Abstract
The UL74 (glycoprotein O [gO])-UL75 (gH)-UL115 (gL) complex of human cytomegalovirus (CMV), known as the gCIII complex, is likely to play an important role in the life cycle of the virus. The gH and gL proteins have been associated with biological activities, such as the induction of virus-neutralizing antibody, cell-virus fusion, and cell-to-cell spread of the virus. The sequences of the two gH gene variants, readily recognizable by restriction endonuclease polymorphism, are well conserved among clinical isolates, but nothing is known about the sequence variability of the gL and gO genes. Sequencing of the full-length gL and gO genes was performed with 22 to 39 clinical isolates, as well as with laboratory strains AD169, Towne, and Toledo, to determine phylogenetically based variants of the genes. The sequence information provided the basis for identifying gL and gO variants by restriction endonuclease polymorphism. The predicted gL amino acid sequences varied less than 2% among the isolates, but the variability of gO among the isolates approached 45%. The variants of the genes coding for gCIII in laboratory strains Towne, AD169, and Toledo were different from those in most clinical isolates. When clinical isolates from different patient populations with various degrees of symptomatic CMV disease were surveyed, the gO1 variant occurred almost exclusively with the gH1 variant. The gL2 variant occurred with a significantly lower frequency in the gH1 variant group. There were no configurations of the gCIII complex that were specifically associated with symptomatic CMV disease or human immunodeficiency virus serologic status. The potential for the gCIII complex to exist in diverse genetic combinations in clinical isolates points to a new aspect that must be considered in studies of the significance of CMV strain variability.
The importance of the infecting strain of cytomegalovirus (CMV) for clinical outcome has been the subject of speculation since it was first recognized that all epidemiologically unrelated clinical isolates of CMV have a unique genetic profile (19). There is a high level of sequence homology, estimated to be 80%, but individual CMV isolates can be readily differentiated by restriction enzyme analysis of full-length genomic DNA. CMV clinical isolates can also be grouped by immunologic assays, such as neutralization kinetics (48, 51). Clinical isolates vary in their ability to mediate myelosuppression in vitro (38) or replicate in either endothelial (34) or smooth-muscle cells in vitro (52). Whether there is a genetic basis for these biological differences among virus isolates is unknown.
Comparative genetic analysis of CMV strains and isolates is still in its early stages and is limited primarily by the size and complexity of the viral genome. Although the complete genome of laboratory strain AD169 is known, as many as 22 additional virus genes were recently found in low-passage clinical isolates that are missing in the widely used laboratory strains AD169 and Towne (5). The variability in individual genes of low-passage isolates of CMV is beginning to be documented. The genes for UL55 (glycoprotein B [gB]) and UL75 (gH) code for two of the immunologically dominant envelope glycoproteins of CMV (7-9) involved in the induction of virus-neutralizing antibody. Based on variability at the cleavage site of the 169-kDa polyprotein, four major variants of the gB gene have been determined. Additional sites of variability have been described at restricted regions of the N and C termini of the gene product (14, 31). Based on sequence analysis of gH from clinical isolates, which revealed variability at the N terminus, two major variants can be defined for the gH gene. Differences among strains have also been reported for UL11 (17), UL144 (30), and exon 3 of the immediate-early gene (15, 54).
The gCIII viral-envelope complex consists of UL74 (gO), UL75 (gH), and UL115 (gL) (13, 20-22, 28). The gH component induces virus-neutralizing antibody (37) and facilitates penetration of CMV into host cells (24-26). gL is necessary for transport of the gH glycoprotein to the cell surface (23, 42). The disulfide-bonded, tripartite gCIII complex is displayed on the surface of infected cells and ultimately becomes part of the virion (44, 45). Positional homologues of the UL74 gene exist among other herpesviruses (21), but the function of the gO gene product is unknown.
In view of the importance of the viral-envelope glycoprotein complexes for both replication and induction of a host immune response, one might predict a high level of genetic conservation. Studies from many laboratories have confirmed the consistent detection of two closely related variants of the gH gene; however, nothing is known about the genetic variability of the individual gL and gO genes among isolates. Moreover, we know nothing about the genetic configuration of the gCIII complex among clinical isolates. Therefore, in conjunction with an analysis of the CMV gH genes, we have phylogenetically classified the variants of the gL and gO genes that were amplified from low-passage clinical isolates, samples from CMV-infected individuals, and laboratory strains of CMV. We have studied the distribution of these three genes that code for the protein components of the gCIII complex components in different patient groups who are at high risk for symptomatic CMV infection.
MATERIALS AND METHODS
Viruses.
Blood buffy coat, urine, and bronchoalveolar lavage samples were obtained from human immunodeficiency virus (HIV)-positive and HIV-negative immunosuppressed patients. Vitreous humor samples from HIV-positive patients have been arbitrarily designated with a DM prefix. Commercially obtained MRC5 and human fibroblast (HF) cells were used for all CMV cultures. Clinical isolates were passaged fewer than five times and stored at −70°C. Laboratory strains AD169, Towne, and Toledo had been passaged for an unknown number of times in fibroblast cultures.
Sequence analysis.
Proteinase K-digested samples were amplified by PCR using the primers listed in Table 1. For amplification, samples were diluted 1:10 to 1:100 in 100 μl of a master mix-sample solution. All master mixes contained 1× PCR buffer, 2.5 mM MgCl2, 150 μM concentrations of each deoxynucleoside triphosphate (dNTP), 2.5 U of Taq polymerase (Life Technologies, Gaithersburg, Md.), and 20 pmol each of upper and lower primers (Operon Technologies, Alameda, Calif.). The cycling parameters were 95°C for 30 s, followed by 40 cycles of 94°C for 15 s, 55°C for 20 s, and 70°C for 2 min, followed by a 10-min hold at 72°C.
