Abstract
Binding of herpes simplex virus (HSV) envelope glycoprotein D (gD) to a cell surface receptor is an essential step of virus entry. We recently determined the crystal structure of gD bound to one receptor, HveA. HveA is a member of the tumor necrosis factor receptor family and contains four characteristic cysteine-rich domains (CRDs). The first two CRDs of HveA are necessary and sufficient for gD binding. The structure of the gD-HveA complex reveals that 17 amino acids in HveA CRD1 and 4 amino acids in HveA CRD2 directly contact gD. To determine the contribution of these 21 HveA residues to virus entry, we constructed forms of HveA mutated in each of these contact residues. We determined the ability of the mutant proteins to bind gD, facilitate virus entry, and form HveA oligomers. Our results point to a binding hot spot centered around HveA-Y23, a residue that protrudes into a crevice on the surface of gD. Both the hydroxyl group and phenyl group of HveA-Y23 contribute to HSV entry. Our results also suggest that an intermolecular β-sheet formed between gD and HveA residues 35 to 37 contributes to binding and that a C37-C19 disulfide bond in CRD1 is a critical component of HveA structure necessary for gD binding. The results argue that CRD2 is required for gD binding mainly to provide structural support for a gD binding site in CRD1. Only one mutant, HveA-R75A, exhibited enhanced gD binding. While some mutations influenced complex formation, the majority did not affect HSV entry, suggesting that most contact residues contribute to HveA receptor function collectively rather than individually. This structure-based dissection of the gD-HveA binding site highlights the contribution of key residues within HveA to gD binding and HSV entry and defines a target region for the design of small-molecule inhibitors.
In humans, herpes simplex virus (HSV) typically causes mucosal lesions and then spreads to the peripheral nervous system, where it establishes latent infections in sensory neurons. HSV encodes at least 11 membrane glycoproteins, and four of these (gD, gH, gL, and gB) are essential for entry of virions into mammalian cells. Expression of these four glycoproteins alone can facilitate cell fusion (4, 33, 37, 41, 44).
The current model for virus entry posits the following series of events. Initially gC and/or gB binds cell surface heparan sulfate proteoglycans (41). Although binding of gC is a high-affinity interaction (KD = 10−8 M) (38), it is not essential for virus entry (41, 42). This gC binding step is followed by binding of gD to one of several cell surface receptors. This essential step leads to pH-independent membrane fusion of the viral envelope with the cell plasma membrane, a process facilitated by gD, gH, gL, and gB (41). Some experiments suggest that the gH/gL heterodimer and/or gB is the viral fusogen, but how their fusogenic activity is triggered by the gD-receptor interaction is unknown (18, 22).
Several cellular receptors for HSV have been identified through expression cloning and homology searches, including herpesvirus entry mediator A (HveA), nectin-1 (HveC), nectin-2 (HveB), and a modified form of heparan sulfate (3-OST-3) (5, 41). The focus of this study is on HveA (also called HVEM, TNFRSF-14, ATAR, and TR2), a member of the tumor necrosis factor receptor (TNFR) protein family (24, 28, 32). HveA has four cysteine-rich domains (CRDs) that are characteristic of the TNFR family (Fig. 1 and 2A). HveA binds directly to HSV gD and mediates entry of most HSV-1 and HSV-2 strains (32, 47).
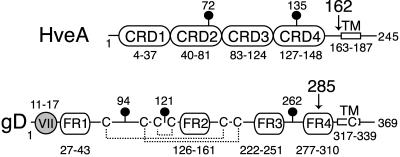
FIG. 1.
Diagram of full-length HveA and gD. The amino acid numbers begin with the first amino acid in the mature protein after signal sequence cleavage. The positions of N-glycosylation sites (lollipops) and transmembrane regions (TM) are indicated. The HveA amino acids comprising each of the four CRDs are labeled. The gD amino acids comprising each of four defined functional regions (FR) and the group VII MAb epitope (gray circle) are labeled. The disulfide bond pattern (dotted lines) and locations of cysteines (C) within gD are indicated. Arrows indicate the sites of truncation for the proteins used to solve the crystal structure.
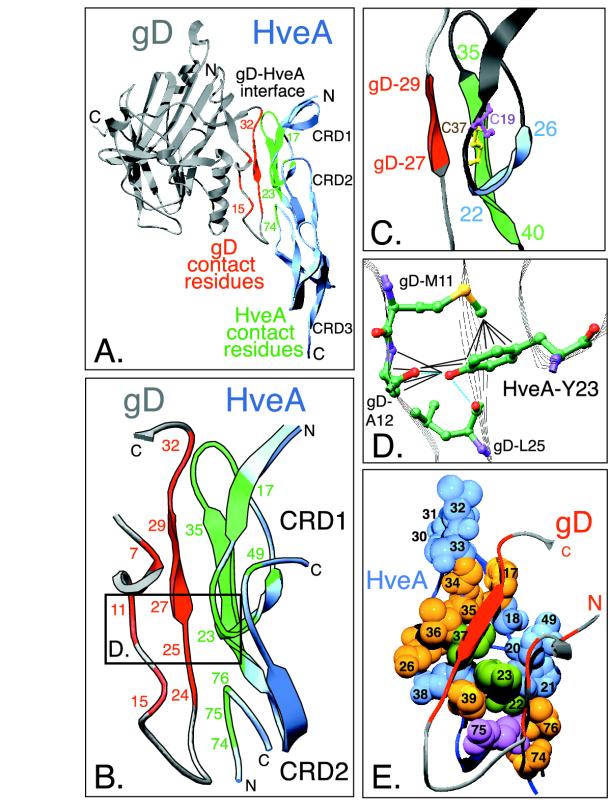
FIG. 2.
Highlighted regions within the gD-HveA crystal structure. (A) Ribbon diagram of the crystal structure of HveA bound to gD. The N- and C-terminal residues observed in the crystal structure and the location of HveA CRDs are indicated. The HveA molecule is shown partially in blue, and the HveA contact residues found within CRD1 and CRD2 are shown in green. The gD molecule is shown in gray, with the contact residues located in the N-terminal loop of gD shown in red. Contact residues were defined as amino acids containing atoms that come within 4 Å of the partner molecule (see Table 1). Some of the contact residues are numbered for reference. (B) An enlarged view of the gD-HveA interface shown in the same orientation as in panel A. HveA contact residues are displayed in green, and gD contact residues are red. The boxed area indicates a region shown at higher magnification in panel D. (C) Magnification of a disulfide bond within HveA CRD1 and three β-strands within the gD-HveA binding site. HveA-C37 (yellow) forms a disulfide bond with HveA-C19 (purple). Residues 35 to 37 within a β-strand of HveA (residues 35 to 40, shown in green) form hydrogen bonds with a short β-strand on gD (gD residues 27 to 29, shown in red) to form an intermolecular, antiparallel β-sheet. This augments a two-stranded β-sheet in HveA CRD1 formed by residues 22 to 26 (blue) and residues 35 to 40 (green). (D) Interactions between HveA-Y23 and gD amino acids. Carbon (green), oxygen (red), nitrogen (purple), and sulfur (yellow) atoms are shown. The blue dotted lines indicate hydrogen bonds between HveA-Y23 and gD. The black lines indicate other interactions, defined as distances of 4 Å or less (see Table 1). The hydroxyl group of HveA-Y23 interacts with gD-A12 and gD-L25. The phenyl ring of HveA-Y23 interacts with gD-M11 and gD-A12. (E) Space-filling model of the gD binding site on the surface of HveA. The gD-HveA interface from panel B is rotated so that the N-terminal loop of gD lies on top of the HveA binding surface. The N-terminal loop of gD is shown as a gray ribbon, with the gD contact residues colored red. The HveA contact residues are numbered. The space-filling spheres represent atoms of HveA contact residues from category 1 (blue), category 2 (purple), category 3 (yellow), and category 4 (green) (see Table 1). HveA-Y23 (green) is located at the center of the interface.
Biochemical studies showed that CRD1 and CRD2 of HveA are necessary and sufficient to bind gD (46). A truncated form of HveA containing both CRD1 and CRD2 binds gD and blocks entry of HSV, but forms of HveA consisting of CRD1 or CRD2 alone neither bind gD nor block virus entry. Furthermore, a monoclonal antibody (MAb), CW3, that recognizes a conformational epitope located on CRD1 blocks gD binding to HveA and partially blocks HSV entry (46). This observation led to the suggestion that gD interacts primarily with CRD1. It was hypothesized that the requirement of CRD2 for gD binding was due either to the presence of a few critical binding residues within CRD2 or to an indirect effect of CRD2 on the presentation of a gD binding site within CRD1.
In a series of structure-function studies with purified forms of gD and HveA proteins, we found that the ectodomain of gD truncated at residue 306, gD(306t), binds HveA with a KD of 3.2 × 10−6 M. A shorter form of gD truncated at residue 285, gD(285t), binds HveA with a 100-fold-higher affinity (KD = 3.7 × 10−8 M) and blocks infectivity 50 to 100 times better than the lower-affinity form (35a, 48).
