Abstract
Fluorescently labeled oligodeoxyribonucleotides containing a single tract of four successive guanines have been used to study the thermodynamic and kinetic properties of short intermolecular DNA quadruplexes. When these assemble to form intermolecular quadruplexes the fluorophores are in close proximity and the fluorescence is quenched. On raising the temperature these complexes dissociate and there is a large increase in fluorescence. These complexes are exceptionally stable in potassium-containing buffers, and possess Tm values that are too high to measure. Tm values were determined in sodium-containing buffers for which the rate of reannealing is extremely slow; the melting profiles are effectively irreversible, and the apparent melting temperatures are dependent on the rates of heating. The dissociation kinetics of these complexes was estimated by rapidly increasing the temperature and following the time-dependent changes in fluorescence. From these data we have estimated the half-lives of these quadruplexes at 37°C. Addition of a T to the unlabeled end of the oligonucleotide increases quadruplex stability. In contrast, addition of a T between the fluorophore and the oligonucleotide leads to a decrease in stability.
INTRODUCTION
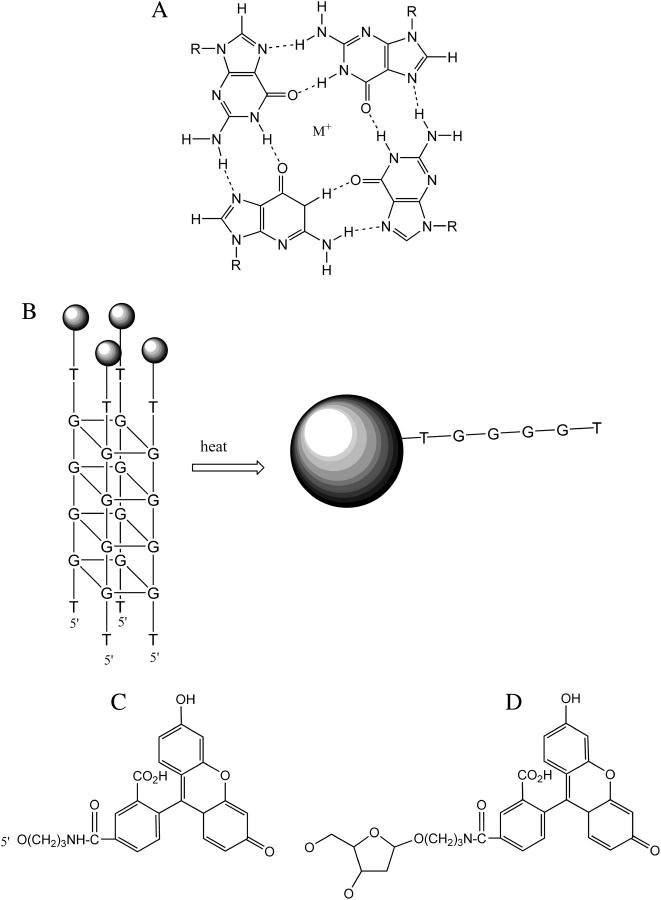
Guanine-rich DNAs can assemble to form four-stranded structures, which are based on stacks of square-planar arrays of G-quartets (1–4). The G-quartets consist of four guanines that are linked by Hoogsteen type base pairing (Fig. 1 A). Monovalent cations are selectively bound in the central cavity between the G-quartets, and these structures are specifically stabilized by potassium; sodium produces less stable complexes, whereas lithium inhibits assembly (5,6). G-quadruplexes can be formed by the intermolecular association of four DNA strands (5,7,8), by the dimerization of sequences that contain two G-tracts (9,10) or by the intramolecular folding of one strand containing at least four G-tracts (11–15). These structures are highly polymorphic and the strand polarity and glycosidic torsion angles are affected by the nature of the cations, the connecting loops, the capping bases, and the presence of other bases (1). As an example, the intramolecular complex formed by the human telomeric repeat [(GGGTTA)n] has been shown to adopt an antiparallel structure in the presence of sodium (determined by NMR) (13) or a parallel structure in the presence of potassium (determined by x-ray crystallography) (14).
FIGURE 1.
(A) Chemical structure of the G-quartet. M+ represents a monovalent cation. (B) Schematic representation of the melting of a fluorescently labeled intermolecular quadruplex. The circles represent the fluorescent group, which is self-quenched in the associated complex. (C and D) Chemical structures of FAM and dR-FAM.
Within the last 10 years there has been considerable interest in these structures as G-rich sequences with the potential to form quadruplexes have been found in a number of important biological processes. In particular, telomeric sequences consist of highly repeated G-rich sequences such as (GGGTTA)n in humans and other higher organisms, (GGGGTT)n in Tetrahymena, and (GGGGTTTT)n in Oxytrichia. Quadruplexes have also been implicated in the control regions of some oncogenes, especially c-myc (16,17), immunoglobulin switch regions (3), the retinblastoma susceptibility gene (18), the FMR-1 gene (19), the chicken β-globin gene (20), and the insulin gene (21). In addition, several synthetic aptamers are known to be based around a G-quadruplex platform including those targeted to HIV-integrase (22) and thrombin (12).
Intermolecular tetrameric complexes are usually only formed by short oligonucleotides that contain single tracts of contiguous guanines. In these complexes, the four strands are in a parallel arrangement and crystal structures and NMR studies have shown that the bases are arranged in the anti configuration (7,8,23,24). In contrast to these structural studies, much less is known about the kinetics of quadruplex formation, though it is clear that they are very stable, with extremely slow association and dissociation rates (25–27). For example, NMR studies have shown that the G(N1H) protons exchange on a timescale of days to weeks, even when dissolved in D2O (28), in contrast to DNA duplexes for which these protons have half-lives measured in milliseconds The rates of formation of intermolecular quadruplexes are also very slow and often require equilibration times in excess of 24 h, though intramolecular complexes form more rapidly. In one study the intermolecular complex formed by T2G4T2 showed a fourth-order association rate constant of 6 × 104 M−3 s−1 and a dissociation rate of 1.3 × 10−7 s−1 (i.e., a half-life of 60 days) at 37°C (27).
In this work we have examined the kinetic stability of nine intermolecular G-quadruplexes, formed by short oligonucleotides that each contain a single G4 tract. These studies have used the fluorescence melting technique, which we developed previously for studying the stability of triplexes and quadruplexes (29–32). The oligonucleotides each contained a single fluorescein moiety, at either the 3′- or 5′-end. When an intermolecular quadruplex is formed the fluorescence is quenched, due to the close proximity of the four fluorophores. On increasing the temperature the quadruplex melts and there is a large increase in fluorescence as the four strands are separated (Fig. 1 B). These experiments are performed in the Roche LightCycler, which allows for simultaneous determination of up to 32 melting profiles. The dissociation kinetics of these complexes was estimated in similar experiments, using a temperature-jump technique in which the temperature was rapidly increased and the time-dependent changes in fluorescence were measured.