TABLE 1.
Primers used for PCR amplification and sequencing of human CMV
| Gene | Procedure | Primer | Primer sequence (5′ to 3′) | Relative position on genea |
|---|---|---|---|---|
| gO (UL74) | PCR | UL74alg5′ | cag caa aac gac cag aat cag | −87 to −67 |
| UL74alg3′ | tcc taa tgt gat gag acg ac | 150 to 170 | ||
| Sequencing | 74up223 | caa acc aca agg cag acg ga | −45 to −25 | |
| 74up63 | acg gtg cgg ggt ctc ctc ct | −25 to −6 | ||
| 74up330 | gtt tga ttt tta tag tac cca gc | 330 to 352 | ||
| 74up989 | cca agt ata tta acg gca cca | 713 to 733 | ||
| 74up1326 | ccg aac cgc cgt gtc aga at | 1050 to 1069 | ||
| 74lp1731 | gtg gac tat gct taa tgc tct | 54 to 74 | ||
| 74lp5 | acg cac ctt tac agt tta tga a | 79 to 100 | ||
| 74lp251 | ata aaa atc aaa cca caa gt | 323 to 342 | ||
| 74lp314 | ttt ctg agc cgt ttg att at | 386 to 405 | ||
| 74lp406 | agg tgg ggg tct gaa tgt tat | 406 to 426 | ||
| 74lp902 | cgt aac gta aaa gca ggg cgg | 626 to 646 | ||
| 74lp1326 | att ctg aca cgg cgg ttc gg | 1050 to 1069 | ||
| 74lp1411 | att ctg aca cgg cgg ttc gg | 1054 to 1074 | ||
| mRNA analysis | 74 up m | cgc ctt ccc aag tat atta | 705 to 723 | |
| 74lp m | cac cac caa agg cta ttg agg | 1458 to 1478 | ||
| Amplification of DNA for restriction digests | 74 up out | cag ctt cga aaa ccg gcc aaa tac g | 349 to 373 | |
| 74 lo out | aat ata ctt ggg gac gcg aaa ata ga | 699 to 723 | ||
| 74 up in | gct tcg aaa acc ggc caa ata cg | 351 to 373 | ||
| 74 lo in | ata ctt ggg gac gcg aaa tag a | 699 to 720 | ||
| 74 TOW up out | caa ctc cgt aaa ccg gcc aaa t | 337 to 358b | ||
| 74 TOW lo out | ata tac ttg gga acg cgg | 693 to 710b | ||
| 74 TOW up in | ctc cgt aaa ccg gcc aaa tat g | 340 to 361b | ||
| 74 TOW lo in | tac ttg gga acg cgg aat | 690 to 707b | ||
| gL (UL115) | PCR | 115Alg5′ | ctg gct cct tac cgt cac ac | −41 to −20 |
| 115Alg3′ | ctc ggg cgg aca cag ata gc | 924 to 943 | ||
| Sequencing | 115SUp2 | gtg ctg ttg gac gat gct ttc | 295 to 314 | |
| 115SUp3 | tac agc gag tgc ggc gat gg | 421 to 440 | ||
| 115SLp1 | cat cgc cgc act cgc tgt ag | 420 to 439 | ||
| 115SLp2 | aag aga gac ggt ggc acc ag | 547 to 566 | ||
| 115SLp3 | cag aga gcc ttt att atc ag | 843 to 862 | ||
| Amplification of DNA for restriction digests | 115 up out | ttg atg tgc cgc cgc ccg gat t | −3 to 19 | |
| 115 lo out | gca cca gct cga agc cta ac | 534 to 553 | ||
| 115 up in | atg tgc cgc cgc ccg gat t | 1 to 19 | ||
| 115 lo in | cca gct cga agc cta ac | 534 to 550 |
Positions are relative to those of sequences from strain AD169 (GenBank accession no. X046066). The positions for the 5′ or upper primers are relative to the first base pair of the atg start codon; those for the 3′ or lower primers are relative to the last base pair of the stop codon of the gene.
Positions of primers designated TOW are relative to the positions of the sequences from the Towne strain.
Following amplification of the full-length gene, PCR products were purified (QIAquick PCR purification kit; Qiagen, Hilden, Germany) and 100 ng per reaction was used for sequencing. For sequencing reactions, 10 μl of each sample was used with dRhodamine dye-labeled dideoxy terminators (Applied Biosystems, Foster City, Calif.) and 5 pmol of each of the corresponding sequencing primers. The sequencing reactions were then analyzed on an ABI model 377 instrument (Applied Biosystems). Data were assembled manually, proofread, and edited using ABI sequence analysis software. Nucleotide and amino acid deviations were determined by comparison with the AD169 consensus sequence. Multiple sequencing of laboratory strains and patient samples was performed in order to validate the reproducibility of the sequence determination as well as to rule out laboratory contamination.
Phylogenetic analysis.
DNA sequences of the gL and gO genes were aligned and analyzed using PAUP version 4.0 software (16) for analysis of relationships by the generation of phylogenetic trees. The radially described phylogenetic trees were based on neighbor-joining methods with a maximum-likelihood-based distance. For each gene, a single, unrooted tree with 100 bootstrap values was generated.
Restriction digests.