We recently crystallized the complex between gD(285t) and the ectodomain of HveA(CRD1-4) (7, 8). This crystallization was probably favored by the high affinity of gD(285t) for HveA. The structure of gD(285t) bound to HveA(CRD1-4) shows that the gD core has a V-like immunoglobulin fold (Fig. 2A). This core is flanked by a long C-terminal extension and an N-terminal hairpin structure that comprises the entire HveA binding site. The structure of HveA is similar to that of other members of the TNFR family, especially within CRD1 and CRD2 (8, 34).
In accordance with the biochemical data, the crystal structure reveals that gD makes direct contact with amino acids in both CRD1 and CRD2 of HveA, but the majority of contacts lie within CRD1. All of the HveA amino acids that contact gD are within residues 17 to 26 and 30 to 39 of CRD1 and residues 49 and 74 to 76 of CRD2 (Table 1, Fig. 2B). At the gD-HveA interface, an intermolecular antiparallel β-sheet is formed between a β-strand consisting of HveA residues 35 to 37 and a short β-strand in gD consisting of residues 27 to 29 (Fig. 2C). The gD-HveA interface centers around the phenol ring of HveA-Y23, which protrudes into a crevice on the surface of gD (Fig. 2D and E) (8).
TABLE 1.
HveA contact residues
| HveA contact residue | No. of interactionsa
|
gD residue(s) contacted | Effect of mutationb on:
|
Cate- gory | ||
|---|---|---|---|---|---|---|
| Side chain | Main chain | gD binding | HSV entry | |||
| K18 | 1 | 1 | A7, T29 | Wild type | Wild type | 1 |
| S20 | 4 | 1 | A7, M11 | Wild type | Wild type | 1 |
| P21 | 1 | P14 | Wild type | Wild type | 1 | |
| G30 | 2 | P32 | Wild type | Wild type | 1 | |
| E31 | 1 | P32 | Wild type | Wild type | 1 | |
| L32 | 3 | P31 | Wild type | Wild type | 1 | |
| T33 | 3 | P31, P32 | Wild type | Wild type | 1 | |
| E38 | 4 | 2 | D26, V24 | Wild type | Wild type | 1 |
| L49 | 1 | A7 | Wild type | Wild type | 1 | |
| R75 | 5 | 6 | P14, N15 | Increased | Wild type | 2 |
| P17 | 1 | 3 | T29, D30 | Reduced | Wild type | 3 |
| K26 | 4 | D26 | Reduced | Wild type | 3 | |
| G34 | 6 | T29, D30, L28 | Reduced | Wild type | 3 | |
| T35 | 1 | 11 | T29, L28, Q27 | Reduced | Wild type | 3 |
| V36 | 2 | Q27 | Reduced | Wild type | 3 | |
| P39 | 4 | V24, L25 | Reduced | Wild type | 3 | |
| S74 | 2 | 5 | N15 | Reduced | Wild type | 3 |
| T76 | 2 | 1 | P14, N15 | Reduced | Wild type | 3 |
| G22 | 1 | P14 | Negative | Negative | 4 | |
| Y23 | 14 | M11, A12, L25 | Negative | Negative | 4 | |
| C37 | 2 | 12 | M11, D26, Q27, T29 | Negative | Negative | 4 |
An interaction was defined as an atom on an HveA residue coming within 4 Å of an atom on a gD residue. Side chain interactions were defined as interactions involving atoms on an HveA residue that are directly eliminated by alanine substitution, i.e., atoms downstream from the Cβ of the residue. Main chain interactions were defined as interactions involving atoms not eliminated by alanine substitution, i.e., atoms upstream from and including the Cβ.
Each contact residue was replaced with alanine individually, and the mutant proteins were assessed for ability to bind gD and mediate HSV entry.
The gD contact residues are contained within only two short segments of gD (residues 7 to 15 and 24 to 32) in the N-terminal hairpin loop. Some of these gD contact residues lie within a previously described epitope (residues 11 to 17) for neutralizing group VII MAbs that block binding of gD to HveA (Fig. 1) (35). Other gD contact residues lie within the previously described functional region 1 (FR1) of gD (residues 27 to 43) (Fig. 1) (10). Mutations in FR1 prevent gD from binding HveA and prevent HSV from infecting cells (10, 27, 48).
In this study, we examined the contribution of each HveA contact residue to gD binding, virus entry, and HveA oligomerization. By use of site-directed mutagenesis, we changed each HveA contact residue to alanine and examined the phenotypes of the mutated gD receptors. Our results show that HveA-Y23 plays a key role in gD binding and HSV entry. Three mutations in CRD1 eliminate HveA function as a gD and HSV entry receptor. The results suggest that CRD2 is required for gD binding mainly to provide structural support for a gD binding site present in CRD1. Although eight mutants showed reduced gD binding and one mutant, HveA-R75A, exhibited enhanced binding, mutation of the majority of contact residues had little effect on the ability of HveA to act as an entry receptor. This result suggests that the majority of contact residues contribute to binding collectively rather than individually.
MATERIALS AND METHODS
Cells and viruses.
B78-H1 mouse melanoma cells (30) and African green monkey kidney (Vero) cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal calf serum (FCS). 293T cells were grown in DMEM supplemented with 10% FCS. HSV-1 hrR3 carries lacZ under the ICP6 promoter and was kindly provided by S. K. Weller (21). HSV Rid1 TK12 carries lacZ under the ICP4 promoter and was kindly provided by P. G. Spear (32). HSV Rid1 TK12 was purified on a sucrose gradient (23).
PAbs and MAbs.
Rabbit serum R7 polyclonal antibody (PAb) was raised against HSV-2 gD and cross-reacts with HSV-1 gD (25). Rabbit serum R140 PAb was raised against HveA(200t) (43). MAb CW3 was raised against HveA(200t) (46). R166 PAb was raised against nectin-1 [HveC(346t) lacking a His tag] (26) and was used here as a negative control. Immunoglobulin G (IgG) was purified from rabbit serum or mouse ascites fluid with HiTrap protein G 1-ml columns (Amersham Phamacia Biotech). R140 PAb and R166 PAb IgGs were directly labeled with Alexa Fluor 488 with the Alexa Fluor 488 protein labeling kit (Molecular Probes).
Production and purification of HSV-1 gDt.
Procedures for production and purification of gD(285t) expressed by recombinant baculovirus-infected insect cells have been described elsewhere (39).
Construction of mutant HveA molecules.
Plasmid pBec10 (32) was used as a template for PCR amplification of HveA. An upstream primer (CGGCGAAGCTTGAGGCATGGAGCCT) and downstream primer (GGCGCTCGAGTCTGTGGGTCAGTGGT) amplified the full-length sequence of wild-type HveA. Using the restriction digest sites in italic, the HveA fragment was digested and ligated with DNA obtained from pcDNA3.1 to generate plasmid pSC386.
The QuikChange site-directed mutagenesis kit (Stratagene Cloning Systems) was used to generate mutant HveA constructs as recommended by the manufacturer. Briefly, primers designed to mutate specific HveA residues to alanine were used to PCR amplify the entire pSC386 plasmid. The reaction products were then treated with DpnI to digest methylated template DNA and used to transform competent bacteria (Escherichia coli XL1-Blue cells [Stratagene] or One Shot Top10F′ cells [Invitrogen]). The mutant HveA sequences were confirmed by sequencing the entire gene. The plasmids were named as follows: HveA-P17A (pSC419), HveA-K18A (pDL433), HveA-C19A (pDL458), HveA-S20A (pSC391), HveA-P21A (pSC410), HveA-G22A (pDL438), HveA-Y23A (pSC411), HveA-Y23F (pDL456), HveA-K26A (pDL434), HveA-G30A (pSC412), HveA-E31A (pDL435), HveA-L32A (pSC403), HveA-T33A (pSC399), HveA-G34A (pSC401), HveA-T35A (pSC393), HveA-V36A (pDL436), HveA-C37A (pSC418), HveA-E38A (pSC413), HveA-P39A (pSC414), HveA-L49A (pDL429), HveA-S74A (pSC400), HveA-R75A (pSC392), HveA-T76A (pSC394), and HveA-74/75/76A (pDL460).
Transfection.
B78-H1 cells were plated on 60-mm plates or six-well plates and transfected with endotoxin-free preparations (Qiagen) of the HveA plasmids with GENEPorter as recommended by the manufacturer (Gene Therapy Systems). For 60-mm plates, a total of 6 μg of DNA with 41 μl of GENEPorter was used. For six-well plates, 2.7 μg of DNA and 18 μl of GENEPorter were used per well. Cells were exposed to the DNA-GENEPorter mixture for 3 h before an equal volume of DMEM containing 20% FCS was added and left on the cells overnight at 37°C. For sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), 293T cells were plated in 48-well plates and transfected similarly, with 0.5 μg of DNA and 2.5 μl of GENEPorter per well.