MATERIAL AND METHODS
Oligodeoxyribonucleotides
Oligodeoxyribonucleotides were purchased from Eurogentec (Southampton, UK) and were synthesized on an Applied Biosystems 394 DNA/RNA synthesizer on either the 0.2- or 1-μmole scale. These were purified by high-performance liquid chromatography. The sequences of the oligonucleotides used in this work are shown in Table 1. Fluorescein was incorporated at the 5′- end of the oligonucleotides using either FAM (FAM-GGGGT) or dR-FAM (dR-FAM-GGGGT, dR-FAM-TGGGGT, dR-FAM-TGGGG), or at the 3′-end using dR-FAM (TGGGGT-dR-FAM, GGGGT-dR-FAM, TGGGG-dR-FAM, pGGGGT-dR-FAM, GGGG-dR-FAM). The chemical structures of FAM and dR-FAM are shown in Fig. 1, C and D.
TABLE 1.
Apparent Tm values for the intermolecular quadruplexes obtained at different heating rates
|
Tm (°C)
|
|||||
|---|---|---|---|---|---|
| Oligonucleotide | 6°C/min | 1°C/min | 0.5°C/min | 0.2°C/min | 0.05°C/min |
| FAM-GGGGT | 74.5 ± 0.1 (72.4) | 69.6 ± 0.1 | 66.4 ± 0.9 | 62.9 ± 0.1 | 57.5 ± 0.1 |
| dR-FAM-GGGGT | 82.4 ± 0.5 (78.4) | 76.2 ± 0.1 | 73.5 ± 0.7 | 69.8 ± 0.2 | 65.4 ± 0.1 |
| dR-FAM-TGGGGT | 70.0 ± 0.2 (69.0) | 64.1 ± 0.2 | 61.6 ± 0.5 | 58.4 ± 0.1 | 53.3 ± 0.1 |
| dR-FAM-TGGGG | 61.9 ± 0.1 (62.0) | 54.6 ± 0.1 | 52.1 ± 0.4 | 47.9 ± 0.2 | 41.3 ± 0.1 |
| TGGGGT-dR-FAM | 66.4 ± 0.2 (64.8) | 60.8 ± 0.4 | 58.3 ± 0.3 | 54.6 ± 0.4 | 50.6 ± 0.1 |
| GGGGT-dR-FAM | 60.7 ± 0.1 (56.8) | 50.4 ± 0.1 | 46.9 ± 1 | 41.6 ± 0.2 | 37.2 ± 0.5 |
| TGGGG-dR-FAM | 83.2 ± 0.2 (81.5) | 76.6 ± 0.2 | 74.4 ± 0.2 | 70.4 ± 0.4 | 65.1 ± 0.6 |
| pGGGGT-dR-FAM | 54.4 ± 0.5 (51.2) | 47.4 ± 0.4 | 44.1 ± 1.2 | 40.4 ± 0.2 | < 35 |
| GGGG-dR-FAM | 64.6 ± 0.1 (67.5) | 58.0 ± 0.1 | 56.1 ± 0.3 | 52.4 ± 0.2 | 48.0 ± 0.2 |
All experiments were performed in 10 mM sodium phosphate, pH 7.4 (I = 0.026). The figures in parentheses in the second column (6°C/min) are apparent melting temperatures derived from simulated melting profiles using the kinetic parameters shown in Table 3.
Buffers
Reactions were performed in sodium phosphate buffer, pH 7.4 (10–250 mM, as indicated) or 5 mM potassium phosphate buffer, pH 7.4. These buffers were prepared as described in Sambrook et al. (33), so that the ionic strength (I) of 10 mM buffer is 0.026.
Fluorescence melting
Fluorescence melting curves were determined using a Roche LightCycler as previously described (29), with excitation and detection wavelengths of 488 nm and 520 nm, respectively. The final oligonucleotide concentration was usually 0.25 μM, in a total volume of 20 μL. Samples were first denatured by heating from 30 to 95°C at a rate of 0.1°C/s (first melt). These were maintained at 95°C for 5 min and annealed by cooling to 30°C at 0.1°C/s. The fluorescence was recorded during both the melting and annealing phases. In some experiments the samples were evaporated to dryness after cooling to 30°C, redissolved in 20 μl water and melted again. Although the slowest heating rate for the LightCycler is 0.1°C/s, curves at lower rates of heating were obtained by raising the temperature in 1°C increments, leaving the samples for long times (between 1 and 20 min) between each temperature rise.
Dissociation kinetics
The dissociation kinetics of these intermolecular complexes were determined by rapidly increasing the temperature and following the subsequent slow changes in fluorescence as the system relaxes to a new equilibrium (32), in a manner similar to that of classical temperature-jump kinetics. In these experiments, the temperature was first increased slowly (0.1°C/s) to a point ∼10°C below the Tm, and the sample was maintained at this temperature for at least 3 min. The temperature was then rapidly increased (20°C/s) to a point 10–20°C higher and the fluorescence was recorded over the next 1.5 min as the system relaxed to a new equilibrium. Although the fast heating phase should yield a theoretical dead-time of 0.5–1 s, we did not analyze any of the data recorded in the first 2 s after initiating the temperature rise.
Data analysis
Tm values were determined from the first derivatives of the melting profiles using the Roche LightCycler software. Dissociation rate constants (k) were determined from the temperature-jump data by fitting the time-dependent increases in fluorescence with a simple exponential function. These values were determined at five or six different final temperatures. Arrhenius plots [ln(k) vs. 1/T] of these dissociation constants were prepared, from which the activation energy Ea, the dissociation rate constant k, and the half-life life at 37°C were calculated.
