Abstract
Titin is the main determinant of passive muscle force. Physiological extension of titin derives largely from its PEVK (Pro-Glu-Val-Lys) domain, which has a different length in different muscle types. Here we characterized the elasticity of the full-length, human soleus PEVK domain by mechanically manipulating its contiguous, recombinant subdomain segments: an N-terminal (PEVKI), a middle (PEVKII), and a C-terminal (PEVKIII) one third. Measurement of the apparent persistence lengths revealed a hierarchical arrangement according to local flexibility: the N-terminal PEVKI is the most rigid and the C-terminal PEVKIII is the most flexible segment within the domain. Immunoelectron microscopy supported the hierarchical extensibility within the PEVK domain. The effective persistence lengths decreased as a function of ionic strength, as predicted by the Odijk-Skolnick-Fixman model of polyelectrolyte chains. The ionic strength dependence of persistence length was similar in all segments, indicating that the residual differences in the elasticity of the segments derive from nonelectrostatic mechanisms.
INTRODUCTION
One of the main determinants of muscle elasticity is the filamentous intrasarcomeric protein titin ((1); also called connectin (2)), a 3.0–3.7 MDa protein (for recent reviews, see Tskhovrebova and Trinick, 2003 (3), Granzier and Labeit, 2004 (4), and Miller et al., 2004 (5)) that spans the half sarcomere. Titin is anchored to the Z- and M-lines and is attached to the thick filaments of the A-band (6). The I-band section of the molecule is constructed of serially linked immunoglobulin (Ig)-like domains (proximal and distal tandem Ig regions) interspersed with unique sequences including a proline (P)-, glutamate (E)-, valine (V)-, and lysine (K)-rich PEVK domain (7). Upon stretching the sarcomere, passive force is generated by the extension of the I-band segment of titin. The extension of titin's I-band section occurs as a series of consecutive events (8): the extension of the tandem Ig segment is followed by the extension of the PEVK domain (9,10) and by the N2-B unique sequence in cardiac muscle (11,12). In different muscle types, different length isoforms of titin are expressed (7,13). Cardiac muscle contains the shortest titin isoform (N2-B) with a ∼180-residue-long PEVK domain (7,13). By contrast, in soleus muscle the PEVK segment is 2174 residues long (7,13).
The PEVK domain of titin has been suggested to acquire a random structure due to the preponderance of prolines and charged residues (7). Indeed, early immunoelectron microscopic analysis has shown that the PEVK domain probably behaves as a quasi-unfolded, random protein chain (10). Recent structural experiments have suggested that the PEVK domain may also contain left-handed polyproline helices (14). Furthermore, a repetitive motif structure of PEVK has been demonstrated based on sequence analysis (15). Two main motifs were identified in the PEVK sequence: a), PPAK motifs (or PEVK repeats (16)), and b), polyE motifs. PPAK motifs are ∼28-residue-long sequences which begin most often with the amino acids PPAK. PolyE motifs contain a preponderance of glutamate. Based on NMR and circular dichroism spectroscopic data, Ma and Wang recently suggested that the PEVK domain has a malleable structure which is capable of transition between various conformational states: polyproline helix, β-turn, and unordered coil (17). However, whether and how the local motif structure influences local elasticity remained unresolved.
In this work, we explored the mechanical property along skeletal PEVK by applying a multifaceted approach: heterologous expression of various skeletal PEVK segments, single-molecule force spectroscopy experiments, and immunoelectron microscopy (IEM). We find that the PEVK domain displays a spatially hierarchical arrangement of local elasticity: the N-terminal region is the most rigid and the C-terminal region is the most flexible.
MATERIALS AND METHODS
Cloning, expression, and purification of human skeletal PEVK
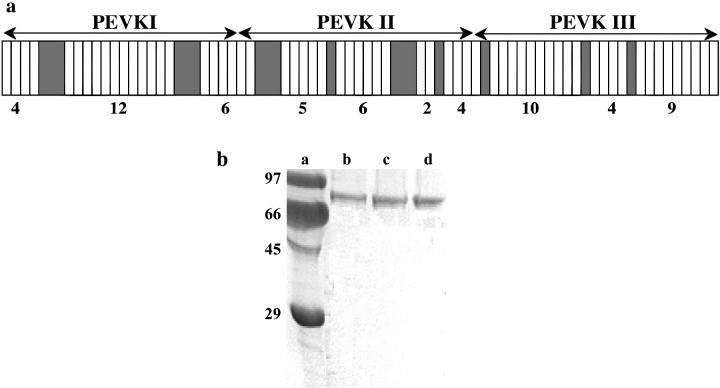
The human skeletal-muscle cDNA library was a generous gift of Dr. Siegfried Labeit. The entire skeletal PEVK domain (largest, m. soleus isoform) was expressed in three contiguous segments (18), each corresponding to ∼1/3 (∼700 residues) of the PEVK length: N-terminal (PEVKI), middle (PEVKII), and C-terminal (PEVKIII). The nucleotide sequence boundaries of the PEVK segments, based on GenBank accession No. X90569 (version X90569.1; Labeit and Kolmerer (7)) are as follows: PEVKI 16852-19074 (AA: 5618-6358), PEVKII 19075-21192 (AA: 6359-7064), and PEVKIII 21193-23373 (AA: 7065-7791). The arrangement of PEVK segments along skeletal PEVK is shown in Fig. 1 a. Each of the segments was cloned into pET-28a vector (Novagen, Darmstadt, Germany) between NheI and XhoI sites introduced independently with polymerase chain reaction (PCR) by using specific oligonucleotides. To the C-terminus of the recombinant proteins, two vicinal cysteins were added for subsequent single-molecule manipulation. Proteins were expressed in Escherichia coli (BL21(DE3)pLysS). His6-tagged (on N-terminus) proteins were purified on Ni2+-NTA columns under native conditions following manufacturer's instructions (Qiagen, Hilden, Germany) and further purified on a Sephadex G-25 column (Sigma-Aldrich, St. Louis, MO). The His6-tag was subsequently used for capturing the PEVK segment's end specifically. The electrophoretogram of the purified PEVK segments is shown in Fig. 1 b. As evidenced by the gel profile, the protein samples were of high purity and devoid of degradation products.
FIGURE 1.
(a) Motif layout of the skeletal-muscle titin PEVK domain (15). White rectangles refer to PPAK motifs and shaded ones to polyE motifs. The boundaries of the PEVK segments expressed and studied in this work are indicated by lines with arrows on both ends. (b) SDS-PAGE pattern of the expressed and purified PEVK segments and fragments. Lanes (a) molecular weight standard, (b) PEVKI, (c) PEVKII, and (d) PEVKIII.