Immunofluorescence assay.
B78-H1 cells transfected with HveA plasmids were trypsinized 24 h posttransfection, seeded on glass coverslips, and grown overnight at 37°C. Cells were fixed in 3% paraformaldehyde overnight at 4°C. The cells were rinsed with phosphate buffered saline (PBS) and incubated with 50 mM NH4Cl for 10 min at 25°C to quench the remaining paraformaldehyde. Fixed cells were incubated with 0.5% bovine serum albumin in PBS (BSA-PBS) for 30 min at 25°C and then labeled with 5 μg/ml of Alexa Fluor 488-conjugated R140 PAb IgG or R166 PAb IgG (as a negative control) diluted in BSA-PBS for 1 h at 25°C. The cells were rinsed three times with PBS and once with water before being mounted on slides in ProLong Antifade (Molecular Probes). Slides were examined with a Nikon Eclipse E600 microscope. Typically a 60× objective lens was used to view the cells.
Flow cytometry.
B78-H1 cells transfected with HveA plasmids were trypsinized 24 h posttransfection, reseeded on fresh plates, and grown overnight at 37°C. Cells were detached with 0.02% (wt/vol) disodium EDTA in PBS (Versene; Gibco-BRL) and fixed in 1% paraformaldehyde overnight at 4°C. The cells were pelleted, rinsed with DMEM containing 5% FCS (DMEM-5% FCS), and resuspended in DMEM-5% FCS containing 50 μg/ml of Alexa Fluor 488-conjugated R140 PAb IgG or R166 PAb IgG (as a negative control). After DMEM-5% FCS and PBS washes, cells were resuspended in 1% paraformaldehyde.
CELISA.
To detect HveA cell surface expression and gDt binding to cells, we used a modification of a published cellular enzyme-linked immunosorbent assay (CELISA) method (19, 31). B78-H1 cells transfected with HveA plasmids were trypsinized 24 h posttransfection and replated on 96-well plates pretreated with 0.2% gelatin. Cells were grown to confluence overnight and either fixed in paraformaldehyde or exposed to serial dilutions of gDt prepared in DMEM-5% FCS for 1 h at 4°C, washed three times with PBS supplemented with 900 μM CaCl2 and 500 μM MgCl2, and then fixed in 3% paraformaldehyde. Cells were incubated with 50 mM NH4Cl for 10 min at 25°C to quench the remaining paraformaldehyde and then rinsed twice with PBS. Cells were incubated for 1 h at 25°C with either 10 μg/ml of PAb R7 IgG to detect gDt binding or serial dilutions of R140 PAb IgG or CW3 MAb IgG to detect HveA cell surface expression. All antibodies were prepared in DMEM-5% FCS. Cells were washed with PBS three times and incubated with horseradish peroxidase-conjugated goat anti-mouse or anti-rabbit Ig antibodies for 30 min at 25°C. Following another three PBS washes, cells were rinsed with 20 mM citrate buffer (pH 4.5). ABTS peroxidase substrate (Moss, Inc.) was added, the absorbance at 405 nm was recorded at multiple time points with a microtiter plate reader, and the mean slopes were recorded.
Virus entry assay.
To detect HSV entry mediated by HveA, we used a modification of a published entry assay (36). Briefly, B78-H1 cells transfected with HveA plasmids as above were plated in 96-well plates and grown to confluence overnight at 37°C. Serial dilutions of HSV-1 hrR3 were added, and the cells were incubated for 60 min at 4°C and then shifted to 37°C. After 6 h, the cells were washed with PBS and lysed in DMEM containing 0.5% NP-40. β-Galactosidase activity in the lysate was measured by adding substrate (chlorophenol red-β-d-galactopyranoside; Boehringer Mannheim) and measuring absorbance at 570 nm with a microtiter plate reader.
SDS-PAGE analysis.
293T cells were lysed 24 h posttransfection and harvested in lysis buffer (10 mM Tris, 150 mM NaCl, 10 mM EDTA, 1% NP-40, 0.5% sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride [pH 8]). Proteins were separated by SDS-PAGE in precast Tris-glycine gels (Novex) under nondenaturing, nonreducing conditions (12). Following SDS-PAGE, the proteins were transferred to nitrocellulose and incubated in PBS containing 5% nonfat dry milk and 0.2% Tween 20 (blocking solution). Blots were reacted with R140 PAb and incubated with secondary antibody (goat anti-mouse or goat anti-rabbit immunoglobulin) coupled to horseradish peroxidase. Blots were washed and visualized by exposure to film after the addition of chemiluminescent substrate (ECL; Amersham).
RESULTS
Production and expression of HveA mutants.
To determine the contribution to receptor function of the 21 HveA amino acids that contact gD, we individually mutated each of these HveA contact residues. HveA contact residues were determined by use of the crystal structure of HveA bound to gD (PDB identification number 1JMA) and were defined as those amino acids containing atoms located within 4 Å of atoms of the gD structure (Table 1; Fig. 2B). Site-directed mutagenesis was used to substitute alanine for each of the HveA contact residues. This amino acid was chosen as a generic substitute because its presence removes the majority of the original residue's side chain atoms. Alanine is also commonly found in many secondary structures in both exposed and buried positions (45). The final plasmid constructs were cloned and sequenced, and the phenotypes of the mutated receptors in cells were compared to wild-type HveA.
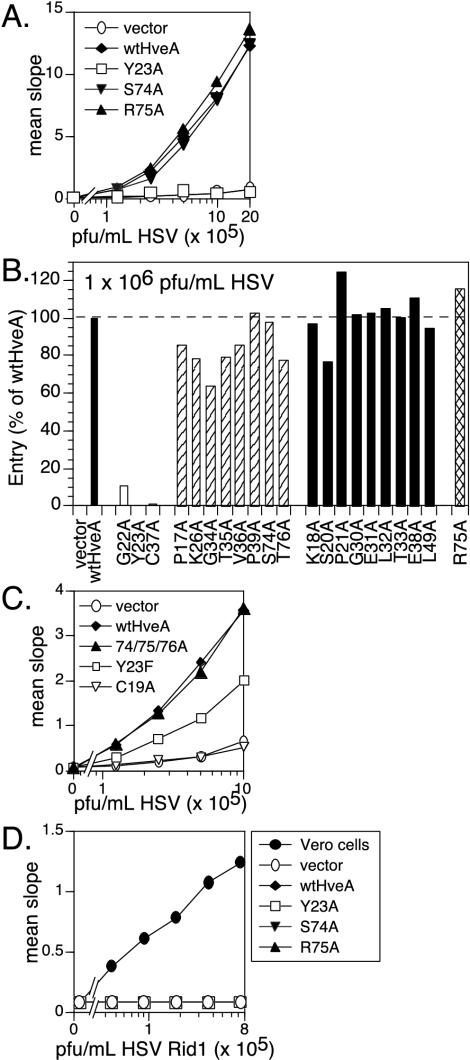
Surface expression was used as a first indication that each of the mutant HveA molecules was properly folded and transported to the plasma membrane. Plasmids carrying each of the HveA mutants were transfected into B78-H1 cells. This mouse melanoma cell line does not express any gD receptors, but it can be made permissive for HSV infection by transfection with a gD receptor (30). Transfected cells were fixed with paraformaldehyde, stained with a fluorescently labeled anti-HveA antibody (R140), and visualized by immunofluorescence microscopy. R140 is a polyclonal antibody that was raised against the purified full ectodomain of HveA (CRD1-4; Fig. 1) (43). Figure 3A shows immunofluorescence assay results for wild-type HveA and three representative HveA mutants: HveA-R75A, HveA-S74A, and HveA-Y23A. Throughout this paper, wild-type HveA and these three mutants will be used as representatives of mutant phenotype categories 1, 2, 3, and 4, respectively (Table 1). These three forms of HveA as well as the other 18 HveA mutants were expressed on the cell surface, as judged by immunofluorescence assay.
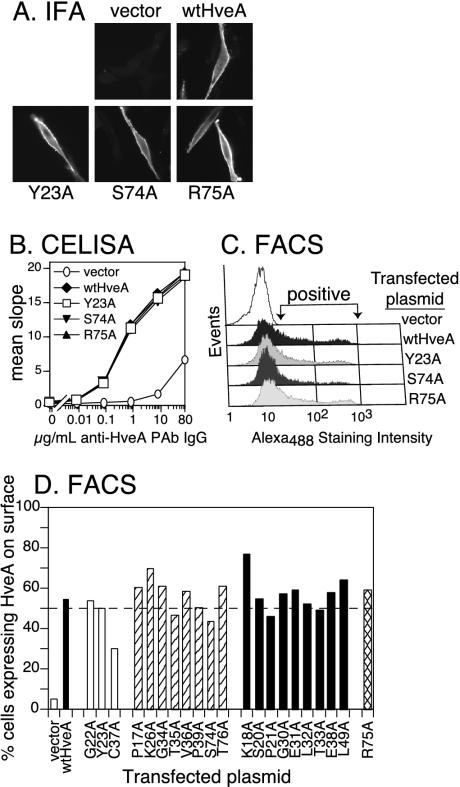
FIG. 3.