The two gH variants were identified by restriction digestion with HhaI by using published methods (53). A single digestion with HpaII was used to characterize the four gO variants by using restriction fragment length polymorphism (RFLP). Double digestion with BamII and HgaI was used to identify the four gL variants. For the gO and gL genes, restriction digest products were analyzed on 2 and 3% Metaphor agarose gels (BME Products, Rockland, Maine), respectively.
mRNA analysis.
mRNA was extracted from cultured fibroblast cells infected with various strains of CMV by using a commercial kit (Direct mRNA mini kit; Qiagen) and analyzed by reverse transcription-PCR (OneStep RT-PCR kit; Qiagen) using the primers shown in Table 1. Samples were diluted 1:3 before 1 μl was added to a final volume of 50 μl of a master mix-sample solution. All master mixes contained 1× PCR buffer, 2.5 mM MgCl2, 1× Q solution, 1.6 mM concentrations of each dNTP, 2 μl of OneStep RT-PCR enzyme mix, and 30 pmol each of upper and lower primers (Operon Technologies). The cycling parameters for each strain were 50°C for 30 s and 95°C for 30 s, followed by 35 cycles of 94°C for 15 s, 53°C for 20 s, and 72°C for 2 min, followed by a 10-min hold at 72°C.
After completion of the cycling, 2 μl of PCR product was added to a final volume of 100 μl of second-round master mix-sample solution. All master mixes contained 1× PCR buffer, 2.5 mM MgCl2, 600 μM concentrations of each dNTP, 2.5 U of Taq DNA polymerase (Life Technologies), and 10 pmol each of upper and lower primers (Operon Technologies). The cycling parameters for each strain were 94°C for 2 min, followed by 35 cycles of 94°C for 15 s, 53°C for 20 s, and 72°C for 2 min, followed by a 10-min hold at 72°C. Following PCR amplification, ethidium bromide-stained products were analyzed based on band migration through a 3% agarose gel (SeaKem LE agarose; FMC BioProducts, Rockland, Maine).
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper are available from the GenBank nucleotide sequence database. The accession numbers are AF530165 to AF530190 for the gL gene and AF531315 to AF531356 for the gO gene.
RESULTS
Phylogenetic assessment of relationship between intragenic variants.
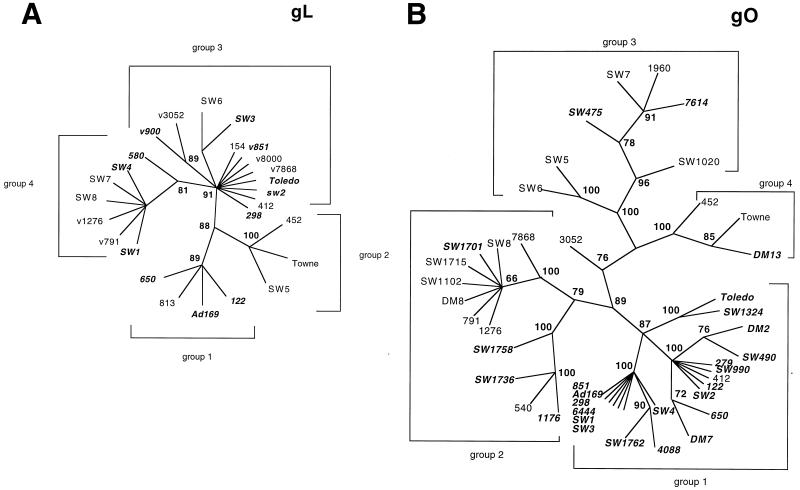
For the gO gene, full-length gene sequences from 40 clinical isolates and three laboratory strains (Towne, Toledo, and AD169) were analyzed, and for the gL gene, full-length gene sequences from 23 clinical isolates and the three laboratory strains were analyzed. The analysis of the phylogenetic relationships for intragenic variants of the gL and gO genes is depicted in Fig. 1. Bootstrap values of greater than 60% are shown at the major nodes of the phylogenetic trees. The groupings were established by using the first level of significant divergence, based on bootstrap values, from the main node of the tree. Four major phylogenetic groups were designated for both the gL and gO genes. For each gene, the group containing the AD169 laboratory strain was designated as group 1. For the gL gene, all isolates and laboratory strains could be readily grouped, with only a single strain (580) diverging significantly from its group 4 cohorts. gL group 3 showed the greatest degree of heterogeneity, with three potential subgroups.
FIG. 1.
Phylogenetic relationships of intragenic variants of UL115 (gL) (A) and UL74 (gO) (B) of CMV. The unrooted radial trees were generated using neighbor-joining methods with a maximum-likelihood-based distance. For each gene, a single tree with 100 bootstrap values was generated. The bootstrap values, indicating the estimated percentage of the frequency of relatedness, are shown in boldface type at the junctions of the major nodes. The individual groups represent clinical isolates derived from both allografts and HIV-infected patients. Laboratory strains AD169, Towne, and Toledo are indicated, and the human CMV strains with the gH1 variant are indicated with italic type.
The highly branched phylogenetic tree for the gO gene reflected a high level of intragenic sequence variability. Four phylogenetic groups could be identified, with only one clinical strain, 3052, not readily fitting into any one of the four groups. While 3052 was intermediate between groups 2 and 3, it did not appear to be a recombinant. Strain 3052 was not included in subsequent analyses of the distribution of gene variants in patients because of its indeterminate genotype.
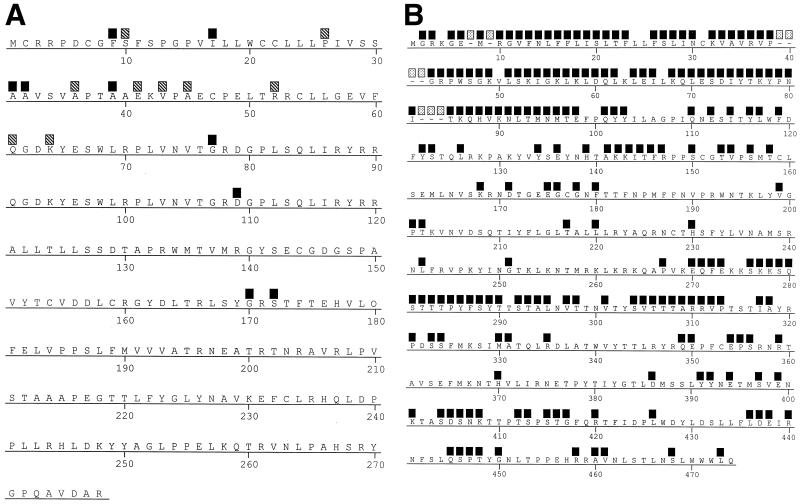
Regions of major variability in amino acids within consensus sequences of gO and gL.
