Abstract
The Wnt signaling pathway plays an important role in neural cell development and function. The key components of this pathway, β-catenin and its partner TCF-4/LEF-1, exert their effects on transcription by entering the nuclei, where they associate with the TCF-4/LEF-1 DNA motif positioned in the promoters of several important genes. Here we examined the role of TCF-4 upon transcription of the human immunodeficiency virus type 1 (HIV-1) promoter in human astrocytic cells. Our results showed that expression of TCF-4 in human astrocytic cells (U-87MG cells) decreased the basal and Tat-mediated transcription of the HIV-1 long terminal repeat (LTR). Results from promoter deletion studies revealed that the promoter sequence of the LTR with no classical binding motif for TCF-4/LEF-1, which spans positions −80 to +80 of the LTR, remained responsive to down-regulation by TCF-4. Noticeably, removal of the sequences between positions −80 and −68 decreased the negative effect of TCF-4 on viral gene transcription. A mutant variant of TCF-4 with no binding site for β-catenin was able to down-regulate LTR transcription, suggesting that β-catenin may not be directly involved in the observed regulatory events. Results from the glutathione S-transferase pull-down assay as well as the combined immunoprecipitation and Western blot analysis of protein extract from U-87MG cells revealed an interaction of Tat with TCF-4. Subcellular examination of TCF-4 and Tat in cells expressing either protein alone showed a predominantly nuclear accumulation of these proteins. However, in cells which coexpressed both TCF-4 and Tat, significant levels of these proteins were found in the cytoplasm. All together, these observations provide evidence for the cooperative interaction of TCF-4, the important transcription factor of the Wnt pathway, with Tat; this interaction may determine the level of viral gene transcription in human astrocytic cells.
Control of human immunodeficiency virus type 1 (HIV-1) gene transcription requires the participation of a variety of cellular proteins which collaborate with viral regulatory factors such as Tat and Vpr (4, 6, 9, 12, 18, 22). Most, if not all, of the cellular regulatory proteins have the capacity to interact with the viral promoter sequence spanning the long terminal repeat (LTR). Further, the interaction of several proteins with each other but without direct binding to the LTR DNA sequence seems to be essential for optimizing viral gene transcription (15). Some of these proteins appear to be ubiquitously expressed in a broad range of cells, whereas others, such as NF-κB, are inducible and their activity is modulated by environmental stimuli (10, 21). During the last decade, our research has been centered on the modulation of LTR transcription in human astrocytic cells, with emphasis on the cooperative interaction between cellular factors and viral proteins. Recently, we paid attention to the Wnt signaling pathway, as several studies pointed to the involvement of this signaling pathway in a variety of biological events, including neural cell fate (1, 13). The most studied participant of this pathway, β-catenin, is usually found in the cytoplasm, where it can be phosphorylated by glycogen synthase kinase 3 and is subjected to proteolysis by ubiquitination (14). Stabilization of β-catenin via mutation in its phosphorylation sites, which are positioned in exon 3 (17), and/or via its association with other cytoplasmic proteins provides the opportunity for this key component of the Wnt pathway to associate with cytoplasmic transcription factors such as cytoplasmic transcription factor 4 (TCF-4), also called lymphoid enhancer binding factor 1 (LEF-1) (8, 13). The β-catenin:TCF-4/LEF-1 complex, in turn, enters the nuclei, where, by associating with the TCF-4/LEF-1 consensus DNA binding site, it stimulates transcription of the corresponding promoter(s) (2). In this study, we examined the effect of the Wnt pathway on the basal and Tat-induced transcription of HIV-1 and investigated Tat interaction with Wnt factors in human astrocytic cells.
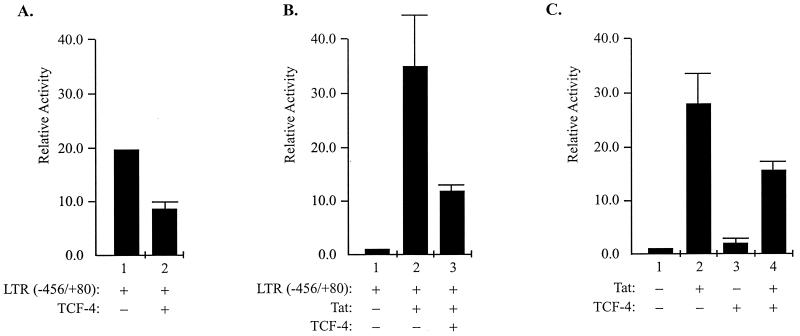
In the first series of experiments, cells from the U-87MG human astrocytic cell line were transfected with the LTR-chloramphenicol acetyltransferase (CAT) reporter plasmid in the absence and presence of plasmids expressing TCF-4. As shown in Fig. 1A, ectopic expression of TCF-4 decreased viral promoter activity (compare lane 1 to lane 2). A similar series of experiments was performed in the presence of Tat, the potent viral early transactivator whose expression is essential for the productive replication of HIV-1 in infected cells (16). As shown in Fig. 1B, the extent of Tat-mediated activation of LTR transcription was smaller in cells expressing TCF-4 (compare lanes 2 and 3). These observations are not restricted to U-87MG cells, as results from the transfection of primary cultures of human astrocytes revealed similar information (data not shown). Furthermore, overexpression of TCF-4 caused a noticeable decrease in the transcriptional activation by Tat of the integrated copy of the LTR-CAT sequence in the cellular gene (Fig. 1C). This observation suggests that TCF-4 is able to down-regulate transcription of the episomal and the integrated DNA containing the HIV-1 LTR sequence. To further investigate the effect of TCF-4 on the transcription of the HIV-1 promoter, we created and utilized various LTR mutants encompassing the DNA sequences between positions −117 and +80, −80 and +80, −68 and +80, and −117 and +3; all of these mutants respond, albeit to various degrees, to Tat activation in U-87MG cells (20). Results from transfection studies revealed that expression of TCF-4 diminished the level of Tat activation of the viral promoter containing the sequences spanning positions −117 to +80 and −80 to +80. Under similar conditions, TCF-4 showed a lesser effect on Tat-mediated LTR transcription, suggesting that the sequences between −80 and −68 may play an important role in the TCF-4-mediated suppression of Tat-induced LTR activation. The absence of a TCF/LEF binding site within this region of the LTR implies that TCF-mediated inactivation of the function of Tat on the LTR may be an indirect event which is independent from the association of TCF-4 with the DNA sequence.