Expression of HveA mutant proteins on transfected cells. B78-H1 cells were transiently transfected with plasmids carrying each of the HveA mutants and evaluated for the expression of HveA after 48 h. (A) Cells were stained with fluorescently labeled anti-HveA PAb IgG and visualized by immunofluorescence microscopy to detect cell surface expression. Results for the transfection of wild-type HveA (wtHveA) and three representative HveA mutants are shown. (B) HveA expression was quantitated by CELISA. Cells were plated on 96-well plates and titrated with anti-HveA PAb IgG (R140) to estimate the level of cell surface expression. Sample data are shown for wild-type HveA and three representative HveA mutants. (C) Cells were stained with fluorescently labeled (Alexa Fluor 488) anti-HveA PAb IgG and analyzed by flow cytometry (FACS). Sample data for wild-type HveA and three representative HveA mutants show the level and range of positive staining. (D) Flow cytometry data for all of the HveA mutants are shown to indicate the percentage of cells staining positive for HveA. The dotted line represents 50% of the cells staining positive for HveA. Mutants were divided into four categories based on their ability to bind gD (see Fig. 4 and Table 1).
Two methods were used to quantitate the relative level of HveA on the cell surface. In the first, expression was measured by CELISA (19). B78-H1 cells were transiently transfected with each of the HveA mutants. The transfected cells were fixed with paraformaldehyde and incubated with various concentrations of R140, followed by secondary antibody conjugated to horseradish peroxide. Signal levels were essentially the same regardless of whether the cells were fixed before or after the addition of antibody (data not shown). Data are shown for wild-type HveA and the three representative HveA mutants (Fig. 3B). HveA expression by cells transfected with plasmids carrying these three mutants was identical to that of cells transfected with the wild-type HveA plasmid. Similar CELISA results were obtained for the other 18 HveA mutants (data not shown).
As a second method, flow cytometry was used to determine both the level and percentage of transiently transfected B78-H1 cells expressing the HveA mutant proteins. Transiently transfected B78-H1 cells were fixed with paraformaldehyde and stained with fluorescently labeled R140 to detect HveA surface expression. Data for wild-type HveA and the three representative HveA mutants showed that signal intensity varied over a broad range (Fig. 3C). For wild-type HveA and 20 of the HveA mutants, approximately 50% of the cells displayed detectable levels of HveA (Fig. 3D). For HveA-C37A, only about 30% of cells stained positive for cell surface expression of the receptor. Thus, flow cytometry profiles similar to those for wild-type HveA were observed for 20 of the 21 mutants.
Binding of gD to HveA mutants.
We used CELISA to determine the ability of gD to bind to the various HveA mutant proteins expressed on the cell surface. Various concentrations of gD(285t) were added to B78-H1 cells that had been transfected with plasmids expressing the HveA mutants. gD binding was detected with an anti-gD polyclonal antibody. By use of this approach, the 21 HveA mutants were initially categorized based on their gD binding phenotype. Sample data for wild-type HveA and three HveA mutants are shown (Fig. 4A). No gD bound to cells transfected with HveA-Y23A. gD bound less well to cells transfected with HveA-S74A than it did to those transfected with wild-type HveA. We were surprised to find that gD showed enhanced binding to cells transfected with HveA-R75A.
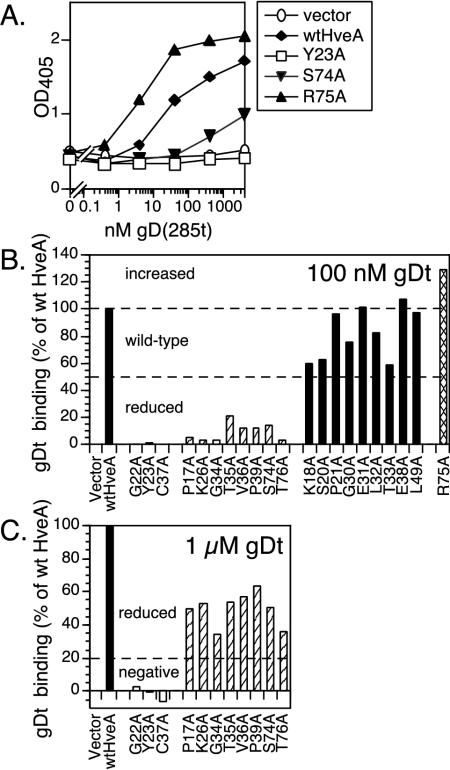
FIG. 4.
Binding of a truncated form of gD, gD(285t), to HveA proteins expressed on transfected cells. Transiently transfected B78-H1 cells expressing wild-type HveA and the HveA mutants were seeded on 96-well plates, incubated with different concentrations of gDt, washed, and probed with an anti-gD PAb to detect the level of gDt binding. (A) Sample data are shown for wild-type HveA (wtHveA) and three representative HveA mutants. Assays detecting HveA expression were run in parallel (Fig. 3). (B) Data for all of the HveA mutant proteins binding gDt at one concentration, 100 nM gD(285t), are shown to illustrate the basis for the separation of the HveA mutants into reduced (striped bars), wild-type (black bars), and increased (crosshatched bars) binding categories. gDt binding is expressed as a percentage of gDt binding to wild-type HveA. Results from one representative experiment are shown. (C) Data for HveA mutants binding gDt at a higher concentration, 1 μM gD(285t), are shown to illustrate the basis for the separation of the HveA mutants into negative (white bars) and reduced (striped bars) categories.
To categorize the gD binding phenotypes of all the HveA mutants (Table 1), we calculated the amount of gD bound to each mutant as a percentage of the amount of gD bound to wild-type HveA by using two different gD concentrations. At 100 nM gDt, nine HveA mutants bound gD at wild-type levels (category 1), one bound gD at increased levels (category 2), and 11 bound gD at reduced levels (Fig. 4B). Of the 11 mutants showing reduced binding at 100 nM gDt, 8 bound at reduced levels when the gDt concentration was increased to 1 μM (category 3), while the remaining 3 mutants did not bind gD at all (category 4) (Fig. 4C). For each HveA mutant, these experiments were repeated at least three times, and the pattern of gD binding remained consistent.
The three HveA mutations that led to a complete loss of gDt binding are located in CRD1 (HveA-Y23A, HveA-G22A, and HveA-C37A). The eight mutations that produced reduced gDt binding are distributed in both CRD1 (HveA-P17A, HveA-K26A, HveA-G34A, HveA-T35A, HveA-V36A, and HveA-P39A) and CRD2 (HveA-S74A and HveA-T76A). The mutation that resulted in significantly increased gDt binding, HveA-R75A, is in CRD2. The remaining nine mutations had only modest effects on gDt binding and are located in both CRD1 (HveA-K18A, HveA-S20A, HveA-P21A, HveA-G30A, HveA-E31A, HveA-L32A, HveA-T33A, and HveA-E38A) and CRD2 (HveA-L49A).
These results indicate that mutations in some contact residues negatively affect HveA binding to gD more than others, i.e., some contact residues are more critical for gD binding than are others. Some of these phenotypes are readily understood based on the structure of the gD-HveA complex. For example, in the crystal structure, HveA-Y23 lies at the center of the interface between gD and HveA, protrudes into a pocket on the surface of gD, and makes extensive contacts to seven atoms of gD (Fig. 2D and E, Table 1). The loss of gD binding by HveA-Y23A is most likely due to the loss of direct gD interactions with the Y23 side chain. In contrast, HveA-R75 makes multiple contacts with gD, and we might have expected mutation to alanine to reduce gD binding. The observed increased gD binding by HveA-R75A suggests that the side chain of HveA-R75 may instead hinder complex formation.
HveA mutants mediating entry of HSV.
We wanted to determine whether the observed changes in gD binding correlate with the ability of HveA to function as an entry receptor. B78-H1 cells were transiently transfected with plasmids expressing each of the 21 HveA mutants and infected with HSV carrying lacZ under the control of the ICP6 promoter. After 6 h of infection, cells were lysed, and the level of β-galactosidase activity was determined and used as a measure of entry (32). The HveA mutants fell into two groups, those that mediated HSV entry and those that did not. Data for wild-type HveA and the three representative HveA mutants are shown in Fig. 5A. HveA-R75A and HveA-S74A were as effective as wild-type HveA for HSV entry, while HveA-Y23A was nonpermissive.
FIG. 5.