Preparation of chemically modified glass slides for single-molecule manipulation
For specific handling of the N-terminal, His6-tagged end of the PEVK segments we used glass slides covered with Ni-NTA (19). Briefly, microscope slides (J. Melvin Freed Brand, Sigma-Aldrich) were cleaned by sonication in ethanol and then in acetone for 20 min and by a subsequent incubation for 20 min in concentrated HNO3 and then in 6N KOH. Between each step, the slides were rinsed three times with MilliQ water (0.2 μm filtered, >18.2 Mohm × cm). Before further use, the slides were dried in a stream of clean N2 gas. For chemical activation, the slides were first incubated for 12 h in toluene vapor containing 2% Glymo (3-glycidiloxypropyl-trimethoxisylane; Sigma-Aldrich), then washed with distilled water and dried. Second, the glymo-covered slides were incubated in 0.01 M Na2CO3 (pH 10), containing 2% (wt/vol) N-(5-amino 1-carboxypenthyl)-iminodiacetic acid (NTA; Dojindo, Tokyo, Japan) for 16 h at 60°C, then washed with distillated water and dried. For activation with Ni, the slides were incubated in assay buffer (AB) buffer (25 mM imidazole-HCl, pH 7.4, 0.2 M KCl, 4 mM MgCl2, 1 mM EGTA, 0.01% NaN3, 1 mM DL-Dithiothreitol, 20 μg/ml leupeptin, 10 μM E-64) containing 10 mM NiCl2 and 5 mM glycine (pH 8.0) for 2 h at room temperature. The slides were washed and stored in distilled water until further use.
Single-molecule force spectroscopy
PEVK segments, held specifically at their ends via genetically engineered His6-tag and vicinal-cysteine handles, were mechanically stretched by using an atomic force microscope (AFM) dedicated for molecular manipulation (MFP1D; Asylum Research, Santa Barbara, CA). The AFM was mounted on a custom-built, low-profile inverted light microscope. PEVK segments, diluted in AB buffer were allowed to bind to the tip of a gold-coated AFM cantilever (Bio-lever; Olympus, Tokyo, Japan) for 10 min. To avoid aggregation, the buffer contained 0.2% Tween-20. Unbound molecules were washed away with AB buffer. The PEVK-coated cantilever was brought gently near the Ni-NTA-coated glass slide. The cantilever approach to the surface was monitored by following the amplitude of the thermally driven cantilever oscillation. Once a contact, via the PEVK segment, was made between the surface and the tip, the cantilever was pulled away from the surface at a constant rate (∼500 nm/s typical cantilever base velocity) to stretch the captured molecule. Previously, the elasticity of the cardiac PEVK was measured (20) by mechanically manipulating a recombinant polyprotein composed of tandemly arranged titin I27 (7) and PEVK domains. Although the polyprotein approach is often considered superior to the direct manipulation of individual molecules (21), identical results were obtained with these two methods in a recent work on a skeletal PEVK fragment (22). Considering the added difficulties in expressing a polyprotein containing modules as large as the PEVK segments studied here, in this work we preferred the direct manipulation of individual molecules. To ensure that only single strands were considered in our analysis, we employed the following experimental strategies: a), protein concentration was kept low (10–100 μg/ml) during incubation on the gold-coated cantilever tip, b), 0.2% Tween-20 was added in the buffer to reduce aggregation and nonspecific surface-protein interaction, c), data displaying sudden force drops back to the baseline were omitted, and d) only data that displayed the expected contour length in repeated mechanical cycles were considered.
Force versus displacement curves were collected in repeated stretch and release cycles. Force was obtained from the bending and the stiffness of the cantilever. Cantilever stiffness (κ) was obtained by calibration with the thermal method (23). Typical cantilever stiffness was ∼6 pN/nm. The force versus displacement curves were corrected for several factors to obtain force versus molecular end-to-end length (24): 1), the zero-length, zero-force data point was obtained from the force response that corresponded to the cantilever tip reaching (or departing from) the substrate surface; 2), forces (F) were corrected for baseline slope obtained from the force response of the displaced but unloaded cantilever; and 3), the end-to-end length (z) of the tethered molecule was calculated by correcting the cantilever base displacement (s) with cantilever bending as
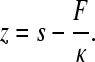 |
(1) |
Analysis of force data
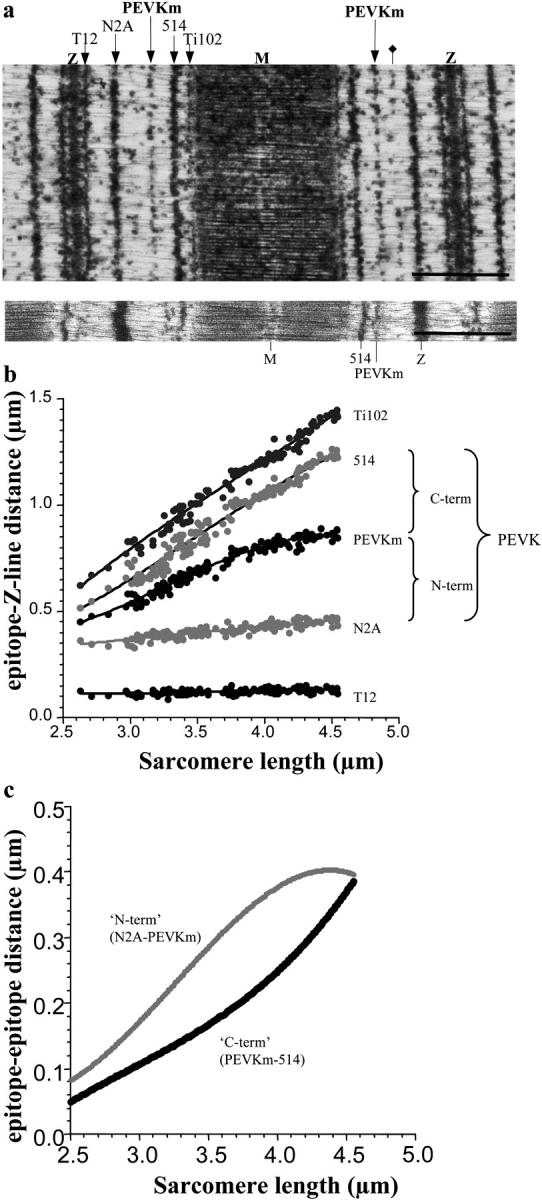
Force data were compared with the wormlike chain (WLC) equation (25,26)
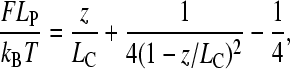 |
(2) |
where LP and LC are the effective persistence length and the contour length of the tethered molecule, respectively. The experimental curves were fitted with the WLC equation by using a nonlinear least squares fit (Marquardt-Levenberg). Nonspecifically captured short tethers were excluded from the analysis based on a comparison of Lc with the contour length predicted from sequence. Typical force range for the WLC fit was 0–50 pN.
