the following filament “life cycle”: a daughter filament is created as a branch on a mother filament. Its barbed end then becomes capped. Next, the daughter filament dissociates from the Arp2/3 complex on the mother filament. Finally, it depolymerizes.
The steady-state properties implied by this life cycle can be obtained by writing the time rate of change of the total number of filaments N as a sum of creation and destruction terms (9):
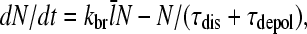 |
(4) |
where 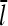 is the average number of subunits per filament. The first term on the right-hand side follows from the definition of kbr as a branching rate per filament subunit. The second term states that the destruction of a filament involves first dissociation from the mother filament, which takes time τdis, and subsequently depolymerization, which takes time τdepol. Thus, the steady-state condition for Gc, dN/dt = 0, implies that
is the average number of subunits per filament. The first term on the right-hand side follows from the definition of kbr as a branching rate per filament subunit. The second term states that the destruction of a filament involves first dissociation from the mother filament, which takes time τdis, and subsequently depolymerization, which takes time τdepol. Thus, the steady-state condition for Gc, dN/dt = 0, implies that
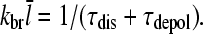 |
(5) |
This result uniquely determines Gc. The left-hand side increases with [G], because kbr increases according to Eq. 1, and 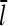 will be enhanced by the increasing on-rate. The right-hand side decreases with increasing [G], because the increasing value of
will be enhanced by the increasing on-rate. The right-hand side decreases with increasing [G], because the increasing value of 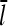 and decreasing off-rate cause τdepol to increase. Therefore, there is only one value of [G] at which the right- and left-hand sides are equal.
and decreasing off-rate cause τdepol to increase. Therefore, there is only one value of [G] at which the right- and left-hand sides are equal.
One might ask whether or not this analysis gives a constant number of subunits in filaments (the total number of polymerized subunits), as it must for a system in steady state. In fact, both the number of subunits in filaments and the number of filaments are constant. The method underlying the analysis is the calculation of a steady-state filament length distribution, in which both of these quantities are constant. We employ the state-state condition for the number of filaments because it gives the simplest mathematical formulation.
Of the four variables in Eq. 5, the [G]-dependence of kbr is given by Eq. 1, and τdis is taken to be independent of [G]. However, the [G]-dependences of 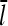 and τdepol are more complex, and are calculated in the next subsection.
and τdepol are more complex, and are calculated in the next subsection.
Dependence of l̄ and τdepol on [G]
In the presence of barbed- and pointed-end capping, there are four species of filaments: uncapped, barbed-end capped, pointed-end capped, and capped at both ends. A previous analysis of the filament length distribution of uncapped filaments (14) showed that this distribution decays exponentially as a function of length, and the average filament length is inversely proportional to the difference between the on- and off-rates. Here, we generalize this analysis by calculating 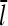 with a kinetic model containing transitions between both different filament lengths and different capping states. To keep the subsequent calculation of Gc from becoming too complicated, we treat only the two barbed-end capping states explicitly; the pointed end is taken to be in an average capping state defined by ηP, the probability of its being uncapped.
with a kinetic model containing transitions between both different filament lengths and different capping states. To keep the subsequent calculation of Gc from becoming too complicated, we treat only the two barbed-end capping states explicitly; the pointed end is taken to be in an average capping state defined by ηP, the probability of its being uncapped.
The two filament populations are 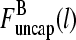 and
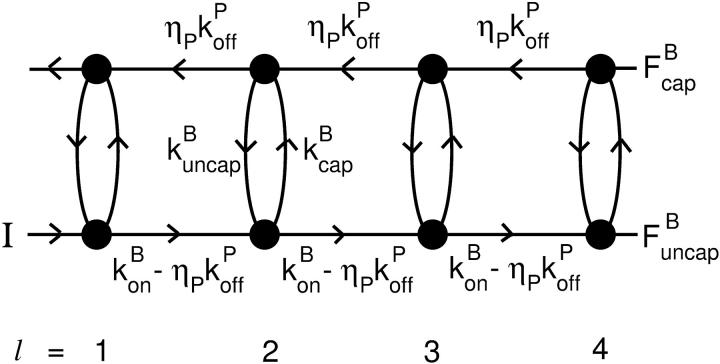
and 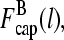 which describe the numbers of filaments of length l, uncapped and capped at the barbed end, respectively. The critical nucleus size of actin is several subunits (15,16), and in principle only l-values greater than or equal to this critical size should be included. However, because actin filament lengths are typically 100 subunits or more, the results are not changed substantially if l-values down to 1 are allowed; this simplifies the calculations. At first, we ignore the time τdis during which the filament is attached to its mother filament. The rate equations correspond to the flow chart in Fig. 1:
which describe the numbers of filaments of length l, uncapped and capped at the barbed end, respectively. The critical nucleus size of actin is several subunits (15,16), and in principle only l-values greater than or equal to this critical size should be included. However, because actin filament lengths are typically 100 subunits or more, the results are not changed substantially if l-values down to 1 are allowed; this simplifies the calculations. At first, we ignore the time τdis during which the filament is attached to its mother filament. The rate equations correspond to the flow chart in Fig. 1:
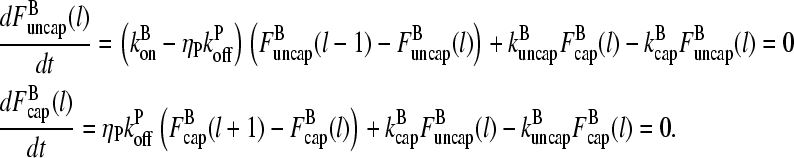 |
(6) |
FIGURE 1.
Schematic of rate equation model for two capping-state system (Eq. 6). Uncapped filaments 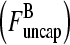 enter at the point labeled I and grow at a rate
enter at the point labeled I and grow at a rate 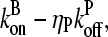 until they become capped. Capped filaments
until they become capped. Capped filaments 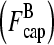 shrink at a rate
shrink at a rate 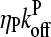 or become capped. Capping and uncapping rates are
or become capped. Capping and uncapping rates are 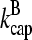 and
and 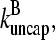 respectively.
respectively.
Here 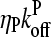 is the average off-rate at a filament pointed end, and
is the average off-rate at a filament pointed end, and 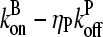 is the average net on rate for a filament whose barbed end is uncapped. By treating only barbed-end polymerization and pointed-end depolymerization, these equations ignore fluctuations resulting from cancellation of polymerization with depolymerization at each end. The very high affinities of capping proteins (17) imply that in most cases ηB < ηP, so [G] will not be too close to
is the average net on rate for a filament whose barbed end is uncapped. By treating only barbed-end polymerization and pointed-end depolymerization, these equations ignore fluctuations resulting from cancellation of polymerization with depolymerization at each end. The very high affinities of capping proteins (17) imply that in most cases ηB < ηP, so [G] will not be too close to 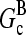 ; this implies that the barbed-end fluctuations will be small. The pointed-end fluctuations are less important because the filament length is set during the extension phase, when barbed-end growth dominates.