HveA mutant proteins mediating HSV entry into transfected cells. Transiently transfected B78-H1 cells expressing the HveA mutant proteins were seeded on 96-well plates, incubated with an HSV β-galactosidase reporter virus for 6 h, and assayed for β-galactosidase activity as a measure of virus entry. (A) Sample data show the ability of wild-type HveA (wtHveA) and three representative HveA mutants to mediate entry of increasing amounts of HSV. Assays detecting HveA expression and gDt binding were run in parallel (Fig. 3 and 4). (B) Data show the original panel of 21 HveA mutants mediating entry at one concentration of HSV. Mutants were divided into the categories based on gD binding phenotype (see Fig. 4 and Table 1). Data are expressed as a percentage of entry mediated by wild-type HveA. Results from one representative experiment are shown. (C) The ability of additional HveA mutants to mediate entry of increasing amounts of HSV is shown. (D) Sample data show the inability of wild-type HveA and three representative HveA mutants to mediate entry of increasing amounts of a mutant β-galactosidase reporter virus, HSV Rid1, after 6 h. Vero cells were included as a positive control for HSV Rid1 entry.
A comparison of entry data for all of the HveA mutants at a single HSV concentration is shown in Fig. 5B. The three HveA mutants that did not bind gD, HveA-Y23A, HveA-G22A, and HveA-C37A, were not permissive for HSV entry (Table 1). All of the remaining HveA mutants were able to mediate HSV entry at or near wild-type levels, regardless of their level of gD binding. Thus, the effect of mutations on HveA function as a viral receptor was less apparent in the entry assay than in the gD binding assay. Some subtle effects of mutation on entry were noted, but the differences were small.
Additional HveA mutations.
We constructed an additional series of HveA mutants to address questions raised by the previous results. HveA expression levels were determined by CELISA, and the ability of the mutants to mediate HSV entry was determined with the protocol described previously.
(i) HveA-Y23.
We have shown that HveA-Y23 is critical for HveA function as a gD receptor. This amino acid interacts with gD through its hydroxyl group as well as its phenyl ring (Fig. 2D). To address the relative importance of the hydroxyl group and phenyl ring of HveA-Y23, we constructed HveA-Y23F. CELISA data indicate that HveA-Y23F is expressed on cells at wild-type levels (data not shown). We found that HveA-Y23F permits entry of HSV, although the efficiency of entry is reduced approximately 50% compared to wild-type HveA (Fig. 5C). The properties of HveA-Y23A and HveA-Y23F indicate that while the hydroxyl group is not required for HSV entry, both the phenyl ring and the hydroxyl group of HveA-Y23 contribute to HveA function as an entry receptor.
(ii) HveA-C37.
The inability of HveA-C37A to bind gD could be due to either the loss of direct contacts between HveA-C37 and gD or the loss of a disulfide bond between HveA-C37 and its partner HveA-C19 (Fig. 2C). To address this issue, we constructed the mutant HveA-C19A. HveA-C19A was expressed on cells at wild-type levels (data not shown) but did not permit entry of HSV (Fig. 5C). We conclude that the failure of HveA-C37A to bind gD and mediate HSV entry is due to the loss of a disulfide bond between HveA-C37 and HveA-C19 that likely affects HveA conformation.
(iii) HveA CRD2.
Mutations of contact residues in CRD2 increased (HveA-R75A), decreased (HveA-S74A, HveA-T76A), or did not affect (HveA-L49A) gD binding, but none eliminated HSV entry. We wanted to determine whether HveA function would be affected when three contiguous CRD2 residues were mutated in the same molecule. We constructed the HveA triple mutant HveA-74/75/76A, containing alanine substitutions for HveA-S74, -R75, and -T76. HveA-74/75/76A was expressed on the cell surface at wild-type levels and functioned as an HSV receptor as well as wild-type HveA did (Fig. 5C). Thus, none of these three CRD2 residues that contact gD are absolutely required for HveA function in HSV entry.
Do any of the HveA mutants mediate entry of HSV Rid1?
HSV Rid1 is a virus with a single amino acid mutation in gD (Q27P). This virus is unable to use human HveA for entry, and gD from this virus exhibits a 10-fold higher affinity for a second HSV receptor, nectin-1 (16, 20, 27, 32). The gD-HveA crystal structure suggests that the Rid1 mutation might distort the intermolecular antiparallel β-sheet formed between HveA-T35, HveA-V36, and HveA-C37 and gD residues 27 to 29 (8).
We asked whether any of the original panel of 21 HveA mutations were able to compensate for the HSV Rid1 entry defect. Dilutions of HSV-Rid1 carrying lacZ under control of the thymidine kinase promoter (32) were added to B78-H1 cells that had been previously transfected with each of the HveA mutants. β-Galactosidase activity was determined as a measure of entry after 6 h. As a positive control, Vero cells, which are permissive for HSV Rid1, were also infected with the virus. Results for wild-type HveA and three representative mutants are shown in Fig. 5D. Neither wild-type HveA nor any of the HveA mutants were able to mediate entry of HSV Rid1 (data not shown).
HveA oligomerization.
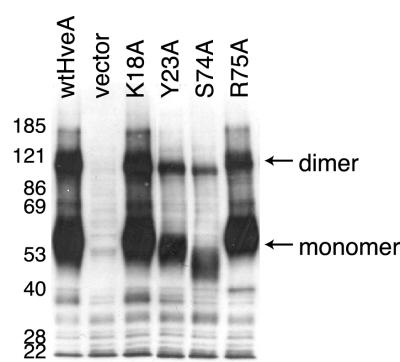
Many members of the TNFR family oligomerize in the absence of ligand via pre-ligand association domains (PLADs) present in CRD1 (9). It has been proposed that TNFR trimerization is required for ligand binding and subsequent signaling events in the cell. The TNFR-1 ectodomain crystallizes as a dimer with multiple monomer-monomer contacts in CRD1 (34). The crystal structure of the ligand-receptor tumor necrosis factor alpha-TNFR-1 complex revealed a tumor necrosis factor alpha trimer bound to three receptors, with no direct contacts among the three receptors (1). In contrast, the crystal structure of the gD(285t)-HveA complex shows a monomer of HveA bound to a monomer of gD. However, nondenaturing Western blot studies and gel filtration analyses indicate that HveA forms oligomers via CRD1, and several biochemical assays indicate that gD is a dimer (32, 46, 48).
Thus, it is possible that mutations in HveA may disrupt HveA oligomerization and thereby affect gD binding and HSV entry. To address this possibility, plasmids carrying the original 21 HveA mutants were transfected into 293T cells to obtain sufficient amounts of receptor for Western blotting. Cell extracts were electrophoresed on SDS gels under nondenaturing conditions (12), and Western blots were probed with R140, a polyclonal antibody that recognizes both monomeric and dimeric forms of HveA. All of the HveA mutants were expressed in 293T cells. Sample data for wild-type HveA and four selected HveA mutants are shown in Fig. 6. The monomer form of wild-type HveA was present at 55 kDa, and a dimer form of HveA was present at 110 kDa. The amount of HveA in these cell extracts was not equilibrated, and differences in signal levels do not indicate differences in levels of expression.
FIG. 6.
Ability of HveA mutant proteins to oligomerize. Extracts of 293T cells transfected with HveA mutants were run on SDS-PAGE under nondenaturing conditions, transferred to nitrocellulose, and probed with an anti-HveA PAb. Results for wild-type HveA (wtHveA) and four selected HveA mutant proteins are shown. The positions of molecular size markers are shown (in kilodaltons), along with the expected positions of the HveA monomer and dimer.
Each of the mutant forms of HveA was detected in both monomeric and dimeric forms to the same extent as the wild type, including HveA-K18A. An alignment of HveA with TNFR-1 shows that HveA-K18 is homologous to TNFR-K32. In contrast to HveA-K18A, the mutation TNFR-K32A prevented TFNR-1 oligomerization (9). The apparently reduced molecular weight of HveA-S74A (Fig. 6) is likely due to the fact that this mutation eliminates an N-glycosylation site at HveA-N72. Loss of this N-glycosylation does not prevent the receptor from mediating HSV entry (Fig. 5A). The other 17 HveA mutants also exhibited a normal pattern of oligomerization (data not shown). These results indicate that the inability of HveA-G22A, HveA-Y23A, and HveA-C37A to bind gD and mediate HSV entry is probably not due to defects in dimerization. The gD binding site and the HveA dimerization site in CRD1 are not the same.
DISCUSSION
The crystal structure of the gD-HveA complex shows that gD directly contacts residues in both CRD1 and CRD2 of HveA. In this study, we used site-directed mutagenesis to determine the contribution of each of the HveA contact residues to gD binding, HSV entry, and HveA oligomerization. We found that some mutations had a greater effect on HveA function than did others. Thus, the HveA contact residues do not contribute equally to gD binding. We determined that HveA-Y23 is critical for gD binding. Twenty-one HveA contact residue mutants were classified into four categories (Table 1). Nine mutants displayed wild-type HveA function (category 1); one bound gDt at increased levels and permitted HSV entry (category 2); eight bound gDt at reduced levels but permitted HSV entry (category 3); and three did not bind gDt or mediate HSV entry (category 4).