By incorporating the data from both the laboratory and clinical strains, consensus amino acid sequences were generated for both gL and gO (Fig. 2). The major amino acid variability observed for gL among the strains occurred within the first 45 amino acids, with only four amino acid changes occurring in the remainder of the protein. Most of the variability in the consensus sequence was due to variability in phylogenetic group 3, as shown by the hatched boxes in Fig. 2A. Amino acid variability that was common to all four groups, as shown by the solid boxes in Fig. 2, was concentrated within the first 32 amino acids of the protein. However there were four single changes prior to amino acid 172. After that point, there was total conservation of sequence among all strains. The overall variability of the amino acid consensus, when compared to that of gL from AD169, was less than 2%.
FIG. 2.
Positional variability in gL (UL115) (A) and gO (UL74) (B) from genetic variants of CMV relative to a computer-generated consensus sequence (MegAlign; DNASTAR, Madison, Wis.). The positions of sequence variability among the products of all four variants are indicated by solid boxes. In panel A, the hatched boxes indicate positions of variability from the gL consensus sequence due only to changes in amino acids of gL group 3. In panel B, the positions of deletions in the gO groups are indicated by speckled boxes.
gO displayed a high level of sequence variability among phylogenetic groups. Much of the variability, particularly at the N terminus, was the result of deletions in the sequences from clinical isolates. Variability was profound in the first 98 amino acids and again between amino acids 270 and 313. There were areas of almost total conservation between amino acids 180 and 269, and only sporadic changes were observed between amino acids 314 and 390. Focal areas of variability occurred regularly after amino acid 390, approaching the C terminus of the molecule. The sequence variability among strains, including both deletions and amino acid changes, was 46% when compared to the 466 amino acids of gO from AD169. The deletions in the sequences from the clinical isolates resulted in predicted gene products ranging from 457 to 466 amino acids. A single strain, 3052, had gO with 472 amino acids.
Only epidemiologically related strains had total sequence identity for either gL or gO. Specifically, two isolates from the same individual, cultured within 1 month of each other, had identical sequences for both gL and gO. In addition, isolates from twins with neonatal CMV infection yielded strains with identical sequences (data not shown).
Genetic linkages among components of the gCIII complex.
The initial analysis included only those isolates for which the full-length sequences of the gL and gO genes were known. Nineteen of the 26 isolates with the gH1 variant (strain 3052 excluded) also had the gO1 variant, 4 had gO2, 2 had gO3, and 1 had gO4. For the 16 isolates with the gH2 variant, 1 had gO1, 8 had gO2, 5 had gO3, and 2 had gO4. The occurrence of the gO1 variant in the isolates with gH1 was significant (P = 0.0008, χ2 analysis with Yates' correction). The distribution of the four gL variants was similar within both the group with gH1 (3, 0, 6, and 2 isolates with each of the four respective gL variants) and the group with gH2 (1, 0, 6, and 4 isolates).
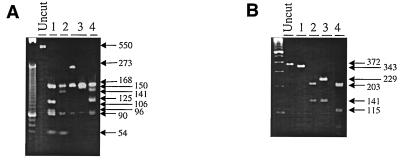
To simplify the analysis of the sequences available for the study of any possible variant among all three genes coding for the gCIII complex, 84 isolates from 81 patients, along with the three laboratory strains, were analyzed by restriction endonuclease polymorphism after digestion with a series of enzymes. The primers used, which were designed to amplify regions of each gene that would produce unique fragments after restriction endonuclease digestion, are listed in Table 1. A single set of primers was adequate for amplifying a region of the gL gene that could be digested for identification of the four variants. However, for the gO gene, the primers designed for the AD169 template did not readily amplify gO4. Therefore, a separate set of primers was used based on a region of gO4 from the Towne strain (Table 1). The predicted gL and gO variants were readily differentiated by characteristic RFLP patterns (Fig. 3). Twenty-four samples were compared for sequence agreement with the predicted RFLP pattern for the gL gene, with all isolates producing a pattern that was consistent with the predicted digestion products. For the gO gene, 36 of 38 samples with known sequences gave the expected RFLP patterns, based on computer simulation of the digest. Two of the 38 clinical strains gave results inconsistent with the predicted pattern, resulting in an “untypeable” genotype designation. The untypeable variants were not included in any analysis.
FIG. 3.
RFLP of the gL and gO variants. A 550-bp fragment of the gL gene and a 372-bp fragment of the gO gene were amplified using the primers listed in Table 1. The amplified fragments are shown in the lanes labeled “Uncut.” (A) Double digestion with BamII and HgaI was used to identify the four gL (UL115) variants. Lanes: 1, gL1; 2, gL2; 3, gL3; 4, gL4. (B) A single digestion with HpaII was used to characterize the four gO (UL74) gene products. Lanes: 1, gO1; 2, gO2; 3, gO3; 4, gO4. The restriction digest products for the gO and gL genes were analyzed on 2 and 3% Metaphor agarose gels, respectively. Arrows indicate the molecular sizes of the fragments (in base pairs).
The distribution of the gH, gL, and gO variants within the gCIII complex of 87 different strains of CMV is shown in Table 2. When the frequency of occurrence of gO1 within the gH1 group, regardless of the gL variant, was compared to that within the gH2 group, the difference was highly significant (P = 0.00001). Within the gH1 group, the frequency of occurrence of gL2 in combination with any gO variant was significantly reduced (P = 0.01). Finally, the gO2-gL4 configuration was missing entirely from the gH1 group but was significantly represented within the gH2 group (P = 0.03).