; this implies that the barbed-end fluctuations will be small. The pointed-end fluctuations are less important because the filament length is set during the extension phase, when barbed-end growth dominates.
The rate equations are difference equations with constant coefficients. Because there are two first-order equations, there are two linearly independent solutions. The standard approach to solving such equations is to search for solutions of the form
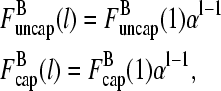 |
(7) |
where α is a constant. If two distinct values are obtained for α that lead to solutions of the difference equations, then these are the only solutions (18). Inserting Eq. 7 into Eq. 6, and dividing by αl−1, gives
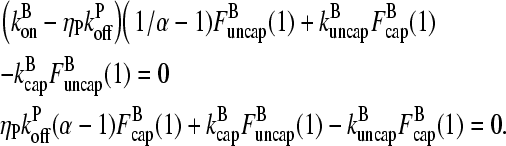 |
(8) |
Because the average filament contains many subunits, the filament populations will decay slowly with l, which means that α is close to unity. Therefore, we assume that 1 − α ≡ δ ≪ 1, and thus 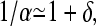 so that
so that
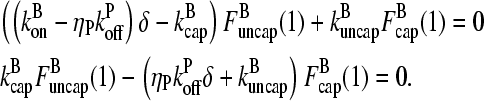 |
(9) |
This system of equations will have nonzero solutions for 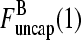 and
and 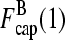 only if the determinant of the matrix
only if the determinant of the matrix
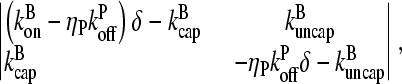 |
(10) |
vanishes. The solutions are readily seen to be δ = 0 and
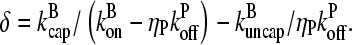 |
(11) |
Because they are distinct, we have found all of the solutions of the problem. The δ = 0 solution is unphysical because it leads to an infinite number of filaments.
For the average filament length, we then obtain:
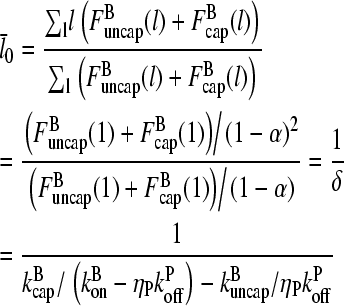 |
(12) |
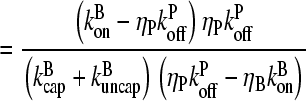 |
(13) |
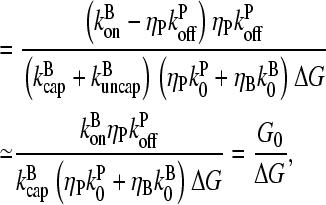 |
(14) |
where 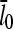 is the value of
is the value of 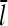 for vanishing τdis,
for vanishing τdis,
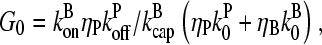 |
(15) |
and ΔG = Gtr − [G] is the deviation of [G] from Gtr, which is caused by branching. The approximations made in the last equation, ignoring 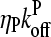 relative to
relative to 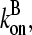 and
and 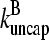 relative to
relative to 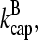 are justified because the high affinity of capping proteins ensures that
are justified because the high affinity of capping proteins ensures that 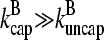 ; as mentioned above, Gc is not too close to
; as mentioned above, Gc is not too close to 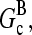 which implies that
which implies that 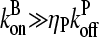 . The 1/ΔG dependence of
. The 1/ΔG dependence of 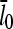 is consistent with the previous result (14) that
is consistent with the previous result (14) that 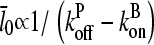 for uncapped filaments, because
for uncapped filaments, because 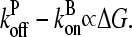
Increasing the branching rate will stimulate polymerization, thereby increasing ΔG and decreasing 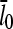 . Thus, the effect of branching on
. Thus, the effect of branching on 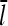 is mediated by its effect on ΔG, rather than being direct. Under Discussion, we will show that
is mediated by its effect on ΔG, rather than being direct. Under Discussion, we will show that 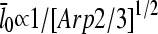 for side branching and
for side branching and 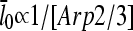 for end branching, and evaluate the consequences of these dependences.
for end branching, and evaluate the consequences of these dependences.
The average filament lifetime after debranching, τdepol, can be obtained from the same rate equations (still taking τdis = 0). We envisage a flow I of newly created filaments with free barbed ends entering at the point 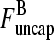 (1); defining τdepol by dN/dt = I − N/τdepol, we have τdepol = N/I in the steady state. Balancing the currents coming in and out of the entry point
(1); defining τdepol by dN/dt = I − N/τdepol, we have τdepol = N/I in the steady state. Balancing the currents coming in and out of the entry point 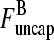 (1) (cf. Fig. 1) gives
(1) (cf. Fig. 1) gives
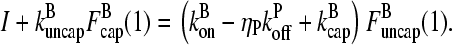 |
(16) |
This equation can be solved for 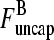 (1) by noting that the ratio
(1) by noting that the ratio 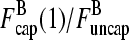 (1) is fixed by the eigenvector of the matrix (10) corresponding to the nonzero solution for δ. Straightforward calculation shows that this eigenvector satisfies
(1) is fixed by the eigenvector of the matrix (10) corresponding to the nonzero solution for δ. Straightforward calculation shows that this eigenvector satisfies
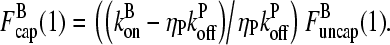 |
(17) |
Thus
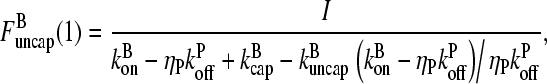 |
(18) |
so that
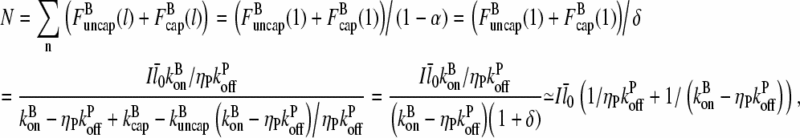 |
(19) |
where the last step holds because δ ≪ 1. Then
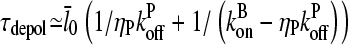 |
(20) |
Again using the fact that 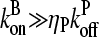 in most cases, we have
in most cases, we have
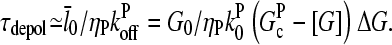 |
(21) |
For τdis ≠ 0, 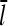 increases by an amount
increases by an amount 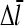 equal to the number of subunits that are added to the filament's barbed end before it detaches from the mother filament:
equal to the number of subunits that are added to the filament's barbed end before it detaches from the mother filament:
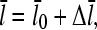 |
(22) |
where
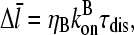 |
(23) |
and τdepol does not change with τdis, because it describes the lifetime after dissociation.