IEM
IEM methods were published earlier (10,27–29). Briefly, skinned fibers were stretched, held at fixed length (as above), then, after a predetermined hold period, they were fixed for 20 min by replacing the relaxing solution with freshly prepared 3% para-formaldehyde in PBS (phosphate-buffered saline). Fibers were then washed, blocked with BSA (bovine serum albumin), and washed and incubated for ∼24 h with anti-titin antibodies in PBS/BSA. The following antibodies were used (see also Fig. 5 a): anti-I2/I3 (T12), anti-I80/I81 (N2A), anti-C-terminal PEVK residues (4596–4606 of human cardiac titin; 514), and anti-I111/I112 (Ti-102). For additional information on T12, see Fürst et al., 1988 (6) and Sebestyen et al., 1995 (30); for 514 and Ti-102, see Trombitás et al., 1998 (10); and for all other antibodies, see Bang et al. (16). Fibers were then washed, labeled with secondary antibody, washed, fixed with glutaraldehyde/tannic acid, stained with OsO4, and embedded in araldite. Ultrathin sections were stained with potassium permanganate and lead citrate and observed with a JEOL 1200 electron microscope (JEOL, Tokyo, Japan). Mid-Z-line to midepitope distances were measured from electron micrographs after high-resolution scanning and digital image processing using custom-written macros for the image analysis program NIH image (v. 1.6, Wayne Rasband, National Institutes of Health). For spatial calibration, the electron microscope's magnification was used.
FIGURE 5.
IEM of soleus muscle using different anti-titin antibodies. (a, upper panel) Immunoelectron microscopic image of a soleus muscle sarcomere stretched to an SL of 3.8 μm. Scale bar 1 μm. The fiber was labeled with a cocktail of T12, N2A, 514, and Ti102 antibodies (indicated above the figure). PEVKm indicates the extra epitope labeled with the 514 antibody. The additional 514-labeled weak epitope is labeled with a diamond. (Lower panel) Immunoelectron microscopic image of a soleus muscle sarcomere labeled with the 514 antibody only. Scale bar 1 μm. (b) Z-line to epitope distance as a function of SL. (c) Segmental extensibility within the PEVK domain (as deduced from the epitope to epitope distances as indicated) as a function of SL.
Theory and calculations
Considering that the PEVK domain contains charged residues and therefore behaves as a polyelectrolyte chain, its effective persistence length (LP) is influenced by electrostatic interactions between the chain-associated charges. Accordingly, based on the Odijk-Skolnick-Fixman (OSF) theory (31), like charge interactions tend to increase LP by limiting the thermally driven bending motions. LP is the sum of a purely elastic (L0) and an electrostatic (Le) term
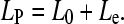 |
(3) |
The electrostatic term (Le) is given by
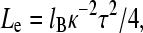 |
(4) |
where lB is the Bjerrum length (distance at which the charges have an interaction energy equivalent to the thermal energy, kBT; lB = 0.7 nm in water at room temperature), τ is line charge density (inverse of mean intercharge distance along the chain), and κ−1 is the Debye screening length:
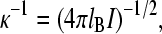 |
(5) |
where I is ionic strength of the solution. To obtain L0 and τ, the LP versus κ−1 data were fitted with Eq. 4. τ was also estimated for the stretched limit by calculating the glutamate-glutamate and lysine-lysine mean nearest neighbor distances (dr) along the PEVK sequence as
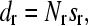 |
(6) |
where Nr is the distance between nearest neighbors of corresponding glutamates or lysines along the sequence expressed in number of residues, and sr is residue spacing (0.38 nm (32)). The relative fractional extension of the tandem Ig region and the PEVKI, PEVKII, and PEVKIII segments were calculated based on the principle of equivalence of forces within a serially linked mechanical system derived from the WLC equation (Eq. 2):
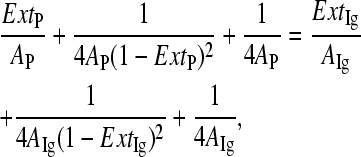 |
(7) |
where ExtP and ExtIg are the fractional extensions, and Ap and AIg are the persistence lengths of the given PEVK segment and the tandem Ig region, respectively. For the persistence length of the tandem Ig region, we used 15 nm (33). The length of a simulated relaxed sarcomere (SL) was calculated from the relative end-to-end distances of the various titin regions as
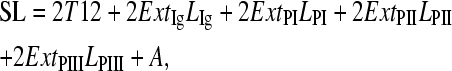 |
(8) |
where T12 is the inextensible Z-line to T12 epitope distance (0.1 μm (34)), A is the A-band width (1.6 μm), and Ext and L are the fractional extension and contour length of the respective regions (Ig = tandem Ig, P1 = PEVKI segment, PII = PEVKII segment, and PIII = PEVKIII segment). The contour length of the tandem Ig region (combined proximal and distal tandem Ig regions) was 0.465 μm (93 Ig domains × 5 nm domain spacing). The contour lengths of the PEVK segments were calculated from their sequence (PEVKI = 0.281 μm, PEVKII = 0.268 μm, and PEVKIII = 0.276 μm).
RESULTS
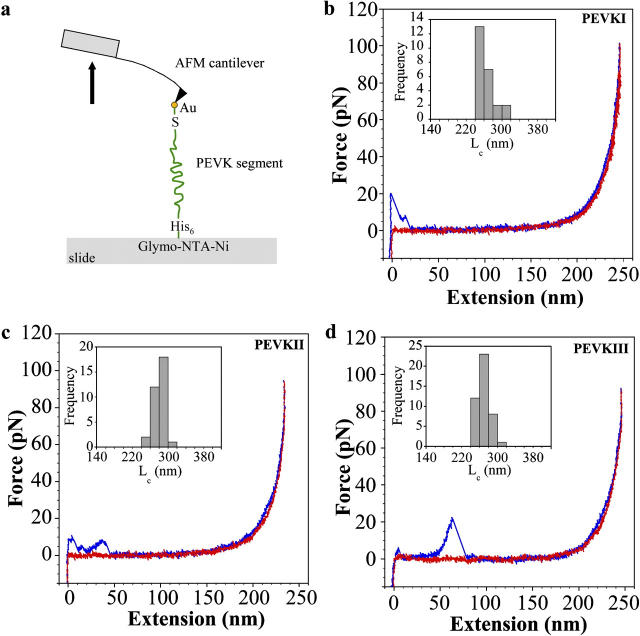
Regional elasticity within the PEVK domain of the largest, soleus isoform of titin was explored by mechanically manipulating its recombinant subsegments (PEVKI, II, and III) using single-molecule force spectroscopy (Fig. 2 a). Representative force versus extension curves of the PEVK segments, measured at a stretch rate of ∼500 nm/s, are shown in Fig. 2, b–d. The force data obtained during the stretch (blue) and release (red) phase of the mechanical cycle overlapped for a wide range (100–1000 nm/s) of stretch rate (except for the initial part of stretch where tip-surface interactions occur). The nonlinear release force curves were fitted with the WLC model, based on which the effective persistence length (LP) and the contour length (LC) of the chain were obtained. The histograms of LC are shown in the insets of Fig. 2, b–d. The LP distribution of the PEVK segments at an ionic strength of 167 mM, obtained from many stretch and release experiments, is shown in Fig. 3. From the distribution, it can be observed that the mean LP of the PEVK segments are different. PEVKI has the longest LP (1.43 nm ± 0.19 mean ± SE), followed by PEVKII (1.01 nm ± 0.08 mean ± SE) and PEVKIII (0.71 nm ± 0.05 mean ± SE). Unpaired t-tests revealed that the differences between the mean persistence lengths are significant (PEVKI and PEVKII p = 0.05, PEVKII and PEVKIII p = 0.002).