RESULTS
Fluorescence melting curves
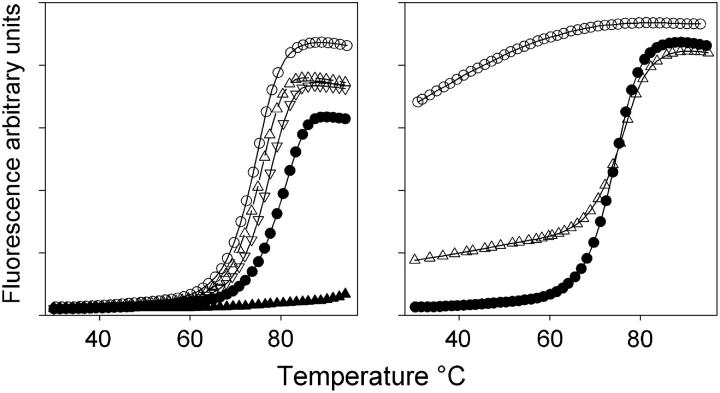
We have examined the stability of a range of intermolecular quadruplexes that are generated by short oligonucleotides containing a single G4-tract. Fluorescein is attached to either the 5′-end of these sequences as FAM or dR-FAM or at the 3′-end as dR-FAM. The sequences of these oligonucleotides are shown in Table 1. In initial experiments fluorescence melting curves for these oligonucleotides were determined at a heating rate of 0.1°C/s, in different concentrations of sodium or potassium. Typical melting profiles for FAM-GGGGT under these conditions are shown in the lefthand panel of Fig. 2. It can be seen that, as expected, there is a large temperature-dependent increase in the fluorescence signal. At low temperatures the oligonucleotide assembles so that the fluorophores are in close proximity and the fluorescence is quenched. When the complex dissociates, a large distance separates the fluorophores, and the fluorescence increases. It can be seen that raising the sodium concentration increases the Tm with values of 74.5, 75.7, 77.7, and 81.2°C at buffer concentrations of 10, 50, 100, and 250 mM, respectively (I = 0.026, 0.13, 0.26, and 0.65). The complex is extremely stable in potassium-containing buffers and the melting temperature was above 95°C even with 5 mM potassium. Because of the extreme stability of these complexes all the experiments described below were performed in 10 mM sodium phosphate buffer (I = 0.026). In contrast, there was only a small decrease in the fluorescence signal when these dissociated complexes were cooled, suggesting that they had not reannealed (Fig. 2, right). It is known that the formation of intermolecular quadruplexes is a very slow process taking from hours to days, and requires high concentrations of DNA in a tetramolecular reaction (9). Under these conditions, with low oligonucleotide concentrations (0.25 μM), reannealing was not promoted by using slower cooling rates, or by leaving the samples at room temperature for over 24 h. However, the melting curve could be regenerated by evaporating the sample to dryness (thereby increasing the effective strand concentration) and redissolving it in the same volume of water (Fig. 2, right). This second melting curve has the same Tm value as the first, though there is a small increase in the baseline fluorescence, suggesting that some of the oligonucleotide had not reassociated into a quadruplex.
FIGURE 2.
(Left) Fluorescence melting curves for FAM-GGGGT measured in different concentrations of sodium phosphate buffer, pH 7.4 (○, 10 mM; ▵, 50 mM; ∇, 100 mM; •, 250 mM), or 5 mM potassium phosphate, pH 7.4 (▴). (Right) Fluorescence melting and annealing curves for FAM-GGGGT measured in 10 mM sodium phosphate buffer, pH 7.4. (•) First melt, (○) reannealing of this denatured complex, and (▵), melting curve after drying the melted samples and redissolving in the same volume of water. In each case, the temperature was changed at 0.1°C/s.
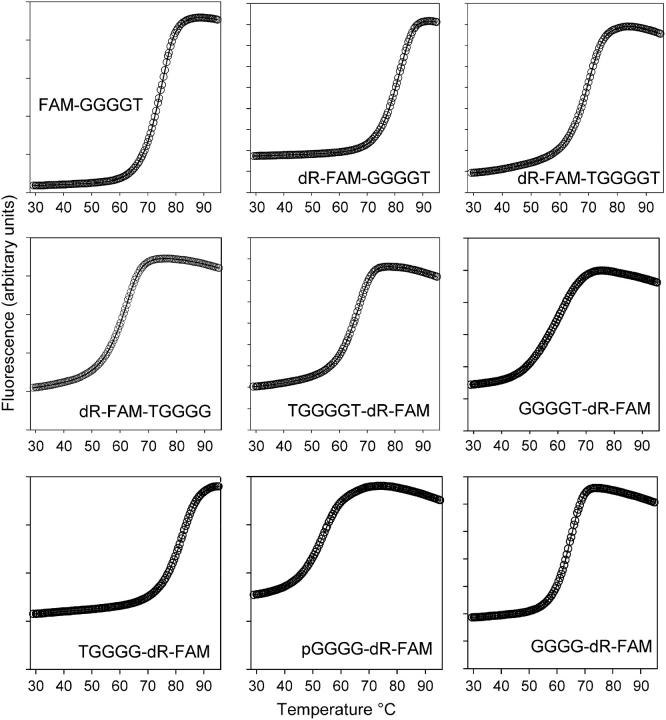
Fluorescence melting curves for all the oligonucleotides studied, determined in 10 mM sodium phosphate buffer (pH 7.4), are shown in Fig. 3. Each oligonucleotide shows a clear monophasic increase in fluorescence as the temperature is increased, which indicates melting of the intermolecular quadruplex. In each case this is followed by a small decrease in fluorescence at high temperatures, which is most evident for the complexes with lower Tms. This has been noted in previous studies with molecular beacons or fluorescence resonance energy transfer and is caused by the small temperature dependence of the fluorescence of fluorescein.
FIGURE 3.
Fluorescence melting curves for the different intermolecular quadruplexes. These were determined in 10 mM sodium phosphate buffer, pH 7.4 (I = 0.026), at a heating rate of 0.1°C/s.
Since these intermolecular complexes do not reanneal when the samples are cooled, it is clear that the melting curves cannot be in thermodynamic equilibrium. Instead of representing the usual thermodynamic equilibrium between folded and unfolded forms, these curves are dominated by the slow dissociation kinetics. As the temperature increases the complexes begin to dissociate, in what is essentially an irreversible reaction with no reassociation of the complex, as shown above. This dissociation is extremely slow and at each temperature the fluorescence would continue to rise if the samples were incubated for longer periods (see below). We would therefore expect the profiles to shift to lower temperatures at slower heating rates. These experiments were therefore repeated at different rates of heating. The apparent Tm values obtained from all these melting curves are summarized in Table 1, in which it can be seen that, as expected, all the values move to lower temperatures at slower heating rates.
Further evidence for the nonequilibrium conditions was obtained by examining the concentration dependence of the melting temperature. Although the Tm of a reversible tetramolecular reaction should be strongly concentration-dependent, we observed that the Tm of FAM-GGGGT was not affected by adding a 200-fold excess of unlabeled dTGGGGT (Fig. S1 of Supplementary Material). This lack of dependence of Tm on oligonucleotide concentration is only possible if the process is essentially irreversible, with no contribution from the concentration-dependent reassociation reaction.