Self-consistent calculation of Gc
We now solve Eq. 5 using the [G]-dependences derived above, at first ignoring τdis. We express Gc in terms of ΔG: Gc = Gtr − ΔG. Inserting Eqs. 1, 14, 15, and 21 into Eq. 5 yields
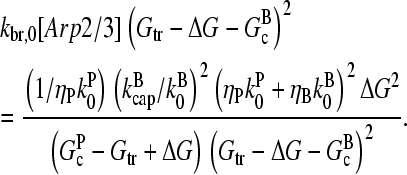 |
(24) |
This equation gives rise to a fifth-order polynomial equation for ΔG, which can be solved numerically. However, our main purpose is to obtain an analytic result that can be readily interpreted. For this reason, we develop a simplified expression valid for low branching rates, by ignoring ΔG in comparison with 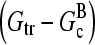 and
and 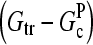 in the terms where they appear together. This yields
in the terms where they appear together. This yields
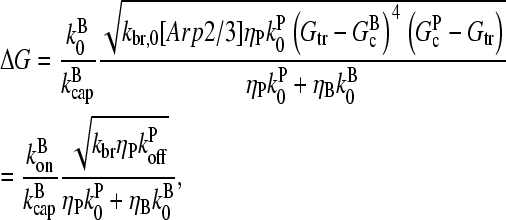 |
(25) |
where kbr, 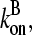 and
and 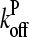 are evaluated at [G] = Gtr.
are evaluated at [G] = Gtr.
The main feature of this result is that ΔG is proportional to 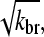 and thus to [Arp2/3]1/2 for small [Arp2/3]. This dependence results from the combination of the
and thus to [Arp2/3]1/2 for small [Arp2/3]. This dependence results from the combination of the 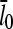 factor in the denominator of the left-hand side of Eq. 5, and the proportionality of τdepol on its right-hand side to
factor in the denominator of the left-hand side of Eq. 5, and the proportionality of τdepol on its right-hand side to 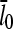 . It is thus robust to deviations from the assumed [G]-dependence of kbr. The physical origin of the
. It is thus robust to deviations from the assumed [G]-dependence of kbr. The physical origin of the 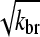 dependence is as follows. For the number of filaments to be constant, each filament must on the average generate one daughter filament before being depolymerized. A doubling of ΔG would correspond to halving the filament length, according to Eq. 14; this would in turn halve the filament lifetime according to Eq. 21. Because the number of daughter filaments formed by a given filament is proportional to its length and its lifetime, a fourfold increase in kbr would be required to keep this number constant. The same argument predicts a linear dependence of ΔG on kbr for end branching, because the factor of
dependence is as follows. For the number of filaments to be constant, each filament must on the average generate one daughter filament before being depolymerized. A doubling of ΔG would correspond to halving the filament length, according to Eq. 14; this would in turn halve the filament lifetime according to Eq. 21. Because the number of daughter filaments formed by a given filament is proportional to its length and its lifetime, a fourfold increase in kbr would be required to keep this number constant. The same argument predicts a linear dependence of ΔG on kbr for end branching, because the factor of 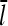 on the left-hand side of Eq. 5 would be absent.
on the left-hand side of Eq. 5 would be absent.
We now include the effects of nonzero τdis, but assume that τdis ≪ τdepol. In Eq. 5, τdis appears directly, and also indirectly through the dependence of 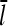 (and thus τdepol) on τdis. Including
(and thus τdepol) on τdis. Including 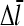 (cf. Eq. 22) in Eq. 5, together with Eq. 21, gives
(cf. Eq. 22) in Eq. 5, together with Eq. 21, gives
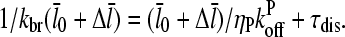 |
(26) |
Using Eq. 14 to express 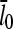 in terms of ΔG, letting kbr and
in terms of ΔG, letting kbr and 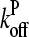 take their values for [G] = Gtr, expanding both sides of Eq. 26 to first order in τdis (recalling that
take their values for [G] = Gtr, expanding both sides of Eq. 26 to first order in τdis (recalling that 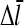 is linearly proportional to τdis), and multiplying both sides by ΔG, we obtain
is linearly proportional to τdis), and multiplying both sides by ΔG, we obtain
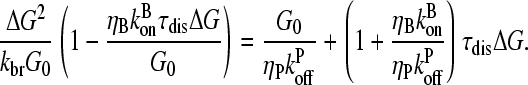 |
(27) |
Solving this equation to first order in τdis, and using Eq. 15, gives
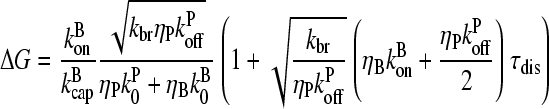 |
(28) |
Thus, increasing τdis increases ΔG, because a filament's pointed end remains capped until it dissociates, and this lowers Gc.
We now analyze the end-branching case briefly. Using Eq. 21, Eq. 5 with τdis = 0 becomes
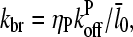 |
(29) |
where kbr is the branching rate per filament. Following the same approach as above for small ΔG, Eq. 25 becomes
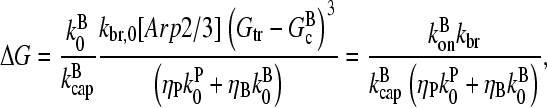 |
(30) |
where kbr and 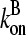 are again evaluated at [G] = Gtr. As expected, ΔG varies linearly with kbr instead of as
are again evaluated at [G] = Gtr. As expected, ΔG varies linearly with kbr instead of as 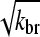 ; at small [Arp2/3], this gives ΔG ∝ [Arp2/3].
; at small [Arp2/3], this gives ΔG ∝ [Arp2/3].
Inclusion of a nonzero τdis in a fashion parallel to that for side branching yields
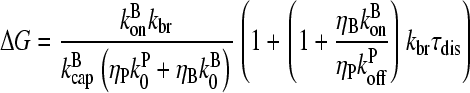 |
(31) |
Comparison of analytic theory with stochastic-simulation results for side branching
The stochastic simulations use a methodology used previously to treat actin filament clusters in solution (9). The coordinates of all filament subunits are stored over time. Isolated filaments nucleate slowly in random directions at random points in space, and subsequent growth, depolymerization, capping, uncapping, branching, and debranching events are treated stochastically. New branches appear on randomly chosen filament subunits, at an angle of 70° to the mother filament (although in bulk polymerization the branch geometry does not affect the filament length or critical concentration). Interactions between subunits on distinct filaments, corresponding to steric exclusion, are ignored because of the low volume fraction of actin at typical in vitro concentrations. Polymerization removes free monomers from solution, and depolymerization replaces them. The simulations treat cubic regions of edge length 5 μm, containing up to ∼150,000 actin monomers. The rate parameters are obtained from previous kinetic fits (7), and are given in Table 1.
TABLE 1.