Mutations that do not affect HveA function (category 1).
gD binding and HSV entry were not greatly affected by alanine substitutions in 9 of the 21 contact residues (Table 1). Most of these residues interact with gD solely through main chain atoms, and thus it is not surprising that substituting alanine for these residues has little effect on HveA function. HveA-S20 and HveA-E38 have multiple side chain interactions with gD (Table 1), yet HveA-S20A and HveA-E38A function as well as wild-type HveA. Thus, analysis of the number of side chain versus main chain interactions for a residue does not always predict the effect of a mutation in that residue on HveA function.
Mutation that increases gDt binding and permits HSV entry (category 2).
HveA-R75A permits HSV entry and binds gDt better than does wild-type HveA. This result suggests that the side chain of HveA-R75 partially interferes with gD binding instead of promoting gD binding, as might be expected from the crystal structure. The negative effect of the HveA-R75 side chain on gD binding may be due to its presence in a region of positively charged gD residues (gD-R16, gD-R18, and gD-K20) and subsequent unfavorable electrostatic interactions across the protein interface. The availability of a presumably high-affinity form of HveA, such as HveA-R75A, may prove valuable for future studies of HveA, just as the high-affinity form of gD, gD(285t), has been fundamental to our studies of gD.
Mutations that reduce gDt binding but permit HSV entry (category 3).
Alanine substitutions in 8 of the 21 contact residues resulted in substantially reduced gDt binding without greatly affecting the level of HSV entry (Table 1). Thus, reduced levels of gD binding do not necessarily affect HveA function in HSV entry. Reduced gD binding may affect the rate of HSV entry, but preliminary experiments suggest that this is not the case (data not shown). Only those mutants that were totally negative for gD binding (category 4) failed to function for HSV entry.
Five of the residues in category 3 (P17, K26, G34, T35, and V36) cluster near a β-strand consisting of gD residues 27 to 29 (Fig. 2E). This β-strand participates in an intermolecular antiparallel β-sheet with HveA residues 35 to 37 and augments a two-stranded β-sheet in HveA CRD1 (Fig. 2C) (8). Results for category 3 mutants suggest that the intermolecular β-sheet is important for gD binding because substituting alanine for any of these five residues diminishes gD binding, possibly by directly or indirectly disrupting this β-sheet.
Mutation of residues that contact gD through side chain atoms resulted in reduced gD binding more frequently than mutation of residues that contact gD through only main chain atoms (Table 1). However, some results for mutants in category 3 were different from what might be predicted from the crystal structure. For example, HveA-V36A shows reduced gD binding despite the fact that only the main chain atoms of HveA-V36 contact gD. The HveA-V36A mutation most likely reduces gD binding indirectly by causing a local change in conformation.
The results for category 2 and 3 mutants suggest that HSV entry tolerates a wide range of affinities between HveA and gD. Similarly, a wide range of affinities between gD and nectin-1 are tolerated for entry of HSV, bovine herpesvirus, and pseudorabies virus (13, 27, 31). Future optical biosensor analysis of gD binding to mutant HveA proteins will define more precisely the binding kinetics and affinity for gD of these HveA mutants.
Why were differences in levels of HSV entry generally more subtle than differences in the levels of gD binding? One possible explanation is that the high levels of HveA expression associated with transient transfection may mask differences in the efficiency of entry mediated by the mutant receptors. Additionally, the interaction between gD and HveA may be different when gD is presented in the context of an intact virion rather than in a soluble form. Studies of influenza virus hemagglutinin (HA) have shown virus infection to be a highly sensitive assay for protein function, in that HA mutants displaying severely diminished fusion capacity were able to facilitate virus entry (14). Binding to an abundant receptor, such as sialic acid in the cases of influenza virus and polyomavirus, may permit the virus to contact multiple receptors, and this avidity may facilitate efficient virus entry. In fact, in the case of polyomavirus, mutants that bind receptor more weakly are more virulent in the host (2).
Mutations in HveA that abolish function (category 4).
We identified four mutations in HveA that result in the complete loss of gD binding and ability to mediate HSV entry. Two of these mutations involve HveA-Y23 (HveA-Y23A and HveA-G22A), and two (HveA-C37A and HveA-C19A) eliminate a disulfide bond in CRD1.
(i) HveA-Y23 is critical for HveA function.
The inability of HveA-Y23A to bind gD was predicted from the gD-HveA crystal structure (8). HveA-Y23 lies at the center of the gD-HveA interface and contacts gD residues on both sides of the N-terminal gD loop (Fig. 2D, E). This amino acid is sandwiched between the side chains of gD-M11 and gD-P14 and makes extensive contacts with seven atoms on three gD residues (Fig. 2D, Table 1). The hydroxyl group of HveA-Y23 is hydrogen bonded to gD-A12 and gD-L25. Previous studies have shown that substitution of gD-L25 with proline permits HSV to overcome gD-mediated interference to virus entry, alters HSV gD receptor usage, and has been postulated to prevent the interaction of gD with HveA (6, 29, 41).
To determine the contribution to HveA function of the hydroxyl group versus the phenyl group of HveA-Y23, we constructed mutant HveA-Y23F. The ability of HveA-Y23F to mediate HSV entry indicates that interactions with the hydroxyl group of HveA-Y23 are not critical for HveA function. This result is supported by the fact that the primate HveA homolog functions as an HSV entry mediator and has phenylalanine in place of tyrosine at position 23 (17). From this result, we predict that gD-M11 is important for gD binding to HveA because gD-M11 interacts with the HveA-Y23 phenyl ring (Fig. 2D) and lies within a known epitope for neutralizing anti-gD MAbs (group VII) (35). However, the reduced capacity of HveA-Y23F to mediate HSV entry indicates the HveA-Y23 hydroxyl group does contribute to HveA function, possibly through the hydrogen bond with gD-L25 (Fig. 2D).
The inability of HveA-G22A to bind gD may be due to a disruption of interactions between the adjacent residue HveA-Y23 and gD. The introduction of an extra methyl group into HveA-G22 may cause a shift in the position of HveA-Y23. Alternatively, replacing glycine with the conformationally more restricted amino acid alanine may cause a structural change in HveA and thereby impair gD binding. This alternative possibility is supported by the observation that G22 is a structurally conserved residue present in many TNFR-like molecules (6, 29, 41).
Studies in other laboratories have indicated that single residues can contribute a large fraction of the binding energy at protein-protein interfaces (3). These residues are located at “hot spots” of binding and are generally found at the center of the interface. A hot spot on HveA for gD binding lies at HveA-Y23. Tyrosine is frequently found in binding hot spots and is three times more likely to be in a hot spot than phenylalanine, suggesting that the ability of tyrosine to hydrogen bond may be important (3).
Structure-based mutagenesis of human growth hormone has shown that a subset of contact residues account for complex formation with the human growth hormone receptor (11, 15). The conclusion that the human growth hormone functional binding epitope is much smaller than the structural epitope suggested that design of smaller hormone mimics may be possible. Likewise, the results of the present study indicate that a small-molecule inhibitor could be designed to prevent gD interactions with the phenol ring of HveA-Y23 (8) and thereby abolish the ability of HveA to permit HSV entry. Two synthetic peptides isolated from phage display libraries have been shown to bind HveA and inhibit gD binding (40). The mechanism of this inhibition is unknown. In future studies we plan to test the ability of these peptides to bind HveA-Y23A, HveA-Y23F, and HveA-G22A. The results may indicate whether the peptides inhibit gD binding by interfering with interactions between HveA-Y23 and gD.
(ii) A disulfide bond in CRD1 is critical for HveA function.
The loss of gDt binding by both HveA-C37A and HveA-C19A is probably due to the loss of a disulfide bond that supports CRD1 structure and the gD binding site (Fig. 2C). However, indirect evidence suggests that HveA-C37A and HveA-C19A retain much of the wild-type HveA conformation. HveA-C37A is expressed on the cell surface and is able to form dimers. Additionally, both HveA-C37A and HveA-C19A are recognized by the MAb CW3 (data not shown), a conformation-dependent MAb that binds HveA CRD1 and blocks HSV entry (46).
An alternative explanation for the loss of gDt binding by both HveA-C37A and HveA-C19A is that the absence of the disulfide bond disrupts an intermolecular β-sheet formed between HveA residues 35 to 37 and gD residues 27 to 29 (Fig. 2C). Mutations in gD-Q27 (Rid mutations) are predicted to disrupt this β-sheet interaction and eliminate an intermolecular hydrogen bond between gD-Q27 and HveA-C37 (8), yielding one explanation for the inability of the HSV Rid viruses to use HveA for entry.
CRD2 contact residues are not critical for HveA function.
Previously, analysis of a series of HveA truncations localized the gD binding domain to CRD1 and CRD2 of HveA (46). These findings were validated by the crystal structure of the gD-HveA complex (8). In the present study, none of the individual mutations in CRD2 contact residues completely eliminated HveA function in gD binding or HSV entry. In fact, even when residues S74, R75, and T76 were all mutated to alanine in the same molecule (HveA-74/75/76A), the receptor still functioned in entry. These results suggest that CRD2 is required for gD binding mainly to provide structural support for the gD binding site in HveA CRD1. The fact that HveA binds gD primarily via residues in CRD1 explains why MAb CW3, an antibody that binds entirely within CRD1, is able to efficiently block gD binding to HveA (46).