Inspection of this table reveals that the various intermolecular quadruplexes have different stabilities. Although the apparent melting temperatures vary with the rate of heating, the order of stability is the same at all the rates of heating. These complexes all contain the same central G4-tract, generating an intermolecular stack of four G-quartets, yet they show a range of different stabilities; dR-FAM-GGGGT and TGGGG-dR-FAM are the most stable, whereas pGGGG-dR-FAM is the least stable. It can be seen that direct attachment of fluorescein at the 5′-end produces a less stable complex than when this fluorophore is attached to deoxyribose (dR-FAM) (compare FAM-GGGGT with dR-FAM-GGGGT). For the 5′-end-labeled oligonucleotides, addition of an extra T to the 3′-end increases the Tm by 8–12°C (compare dR-FAM-TGGGGT with dR-FAM-TGGGG). A similar effect is produced by adding a T to the 5′-end of the 3′-labeled oligonucleotides (compare TGGGGT-dR-FAM with GGGGT-dR-FAM and TGGGG-dR-FAM with GGGG-dR-FAM). This effect must be due to the additional base, rather than the extra phosphate, as pGGGGT-dR-FAM is less stable than GGGG-dR-FAM. In contrast, inserting a T between the fluorophore and the G-tract decreases the stability (compare dR-FAM-GGGGT with dR-FAM-TGGGGT, TGGGGT-dR-FAM with TGGGG-dR-FAM, and GGGGT-dR-FAM with GGGG-dR-FAM). It can be seen that exchanging the fluorophore and T between opposite ends of the oligonucleotides produces complexes with similar stabilities; dR-FAM-GGGGT and TGGGG-dR-FAM have similar Tms, as do dR-FAM-TGGGG and GGGGT-dR-FAM and dR-FAM-TGGGGT and TGGGGT-dR-FAM.
Mixtures of oligonucleotides
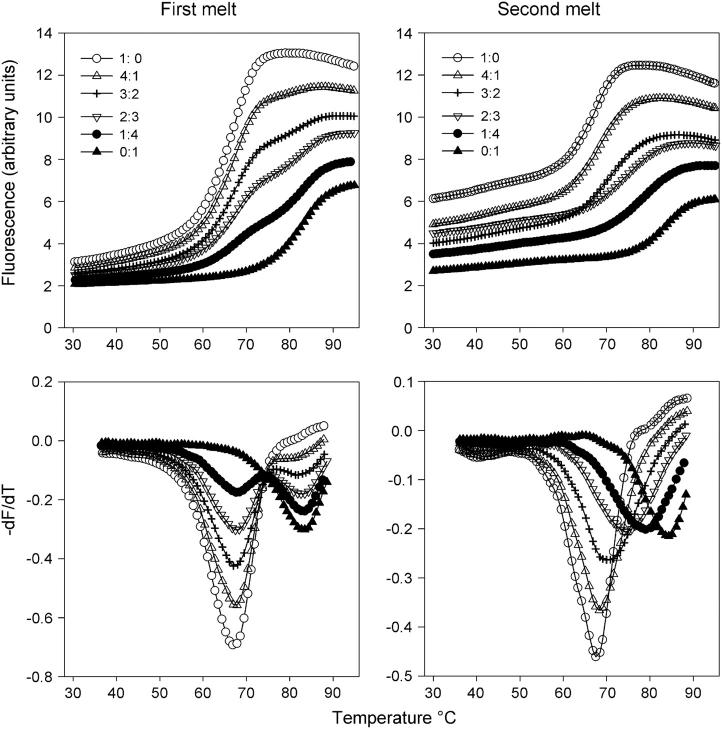
Since these intermolecular complexes have different stabilities we further explored their properties by examining whether the oligonucleotides could combine to form mixed complexes of intermediate stability. In these experiments, pairs of oligonucleotides were mixed together and their melting curves were examined. The results for mixtures of TGGGGT-dR-FAM and TGGGG-dR-FAM are shown in Fig. 4. It can be seen that when the mixture is melted for the first time the melting curves are biphasic (Fig. 4, left) and the first derivatives clearly show two peaks, which correspond to melting of the individual quadruplexes that contain only TGGGGT-dR-FAM or TGGGG-dR-FAM. This is what would be expected, as the constituent oligonucleotides in the two quadruplexes are in very slow exchange and so melt independently of each other. In contrast, single melting transitions were observed after these mixtures had been reannealed by evaporating the mixture to dryness and redissolving in the same volume (Fig. 4, right). This process reassembles the oligonucleotides into different combinations and the presence of a single transition confirms the formation of a mixed quadruplex that contains both strands. The Tm values for other 1:1 mixtures of these oligonucleotides are presented in Table 2 and reveal that the first melt is always biphasic (except in those cases where the two component oligonucleotides have similar Tms). A single monophasic transition is always observed after melting and reannealing. However the width of this transition is greater for the mixtures, suggesting that these curves represent a heterogeneous mixture of complexes that contain different combinations of the two strands (i.e., 4:0, 3:1, 2:2, 1:3, and 0:4). For the mixture of TGGGGT-dR-FAM and TGGGG-dR-FAM (shown in Fig. 4) the widths at the half-maximal dF/dT are 9.9, 10.9, 12.6, 15.1, 14.3, 13.9, and 11.0°C at ratios of 4;0, 4:1, 3:2, 1:1, 2:3, 1:4, and 0:4, respectively. For mixtures of oligonucleotides with very different Tms the transitions become much broader; for example, the mixture of pGGGG-dR-FAM and TGGGG-dR-FAM yield widths of 10.7, 20.5, 25.4, 21.8, 15.7, and 11.9°C at ratios of 4:0, 4:1, 3:2, 2:3, 1:4, and 0:4, respectively. The second melting temperature (Tm2) of the 1:1 mixture is sometimes intermediate to those of the two constituents (dR-FAM-GGGGT + dR-FAM-TGGGGT; TGGGGT-dR-FAM + TGGGG-dR-FAM; GGGGT-dR-FAM + TGGGG-dR-FAM, and pGGGGT-dR-FAM + GGGG-dR-FAM), whereas in other cases it is dominated by the oligonucleotide that produces the most stable complex (FAM-GGGGT + dR-FAM-TGGGG; dR-FAM-GGGGT + dR-FAM-TGGGG; TGGGGT-dR-FAM + GGGGT-dR-FAM; TGGGGT-dR-FAM + pGGGGT-dR-FAM; TGGGG-dR-FAM + pGGGGT-dR-FAM; and TGGGG-dR-FAM + GGGG-dR-FAM). In one combination (GGGGT-dR-FAM + pGGGGT-dR-FAM), the Tm of the mixture is closest to that of the least stable oligonucleotide.
FIGURE 4.