Parameter values
To evaluate Gc, one could gradually ramp up the total actin concentration (the free-actin concentration at the beginning of the simulations) until polymerization begins. However, this procedure gives a very slow convergence of the concentration to Gc, and also gives noisy 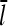 results, because of the small number of filaments present. For this reason, we instead treat a system of 2 μM total actin, in which [CP] and [Arp2/3] have values scaled up by a factor of 100, and their corresponding on-rates
results, because of the small number of filaments present. For this reason, we instead treat a system of 2 μM total actin, in which [CP] and [Arp2/3] have values scaled up by a factor of 100, and their corresponding on-rates 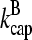 and
and 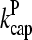 are scaled down by a factor of 100. This results in a “linearized” calculation, in which [CP] and [Arp2/3] are effectively constant during the polymerization runs despite most of the actin being polymerized. Gc and
are scaled down by a factor of 100. This results in a “linearized” calculation, in which [CP] and [Arp2/3] are effectively constant during the polymerization runs despite most of the actin being polymerized. Gc and 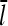 are then obtained as time averages over the last half of each run, and the runs are taken long enough that both properties have stabilized at that point. Additional statistical averaging is performed by repeating each run 10 times, with different starting seeds for the random-number generator. This leads to statistical uncertainties of ∼0.002 μM in Gc and 2% in
are then obtained as time averages over the last half of each run, and the runs are taken long enough that both properties have stabilized at that point. Additional statistical averaging is performed by repeating each run 10 times, with different starting seeds for the random-number generator. This leads to statistical uncertainties of ∼0.002 μM in Gc and 2% in 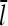 . This procedure gives the same Gc and
. This procedure gives the same Gc and 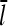 results as the ramping-up procedure, but is more computationally convenient.
results as the ramping-up procedure, but is more computationally convenient.
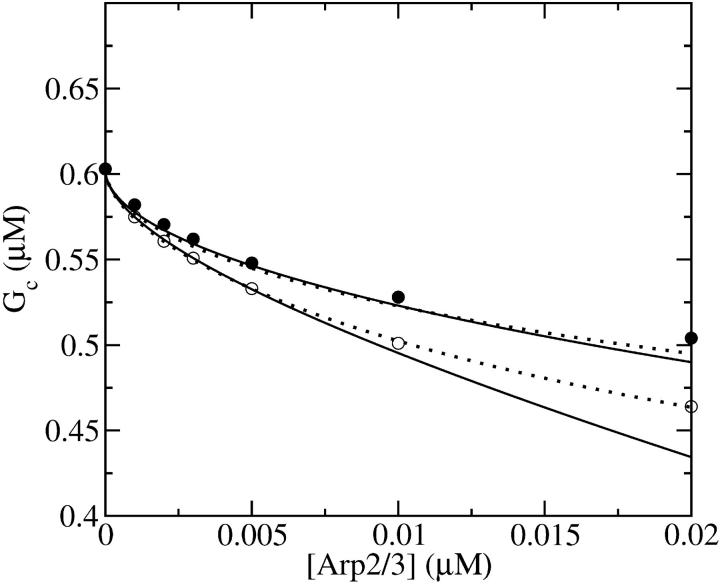
Figs. 2 and 3 show the dependence of Gc on [Arp2/3] for [CP] = 2 nM. Because 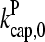 is not precisely known, we perform runs for both
is not precisely known, we perform runs for both 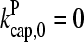 and the value from Table 1, which is plausible but not quantitatively accurate. The results for
and the value from Table 1, which is plausible but not quantitatively accurate. The results for 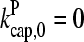 (Fig. 2) are shown for both instantaneous debranching (solid circles) and an in vitro debranching rate of 0.0018 s−1 (open circles) derived (7) from fitting microscopy data for debranching (19). For both cases, the agreement between the analytic result of Eq. 28 and the numerical results is excellent. The full numerical solution of Eq. 24 (dotted lines) gives still closer agreement with the simulations. Fig. 3 shows corresponding results using
(Fig. 2) are shown for both instantaneous debranching (solid circles) and an in vitro debranching rate of 0.0018 s−1 (open circles) derived (7) from fitting microscopy data for debranching (19). For both cases, the agreement between the analytic result of Eq. 28 and the numerical results is excellent. The full numerical solution of Eq. 24 (dotted lines) gives still closer agreement with the simulations. Fig. 3 shows corresponding results using 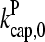 from Table 1; the dashed line denotes Gtr. Again, the results for small [Arp2/3] are quite accurate. At larger [Arp2/3], ΔG (the difference between the dashed line and the simulation points) is overestimated by ∼40%, because the decrease in kbr due to decreasing [G] is not included in our approximate solution of Eq. 24.
from Table 1; the dashed line denotes Gtr. Again, the results for small [Arp2/3] are quite accurate. At larger [Arp2/3], ΔG (the difference between the dashed line and the simulation points) is overestimated by ∼40%, because the decrease in kbr due to decreasing [G] is not included in our approximate solution of Eq. 24.
FIGURE 2.
Effect of increasing Arp2/3 complex concentration [Arp2/3] on critical concentration Gc of actin solution. Capping protein concentration [CP] = 2 nM. Pointed-end capping effects are ignored. (•) Simulation results for τdis = 0. (○) Simulation results for τdis = 0.177 s−1. (Solid lines) Analytic theory (Eq. 28) for τdis = 0 and τdis = 0.177 s−1, respectively. (Dotted lines) Numerical solution of Eq. 28 for τdis = 0 and τdis = 0.177 s−1.
FIGURE 3.
Effect of increasing [Arp2/3] on critical concentration of actin solution with pointed-end capping. [CP] = 2 nM. (•) Simulation results. (Solid line) Analytic theory (Eq. 28) for τdis = 0. (Dashed line) Treadmilling concentration Gtr.