FIGURE 2.
(a) Layout of the molecular mechanics experiment. (b–d) Examples of force-extension curves for PEVKI (b), PEVKII (c), and PEVKIII (d) segments at 167 mM ionic strength. Stretch data are indicated in blue and release data in red. The force curves were acquired at a stretch rate of 500 nm/s. Overlap of stretch and relaxation force data indicates purely elastic (reversible) behavior. (b–d insets) Contour length histograms derived from the force data considered in this work. Mean contour lengths at 167 mM ionic strength were 262.66 nm (±3.97 mean ± SE), 280.99 nm (±2.46 mean ± SE), and 269.48 nm (±1.91 mean ± SE) for PEVKI, II, and III, respectively.
FIGURE 3.
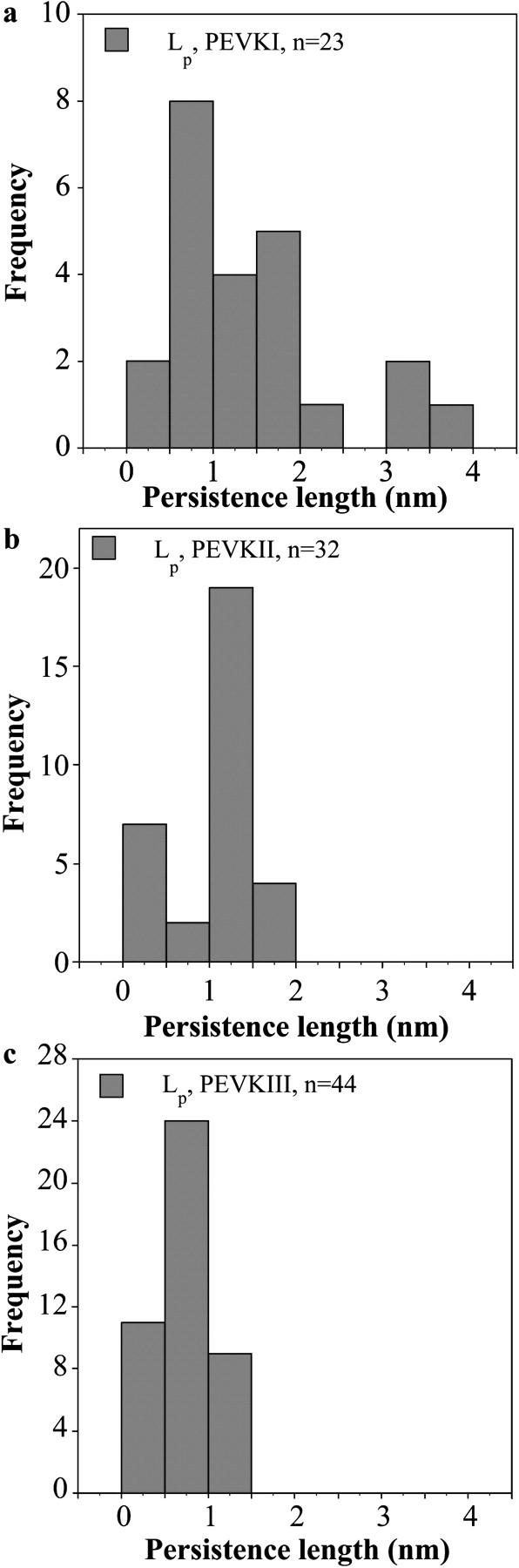
Effective persistence length distribution of PEVK segments at 167 mM ionic strength. Data are shown for PEVKI (a), PEVKII (b), and PEVKIII (c) segments.
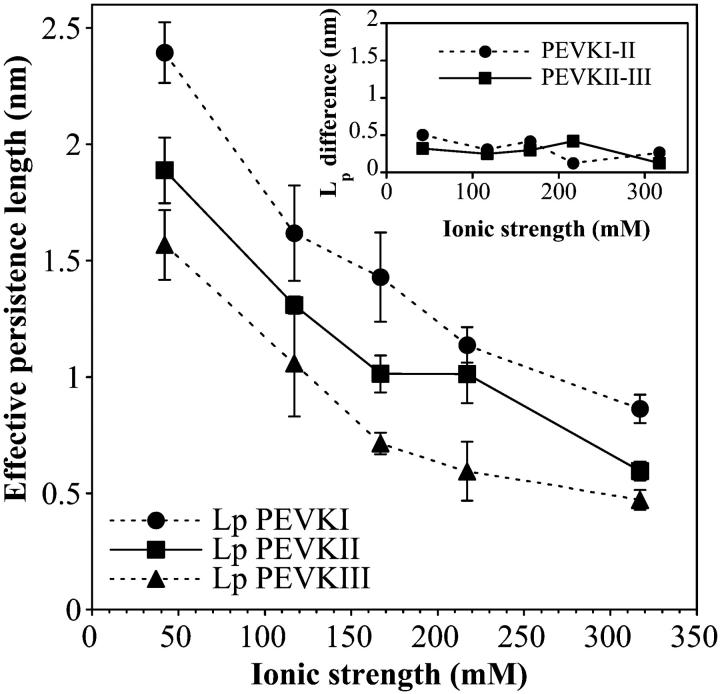
To explore the mechanisms behind the elasticity of the PEVK domain, the PEVK segments were stretched in buffer solutions of different ionic strengths. LP as a function of ionic strength for the different segments is shown in Fig. 4. LP decreases nonlinearly as a function of ionic strength. The decrease of LP with increasing ionic strength is quite large. As an example, LP of PEVKI drops from 2.39 nm (± 0.13 mean ± SE) at 42 mM ionic strength to 0.86 nm (± 0.06 mean ± SE) at 317 mM ionic strength. The order of LP among the PEVK segments is unchanged across the entire ionic strength range studied. The difference between the LP of the PEVK segments is relatively constant throughout the entire ionic strength range and persists even at high ionic strength (317 mM; Fig. 4, inset).