Fluorescence melting curves for mixtures of oligonucleotides TGGGGT-dR-FAM and TGGGG-dR-FAM. These were determined in 10 mM sodium phosphate buffer, pH 7.4, at a heating rate of 0.1°C/s. The left-hand panels show the results for the first melt, whereas the right-hand panels show the melting transitions obtained after melting and annealing the mixtures. The various curves correspond to different ratios of TGGGGT-dR-FAM to TGGGG-dR-FAM, as indicated. The top panels show the melting curves, whereas the bottom panels show their first derivatives.
TABLE 2.
Melting temperatures for intermolecular quadruplex produced with 1:1 mixtures of oligonucleotides
| Oligonucleotides | Tm1 (°C) | Tm2 (°C) | |
|---|---|---|---|
| FAM-GGGGT + dR-FAM-GGGGT | 77.5 ± 0.5 | 81.4 ± 0.2 | 81.8 ± 0.3 |
| FAM-GGGGT +dR-FAM-TGGGGT | 72 ± 0.1 | 74.3 ± 0.6 | |
| FAM-GGGGT + dR-FAM-TGGGG | 75.5 ± 0.4 | 63.5 ± 0.6 | 73.4 ± 0.3 |
| dR-FAM-GGGGT + dR-FAM-TGGGGT | 81.9 ± 0.7 | 71.7 ± 0.3 | 76.6 ± 0.3 |
| dR-FAM-GGGGT + dR-FAM-TGGGG | 82.6 ± 0.4 | 62.9 ± 0.9 | 78.7 ± 0.2 |
| dR-FAM-TGGGGT + dR-FAM-TGGGG | 70.2 ± 0.6 | 64.5 ± 1.8 | 69.9 ± 0.1 |
| TGGGGT-dR-FAM + GGGGT-dR-FAM | 66.4 ± 0.8 | 55.6 ± 1.4 | 65.2 ± 0.5 |
| TGGGGT-dR-FAM + TGGGG-dR-FAM | 67.3 ± 0.3 | 83.3 ± 0.2 | 75.0 ± 0.7 |
| TGGGGT-dR-FAM + pGGGGT-dR-FAM | 67.2 ± 0.2 | 55.9 ± 1.2 | 66.1 ± 0.1 |
| TGGGGT-dR-FAM + GGGG-dR-FAM | 66.7 ± 0.2 | 68.8 ± 0.1 | |
| GGGGT-dR-FAM + TGGGG-dR-FAM | 62.7 ± 0.5 | 83.3 ± 0.2 | 72.1 ± 3.8 |
| GGGGT-dR-FAM + pGGGGT-dR-FAM | 61 ± 2 | 55.2 ± 0.8 | 53.2 ± 0.4 |
| GGGGT-dR-FAM + GGGG-dR-FAM | 64.9 ± 0.7 | 63.3 ± 3.3 | |
| TGGGG-dR-FAM + pGGGGT-dR-FAM | 83.7 ± 1 | 58.1 ± 6 | 78.9 ± 1.6 |
| TGGGG-dR-FAM + GGGG-dR-FAM | 83.7 ± 0.5 | 66.3 ± 0.5 | 78.8 ± 0.2 |
| pGGGGT-dR-FAM + GGGG-dR-FAM | 55.2 ± 0.3 | 66.4 ± 0.2 | 59.1 ± 0.6 |
All the experiments were performed in 10 mM sodium phosphate, pH 7.4 (I = 0.026), at a heating rate of 0.1°C/s. Tm1 corresponds to the first melting curve obtained after simply mixing the oligonucleotides; in most cases, this produces a biphasic melting curve and in these cases the Tms for both transitions are shown. Tm2 corresponds to the melting curve obtained after melting and reannealing the mixture.
Temperature-jump kinetics
The melting curves presented above are clearly dominated by the slow kinetic parameters, and thermodynamic data cannot be derived from these as they are not at equilibrium. We therefore sought to directly analyze the slow dissociation kinetics for these complexes, using a modification of the temperature-jump technique that we have previously used for assessing triplex stability (32). In these experiments the samples were first heated at 0.1°C/s, until the required temperature was reached. The temperature was then increased by 10°C at the fastest rate of heating (20°C/s) and the time-dependent changes in fluorescence were monitored. These kinetic profiles were then fitted with single exponential curves allowing us to estimate the dissociation rate constant at each temperature.
Representative temperature-jump experiments with oligonucleotide TGGGGT-dR-FAM are shown in Fig. 5. Although the rate of relaxation to the new equilibrium should be a function of both the association and dissociation rate constants, we have ignored the association rate as this is extremely slow, evidenced by the lack of reassociation (described above), and the observation that the melting temperatures are independent of oligonucleotide concentration. These data were then used to construct Arrhenius plots, which are shown in Fig. 6 for all the oligonucleotides studied. Activation energies were determined from the slopes of the lines and from these and the preexponential factors we estimated the dissociation rate constants (k37) and half-lives (t½) at 37°C. A similar procedure was used by Mergny et al. (27), who also ignored the association reaction and extrapolated Arrhenius plots to obtain kinetic parameters at 37°C. These kinetic parameters are summarized in Table 3. It can be seen that these intermolecular quadruplexes, which have very similar structures, have widely differing dissociation half-lives at 37°C that vary between ∼10 min and 600 h. The most stable complex is formed by TGGGG-dR-FAM, with an estimated half-life of 600 h at 37°C, whereas the least stable is pGGGGT-dR-FAM, with a half-life of only 10 min. These activation energies are all around 200 kJ mol−1, and are similar to those previously reported by Mergny et al. (27) at an ionic strength of 0.11. The Ea values for GGGGT-dR-FAM and pGGGG-dR-FAM are much lower and correlate well with the trend observed in the Tm determinations.
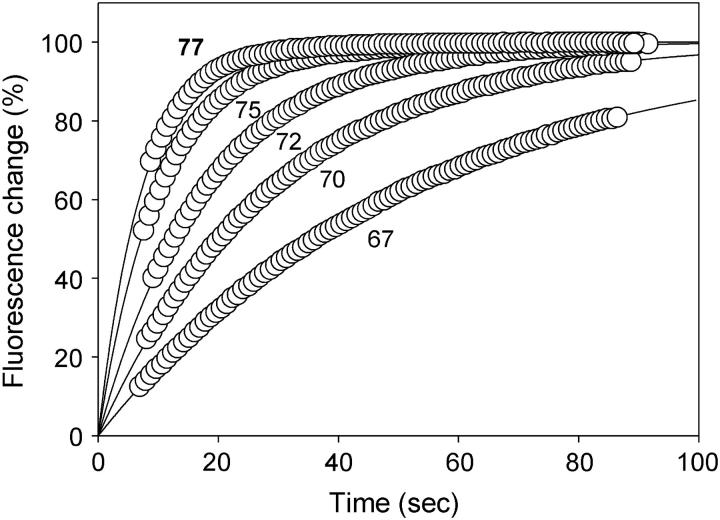
FIGURE 5.