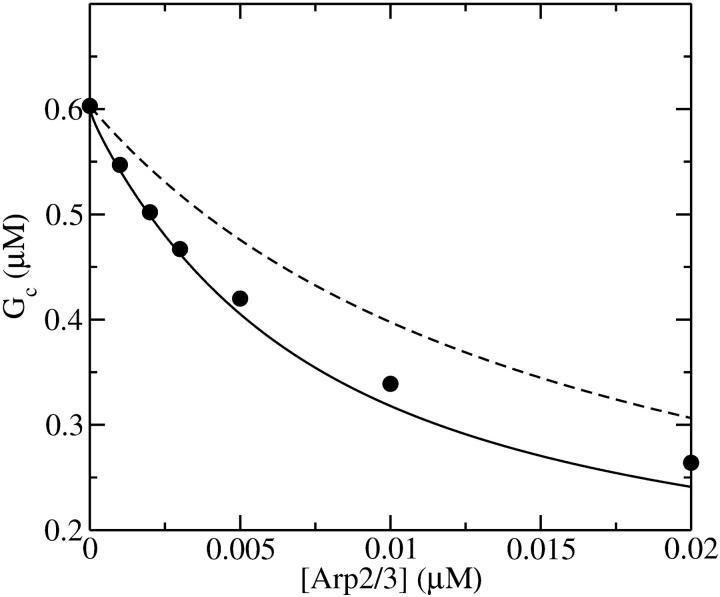
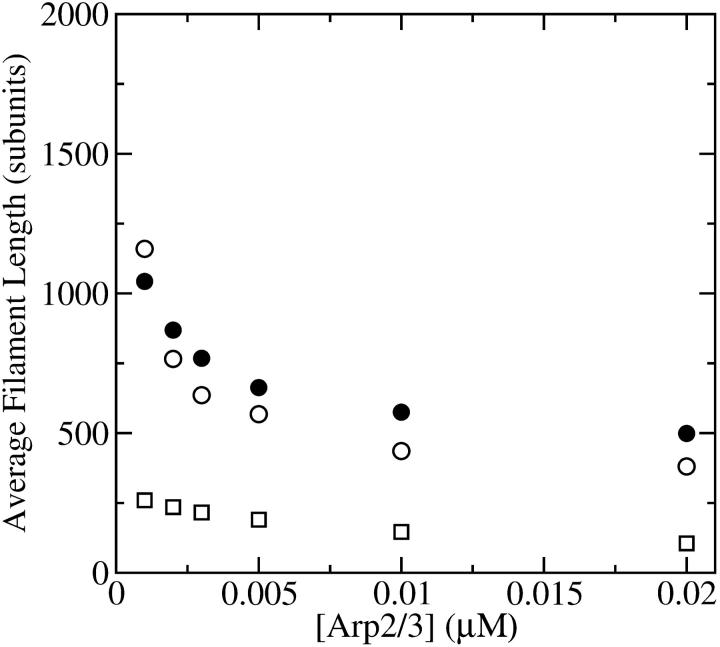
Figs. 4 and 5 compare the analytic theory with simulation results for the dependence of 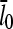 on [Arp2/3], again using
on [Arp2/3], again using 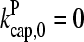 as well as the value from Table 1. To correspond as closely as possible to an experiment in which Gc is measured, at each value of [Arp2/3] we use the simulation value of Gc when calculating the on and off rates entering
as well as the value from Table 1. To correspond as closely as possible to an experiment in which Gc is measured, at each value of [Arp2/3] we use the simulation value of Gc when calculating the on and off rates entering 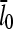 . It is seen that the agreement between the theory and simulations is close for both values of
. It is seen that the agreement between the theory and simulations is close for both values of 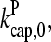 with errors of <25%. The squares in Figs. 4 and 5 give the values of
with errors of <25%. The squares in Figs. 4 and 5 give the values of 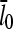 that are obtained in the absence of uncapping effects. The significance of the uncapping effects for filament lengths is treated more fully under Discussion.
that are obtained in the absence of uncapping effects. The significance of the uncapping effects for filament lengths is treated more fully under Discussion.
FIGURE 4.
Effect of increasing [Arp2/3] on average filament length of actin solutions without pointed-end capping. (•) Simulation results. (○) Analytic theory (Eq. 13) using critical concentrations obtained from simulations. (□) Average filament length lcap (Eq. 37) in absence of uncapping.
FIGURE 5.
Effect of increasing [Arp2/3] on average filament length of actin solutions, with pointed-end capping. (•) Simulation results. (○) Analytic theory (Eq. 13) using critical concentrations obtained from simulations. (□) Average filament length lcap (Eq. 37) in absence of uncapping.
Critique of model
Here, we discuss the significance of the main approximations made in our model, namely the neglect of filament severing and the possibility that kbr, 0 decays over time as filaments age.
Severing
Severing is closely analogous to side branching, in that new filaments are generated from an existing filament at a rate proportional to the filaments' length; the rates are thus both defined per filament subunit per second. The spontaneous severing rate is believed to be ∼10−8 s−1 (20). A rough estimate of the relative importance of severing can be obtained by evaluating the value of [Arp2/3] at which the severing rate equals the value of kbr. If we take a typical value of [G] to be 0.4 μM, roughly halfway between 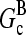 and
and 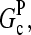 straightforward calculations based on Eq. 1 and the rate constant in Table 1 show that [Arp2/3] = 0.17 nM when kbr = 10−8 s−1. On the scale of Figs. 2–5, this value is essentially at the origin. Therefore, spontaneous severing may safely be ignored. However, several actin-binding proteins, such as ADF/cofilin, are known to accelerate filament severing. Their effects will be significant if the rate of generation of new filaments by severing is comparable to that from branching. In cells, the severing rate is probably less than the branching rate at the cell membrane. If it were not, filaments would sever in the time it takes them to move a branch spacing, and the cortical actin network would contain only one layer of branches; observed cortical actin networks contain many layers. Thus, the effects of severing should not greatly affect our subsequent discussion of branching at membranes.
straightforward calculations based on Eq. 1 and the rate constant in Table 1 show that [Arp2/3] = 0.17 nM when kbr = 10−8 s−1. On the scale of Figs. 2–5, this value is essentially at the origin. Therefore, spontaneous severing may safely be ignored. However, several actin-binding proteins, such as ADF/cofilin, are known to accelerate filament severing. Their effects will be significant if the rate of generation of new filaments by severing is comparable to that from branching. In cells, the severing rate is probably less than the branching rate at the cell membrane. If it were not, filaments would sever in the time it takes them to move a branch spacing, and the cortical actin network would contain only one layer of branches; observed cortical actin networks contain many layers. Thus, the effects of severing should not greatly affect our subsequent discussion of branching at membranes.
Nonuniformity of branch distribution
Recent real-time fluorescence-microscopy studies of branching (10,11) have shown that although branches can occur everywhere along a filament, they are more frequent near the barbed end. To account for this, a modification of the side-branching model, in which the capacity of filament subunits to form new side branches diminishes as filaments age, has been proposed (10,11). This suggestion has been supported by an analysis of polymerization data by kinetic simulations (7), which gave an aging time of ∼110 s. We have not found a simple way of including this effect in our analytic theory. However, we have performed a few simulation runs using a parameter set in which kbr,0 decays with filament age, and a correspondingly larger starting value of kbr,0 is used to compensate for the reduced branching of older filaments. The calculated Gc values are within 0.02 μM of those obtained without aging. This suggests that increasing the starting kbr,0 accounts fairly accurately for the effects of aging in vitro. If branching were highly localized near the barbed end, terms treating the interaction of Arp2/3 complex and capping protein would be needed in the model. However, the real-time branching studies do not provide evidence for such a high degree of localization.
In cells, aging at the in vitro rate would not affect branching significantly, because filament subunits branch mainly when they are in close proximity to the cell membrane. At typical intracellular on-rates (21), a subunit will remain in such close proximity for <1 s, much less than the in vitro aging time. However, if aging is greatly accelerated by actin-binding proteins such as ADF/cofilin, the effects could be important.
We also note that the calculation of the filament length at a given value of [G] is not affected by the aging, because the length depends only on the on- and off-rates.
DISCUSSION
The main results of our analysis are Eqs. 14, 28, and 31. Here we use these results to propose experiments to validate the model treated here, to develop hypotheses regarding actin polymerization in cells, and to design in vitro experiments that shed light on the branching process and allow the measurement of key parameters.