FIGURE 4.
Effective persistence length of PEVK segments as a function of ionic strength. Ionic strength was adjusted by changing the KCl concentration in the solution. (Inset) Differences in the effective persistence lengths of PEVKI and PEVKII, and PEVKII and PEVKIII as a function of ionic strength.
To investigate the in situ extensibility within the PEVK domain, relaxed soleus muscle fibers stretched to different lengths were analyzed with IEM. The N- and C-terminal ends of the PEVK segment were demarcated by the N2A and 514 antibodies, respectively (10). In preparations labeled with only the 514 antibody, we observed a relatively weak but consistently present epitope located in approximately the middle of the PEVK domain in long sarcomeres (Fig. 5 a, lower panel). We named this epitope PEVKm (m for middle). In the 514-labeled fibers, sometimes an additional epitope between PEVKm and N2A also appeared (Fig. 5 a, upper panel), but this was too weak for reliable positional analysis. A representative immunoelectron microscopic image of a sarcomere labeled with the cocktail of antibodies is shown in Fig. 5 a, upper panel. The Z-line to epitope distances as a function of sarcomere length are shown in Fig. 5 b, and the deduced PEVK intradomain extensibilities as a function of SL in Fig. 5 c. As evidenced by the measurements, upon stretching the sarcomere the extension of the N-terminal region of the PEVK domain dominates first. Then, at higher SLs, extension of the C-terminal PEVK region becomes dominant. The observation supports the prediction of spatially hierarchical PEVK extensibility made based on the single-molecule force spectroscopy results.
The origin of the observed effective persistence length differences observed was investigated by comparing the data with the predictions of the OSF theory of polyelectrolyte chains ((31); see Materials and Methods). Fig. 6 a compares the experimentally obtained and theoretically predicted LP as a function of the electrostatic (Debye) screening length. The OSF theory fit the data well in the case of each PEVK segment (PEVKI r = 0.96, PEVKII r = 0.94, and PEVKIII r = 0.96). Based on the OSF fits, the mean line charge density (inverse of the average distance along the chain between charges (31)) of the PEVK segments were the following: PEVKI 1.42 nm−1 (± 0.12 mean ± SE), PEVKII 1.27 nm−1 (± 0.13 mean ± SE), and PEVKIII 1.23 nm−1 (± 0.10 mean ± SE). The residual, purely elastic persistence lengths (L0) of the PEVK segments were PEVKI 0.88 nm (± 0.14 mean ± SE), PEVII 0.67 (± 0.13 mean ± SE), and PEVKIII 0.42 nm (± 0.10 mean ± SE).
FIGURE 6.
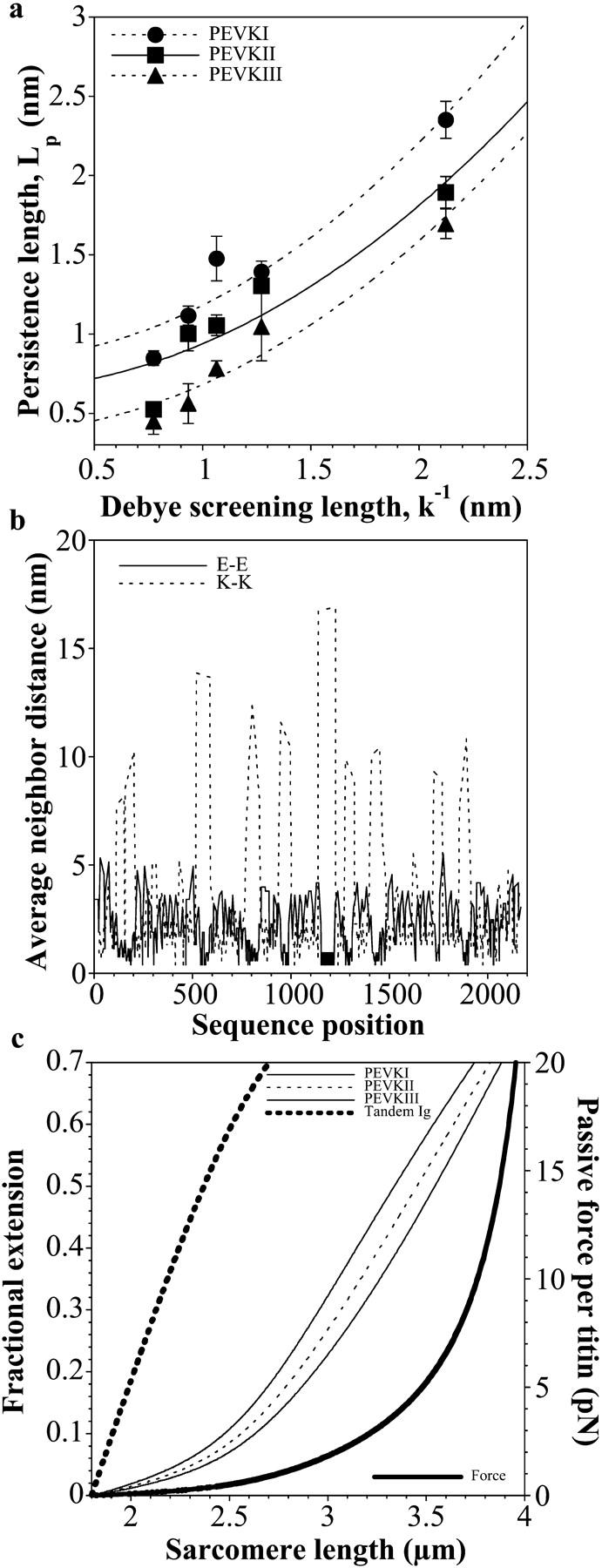
(a) Comparison of PEVK's persistence length with predictions of the OSF theory of polyelectrolyte chains. (b) Glutamate-glutamate (E-E) and lysine-lysine (K-K) average neighbor distance along the PEVK segment. (c) PEVK segment extensibility and passive force as a function of SL. Fractional extension of the PEVK segments was calculated based on the experimentally derived persistence lengths at 167 mM ionic strength.
DISCUSSION
In this work, we investigated the elasticity of the PEVK domain of full-length skeletal-muscle titin by using single-molecule mechanics and immunoelectron microscopic methods. The PEVK domain of the full-length soleus isoform of human titin was expressed in three contiguous, approximately equal-length portions: an N-terminal (PEVKI), a middle (PEVKII), and a C-terminal (PEVKIII) segment (Fig. 1). This approach allowed us not only to describe for the first time, to our knowledge, the elasticity of the entire PEVK domain of skeletal muscle, but also to explore regional differences in the domain's mechanical behavior.