Dissociation profiles for TGGGGT-dR-FAM measured in 10 mM sodium phosphate buffer measured at temperatures 67°C, 70°C, 72°C, 75°C, and 77°C, as indicated. The data have been normalized so that the y axis corresponds to the percentage of the total fluorescence change in each case. For the sake of clarity, only 10% of the data points are plotted for each dataset and the lines show exponential curves, fitted to these data, extrapolated to zero time.
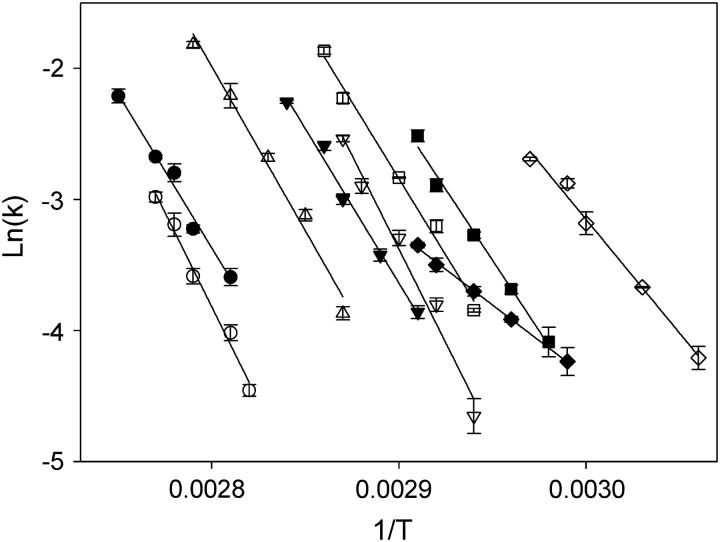
FIGURE 6.
Arrhenius plots for the dissociation rate constants of the different intermolecular G-quadruplexes. (○) TGGGG-dR-FAM; (•) dR-FAM-GGGGT; (▵) FAM-GGGGT; (▾) dR-FAM-TGGGGT; (∇) GGGG-dR-FAM; (♦) GGGGT-dR-FAM; (□) TGGGGT-dR-FAM; (▪) dR-FAM-TGGGG; and (⋄) pGGGGT-dR-FAM.
TABLE 3.
Kinetic parameters for the dissociation of the various intermolecular quadruplexes
| Oligonucleotide | Ea (kJ/mol) | A (s−1) | k37 (s−1) |
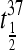 (h) (h) |
|---|---|---|---|---|
| FAM-GGGGT | 202 ± 5 | 4.5 × 1028 | 4.6 × 10−6 | 42 |
| dR-FAM-GGGGT | 202 ± 9 | 1.3 × 1028 | 1.1 × 10−6 | 183 |
| dR-FAM-TGGGGT | 198 ± 5 | 2.2 × 1028 | 1.1 × 10−5 | 17 |
| dR-FAM-TGGGG | 189 ± 8 | 3.7 × 1027 | 5.9 × 10−5 | 3 |
| TGGGGT-dR-FAM | 197 ± 2 | 3.7 × 1028 | 2.5 × 10−5 | 8 |
| GGGGT-dR-FAM | 92 ± 5 | 2.8 × 1012 | 1.1 × 10−3 | 0.18 |
| TGGGG-dR-FAM | 218 ± 8 | 1.8 × 1030 | 3.2 × 10−7 | 606 |
| pGGGGT-dR-FAM | 131 ± 2 | 1.3 × 1019 | 1.2 × 10−3 | 0.16 |
| GGGG-dR-FAM | 211 ± 8 | 3.1 × 1030 | 8.5 × 10−6 | 23 |
The kinetic parameters were derived from Arrhenius plots of the relaxation times obtained from the temperature-jump experiments.
From these kinetic data we simulated the apparent melting curves, as previously described for d(TTGGGGTT) (26). For these calculations we assumed that the oligonucleotides were 100% in the quadruplex form at 30°C and then calculated the fraction that would have progressively dissociated at each temperature using the dissociation rate constants (calculated from the figures given in Table 3) and the heating rate. Reassociation was ignored in this simulation, as we have demonstrated that this is insignificant under these conditions. The predicted apparent Tm values obtained at a heating rate of 6°C/min are shown in parentheses in the first column of Table 1. It can be seen that there is good agreement between the simulated and the observed values, confirming the values obtained for the kinetic constants. The agreement became progressively less at slower rates of heating (not shown) as the assumptions used in the simulation become less appropriate (i.e., no reassociation at all temperatures).
DISCUSSION
The results presented in this article confirm that all these intermolecular quadruplexes are extremely stable, and display very slow rates of dissociation and association. The extremely slow rate of intermolecular quadruplex dissociation has been shown in NMR studies, in which the guanine imino protons exchange with solvent protons on a very slow scale, from hours to days (28). Similar slow kinetics, generating melting profiles that are essentially irreversible, with Tm values that depend on the rate of heating, have previously been noted for other intermolecular quadruplexes (26,27,34). Mergny et al. (34) noted that the melting transition of (dTGGGG)4 was completely irreversible, showing apparent melting temperatures (with 20 μM oligonucleotide at a heating rate of 0.2°C/min) of 88°C in the presence of 10 mM KCl, but failing to melt in the presence of 100 mM KCl. These studies have recently been extended to assess the kinetics of association and dissociation of a wide range of related tetramolecular complexes (27). These authors showed that at low oligonucleotide concentrations association is so slow that the dissociation can be considered an irreversible process. Similarly, Wyatt et al. (26) determined Tm values of 73.4 and 61.0°C for (dTTGGGGTT)4 at heating rates of 1.0 and 0.025°C/min, respectively, making equilibrium measurement impractical. However, other studies—for example, with (dT4G4)4 and (dT4G4T)4 (37) or (dTGGGT)4 (35)—have presented Tm values that were determined at heating rates of 0.5°C/min; clearly, these cannot have been at thermodynamic equilibrium. Other studies have shown that the rate of quadruplex reassociation is extremely slow; for example, the (dTGGGT)4 quadruplex fully refolded only after keeping the sample at 0°C for 20 h in 1 M K+ (25). (dTTGGGGTT)4 reassociated with a fourth-order rate constant of 6 × 104 M−3 s−1 (26), which would extrapolate to a half-life for quadruplex formation of over 107 years under the conditions used in this work (0.25 μM oligonucleotide), though this would decrease to only 10 h with 1 mM oligonucleotide. We demonstrated that reassociation is possible by evaporating the melted complex to dryness, thereby increasing the oligonucleotide concentration and reestablishing the original quadruplex profile. The slow rate of dissociation is further emphasized by the experiments using mixtures of oligonucleotides, in which the strands did not reequilibrate until after the complexes had been melted, allowing them to recombine in different combinations.