TABLE 2.
Distribution of variants of the gH, gL, and gO genes coding for the gCIII complex in clinical isolates and laboratory strains of human CMV
| gL variant | No. of isolates (plus laboratory strain) with gH1 and gO variant:
|
No. of isolates (plus laboratory strain) with gH2 and gO variant:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 2 | 3 | 4 | Mix | 1a | 2 | 3 | 4 | Mix | |
| 1 | 8 (AD169) | 1 | 1 | 0 | 3 | 1 | 2 | 0 | 0 | 3 |
| 2b | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 (Towne) | 0 |
| 3 | 12 (Toledo) | 4 | 1 | 1 | 4 | 1 | 3 | 6 | 1 | 2 |
| 4 | 13 | 0 | 5 | 3 | 0 | 0 | 7c | 6 | 1 | 3 |
| Mix | 3 | 1 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 1 |
P = 0.00001 when the frequency of the association of gO1 with gH1, irrespective of the gL variant, was compared to that with gH2.
P = 0.01 when the frequency of gL2 association with any gO variant within the gH1 variant group was compared to that of any other gL variant.
P = 0.03 when the frequency of gO2-gL4 in the gH2 variant group was compared with that in the gH1 variant group.
Genes for the gCIII complex do not segregate according to patient group.
The gH, gL, and gO genes in isolates from 46 patients with HIV infection and 35 patients who were immunocompromised but HIV negative were analyzed. In the HIV-positive group, 17 isolates with gH1 and 18 isolates with gH2 were characterized. Among the HIV-positive patients, 33 isolates with gH1 and 13 with gH2 were evaluated. There was no segregation of any of the variants, either alone or as members of the gCIII complex, according to HIV status.
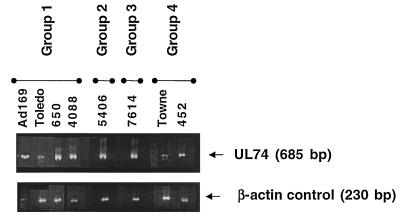
The gO variants of all four phylogenetic groups are transcribed in human fibroblast cells.
Isolates or laboratory strains representing each of the four genetic groups of the gO gene were grown in HF cells for extraction of mRNA (Fig. 4). The presence of specific CMV gO transcripts was evaluated by amplifying a conserved region of the gO gene with the primers listed in Table 1. The amplification of endogenous β-actin mRNA, used as a positive control, was similar in all wells. The gO mRNA was transcribed by all of the CMV isolates, with slight shifts in molecular size resulting from deletions in the sequence of the gene. The β-actin mRNA was downregulated in AD169-infected cells, as has been previously reported (30).
FIG. 4.
Transcription of mRNA by each of the four gO genetic groups of CMV. Human embryonic fibroblast cultures were infected with the indicated strains of CMV, representing each of the four genetic groups. When a cytopathic effect was evident, mRNA was extracted from the infected cultures and reverse transcribed, using the primers listed in Table 1, to generate the expected 685-bp product. The resulting cDNA was electrophoresed in a 3% agarose gel, and bands were visualized by ethidium bromide staining. A 230-bp fragment from β-actin mRNA was amplified as a positive control, and the results are shown in the lower panel.
DISCUSSION
The data in this investigation have shown that there is predictable variation in the genes for the glycoproteins of the gCIII complex of human CMV. In addition to the two known variants for the gH gene, there are four phylogenetically defined groups for the gL, as well as four groups for the gO, gene. These data reveal the large number of gH-gL-gO configurations that may potentially exist in nature.
The magnitude and location of the nucleotide variation that resulted in changes at the amino acid level were different for each gene coding for the complex. It has been reported that the approximately 15% variability in gH is restricted to the first 30 amino acids of the N terminus. In this study, variability in gL was also localized within the first 45 amino acids. There was an approximately 2% difference in amino acid sequences among proteins from different strains. Most of the differences from the consensus sequence were the result of changes in the gL3 variant. The high degree of amino acid conservation in gL likely reflects an essential role in the virus replication process. In support of this idea, gL appears to be essential for virus infectivity (18). The gL homologues of the various herpesviruses are genetically unique; however, it has recently been shown that gL of Epstein-Barr virus (EBV) can be a substitute for gL of varicella-zoster virus, pointing to a high degree of conserved function (29). In contrast to what was observed with gL, variability within gO occurred throughout the entire sequence. Deletions that contributed to the variability among proteins from different strains were restricted to the first 90 amino acids. Between amino acids 1 and 180 and 270 and 313, virtually every amino acid was different among the groups when compared to an overall consensus sequence generated by computer analysis. The variability in gO is unlikely to be an artifact of the tissue culture passage of clinical isolates. All isolates were low passage (passaged fewer than five times in fibroblast cell culture). In addition, when gO sequence determination was performed directly from vitreous humor samples from patients, the sequences were remarkably similar to those from clinical isolates within the same genotypic group that had undergone limited tissue culture passage. The variability in gO is also unlikely to be the result of inadvertent analysis of mixed strains in the HIV-infected patients, since distribution of the gO variants was similar in both HIV-positive and HIV-negative individuals. Transcription of mRNA from each of the four gO variants indicates that genetic variation is unlikely to impede the encoding of a functioning protein. Our data are consistent with those reported in a recently published study of gO variability in which the gO gene from a single clinical isolate was sequenced (34).