The force versus extension curves for each PEVK segment revealed nonlinear elasticity that could be well described with the WLC model (35; Fig. 2). The WLC model describes the polymer chain as a bendable continuum in which thermally excited bending motions evoke the contraction of the chain (reduction of its end-to-end distance) and increase the chain's conformational entropy. The equilibrium conformation depends on the chain's contour length and bending rigidity expressed in terms of persistence length, which is a distance across which the thermally driven bending motions are correlated. The longer the persistence length, the more rigid the chain and the smaller the force required to stretch it to a given relative extension. The contour lengths deduced from the force curves (Fig. 2, b–d insets) indicate that we indeed captured the ends of the PEVK segments. The good fits with the WLC model support previous notions that the PEVK domain can be described as an entropic chain (10,20,36–38). Although in some cases the stretch force data were above the release force data during the nonlinearly rising phase of the curves, a comparison of the persistence lengths obtained from fits to the stretch and release data revealed no significant differences. Such a lack of hysteresis in the force data (Fig. 2, b–d) indicates that the molecular system traverses identical conformational states at each force level during stretch and release, therefore thermodynamic equilibrium is established throughout the mechanical cycle. Similar observations were made on the short (167-residue-long) cardiac PEVK isoform (20,38) and a skeletal PEVK fragment (22). Altogether the entire PEVK domain can be considered an ideal elastic spring that completely recovers the mechanical energy invested in it during stretch.
Although the overall elastic mechanisms appear similar along the PEVK domain, there are regional differences. A comparison of the PEVK segments' persistence lengths reveals a spatial hierarchy of elastic behavior: the N-terminal PEVK segment is the stiffest (longest LP), whereas the C-terminal segment is the most compliant region in the domain (shortest LP). Thus, it is expected that upon stretching the PEVK domain, its N-terminal region extends first, followed by its C-terminal region. We tested this prediction with IEM by following the position of a 514-antibody-based epitope that fortuitously appeared near the middle of the PEVK domain (Fig. 5). Appearance of extra epitopes within the PEVK domain has been shown before (29) and can be attributed to the repetitive motif structure of the domain (15). The extra epitopes are stable as evidenced by the uniform cross-striation appearance of the immunolabel across a wide range of SLs. The IEM experiments confirmed the prediction and revealed that under in situ conditions during sarcomere stretch it is indeed the N-terminal region of the PEVK domain that dominates extensibility initially, followed by the domain's C-terminal region.
A comparison of the ionic strength dependence (in the 47–317 mM range) of LP with the predictions of the OSF theory (31) of polyelectrolyte chains gave good fits for each PEVK fragment. The OSF theory predicts that the interaction between like charges along the polyelectrolyte chain contributes to the polymer's elastic properties by stiffening the chain. These interactions, however, are reduced if the solution ionic strength is increased, due to electrostatic screening. Electrostatic stiffening has previously been hypothesized for the PEVK domain of rabbit psoas muscle using myofibril mechanics experiments (36). Furthermore, recently it was reported that the persistence length of a recombinant skeletal PEVK fragment decreased in the presence of increased calcium concentrations (22), further supporting the idea that interactions between relatively closely spaced like charges significantly influence the elasticity of the PEVK domain. Our results herein provide direct evidence for the polyelectrolyte behavior of the PEVK domain, in which like charge interactions contribute to the effective elastic response. The line charge densities calculated from the OSF fits correspond to average intercharge distances of 0.71 nm (± 0.06 mean ± SE), 0.79 nm (± 0.08 mean ± SE), and 0.81 nm (± 0.07 mean ± SE) for PEVKI, II, and III, respectively. These average distances are comparable to each other, suggesting that the electrostatic contribution to the persistence length of the different PEVK segments is similar. The average intercharge distances calculated from the OSF fits are also comparable to the mean nearest neighbor glutamate-glutamate (E-E) and lysine-lysine (K-K) distances of 0.97 nm (±0.04 mean ± SE) and 1.16 nm (±0.06 mean ± SE), respectively, calculated for a completely stretched chain from the PEVK sequence. The E-E and K-K distances are not uniform along the PEVK sequence, however. The mean E-E and K-K neighbor distance versus PEVK sequence position plot (Fig. 6 b) reveals that islands of short distances, corresponding to polyE and PPAK motifs, respectively (15), are interrupted with regions of long distances. The modular arrangement of high like charge densities adds a complexity to the polyelectrolyte behavior of PEVK, which is why a slight systematic deviation from the OSF fit may be observed despite the good fit (Fig. 6 a). In contrast to our observations, a recent work, in which native titin and a PEVK fragment were manipulated with optical tweezers using microbeads coated with sequence-specific antibodies, reported that the LP of the PEVK domain increased, rather than decreased, with increasing ionic strength (39). It is conceivable that in these experiments, interactions between opposite charges farther apart along the PEVK sequence dominated the mechanics results. These attractive interactions connect points along the chain, thereby shortening the effective contour length (40). During stretch, the interconnections are peeled apart and the contour length is continuously increased. Because of the continuous peeling transition, a reduced LP is observed (40,41), explaining why Leake et al. may have observed short LP at low ionic strength (39). In support of the idea, Leake et al. also observed force hysteresis, indicating that the effective contour length of the PEVK fragment was indeed reduced at the beginning of stretch (39). Thus, the discrepancies between PEVK's ionic strength-dependent persistence lengths observed by Leake et al. and us may be explained with attractive versus repulsive electrostatic interactions, respectively. It is possible that under physiological conditions in the sarcomere, both types of interactions contribute to the mechanical behavior of the PEVK domain. Chain-shortening attractive interactions may be important in the contracted state of the domain at low SLs and may be responsible for the stress relaxation of passive muscle force (40,42). By contrast, repulsive interactions may be more relevant in the partially extended state of the PEVK domain by stiffening its molecular architecture.
The overall line charge densities of the different PEVK segments are comparable, suggesting that the differences between their persistence lengths (Fig. 4) are not of electrostatic origin. Furthermore, there is a more or less constant difference between the LP of the different PEVK segments across the entire ionic strength range studied (Fig. 4, inset) that persists even at high ionic strength, where the charge-charge interactions are efficiently screened. The result indicates that the source of the LP differences is indeed nonelectrostatic. Currently we can only speculate as to what the source of the elasticity difference might be. A possible explanation is that polyproline helices are formed within the PEVK domain, but the density of these helices differs along the PEVK sequence. During stretch, the chain, shortened initially by the presence of the helices, may go through a reversible transition that results in a reduction of apparent persistence length (in a mechanism similar to attractive interactions discussed above). The density of potentially helix-forming prolines along the PEVK sequence is greatest toward the C-terminus, lending support to the idea that prolines might be involved in modulating the local elasticity of the domain.