None of the HveA mutations compensate for the entry deficiency of HSV mutant strain Rid1.
The mutant strain HSV Rid1 carries the single gD mutation Q27P and was selected for its resistance to gD-mediated interference to infection (16). Although human HveA does not serve as an entry receptor for HSV Rid1, the murine HveA homolog has been reported to mediate entry of HSV Rid1 (41), suggesting that conservative changes in HveA can compensate for the block to entry.
Two of the mutants made in this study (HveA-E38A and HveA-L49A) have changes identical to changes present in the murine HveA homolog (24). The inability of these two mutants to mediate HSV Rid1 entry indicates that the E38A and L49A changes are not solely responsible for the ability of murine HveA to mediate HSV Rid1 entry. In fact, none of the mutations introduced in this study resulted in forms of HveA that permit entry of HSV Rid1. Even the increased gD binding of HveA-R75A was insufficient to overcome the HSV Rid1 block to HveA-mediated entry. The ability of murine HveA to mediate HSV Rid1 entry is probably due to multiple changes in the gD binding region.
HveA oligomerization is not affected by the HveA mutations.
Truncations of the HveA ectodomain have indicated that CRD1 plays a major role in HveA oligomerization (46). Many members of the TNFR family oligomerize via a domain present in CRD1 called a PLAD (9). A single alanine substitution at lysine-32 within TNFR-1 CRD1 prevents trimers from forming, suggesting that this residue is a required element of the TNFR-1 PLAD (9). Our Western blot analysis of proteins electrophoresed on nondenaturing gels indicates that a mutation in HveA homologous to TNFR-K32A, HveA-K18A, does not affect HveA oligomerization. Although it is possible that this analysis of HveA in cell extracts does not accurately reflect the state of the HveA on the cell surface, the results suggest that if HveA contains a PLAD, it differs from the PLAD of TNFR-1.
Changes in the ability of some HveA mutants to bind gD and mediate HSV entry cannot be attributed to a lack of HveA dimerization. None of the HveA mutants were deficient in dimerization, indicating that the gD binding site does not directly overlap the HveA dimerization site in CRD1.
Although HveA crystallizes as a monomer bound to gD (8), gel filtration analyses have suggested that a dimer of HveA binds to a monomer of gD (47). HveA mutants that dimerize but do not permit HSV entry, such as HveA-Y23A, could be used to examine whether HveA oligomerization plays a role in entry. If so, HveA-Y23A may dominantly interfere with the function of wild-type HveA during HSV entry. Such studies are anticipated as a follow-up to this initial report.
In summary, we have determined the contribution of specific HveA residues within the gD binding site to HveA function as a gD and HSV entry receptor. HveA-Y23 plays a key role in gD binding and HSV entry due to a large number of interactions with gD. An intermolecular β-sheet formed between gD and HveA residues 35 to 37 also contributes to binding. Our results suggest that the functional gD binding site lies primarily within HveA CRD1. Most of the other HveA contact residues contribute collectively rather than individually to the interaction between HveA and gD. The binding hot spot containing HveA-Y23 may be a good target for the design of an inhibitor of gD-HveA complex formation. Similar mutagenesis studies to examine the properties of gD contact residues are currently under way.
Acknowledgments
We thank S. C. Harrison for generous and critical review of the manuscript; N.W. Fraser, P. G. Spear, and S. K. Weller for reagents; and I. Baribaud, F. C. Bender, T. M. Cairns, C. Krummenacher, R. S. B. Milne, and J. C. Whitbeck for helpful advice.
This research was supported by Public Health Service grant AI-18289 to G.H.C. and R.J.E. from the National Institute of Allergy and Infectious Diseases and grants NS-36731 and NS-30606 to R.J.E. and G.H.C. from the National Institute of Neurological Disorders and Stroke. S.A.C. is a predoctoral trainee supported by grant AI-07325 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Banner, D. W., A. D'Arcy, W. Janes, R. Gentz, H. J. Schoenfeld, C. Broger, H. Loetscher, and W. Lesslauer. 1993. Crystal structure of the soluble human 55 kd TNF receptor-human TNF beta complex: implications for TNF receptor activation. Cell 73:431-445. [DOI] [PubMed] [Google Scholar]
- 2.Bauer, P. H., R. T. Bronson, S. C. Fung, R. Freund, T. Stehle, S. C. Harrison, and T. L. Benjamin. 1995. Genetic and structural analysis of a virulence determinant in polyomavirus VP1. J. Virol. 69:7925-7931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogan, A. A., and K. S. Thorn. 1998. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 280:1-9. [DOI] [PubMed] [Google Scholar]
- 4.Browne, H., B. Bruun, and T. Minson. 2001. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH and gL. J. Gen. Virol. 82:1419-1422. [DOI] [PubMed] [Google Scholar]
- 5.Campadelli-Fiume, G., F. Cocchi, L. Menotti, and M. Lopez. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305-319. [DOI] [PubMed] [Google Scholar]
- 6.Campadelli-Fiume, G., S. Qi, E. Avitabile, L. Foà-Tomasi, R. Brandimarti, and B. Roizman. 1990. Glycoprotein D of herpes simplex virus encodes a domain which precludes penetration of cells expressing the glycoprotein by superinfecting herpes simplex virus. J. Virol. 64:6070-6079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carfi, A., H. Gong, H. Lou, S. H. Willis, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2002. Crystallization and preliminary diffraction studies of the ectodomain of the envelope glycoprotein D from herpes simplex virus 1 alone and in complex with the ectodomain of the human receptor HveA. Acta Crystallogr. D 58:836-838. [DOI] [PubMed] [Google Scholar]
- 8.Carfi, A., S. H. Willis, J. C. Whitbeck, C. Krummenacher, G. H. Cohen, R. J. Eisenberg, and D. C. Wiley. 2001. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol. Cell 8:169-179. [DOI] [PubMed] [Google Scholar]
- 9.Chan, F. K., H. J. Chun, L. Zheng, R. M. Siegel, K. L. Bui, and M. J. Lenardo. 2000. A domain in TNF receptors that mediates ligand-independent receptor assembly and signaling. Science 288:2351-2354. [DOI] [PubMed] [Google Scholar]
- 10.Chiang, H.-Y., G. H. Cohen, and R. J. Eisenberg. 1994. Identification of functional regions of herpes simplex virus glycoprotein gD by with linker-insertion mutagenesis. J. Virol. 68:2529-2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clackson, T., and J. A. Wells. 1995. A hot spot of binding energy in a hormone-receptor interface. Science 267:383-386. [DOI] [PubMed] [Google Scholar]
- 12.Cohen, G. H., V. J. Isola, J. Kuhns, P. W. Berman, and R. J. Eisenberg. 1986. Localization of discontinuous epitopes of herpes simplex virus glycoprotein D: use of a nondenaturing (“native” gel) system of polyacrylamide gel electrophoresis coupled with Western blotting. J. Virol. 60:157-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Connolly, S. A., J. C. Whitbeck, A. H. Rux, C. Krummenacher, S. van Drunen Littel-van den Hurk, G. H. Cohen, and R. J. Eisenberg. 2001. Glycoprotein D homologues in herpes simplex virus type 1, pseudorabies virus, and bovine herpes virus type 1 bind directly to human HveC (nectin-1) with different affinities. Virology 280:7-18. [DOI] [PubMed] [Google Scholar]
- 14.Cross, K. J., S. A. Wharton, J. J. Skehel, D. C. Wiley, and D. A. Steinhauer. 2001. Studies on influenza haemagglutinin fusion peptide mutants generated by reverse genetics. EMBO J. 20:4432-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunningham, B. C., and J. A. Wells. 1993. Comparison of a structural and a functional epitope. J. Mol. Biol. 234:554-563. [DOI] [PubMed] [Google Scholar]
- 16.Dean, H. J., S. S. Terhune, M. Shieh, N. Susmarski, and P. G. Spear. 1994. Single amino acid substitutions in gD of herpes simplex virus 1 confer resistance to gD-mediated interference and cause cell-type-dependent alterations in infectivity. Virology 199:67-80. [DOI] [PubMed] [Google Scholar]
- 17.Foster, T. P., V. N. Chouljenko, and K. G. Kousoulas. 1999. Functional characterization of the HveA homolog specified by African green monkey kidney cells with a herpes simplex virus expressing the green fluorescence protein. Virology 258:365-374. [DOI] [PubMed] [Google Scholar]