Validation of model
Direct validation of the model, for example, by measuring the extent of polymerization as a function of [Arp2/3], is difficult because of the dual functions of Arp2/3 complex in capping pointed ends and generating new branches, and also because of uncertainties in several key rate parameters. However, the dependence of 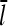 on [Arp2/3] and [CP], for large [CP], can be obtained in a form simple enough to be tested by biochemical measurements. Our calculations show that the criterion of large [CP] is fulfilled if [CP] > 2 nM and [CP] > 4[Arp2/3]. Under these conditions, ηB and thus
on [Arp2/3] and [CP], for large [CP], can be obtained in a form simple enough to be tested by biochemical measurements. Our calculations show that the criterion of large [CP] is fulfilled if [CP] > 2 nM and [CP] > 4[Arp2/3]. Under these conditions, ηB and thus 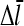 (cf. Eq. 23) are small, so we take
(cf. Eq. 23) are small, so we take 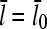 . Combining Eqs. 14, 15, and 25, we then obtain
. Combining Eqs. 14, 15, and 25, we then obtain
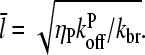 |
(32) |
To calculate 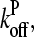 we note that according to Eq.3,
we note that according to Eq.3, 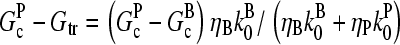 ; if [CP] is large,
; if [CP] is large, 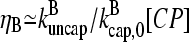 and the
and the 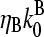 term in the denominator can be ignored, so
term in the denominator can be ignored, so 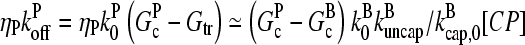 . To calculate kbr, we note that for large [CP], Gtr is close to
. To calculate kbr, we note that for large [CP], Gtr is close to 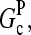 and we thus take
and we thus take 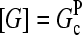 in Eq. 1. Inserting these results into Eq. 32, we obtain
in Eq. 1. Inserting these results into Eq. 32, we obtain
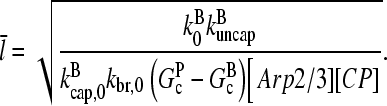 |
(33) |
This result has a simple dependence on [Arp2/3] and [CP]. Although the values of the rate parameters entering Eq. 33 are uncertain, the form of this dependence can be compared directly with experiment. For example, if both [Arp2/3] and [CP] are doubled, the measured filament length should decrease by 50%. If either one or the other is doubled, the length should decrease by 30%.
Effect of [CP] on response to Arp2/3 activation
The strength of the response of an actin solution or cell to Arp2/3 complex activation depends on [CP] in a way that can be estimated from the above results. Considering first the effect of pointed-end capping, the response of the cell (assuming that Arp2/3 complex activation enhances pointed-end capping) can be described by the function dGtr/dηP. This quantity is positive, because an increase in ηP will lead to depolymerization and thus increased Gtr. Using Eq. 3 shows that
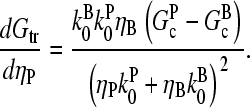 |
(34) |
This function has a maximum at 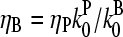 ; because
; because 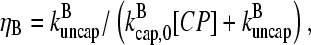 this corresponds to an optimal value of [CP]. To understand the origin of this effect, we note that for very large values of [CP], capping pointed ends causes little incremental polymerization because there are few free barbed ends. For very small values of [CP], so much of the actin is already polymerized that capping pointed ends has little further effect.
this corresponds to an optimal value of [CP]. To understand the origin of this effect, we note that for very large values of [CP], capping pointed ends causes little incremental polymerization because there are few free barbed ends. For very small values of [CP], so much of the actin is already polymerized that capping pointed ends has little further effect.
The response to branching along filament sides in vitro behaves similarly. For small values of [Arp2/3], using the proportionalities 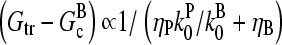 and
and 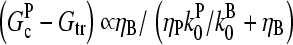 in Eq. 28 yields
in Eq. 28 yields 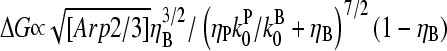 ; taking ηB ≪ 1 in view of the high [CP] binding affinity gives
; taking ηB ≪ 1 in view of the high [CP] binding affinity gives
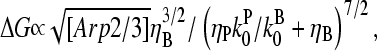 |
(35) |
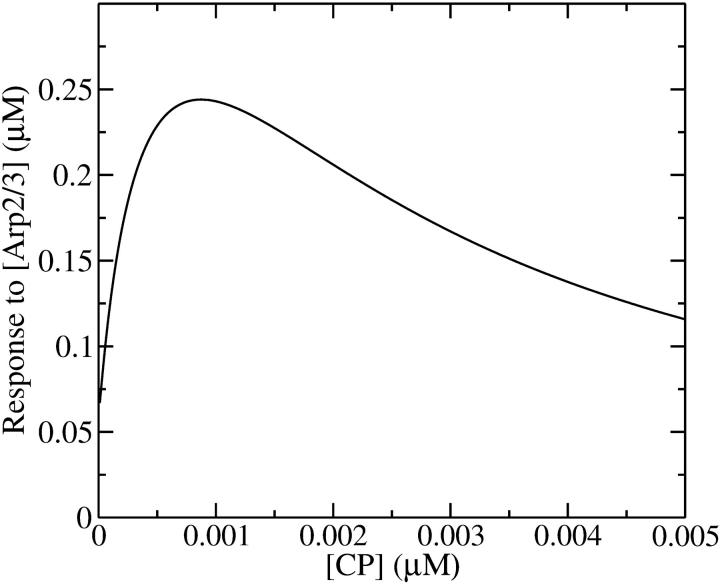
which has a maximum at 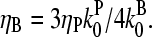 This phenomenon is illustrated in Fig. 6, which shows the change in the polymerized-actin concentration (the opposite of the change in Gc) induced in our side-branching model with pointed-end capping by 0.005 μM Arp2/3 complex, as a function of [CP]. The well-defined maximum at [CP] = 0.0008 μM is consistent with the above expectations. Thus, the response of an actin solution to Arp2/3 complex activation is strongest at an optimal value of [CP].
This phenomenon is illustrated in Fig. 6, which shows the change in the polymerized-actin concentration (the opposite of the change in Gc) induced in our side-branching model with pointed-end capping by 0.005 μM Arp2/3 complex, as a function of [CP]. The well-defined maximum at [CP] = 0.0008 μM is consistent with the above expectations. Thus, the response of an actin solution to Arp2/3 complex activation is strongest at an optimal value of [CP].
FIGURE 6.
Effect of [CP] on response of Gc to [Arp2/3], defined as Gc([Arp2/3] = 0) − Gc([Arp2/3] = 0.005 μM). Pointed-end capping is included.