Based on the experimentally derived persistence lengths of the contiguous segments of the PEVK domain, the elastic response of the entire domain can be calculated, considering that the segments are serially linked. Furthermore, the local extensibilities of the PEVK segments as a function of sarcomere stretch can be predicted (Fig. 6 c). The extension of the PEVK domain is most prevalent in soleus muscle across an SL range of ∼2.5–4.5 μm. In this range, forces up to 20 pN per single titin molecule are generated. The hierarchical extension of the PEVK segments persists across the entire SL range of 2–4 μm. As an example, between ∼2.5 and 4.0 μm, the N-terminal half of the PEVK is ∼1.5-fold more extended than the C-terminal half (Fig. 5 c). What could be a physiological role of the hierarchical intradomain extensibility of PEVK? It is conceivable that the hierarchical PEVK extensibility has a purely mechanical role by providing a unique passive force SL relationship, and the spatial arrangement in the hierarchy is merely coincidental. An intriguing possibility, however, is that the PEVK domain might act as a continuous stretch sensor. Stretch sensing is thought to be a very important but little understood process. Recently it has been suggested that several different regions of titin might play a role in the sensing of stretch in muscle by binding various ligands (4). A simple binding/dissociation may provide only a discrete force-sensing mechanism that reports the absence or presence of a force of a given magnitude. If, however, ligands (e.g., calcium) bound to a long stretch of the titin domain dissociate gradually driven by the domains hierarchical extension, then the state of sarcomeric may be monitored continuously across a wide range of SL. Further research may fully elucidate the functional significance of this spatially hierarchical PEVK extensibility.
Acknowledgments
We thank Dr. Siegfried Labeit for providing the human titin cDNA library and Mark McNabb for help with electron micrographs.
This work was supported by grants from the Hungarian Science Foundation (OTKA T037935), Hungarian Ministry of Education (BIO-110/2002), European Union (HPRN-CT-2000-00091), the South Trans-Danubian Co-operative Research Center to M.S.Z.K., and National Institutes of Health (HL062881/061497/067274) to H.G. M.S.Z.K. is a Howard Hughes Medical Institute International Research Scholar.
P. Bianco's permanent address is Universita' degli Studi di Firenze, LENS, via Nello Carrara 1, 50019 Sesto Fiorentino Florence, Italy.
References
- 1.Wang, K., J. McClure, and A. Tu. 1979. Titin: major myofibrillar components of striated muscle. Proc. Natl. Acad. Sci. USA. 76:3698–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maruyama, K. 1997. Connectin/titin, giant elastic protein of muscle. FASEB J. 11:341–345. [DOI] [PubMed] [Google Scholar]
- 3.Tskhovrebova, L., and J. Trinick. 2003. Titin: properties and family relationships. Nat. Rev. Mol. Cell Biol. 4:679–689. [DOI] [PubMed] [Google Scholar]
- 4.Granzier, H. L., and S. Labeit. 2004. The giant protein titin: a major player in myocardial mechanics, signaling, and disease. Circ. Res. 94:284–295. [DOI] [PubMed] [Google Scholar]
- 5.Miller, M. K., H. Granzier, E. Ehler, and C. C. Gregorio. 2004. The sensitive giant: the role of titin-based stretch sensing complexes in the heart. Trends Cell Biol. 14:119–126. [DOI] [PubMed] [Google Scholar]
- 6.Fürst, D. O., M. Osborn, R. Nave, and K. Weber. 1988. The organization of titin filaments in the half-sarcomere revealed by monoclonal antibodies in immunoelectron microscopy: a map of ten nonrepetitive epitopes starting at the Z line extends close to the M line. J. Cell Biol. 106:1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Labeit, S., and B. Kolmerer. 1995. Titins: giant proteins in charge of muscle ultrastructure and elasticity. Science. 270:293–296. [DOI] [PubMed] [Google Scholar]
- 8.Gautel, M., and D. Goulding. 1996. A molecular map of titin/connectin elasticity reveals two different mechanisms acting in series. FEBS Lett. 385:11–14. [DOI] [PubMed] [Google Scholar]
- 9.Linke, W. A., M. Ivemeyer, N. Olivieri, B. Kolmerer, J. C. Ruegg, and S. Labeit. 1996. Towards a molecular understanding of the elasticity of titin. J. Mol. Biol. 261:62–71. [DOI] [PubMed] [Google Scholar]
- 10.Trombitás, K., M. Greaser, S. Labeit, J. P. Jin, M. Kellermayer, M. Helmes, and H. Granzier. 1998. Titin extensibility in situ: entropic elasticity of permanently folded and permanently unfolded molecular segments. J. Cell Biol. 140:853–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helmes, M., K. Trombitas, T. Centner, M. Kellermayer, S. Labeit, W. A. Linke, and H. Granzier. 1999. Mechanically driven contour-length adjustment in rat cardiac titin's unique N2B sequence: titin is an adjustable spring. Circ. Res. 84:1339–1352. [DOI] [PubMed] [Google Scholar]
- 12.Linke, W. A., D. E. Rudy, T. Centner, M. Gautel, C. Witt, S. Labeit, and C. C. Gregorio. 1999. I-band titin in cardiac muscle is a three-element molecular spring and is critical for maintaining thin filament structure. J. Cell Biol. 146:631–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freiburg, A., K. Trombitas, W. Hell, O. Cazorla, F. Fougerousse, T. Centner, B. Kolmerer, C. Witt, J. S. Beckmann, C. C. Gregorio, H. Granzier, and S. Labeit. 2000. Series of exon-skipping events in the elastic spring region of titin as the structural basis for myofibrillar elastic diversity. Circ. Res. 86:1114–1121. [DOI] [PubMed] [Google Scholar]
- 14.Ma, K., L. Kan, and K. Wang. 2001. Polyproline II helix is a key structural motif of the elastic PEVK segment of titin. Biochemistry. 40:3427–3438. [DOI] [PubMed] [Google Scholar]
- 15.Greaser, M. 2001. Identification of new repeating motifs in titin. Proteins. 43:145–149. [DOI] [PubMed] [Google Scholar]
- 16.Bang, M. L., T. Centner, F. Fornoff, A. J. Geach, M. Gotthardt, M. McNabb, C. C. Witt, D. Labeit, C. C. Gregorio, H. Granzier, and S. Labeit. 2001. The complete gene sequence of titin, expression of an unusual approximately 700-kDa titin isoform, and its interaction with obscurin identify a novel Z-line to I-band linking system. Circ. Res. 89:1065–1072. [DOI] [PubMed] [Google Scholar]