A strong genetic linkage between the gH1 and the gO1 variants is reported. The proximity of the open reading frames for the two genes on the DNA of CMV (UL74 for gO and UL75 for gH) might make this an expected finding. However, it is not clear why this link occurs with the gH1 variant. There were also some combinations of the gCIII complex that were detected with greater or lesser frequency. For example, the gL2 appeared with a significantly lower frequency in the groups with gH1 than did the other gL variants. There was an insufficient number of samples in the gH2 group for a similar analysis. In addition, the gO2-gL4 combination appeared with higher frequency in the gH2 group than in the gH1 group. However, the lack of a genetic linkage between most of the gL variants and either the gH or gO variant is consistent with the high degree of conserved primary sequence in the gL gene. The reasons for the selective occurrence of some combinations could be physical and could depend on folding constraints within the tertiary structure of the complex. Alternatively, different combinations of the gCIII complex may vary in their immunological potential. Some combinations may favor immune evasion, while others may promote a vigorous immune response that favors virus eradication.
The biological consequences of the variability of the gCIII complex are unknown. We do know that the gH-gL complex of herpes simplex virus stimulates neutralizing antibody that protects mice against a lethal challenge dose (35). The genetic homologies of the gH and gL genes among betaherpesviruses suggest a potentially important role for gH and gL in protective immunity for CMV as well as for herpes simplex virus. Should the gCIII complex prove to be more important than its single components in the induction of virus-neutralizing antibody, then it will be important to know whether all genetic variants are similarly susceptible to gCIII antibody to a single virus strain. This issue becomes relevant when considering the data showing that preexisting immunity to CMV does not ensure protection from reinfection (2, 10, 11). There is also some evidence that virus-neutralizing antibody associated with gH variants is strain specific (47). Antibody to gO can also inhibit cell fusion initiated by a fusogenic strain of CMV, AD169 (34). The association of gO with biological activities should stimulate future efforts to understand the biological consequences of strain variability in the gO gene. The variability in the gH, gO, and gL components of the gCIII complex, if proven to be significant for cross-neutralization of virus strains, would most certainly have to be considered in the development of a vaccine against CMV.
The potential for the gCIII complex to exist in different genetic configurations in clinical isolates may have implications for cellular tropism and provide some insight into the mechanism of the pantropism of CMV in vivo. A complex similar to gCIII of CMV has been described for EBV. EBV enters B lymphocytes and epithelial cells by different routes, depending on whether gp42, a genetic homologue of CMV gO, is present. Without gp42, EBV can bind to, but not infect, B lymphocytes (32, 33, 49, 50). Whether gO of CMV has a similar role in virus entry remains to be determined.
Assessment of the significance of the role of the gCIII complex in the biology of CMV must await the introduction of new tools for the evaluation of systems that reflect in vivo virulence. The biology of the widely used laboratory strains of CMV, Towne and AD169, does not reflect that of field isolates (1, 4-6, 27, 36, 40, 41, 43, 46, 52). The clinical condition of human patients is a poor indicator of the virulence of laboratory strains because of the frequent shedding of CMV in the absence of symptomatic disease. Some animal (3, 12) and specialized cell culture (39, 52) models appear to discriminate between the replication of clinical and laboratory strains. Understanding the impact of strain variability will ultimately require direct analysis of experimentally modified recombinant viruses (18). The recombinant viruses, identical in all genetic aspects except for the altered region, can then be studied in either in vivo or in vitro model systems to pinpoint genes that are crucial for virulence as well as for other biological activities that have been attributed to CMV.
Acknowledgments
This work was supported by grants NIH AI 41875 and AmFAR 0253-21-RG (a Gift for Life Research Grant) to L.R.
We are grateful to Sunwen Chou for providing some of the samples from transplant patients and to Todd Margolis for providing vitreous humor samples from HIV patients with CMV retinitis. CMV isolates from allograft recipients were kindly provided by Hiro Margesson and Ann Warford of the Stanford University Hospital Diagnostic Virology Laboratory. The collegiality of T. Merigan and the members of the Stanford Center for AIDS Research is gratefully acknowledged. We thank Darcy Levee, Yvette Girard, and Kristi Cooley for assistance with the DNA sequence analysis and Robert Shafer and Jonathan Eisen for assistance with the phylogenetic analysis.
REFERENCES
- 1.Alderete, J. P., S. Jarrahian, and A. P. Geballe. 1999. Translational effects of mutations and polymorphisms in a repressive upstream open reading frame of the human cytomegalovirus UL4 gene. J. Virol. 73:8330-8337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boppana, S. B., L. B. Rivera, K. B. Fowler, M. Mach, and W. J. Britt. 2001. Intrauterine transmission of cytomegalovirus to infants of women with preconceptional immunity. N. Engl. J. Med. 344:1366-1371. [DOI] [PubMed] [Google Scholar]
- 3.Brown, J. M., H. Kaneshima, and E. S. Mocarski. 1995. Dramatic interstrain differences in the replication of human cytomegalovirus in SCID-hu mice. J. Infect. Dis. 171:1599-1603. [DOI] [PubMed] [Google Scholar]