This work shows that small alterations in the oligonucleotide sequence surrounding the G4-tract, such as the addition of flanking bases, can have large effects on the quadruplex stability. These results show that the location of the fluorophores affects quadruplex stability. Addition of dR-FAM to the 5′-end produces complexes with slightly faster dissociation rates than addition of this group to the 3′-end (compare dR-FAM-GGGGT with TGGGG-dR-FAM), though there is little difference in the Tm values. This presumably occurs because the fluorescein stacks differently on the terminal 5′ and 3′ G-quartets. Direct attachment of fluorescein at the 5′-end produces a slightly less stable complex than attaching this fluorophore to deoxyribose (compare FAM-GGGGT with dR-FAM-GGGGT). Comparison with previous work using similar unlabeled oligonucleotides (34) suggests that addition of the fluorophore also increases quadruplex stability, presumably because this group stacks on the terminal G-quartets. The fluorophore has the greatest effect on quadruplex stability if it is placed directly adjacent to the terminal G-quartet. Although dR-FAM-TGGGGT and TGGGGT-dR-FAM have similar Tms (differing by only 3–4°C), dR-FAM-GGGGT and GGGGT-dR-FAM are markedly different, as are dR-FAM-TGGGG and TGGGG-dR-FAM. Placement of the fluorophore adjacent to an exposed terminal G-quartet therefore appears to affect the Tm in a similar fashion to addition of an extra base. However, this contrasts with a recent study showing that addition of fluorescein only has a small effect on duplex stability (36). This suggests that the fluorescent reporter groups can affect quadruplex stability if they mask the free end of a quadruplex.
The effects of changing the location of fluorophore attachment are much less than the addition of an extra T so long as this does not alter the exposure of the terminal G-quartet. The Tm values of dR-FAM-GGGGT and T-GGGG-dR-FAM differ by <1°C, whereas those of dR-FAM-TGGGGT and TGGGGT-dR-FAM differ by only 3.5°C and those of dR-FAM-TGGGG and GGGG-T-dR-FAM by 1.2°C. These differences are much less pronounced than the changes caused by adding or removing the terminal T residues. The dissociation parameters of each of these oligonucleotide pairs are also similar, differing by factors of 3 (dR-FAM-GGGGT and T-GGGG-dR-FAM) or less (dR-FAM-TGGGGT and TGGGGT-dR-FAM). The kinetics of dR-FAM-TGGGG and GGGG-T-dR-FAM differ by 15-fold, possibly as a result of the exposed G-quartets at either end of the complexes, but this is still much less than the effects of adding a T residue.
Addition of a T at the opposite end of the quadruplex to the fluorophore also increases quadruplex stability (compare dR-FAM-TGGGGT with dR-FAM-TGGGG, TGGGGT-dR-FAM with GGGGT-dR-FAM, and TGGGG-dR-FAM with GGGG-dR-FAM). Again, this must be caused by stacking of this base against the terminal quartet, masking its exposure to solvent. It has previously been shown that addition of a terminal T stabilizes quadruplex structures (37), an effect which is caused by the additional base stacking with possibly some pairing with the terminal G-quartet (38). Addition of a T to the free 3′-end induces a sixfold increase in t½ (compare dR-FAM-TGGGGT with dR-FAM-TGGGG), whereas it appears to have a greater effect at the 5′-end, producing a 30-fold increase in t½ (compare TGGGG-dR-FAM with GGGG-dR-FAM and TGGGGT-dR-FAM with GGGGT-dR-FAM). Addition of a terminal phosphate (pGGGGT-dR-FAM) produces the least stable complex, presumably because this combines exposure of the terminal quartet with charge repulsion from the extra negative charges. Fluorescein appears to have a greater effect than T at the exposed terminus, since insertion of T between the fluorophore and the terminal G produces a decrease in stability (compare dR-FAM-GGGGT with dR-FAM-TGGGGT, TGGGGT-dR-FAM with TGGGG-dR-FAM, and GGGGT-dR-FAM with GGGG-dR-FAM).
These observations are also consistent with the apparent melting temperatures of the mixtures of oligonucleotides obtained after melting and reannealing (Tm2, Table 2). When the melting temperature of the mixture is close to that of one of the oligonucleotides, one of the oligonucleotides is capped by a T at the end opposite the fluorophore (e.g., TGGGGT-dR-FAM), whereas the other, less stable oligonucleotide in the pair has a terminal G (e.g., GGGGT-dR-FAM). It therefore appears that the additional T can stabilize the combined structure by stacking against the terminal G-quartet in the same way as it stabilizes the single oligonucleotide TGGGGT-dR-FAM. For the combination in which the melting temperature is close to that of the least stable partner (GGGGT-dR-FAM + pGGGGT-dR-FAM), the oligonucleotide with a terminal phosphate is placed against a partner with an uncapped 5′-terminal G. However, this effect is not seen with the combination pGGGGT-dR-FAM + GGGG-dR-FAM, which melts at an intermediate temperature.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http:www.biophysj.org. (The concentration dependence of the melting of FAM-GGGGT is available.)
Supplementary Material
Acknowledgments
This work was supported by a grant from the European Union.