This phenomenon should also occur in cells. The stimulation received by a cell is not constant over time. But the characteristic timescale of the polymerization-depolymerization processes is probably not much greater than the filament lifetime τdis + τdepol, which turnover measurements (22) suggest to be ∼20 s. If a stimulus is applied for this length of time or longer, our steady-state analysis may be relevant. New branch formation occurs mainly near filament ends at the cell membrane. Thus, the end branching analysis is appropriate. Eq. 31 yields, in analogy with Eq. 35,
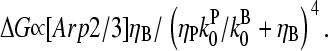 |
(36) |
This has a maximum at 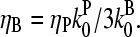 When the pointed-end capping effect is added to branching, we expect the optimal value of
When the pointed-end capping effect is added to branching, we expect the optimal value of 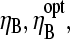 to be between the values obtained for branching and pointed-end capping:
to be between the values obtained for branching and pointed-end capping: 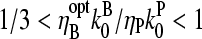 ; using
; using 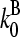 and
and 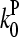 from Table 1 gives
from Table 1 gives 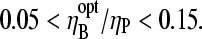 To compare this prediction with cellular values of
To compare this prediction with cellular values of 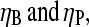 we note that the capping and uncapping rate parameters in Table 1 give binding affinities of 2 nM for Arp2/3 complex at pointed ends, and 0.05 nM for CP at barbed ends. If we assume that [CP] is a few times smaller than [Arp2/3] in cells (21), we obtain
we note that the capping and uncapping rate parameters in Table 1 give binding affinities of 2 nM for Arp2/3 complex at pointed ends, and 0.05 nM for CP at barbed ends. If we assume that [CP] is a few times smaller than [Arp2/3] in cells (21), we obtain 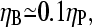 consistent with the above range. Thus, it is possible that the values of [CP] in cells are influenced by their need to respond strongly to Arp2/3 complex stimulation.
consistent with the above range. Thus, it is possible that the values of [CP] in cells are influenced by their need to respond strongly to Arp2/3 complex stimulation.
The existence of an optimal [CP] value for response to branching stimulation may be related to measurements of the motion of Listeria bacteria in pure-protein media (23). These bacteria are partly coated with the ActA protein, which stimulates actin filament branching by activating Arp2/3 complex. The measurements show a velocity maximum as a function of [CP]. Insofar as the motion of the bacterium involves a polymerization response to the branching stimulus provided by activating Arp2/3 complex, the maximum of our calculated polymerization response to Arp2/3 complex activation could be connected with the maximum of the Listeria velocity. Because these experiments used solutions containing ADF/cofilin, we are not able to estimate the optimal value of [CP] for their conditions.
The magnitude of ΔG at cell membranes
The value of kbr to use when applying Eq. 31 to cells is not known. However, the formation of branches roughly every 20 subunits in typical lamellipodial networks (24) suggests that 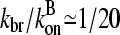 . Concentration estimates available for cells (21) suggest that [G]
. Concentration estimates available for cells (21) suggest that [G] 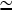 100 μM and [CP]
100 μM and [CP] 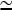 1 μM. Using the rate parameters in Table 1, we obtain
1 μM. Using the rate parameters in Table 1, we obtain 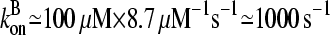 and
and 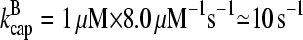 . A lower bound for ΔG is obtained if we take ηB = 1. In the absence of τdis, Eq. 31 then yields
. A lower bound for ΔG is obtained if we take ηB = 1. In the absence of τdis, Eq. 31 then yields 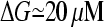 ; inclusion of the τdis term and a more realistic value of ηB would further increase ΔG. The value 20 μM is too large for this theory to accurately predict, but the calculation shows that the effect of branching on Gc is very large. Thus, the branching in the immediate vicinity of the membrane leads to a much lower critical concentration than in the cell interior.
; inclusion of the τdis term and a more realistic value of ηB would further increase ΔG. The value 20 μM is too large for this theory to accurately predict, but the calculation shows that the effect of branching on Gc is very large. Thus, the branching in the immediate vicinity of the membrane leads to a much lower critical concentration than in the cell interior.
End versus side branching in vitro
The dependence of the filament length on [Arp2/3] provides a direct comparison between the predictions of end and side branching. For side branching, combining Eqs. 14, 22, and 28 shows that for sufficiently small [Arp2/3] (where the τdis terms in Eqs. 22 and 28 are small by comparison with the other terms), 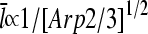 . A similar argument shows that
. A similar argument shows that 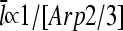 for end branching. Measurements of the dependence of
for end branching. Measurements of the dependence of 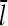 on [Arp2/3] could thus provide useful information with regard to the geometry of new branch formation, supplementing existing analysis based on polymerization kinetics and direct observation of branch formation.
on [Arp2/3] could thus provide useful information with regard to the geometry of new branch formation, supplementing existing analysis based on polymerization kinetics and direct observation of branch formation.
Effect of uncapping on filament lengths
In the absence of barbed-end uncapping effects (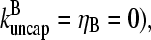 Eq. 13 reduces to
Eq. 13 reduces to
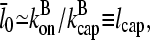 |
(37) |
where we ignore the 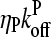 term in comparison with
term in comparison with 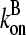 . This is the relation resulting from a simple picture in which a filament grows until it is capped, and then depolymerizes. Dividing the numerator and denominator of Eq. 13 by
. This is the relation resulting from a simple picture in which a filament grows until it is capped, and then depolymerizes. Dividing the numerator and denominator of Eq. 13 by 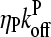 gives
gives
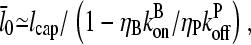 |
(38) |
so that the denominator captures the corrections from uncapping effects. Figs. 4 and 5 compare 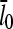 with lcap, and show that even for the rate of only 0.0004 s−1 used here, uncapping can increase
with lcap, and show that even for the rate of only 0.0004 s−1 used here, uncapping can increase 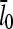 under in vitro conditions by a factor of three or more. The increase in
under in vitro conditions by a factor of three or more. The increase in 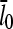 results from intermittent uncapping of filaments, which has been observed in fluorescence microscopy studies of filament growth in vitro (5). The effect on filament lengths seen in these experiments was smaller than that predicted here, probably because the timescale of the experiments was too short for steady state to be reached.
results from intermittent uncapping of filaments, which has been observed in fluorescence microscopy studies of filament growth in vitro (5). The effect on filament lengths seen in these experiments was smaller than that predicted here, probably because the timescale of the experiments was too short for steady state to be reached.
We do not have enough information about rate parameters to evaluate the extent of the uncapping corrections in vivo. However, we can estimate the critical value of 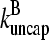 required to cause a substantial increase in
required to cause a substantial increase in 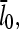 which from Eq. 38 (taking ηB ≪ 1) is
which from Eq. 38 (taking ηB ≪ 1) is 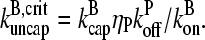 The values of ηP and
The values of ηP and 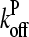 in the cellular environment are not known. However, Eq. 21 implies that
in the cellular environment are not known. However, Eq. 21 implies that 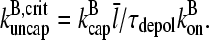 Typical filament lengths in branched networks near cell membranes are a few tenths of a micron, which corresponds to
Typical filament lengths in branched networks near cell membranes are a few tenths of a micron, which corresponds to 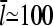 ; as mentioned above, the filament lifetime is ∼20 s, which implies that τdepol < 20 s. Thus, using the above values of
; as mentioned above, the filament lifetime is ∼20 s, which implies that τdepol < 20 s. Thus, using the above values of 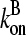 and
and 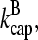 we obtain
we obtain 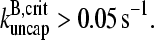 This is much faster than the spontaneous uncapping rate of 0.0004 s−1 given above, consistent with the general expectation that spontaneous uncapping is unimportant in cells. However, cell membranes contain uncapping agents, such PIP2, which might uncap at such high rates.