- 4.Cerboni, C., M. Mousavi-Jazi, A. Linde, K. Soderstrom, M. Brytting, B. Wahren, K. Karre, and E. Carbone. 2000. Human cytomegalovirus strain-dependent changes in NK cell recognition of infected fibroblasts. J. Immunol. 164:4775-4782. [DOI] [PubMed] [Google Scholar]
- 5.Cha, T. A., E. Tom, G. W. Kemble, G. M. Duke, E. S. Mocarski, and R. R. Spaete. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Z., S. Watanabe, and N. Yamaguchi. 1996. Strain-dependent differences in the human cytomegalovirus replication origin. Arch. Virol. 141:13-30. [DOI] [PubMed] [Google Scholar]
- 7.Chou, S. 1992. Comparative analysis of sequence variation in gp116 and gp55 components of glycoprotein B of human cytomegalovirus. Virology 188:388-390. [DOI] [PubMed] [Google Scholar]
- 8.Chou, S. 1992. Molecular epidemiology of envelope glycoprotein H of human cytomegalovirus. J. Infect. Dis. 166:604-607. [DOI] [PubMed] [Google Scholar]
- 9.Chou, S., and K. Dennison. 1991. Analysis of interstrain variation in cytomegalovirus glycoprotein B sequences encoding neutralization-related epitopes. J. Infect. Dis. 163:1229-1234. [DOI] [PubMed] [Google Scholar]
- 10.Chou, S. W. 1989. Neutralizing antibody responses to reinfecting strains of cytomegalovirus in transplant recipients. J. Infect. Dis. 160:16-21. [DOI] [PubMed] [Google Scholar]
- 11.Chou, S. W. 1989. Reactivation and recombination of multiple cytomegalovirus strains from individual organ donors. J. Infect. Dis. 160:11-15. [DOI] [PubMed] [Google Scholar]
- 12.DiLoreto, D., Jr., L. G. Epstein, E. S. Lazar, W. J. Britt, and M. del Cerro. 1994. Cytomegalovirus infection of human retinal tissue: an in vivo model. Lab. Investig. 71:141-148. [PubMed] [Google Scholar]
- 13.Gretch, D. R., B. Kari, L. Rasmussen, R. C. Gehrz, and M. F. Stinski. 1988. Identification and characterization of three distinct families of glycoprotein complexes in the envelopes of human cytomegalovirus. J. Virol. 62:875-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haberland, M., U. Meyer-Konig, and F. T. Hufert. 1999. Variation within the glycoprotein B gene of human cytomegalovirus is due to homologous recombination. J. Gen. Virol. 80:1495-1500. [DOI] [PubMed] [Google Scholar]
- 15.Hebart, H., M. Greif, H. Krause, L. Kanz, G. Jahn, C. A. Muller, and H. Einsele. 1997. Interstrain variation of immediate early DNA sequences and glycoprotein B genotypes in cytomegalovirus clinical isolates. Med. Microbiol. Immunol. 186:135-138. [DOI] [PubMed] [Google Scholar]
- 16.Hillis, D., C. Moritz, and B. Mable. 1996. Molecular systematics, 2nd ed. Sinauer Associates, Inc., Sunderland, Mass.
- 17.Hitomi, S., H. Kozuka-Hata, Z. Chen, S. Sugano, N. Yamaguchi, and S. Watanabe. 1997. Human cytomegalovirus open reading frame UL11 encodes a highly polymorphic protein expressed on the infected cell surface. Arch. Virol. 142:1407-1427. [DOI] [PubMed] [Google Scholar]
- 18.Hobom, U., W. Brune, M. Messerle, G. Hahn, and U. H. Koszinowski. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, E. S., B. A. Kilpatrick, Y. T. Huang, and J. S. Pagano. 1976. Detection of human cytomegalovirus and analysis of strain variation. Yale J. Biol. Med. 49:29-43. [PMC free article] [PubMed] [Google Scholar]
- 20.Huber, M. T., and T. Compton. 1997. Characterization of a novel third member of the human cytomegalovirus glycoprotein H-glycoprotein L complex. J. Virol. 71:5391-5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huber, M. T., and T. Compton. 1998. The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J. Virol. 72:8191-8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huber, M. T., and T. Compton. 1999. Intracellular formation and processing of the heterotrimeric gH-gL-gO (gCIII) glycoprotein envelope complex of human cytomegalovirus. J. Virol. 73:3886-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaye, J. F., U. A. Gompels, and A. C. Minson. 1992. Glycoprotein H of human cytomegalovirus (HCMV) forms a stable complex with the HCMV UL115 gene product. J. Gen. Virol. 73:2693-2698. [DOI] [PubMed] [Google Scholar]
- 24.Keay, S., and B. Baldwin. 1991. Anti-idiotype antibodies that mimic gp86 of human cytomegalovirus inhibit viral fusion but not attachment. J. Virol. 65:5124-5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keay, S., and B. R. Baldwin. 1996. Evidence for the role of cell protein phosphorylation in human cytomegalovirus/host cell fusion. J. Gen. Virol. 77:2597-2604. [DOI] [PubMed] [Google Scholar]
- 26.Keay, S., T. C. Merigan, and L. Rasmussen. 1989. Identification of cell surface receptors for the 86-kilodalton glycoprotein of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 86:10100-10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein, M., K. Schoppel, N. Amvrossiadis, and M. Mach. 1999. Strain-specific neutralization of human cytomegalovirus isolates by human sera. J. Virol. 73:878-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, L., J. A. Nelson, and W. J. Britt. 1997. Glycoprotein H-related complexes of human cytomegalovirus: identification of a third protein in the gCIII complex. J. Virol. 71:3090-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, Q., C. Buranathai, C. Grose, and L. M. Hutt-Fletcher. 1997. Chaperone functions common to nonhomologous Epstein-Barr virus gL and varicella-zoster virus gL proteins. J. Virol. 71:1667-1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lurain, N. S., K. S. Kapell, D. D. Huang, J. A. Short, J. Paintsil, E. Winkfield, C. A. Benedict, C. F. Ware, and J. W. Bremer. 1999. Human cytomegalovirus UL144 open reading frame: sequence hypervariability in low-passage clinical isolates. J. Virol. 73:10040-10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer-Konig, U., M. Haberland, D. von Laer, O. Haller, and F. T. Hufert. 1998. Intragenic variability of human cytomegalovirus glycoprotein B in clinical strains. J. Infect. Dis. 177:1162-1169. [DOI] [PubMed] [Google Scholar]