References
- 1.Simonsson, T. 2001. G-quadruplex DNA structures—variations on a theme. Biol. Chem. 382:621–628. [DOI] [PubMed] [Google Scholar]
- 2.Williamson, J. R. 1994. G-quartet structures on telomeric DNA. Annu. Rev. Biophys. Biomol. Struct. 23:703–730. [DOI] [PubMed] [Google Scholar]
- 3.Keniry, M. A. 2000. Quadruplex structures in nucleic acids. Biopolymers. 56:123–146. [DOI] [PubMed] [Google Scholar]
- 4.Sen, D., and W. Gilbert. 1988. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 334:364–366. [DOI] [PubMed] [Google Scholar]
- 5.Williamson, J. R., M. K. Raghuraman, and T. R. Cech. 1989. Monovalent cation induced structure of telomeric DNA—the G-quartet model. Cell. 59:871–880. [DOI] [PubMed] [Google Scholar]
- 6.Sen, D., and W. Gilbert. 1990. A sodium-potassium switch in the formation of 4-stranded G4-DNA. Nature. 344:410–414. [DOI] [PubMed] [Google Scholar]
- 7.Laughlan, G., A. I. H. Murchie, D. G. Norman, M. H. Moore, P. C. Moody, D. M. J. Lilley, and B. Luisi. 1994. The high-resolution crystal structure of a parallel stranded guanine tetraplex. Science. 265:520–524. [DOI] [PubMed] [Google Scholar]
- 8.Phillips, K., Z. Dauter, A. I. Murchie, D. M. J. Lilley, and B. Luisi. 1997. The crystal structure of a parallel-stranded guanine tetraplex at 0.95 Angstrom resolution. J. Mol. Biol. 273:171–182. [DOI] [PubMed] [Google Scholar]
- 9.Sundquist, W. I., and A. Klug. 1989. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 342:825–829. [DOI] [PubMed] [Google Scholar]
- 10.Keniry, M. A., G. D. Strahan, E. A. Owen, and R. H. Shafer. 1995. Solution structure of the Na+ form of the dimeric guanine tetraplex [d(G3T4G3)]2. Eur. J. Biochem. 233:631–643. [DOI] [PubMed] [Google Scholar]
- 11.Henderson, E., C. C. Hardin, S. K. Walk, I. Tinoco, and E. H. Blackburn. 1987. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell. 51:899–908. [DOI] [PubMed] [Google Scholar]
- 12.Macaya, R. F., P. Schultze, F. W. Smith, J. A. Roe, and J. Feigon. 1993. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl. Acad. Sci. USA. 90:3745–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang, Y., and D. J. Patel. 1993. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1:76–94. [DOI] [PubMed] [Google Scholar]
- 14.Parkinson, G. N., M. P. H. Lee, and S. Neidle. 2002. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 417:876–880. [DOI] [PubMed] [Google Scholar]
- 15.Wang, Y., and D. J. Patel. 1995. Solution structure of the Oxytricha telomeric repeat d[G4(T4G4)3] G-tetraplex. J. Mol. Biol. 251:263–282. [DOI] [PubMed] [Google Scholar]
- 16.Simonsson, T., P. Pecinka, and M. Kubista. 1998. DNA tetraplex formation in the control regions of c-myc. Nucleic Acids Res. 26:1167–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siddiqui-Jain, A., C. L. Grand, D. J. Bearss, and L. H. Hurley. 2002. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA. 99:11593–11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murchie, A. I., and D. M. J. Lilley. 1992. Retinoblastoma susceptibility genes contain 5′ sequences with a high propensity to form guanine-tetrad structures. Nucleic Acids Res. 20:49–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fry, M., and L. A. Loeb. 1994. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl. Acad. Sci. USA. 91:4950–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howell, R. M., K. J. Woodford, M. N. Weitzmann, and K. Usdin. 1996. The chicken beta-globin gene promoter forms a novel “cinched” tetrahelical structure. J. Biol. Chem. 271:5208–5214. [DOI] [PubMed] [Google Scholar]
- 21.Lew, A., W. J. Rutter, and G. C. Kennedy. 2000. Unusual DNA structure of the diabetes susceptibility locus IDDM2 and its effect on transcription by the insulin promoter factor Pur-1/MAZ. Proc. Natl. Acad. Sci. USA. 97:12508–12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jing, N., R. F. Rando, Y. Pommier, and M. E. Hogan. 1997. Ion selective folding of loop domains in a potent anti-HIV oligonucleotide. Biochemistry. 36:12498–12505. [DOI] [PubMed] [Google Scholar]
- 23.Aboul-ela, F., A. I. Murchie, and D. M. J. Lilley. 1993. NMR study of parallel-stranded tetraplex formation by the hexadeoxynucleotide d(TG4T). Nature. 360:280–282. [DOI] [PubMed] [Google Scholar]
- 24.Wang, Y., and D. J. Patel. 1993b. Solution structure of a parallel-stranded G-quadruplex DNA. J. Mol. Biol. 234:1171–1183. [DOI] [PubMed] [Google Scholar]
- 25.Hardin, C. C., M. J. Corregan, D. V. Lieberman, and B. A. Brown. 1997. Allosteric interactions between DNA strands and monovalent cations in DNA quadruplex assembly: thermodynamic evidence for three linked association pathways. Biochemistry. 36:15428–15450. [DOI] [PubMed] [Google Scholar]
- 26.Wyatt, J. R., P. W. Davis, and S. M. Freier. 1996. Kinetics of G-quartet-mediated tetramer formation. Biochemistry. 35:8002–8008. [DOI] [PubMed] [Google Scholar]
- 27.Mergny, J.-L., A. De Cian, A. Ghelab, B. Saccà, and L. Lacroix. 2005. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 33:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheong, C., and P. B. Moore. 1992. Solution structure of an unusually stable RNA tetraplex containing G- and U-quartet structures. Biochemistry. 31:8406–8414. [DOI] [PubMed] [Google Scholar]
- 29.Darby, R. A. J., M. Sollogoub, C. McKeen, L. Brown, A. Risitano, N. Brown, C. Barton, T. Brown, and K. R. Fox. 2002. High throughput measurement of duplex, triplex and quadruplex melting curves using molecular beacons and a LightCycler. Nucleic Acids Res. 30:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Risitano, A., and K. R. Fox. 2003. Stability of intramolecular DNA quadruplexes: comparison with DNA duplexes. Biochemistry. 42:6507–6513. [DOI] [PubMed] [Google Scholar]
- 31.Risitano, A., and K. R. Fox. 2004. Influence of loop size on the stability of intramolecular DNA quadruplexes. Nucleic Acids Res. 32:2598–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.James, P. L., T. Brown, and K. R. Fox. 2003. Thermodynamic and kinetic stability of intermolecular triple helices containing different proportions of C+·GC and T·AT triplets. Nucleic Acids Res. 31:5598–5606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 34.Mergny, J.-L., A.-T. Phan, and L. Lacroix. 1998. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 435:74–78. [DOI] [PubMed] [Google Scholar]
- 35.Petraccone, L., E. Erra, L. Nasti, A. Galeone, A. Randazzo, L. Mayol, G. Barone, and C. Giancola. 2003. Effect of a modified thymine on the structure and stability of [d(TGGGT)]4 quadruplex. Int. J. Biol. Macromol. 31:131–137. [DOI] [PubMed] [Google Scholar]
- 36.Moreira, B. G., Y. You, M. A. Behlke, and R. Owczarzy. 2005. Effects of fluorescent dyes, quenchers, and dangling ends on DNA duplex stability. Biochem. Biophys. Res. Commun. 327:473–484. [DOI] [PubMed] [Google Scholar]
- 37.Lu, M., Q. Guo, and N. R. Kallenbach. 1992. Structure and stability of sodium and potassium complexes of dT4G4 and dT4G4T. Biochemistry. 31:2455–2459. [DOI] [PubMed] [Google Scholar]
- 38.Guo, Q., M. Lu, and N. R. Kallenbach. 1993. Effect of thymine tract length on the structure and stability of model telomeric sequences. Biochemistry. 32:3596–3603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.