- 17.Ma, K., and K. Wang. 2003. Malleable conformation of the elastic PEVK segment of titin: non-cooperative interconversion of polyproline II helix, beta turn and unordered structures. Biochem. J. 374:687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagy, A., P. Cacciafesta, L. Grama, A. Kengyel, A. Málnási-Csizmadia, and M. S. Z. Kellermayer. 2004. Differential actin binding along the PEVK domain of skeletal-muscle titin. J. Cell Sci. 177:5781–5789. [DOI] [PubMed] [Google Scholar]
- 19.Yasuda, R., H. Noji, K. Kinosita Jr., and M. Yoshida. 1998. F1-ATPase is a highly efficient molecular motor that rotates with discrete 120 degree steps. Cell. 93:1117–1124. [DOI] [PubMed] [Google Scholar]
- 20.Li, H., A. F. Oberhauser, S. D. Redick, M. Carrion-Vazquez, H. P. Erickson, and J. M. Fernandez. 2001. Multiple conformations of PEVK proteins detected by single-molecule techniques. Proc. Natl. Acad. Sci. USA. 98:10682–10686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Best, R. B., D. J. Brockwell, J. L. Toca-Herrera, A. W. Blake, D. A. Smith, S. E. Radford, and J. Clarke. 2003. Force mode atomic force microscopy as a tool for protein folding studies. Anal. Chim. Acta. 479:87–105. [Google Scholar]
- 22.Labeit, D., K. Watanabe, C. Witt, H. Fujita, Y. Wu, S. Lahmers, T. Funck, S. Labeit, and H. Granzier. 2003. Calcium-dependent molecular spring elements in the giant protein titin. Proc. Natl. Acad. Sci. USA. 100:13716–13721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutter, J. L., and J. Bechhoefer. 1993. Calibration of atomic-force microscope tips. Rev. Sci. Instrum. 64:1868–1873. [Google Scholar]
- 24.Kellermayer, M. S., C. Bustamante, and H. L. Granzier. 2003. Mechanics and structure of titin oligomers explored with atomic force microscopy. Biochim. Biophys. Acta. 1604:105–114. [DOI] [PubMed] [Google Scholar]
- 25.Bustamante, C., J. F. Marko, E. D. Siggia, and S. Smith. 1994. Entropic elasticity of lambda-phage DNA. Science. 265:1599–1600. [DOI] [PubMed] [Google Scholar]
- 26.Kellermayer, M. S. Z., S. B. Smith, H. L. Granzier, and C. Bustamante. 1997. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science. 276:1112–1116. [DOI] [PubMed] [Google Scholar]
- 27.Granzier, H., M. Helmes, and K. Trombitás. 1996. Nonuniform elasticity of titin in cardiac myocytes: a study using immunoelectron microscopy and cellular mechanics. Biophys. J. 70:430–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trombitás, K., P. H. Baatsen, M. S. Kellermayer, and G. H. Pollack. 1991. Nature and origin of gap filaments in striated muscle. J. Cell Sci. 100(Pt. 4):809–814. [DOI] [PubMed] [Google Scholar]
- 29.Trombitás, K., M. Greaser, G. French, and H. Granzier. 1998. PEVK extension of human soleus muscle titin revealed by immunolabeling with the anti-titin antibody 9D10. J. Struct. Biol. 122:188–196. [DOI] [PubMed] [Google Scholar]
- 30.Sebestyen, M. G., J. A. Wolff, and M. L. Greaser. 1995. Characterization of a 5.4 kb cDNA fragment from the Z-line region of rabbit cardiac titin reveals phosphorylation sites for proline-directed kinases. J. Cell Sci. 108:3029–3037. [DOI] [PubMed] [Google Scholar]
- 31.Hugel, T., M. Grosholz, H. Clausen-Schaumann, A. Pfau, H. E. Gaub, and M. Seitz. 2001. Elasticity of single polyelectrolyte chains and their desorption from solid supports studied by AFM based single molecule force spectroscopy. Macromolecules. 34:1039–1047. [Google Scholar]
- 32.Erickson, H. P. 1994. Reversible unfolding of fibronectin type III and immunoglobulin domains provides the structural basis for stretch and elasticity of titin and fibronectin. Proc. Natl. Acad. Sci. USA. 91:10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higuchi, H., Y. Nakauchi, K. Maruyama, and S. Fujime. 1993. Characterization of beta-connectin (titin 2) from striated muscle by dynamic light scattering. Biophys. J. 65:1906–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Granzier, H., M. Kellermayer, M. Helmes, and K. Trombitás. 1997. Titin elasticity and mechanism of passive force development in rat cardiac myocytes probed by thin-filament extraction. Biophys. J. 73:2043–2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doi, M. and S. F. Edwards. 1986. The Theory of Polymer Dynamics. Clarendon Press, Oxford. 316–318.
- 36.Linke, W. A., M. Ivemeyer, P. Mundel, M. R. Stockmeier, and B. Kolmerer. 1998. Nature of PEVK-titin elasticity in skeletal muscle. Proc. Natl. Acad. Sci. USA. 95:8052–8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linke, W. A., M. Kulke, H. Li, S. Fujita-Becker, C. Neagoe, D. J. Manstein, M. Gautel, and J. M. Fernandez. 2002. PEVK Domain of Titin: An Entropic Spring with Actin-Binding Properties. J. Struct. Biol. 137:194–205. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe, K., P. Nair, D. Labeit, M. S. Kellermayer, M. Greaser, S. Labeit, and H. Granzier. 2002. Molecular mechanics of cardiac titin's PEVK and N2B spring elements. J. Biol. Chem. 277:11549–11558. [DOI] [PubMed] [Google Scholar]
- 39.Leake, M. C., D. Wilson, M. Gautel, and R. M. Simmons. 2004. The elasticity of single titin molecules using a two-bead optical tweezers assay. Biophys. J. 87:1112–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kellermayer, M. S., S. B. Smith, C. Bustamante, and H. L. Granzier. 2001. Mechanical fatigue in repetitively stretched single molecules of titin. Biophys. J. 80:852–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Case, R. B., Y. P. Chang, S. B. Smith, J. Gore, N. R. Cozzarelli, and C. Bustamante. 2004. The bacterial condensin MukBEF compacts DNA into a repetitive, stable structure. Science. 305:222–227. [DOI] [PubMed] [Google Scholar]
- 42.Trombitás, K., Y. Wu, M. McNabb, M. Greaser, M. S. Kellermayer, S. Labeit, and H. Granzier. 2003. Molecular basis of passive stress relaxation in human soleus fibers: assessment of the role of immunoglobulin-like domain unfolding. Biophys. J. 85:3142–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]