- 18.Fuller, A. O., and W. C. Lee. 1992. Herpes simplex virus type 1 entry through a cascade of virus-cell interactions requires different roles of gD and gH in penetration. J. Virol. 66:5002-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geraghty, R. J., C. R. Jogger, and P. G. Spear. 2000. Cellular expression of alphaherpesvirus gD interferes with entry of homologous and heterologous alphaherpesviruses by blocking access to a shared gD receptor. Virology 268:147-158. [DOI] [PubMed] [Google Scholar]
- 20.Geraghty, R. J., C. Krummenacher, R. J. Eisenberg, G. H. Cohen, and P. G. Spear. 1998. Entry of alphaherpesviruses mediated by poliovirus receptor related protein 1 and poliovirus receptor. Science 280:1618-1620. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein, D. J., and S. K. Weller. 1988. Herpes simplex virus type 1-induced ribonucleotide reductase activity is dispensable for virus growth and DNA synthesis: isolation and characterization of an ICP6 lacZ insertion mutant. J. Virol. 62:196-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Handler, C. G., G. H. Cohen, and R. J. Eisenberg. 1996. Cross-linking of glycoprotein oligomers during herpes simplex virus type 1 entry. J. Virol. 70:6076-6082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Handler, C. G., R. J. Eisenberg, and G. H. Cohen. 1996. Oligomeric structure of glycoproteins in herpes simplex virus type 1. J. Virol. 70:6067-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsu, S., I. Solovyev, A. Colombero, R. Elliott, M. Kelley, and W. J. Boyle. 1997. ATAR, a novel tumor necrosis factor receptor family member, signals through TRAF2 and TRAF5. J. Biol. Chem. 272:13471-13474. [DOI] [PubMed] [Google Scholar]
- 25.Isola, V. J., R. J. Eisenberg, G. R. Siebert, C. J. Heilman, W. C. Wilcox, and G. H. Cohen. 1989. Fine mapping of antigenic site II of herpes simplex virus glycoprotein D. J. Virol. 63:2325-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krummenacher, C., I. Baribaud, M. Ponce de Leon, J. C. Whitbeck, H. Lou, G. H. Cohen, and R. J. Eisenberg. 2000. Localization of a binding site for herpes simplex virus glycoprotein D on the herpesvirus entry mediator C by with antireceptor monoclonal antibodies. J. Virol. 74:10863-10872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krummenacher, C., A. V. Nicola, J. C. Whitbeck, H. Lou, W. Hou, J. D. Lambris, R. J. Geraghty, P. G. Spear, G. H. Cohen, and R. J. Eisenberg. 1998. Herpes simplex virus glycoprotein D can bind to poliovirus receptor-related protein 1 or herpesvirus entry mediator, two structurally unrelated mediators of virus entry. J. Virol. 72:7064-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwon, B. S., K. B. Tan, J. Ni, K. O. Oh, Z. H. Lee, K. K. Kim, Y. J. Kim, S. Wang, R. Gentz, G.-L. Yu, J. Harrop, S. D. Lyn, C. Silverman, T. G. Porter, A. Truneh, and P. R. Young. 1997. A newly identified member of the tumor necrosis factor receptor superfamily with a wide tissue distribution and involvement in lymphocyte activation. J. Biol. Chem. 272:14272-14276. [DOI] [PubMed] [Google Scholar]
- 29.Lopez, M., F. Cocchi, L. Menotti, E. Avitabile, P. Dubreuil, and G. Campadelli-Fiume. 2000. Nectin2α (PRR2α or HveB) and nectin2δ are low-efficiency mediators for entry of herpes simplex virus mutants carrying the Leu25Pro substitution in glycoprotein D. J. Virol. 74:1267-1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller, C. G., C. Krummenacher, R. J. Eisenberg, G. H. Cohen, and N. W. Fraser. 2001. Development of a syngenic murine B16 cell line-derived melanoma susceptible to destruction by neuroattenuated HSV-1. Mol. Ther. 3:160-168. [DOI] [PubMed] [Google Scholar]
- 31.Milne, R. S. B., S. A. Connolly, C. Krummenacher, R. J. Eisenberg, and G. H. Cohen. 2001. Porcine HveC, a member of the highly conserved HveC/nectin 1 family, is a functional alphaherpesvirus receptor. Virology 281:315-328. [DOI] [PubMed] [Google Scholar]
- 32.Montgomery, R. I., M. S. Warner, B. J. Lum, and P. G. Spear. 1996. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell 87:427-436. [DOI] [PubMed] [Google Scholar]
- 33.Muggeridge, M. I. 2000. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J. Gen. Virol. 81:2017-2027. [DOI] [PubMed] [Google Scholar]
- 34.Naismith, J. H., T. Q. Devine, T. Kohno, and S. R. Sprang. 1996. Structures of the extracellular domain of the type I tumor necrosis factor receptor. Structure 4:1251-1262. [DOI] [PubMed] [Google Scholar]
- 35.Nicola, A. V., M. Ponce de Leon, R. Xu, W. Hou, J. C. Whitbeck, C. Krummenacher, R. I. Montgomery, P. G. Spear, R. J. Eisenberg, and G. H. Cohen. 1998. Monoclonal antibodies to distinct sites on the herpes simplex virus (HSV) glycoprotein D block HSV binding to HVEM. J. Virol. 72:3595-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Nicola, A. V., S. H. Willis, N. N. Naidoo, R. J. Eisenberg, and G. H. Cohen. 1996. Structure-function analysis of soluble forms of herpes simplex virus glycoprotein D. J. Virol. 70:3815-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng, T., M. Ponce de Leon, H. Jiang, G. Dubin, J. Lubinski, R. J. Eisenberg, and G. H. Cohen. 1998. The gH-gL complex of herpes simplex virus (HSV) stimulates neutralizing antibody and protects mice against HSV type 1 challenge. J. Virol. 72:65-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pertel, P. E., A. Fridberg, M. L. Parish, and P. G. Spear. 2001. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology 279:313-324. [DOI] [PubMed] [Google Scholar]
- 38.Rux, A. H., H. Lou, J. D. Lambris, H. M. Friedman, R. J. Eisenberg, and G. H. Cohen. 2002. Kinetic analysis of glycoprotein C of herpes simplex virus types 1 and 2 binding to heparin, heparan sulfate and complement component C3b. Virology 294:324-332. [DOI] [PubMed] [Google Scholar]
- 39.Rux, A. H., S. H. Willis, A. V. Nicola, W. Hou, C. Peng, H. Lou, G. H. Cohen, and R. J. Eisenberg. 1998. Functional region IV of glycoprotein D from herpes simplex virus modulates glycoprotein binding to the herpes virus entry mediator. J. Virol. 72:7091-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarrias, M. R., J. C. Whitbeck, I. Rooney, L. Spruce, B. K. Kay, R. I. Montgomery, P. G. Spear, C. F. Ware, R. J. Eisenberg, G. H. Cohen, and J. D. Lambris. 1999. Inhibition of herpes simplex virus glycoprotein gD and lymphotoxin-α binding to HveA by peptide antagonists. J. Virol. 73:5681-5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 42.Tal-Singer, R., C. Peng, M. Ponce de Leon, W. R. Abrams, B. W. Banfield, F. Tufaro, G. H. Cohen, and R. J. Eisenberg. 1995. Interaction of herpes simplex virus glycoprotein gC with mammalian cell surface molecules. J. Virol. 69:4471-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Terry-Allison, T., R. I. Montgomery, J. C. Whitbeck, R. Xu, G. H. Cohen, R. J. Eisenberg, and P. G. Spear. 1998. HveA (herpesvirus entry mediator A), a coreceptor for herpes simplex virus entry, also participates in virus-induced cell fusion. J. Virol. 72:5802-5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Turner, A., B. Bruun, T. Minson, and H. Browne. 1998. Glycoproteins gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a Cos cell transfection system. J. Virol. 72:873-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wells, J. A. 1991. Systematic mutational analyses of protein-protein interfaces. Methods Enzymol. 202:390-411. [DOI] [PubMed] [Google Scholar]
- 46.Whitbeck, J. C., S. A. Connolly, S. H. Willis, W. Hou, C. Krummenacher, M. Ponce de Leon, H. Lou, I. Baribaud, R. J. Eisenberg, and G. H. Cohen. 2001. Localization of the gD-binding region of the human herpes simplex virus receptor, HveA. J. Virol. 75:171-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Whitbeck, J. C., C. Peng, H. Lou, R. Xu, S. H. Willis, M. Ponce de Leon, T. Peng, A. V. Nicola, R. I. Montgomery, M. S. Warner, A. M. Soulika, L. A. Spruce, W. T. Moore, J. D. Lambris, P. G. Spear, G. H. Cohen, and R. J. Eisenberg. 1997. Glycoprotein D of herpes simplex virus (HSV) binds directly to HVEM, a member of the tumor necrosis factor receptor superfamily and a mediator of HSV entry. J. Virol. 71:6083-6093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willis, S. H., A. H. Rux, C. Peng, J. C. Whitbeck, A. V. Nicola, H. Lou, W. Hou, L. Salvador, G. H. Cohen, and R. J. Eisenberg. 1998. Examination of the kinetics of herpes simplex virus glycoprotein D binding to the herpesvirus entry mediator, with surface plasmon resonance. J. Virol. 72:5937-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]