This is much faster than the spontaneous uncapping rate of 0.0004 s−1 given above, consistent with the general expectation that spontaneous uncapping is unimportant in cells. However, cell membranes contain uncapping agents, such PIP2, which might uncap at such high rates.
Evaluation of filament end binding properties from measured filament lengths
Equations 22 and 38 show that the filament length is determined by the on- and off-rate parameters, the “open” fractions ηB and ηP, and τdis. For small [Arp2/3], ΔG is small, so that 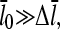 and one can take
and one can take 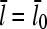 . The rate parameters entering Eq. 38 are known. Therefore, measurements of
. The rate parameters entering Eq. 38 are known. Therefore, measurements of 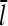 could be used to determine the ratio
could be used to determine the ratio 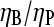 in vitro. Rough estimates of ηB exist, and these could be used to determine ηP, and thus give a new way to evaluate the binding affinity of Arp2/3 complex to pointed ends. With the ratio
in vitro. Rough estimates of ηB exist, and these could be used to determine ηP, and thus give a new way to evaluate the binding affinity of Arp2/3 complex to pointed ends. With the ratio 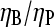 established, one can also evaluate Gtr from Eq. 3, which will be useful below.
established, one can also evaluate Gtr from Eq. 3, which will be useful below.
Evaluation of branching rate parameter from measured ΔG
With Gtr determined as above, ΔG can be obtained from measured values of Gc. If [Arp2/3] is sufficiently small, then ηP in Eq. 28 can be replaced by unity, and the τdis term can be ignored. Further, ηB can be obtained from the above-described measurement of 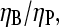 so all of the terms entering ΔG are known except for kbr,0. Therefore, this parameter could be evaluated from the measured ΔG.
so all of the terms entering ΔG are known except for kbr,0. Therefore, this parameter could be evaluated from the measured ΔG.
Acknowledgments
I appreciate informative conversations with John Cooper and David Sept, and a careful reading of the article by Frank Brooks.
This work was supported by the National Science Foundation under grant DMS-0240770.
References
- 1.Bray, D. 2001. Cell Movements: From Molecules to Motility. Garland Publishing, New York, NY.
- 2.Pollard, T. D., and G. G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–456. [DOI] [PubMed] [Google Scholar]
- 3.Higgs, H. N., and T. D. Pollard. 2001. Regulation of actin filament formation through Arp2/3 complex. Annu. Rev. Biochem. 70:649–676. [DOI] [PubMed] [Google Scholar]
- 4.Wear, M. A., and J. A. Cooper. 2004. Capping protein: new insights into mechanism and regulation. Trends Biochem. Sci. 29:418–428. [DOI] [PubMed] [Google Scholar]
- 5.Blanchoin, L., K. J. Amann, H. N. Higgs, J.-P. Marchand, D. A. Kaiser, and T. D. Pollard. 2000. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins. Nature. 404:1007–1011. [DOI] [PubMed] [Google Scholar]
- 6.Pantaloni, D., R. Boujemaa, D. Didry, P. Gounon, and M.-F. Carlier. 2000. The Arp2/3 complex branches filament barbed ends: functional antagonism with capping proteins. Nat. Cell Biol. 2:385–391. [DOI] [PubMed] [Google Scholar]
- 7.Carlsson, A. E., M. A. Wear, and J. A. Cooper. 2004. End versus side branching by Arp2/3 complex. Biophys. J. 86:1074–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mogilner, A., and L. Edelstein-Keshet. 2002. Regulation of actin dynamics in rapidly moving cells: a quantitative analysis. Biophys. J. 83:1237–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlsson, A. E. 2004. Structure of autocatalytically branched actin solutions. Phys. Rev. Lett. 92:238102. [DOI] [PubMed] [Google Scholar]
- 10.Amann, K. J., and T. D. Pollard. 2001. The Arp2/3 complex nucleates actin filament branches from the sides of pre-existing filaments. Nat. Cell Biol. 3:306–310. [DOI] [PubMed] [Google Scholar]
- 11.Ichetovkin, I., W. Grant, and J. Condeelis. 2002. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 12:79–84. [DOI] [PubMed] [Google Scholar]
- 12.Fujiwara, I., S. Suetsugu, S. Uemura, T. Takenawa, and S. Ishiwata. 2002. Visualization and force measurement of branching by Arp2/3 complex and N-WASP in actin filament. Biochem. Biophys. Res. Commun. 293:1550–1555. [DOI] [PubMed] [Google Scholar]
- 13.Berg, H. C. 1993. Random Walks in Biology, Chapt. 3. Princeton University Press, Princeton, NJ.
- 14.Howard, J. 2001. Mechanics of Motor Proteins and the Cytoskeleton. Sinauer Associates, Sunderland, MA.
- 15.Pollard, T. D., and J. A. Cooper. 1986. Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu. Rev. Biochem. 55:987–1035. [DOI] [PubMed] [Google Scholar]
- 16.Buzan, J. M., and C. Frieden. 1996. Yeast actin: polymerization kinetic studies of wild type and a poorly polymerizing mutant. Proc. Natl. Acad. Sci. USA. 93:91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooper, J. A., M. C. Hart, T. S. Karpova, and D. A. Shafer. 1999. Capping protein. In Guidebook to the Cytoskeletal and Motor Proteins. T. Kreis and R. Vale, editors. Oxford University Press, New York, NY. 62–64.
- 18.Bender, C. M., and S. A. Orszag. 1978. Advanced Mathematical Methods for Scientists and Engineers. McGraw-Hill, New York, NY.
- 19.Weaver, A. M., A. V. Karginov, A. W. Kinley, S. A. Weed, Y. Li, J. T. Parsons, and J. A. Cooper. 2001. Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 11:370–374. [DOI] [PubMed] [Google Scholar]
- 20.Sept, D., J. Xu, T. D. Pollard, and J. A. McCammon. 1999. Annealing accounts for the length of actin filaments formed by spontaneous polymerization. Biophys. J. 77:2911–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollard, T. D., L. Blanchoin, and R. D. Mullins. 2000. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29:545–576. [DOI] [PubMed] [Google Scholar]
- 22.Watanabe, N., and T. J. Mitchison. 2002. Single-molecule speckle analysis of actin filament turnover in lamellipodia. Science. 295:1083–1086. [DOI] [PubMed] [Google Scholar]
- 23.Loisel, T. P., R. Boujemaa, D. Pantaloni, and M.-F. Carlier. 1999. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 401:613–616. [DOI] [PubMed] [Google Scholar]
- 24.Svitkina, T. M., and G. G. Borisy. 1999. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 145:1009–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higgs, H. N., L. Blanchoin, and T. D. Pollard. 1999. Influence of the C terminus of Wiskott-Aldrich syndrome protein WASp and the Arp2/3 complex on actin polymerization. Biochemistry. 38:15212–15222. [DOI] [PubMed] [Google Scholar]
- 26.Pollard, T. 1986. Rate constants for the reactions of ATP- and ADP-actin with the ends of actin filaments. J. Cell Biol. 103:2747–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]