- 32.Miller, N., and L. M. Hutt-Fletcher. 1992. Epstein-Barr virus enters B cells and epithelial cells by different routes. J. Virol. 66:3409-3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Molesworth, S. J., C. M. Lake, C. M. Borza, S. M. Turk, and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J. Virol. 74:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paterson, D. A., A. P. Dyer, R. S. Milne, E. Sevilla-Reyes, and U. A. Gompels. 2002. A role for human cytomegalovirus glycoprotein O (gO) in cell fusion and a new hypervariable locus. Virology 293:281-294. [DOI] [PubMed] [Google Scholar]
- 35.Peng, T., M. Ponce-de-Leon, H. Jiang, G. Dubin, J. M. Lubinski, R. J. Eisenberg, and G. H. Cohen. 1998. The gH-gL complex of herpes simplex virus (HSV) stimulates neutralizing antibody and protects mice against HSV type 1 challenge. J. Virol. 72:65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinnan, G., M. Deiry, A. Rook, W. Frederick, J. Epstein, J. Manischewitz, L. R. Jackson, K. Mittal, S. Plotkin, and M. Hilleman. 1984. Comparative virulence and immunogenicity of the Towne strain and a nonattenuated strain of cytomegalovirus. Ann. Intern. Med. 101:478-483. [DOI] [PubMed] [Google Scholar]
- 37.Rasmussen, L. E., R. M. Nelson, D. C. Kelsall, and T. C. Merigan. 1984. Murine monoclonal antibody to a single protein neutralizes the infectivity of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 81:876-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simmons, R., K. Kaushansky, and B. Torok-Storb. 1990. Mechanisms of cytomegalovirus-mediated myelosuppression: perturbation of stromal cell function versus direct infection of myeloid cells. Proc. Natl. Acad. Sci. USA 87:1386-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sinzger, C., A. L. Bissinger, R. Viebahn, H. Oettle, C. Radke, C. A. Schmidt, and G. Jahn. 1999. Hepatocytes are permissive for human cytomegalovirus infection in human liver cell culture and in vivo. J. Infect. Dis. 180:976-986. [DOI] [PubMed] [Google Scholar]
- 40.Sinzger, C., and G. Jahn. 1996. Human cytomegalovirus cell tropism and pathogenesis. Intervirology 39:302-319. [DOI] [PubMed] [Google Scholar]
- 41.Sinzger, C., J. Knapp, B. Plachter, K. Schmidt, and G. Jahn. 1997. Quantification of replication of clinical cytomegalovirus isolates in cultured endothelial cells and fibroblasts by a focus expansion assay. J. Virol. Methods 63:103-112. [DOI] [PubMed] [Google Scholar]
- 42.Spaete, R. R., K. Perot, P. I. Scott, J. A. Nelson, M. F. Stinski, and C. Pachl. 1993. Coexpression of truncated human cytomegalovirus gH with the UL115 gene product or the truncated human fibroblast growth factor receptor results in transport of gH to the cell surface. Virology 193:853-861. [DOI] [PubMed] [Google Scholar]
- 43.Srivastava, R., M. Curtis, S. Hendrickson, W. H. Burns, and J. D. Hosenpud. 1999. Strain specific effects of cytomegalovirus on endothelial cells: implications for investigating the relationship between CMV and cardiac allograft vasculopathy. Transplantation 68:1568-1573. [DOI] [PubMed] [Google Scholar]
- 44.Theiler, R. N., and T. Compton. 2001. Characterization of the signal peptide processing and membrane association of human cytomegalovirus glycoprotein O. J. Biol. Chem. 276:39226-39231. [DOI] [PubMed] [Google Scholar]
- 45.Theiler, R. N., and T. Compton. 2002. Distinct glycoprotein O complexes arise in a post-Golgi compartment of cytomegalovirus-infected cells. J. Virol. 76:2890-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torok-Storb, B., M. Boeckh, C. Hoy, W. Leisenring, D. Myerson, and T. Gooley. 1997. Association of specific cytomegalovirus genotypes with death from myelosuppression after marrow transplantation. Blood 90:2097-2102. [PubMed] [Google Scholar]
- 47.Urban, M., W. Britt, and M. Mach. 1992. The dominant linear neutralizing antibody-binding site of glycoprotein gp86 of human cytomegalovirus is strain specific. J. Virol. 66:1303-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Waner, J. L., and T. H. Weller. 1978. Analysis of antigenic diversity among human cytomegaloviruses by kinetic neutralization tests with high-titered rabbit antisera. Infect. Immun. 21:151-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, X., and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus lacking glycoprotein gp42 can bind to B cells but is not able to infect. J. Virol. 72:158-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, X., W. J. Kenyon, Q. Li, J. Mullberg, and L. M. Hutt-Fletcher. 1998. Epstein-Barr virus uses different complexes of glycoproteins gH and gL to infect B lymphocytes and epithelial cells. J. Virol. 72:5552-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weller, T. H., J. L. Waner, D. R. Hopkins, and E. N. Allred. 1978. Neutralization kinetic studies with genital cytomegalovirus isolates, an antigenically variable group. IARC Sci. Publ. 24:177-184. [PubMed] [Google Scholar]
- 52.Woodroffe, S. B., J. Hamilton, and H. M. Garnett. 1997. Comparison of the infectivity of the laboratory strain AD169 and a clinical isolate of human cytomegalovirus to human smooth muscle cells. J. Virol. Methods 63:181-191. [DOI] [PubMed] [Google Scholar]
- 53.Zipeto, D., S. Morris, C. Hong, A. Dowling, R. Wolitz, T. C. Merigan, and L. Rasmussen. 1995. Human cytomegalovirus (CMV) DNA in plasma reflects quantity of CMV DNA present in leukocytes. J. Clin. Microbiol. 33:2607-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zweygberg Wirgart, B., M. Brytting, A. Linde, B. Wahren, and L. Grillner. 1998. Sequence variation within three important cytomegalovirus gene regions in isolates from four different patient populations. J. Clin. Microbiol. 36:3662-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]