FIG. 1.
Effect of TCF-4 on transcription of the HIV-1 LTR. (A) Human U-87MG astrocytic cells were transfected with 1 μg of a reporter plasmid containing the entire sequence from nucleotides −456 to +80 of the LTR (−456/+80) alone (bar 1) or together with 5 μg of TCF-4 expression plasmid (as described in reference 7). After 48 h, the level of CAT gene expression was determined by CAT enzymatic assay (7). Bar graphs depict the means of relative transcription activities ± standard deviations of triplicate determinations. Transcriptional activity was determined by the level of conversion of [14C]chloramphenicol to its acetylated forms as measured by scintillation counting of the cells on the thin-layer chromatography plates. The value obtained for cells transfected with LTR-CAT alone (bar 1) was used as a baseline and given an arbitrary value of 1. (B) Cells were transfected with the LTR (−456/+80) and/or TCF-4 as described for panel A with or without the addition of 0.5 μg of Tat expression plasmid as shown. The relative levels of transcription activity were determined as described for panel A. (C) HeLa cells containing integrated copies of LTR-CAT were transfected with plasmids expressing Tat and TCF-4 as described for panel A. In all transfections, the amount of DNA was kept constant (15 μg per 60-mm-diameter plate) by adding vector control plasmid DNA to the transfection mixture. Transfection efficiency was controlled by including a plasmid expressing green fluorescent protein and testing its expression in the transfected cells after 24 h.
As the region between −80 and −68 is composed of GC-rich sequences, one may envision a role for GC-rich binding proteins such as Sp1 in the observed regulatory event. In order to examine the importance of the Tat-responsive transactivation response (TAR) elements that are located between positions +1 and +80, we utilized an LTR promoter construct containing sequences between positions −117 and +3. As shown in Fig. 2D, the level of TAR-independent activation of the LTR by Tat in glial cells was noticeably decreased when cells were cotransfected with TCF.
FIG. 2.
Regulation of mutant LTR transcription by TCF-4. U-87MG cells were transfected with 1 μg of the reporter CAT DNA plasmid containing nucleotide sequences from positions −117 to +80 (A), −80 to +80 (B), −68 to +80 (C), and −117 to +3 (D). In cotransfection assays, 5 μg of each of the TCF-4 expressor plasmids was used. The levels of CAT activity were determined after 48 h, and the relative transcription activities of the reporter plasmids were identified and presented as described for Fig. 1.
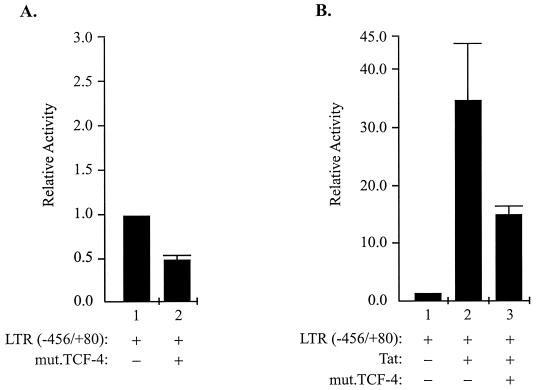
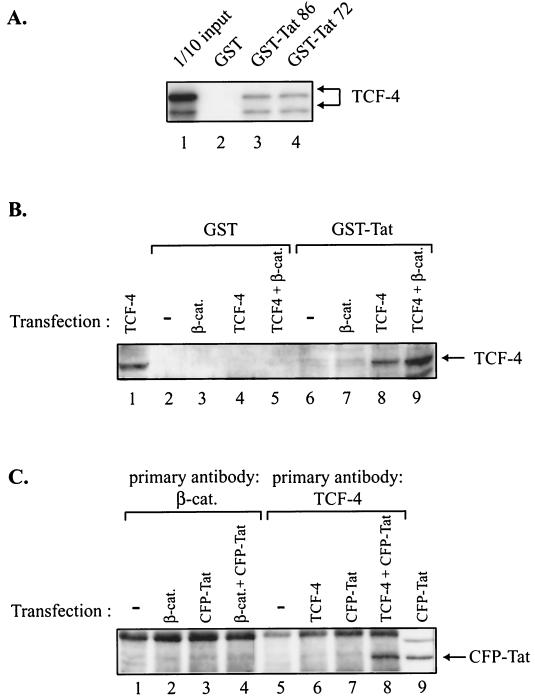
To determine whether the association of TCF-4 with its well-studied partner, β-catenin, is important in the regulation of the basal and Tat-mediated transcription of the LTR, a mutant TCF-4 with a deletion in the β-catenin binding site was utilized in transfection experiments. As shown in Fig. 3, mutant TCF-4 also showed a negative effect on the basal transcription of the LTR (Fig. 3A) and was able to suppress the level of LTR transcriptional activation by Tat (Fig. 3B) in U-87MG cells. These observations suggest that the association of TCF-4 with β-catenin may not be directly involved in the observed down-regulation of the LTR by TCF; the ability of TCF-4 to suppress Tat activity in the absence and presence of TAR raised the possibility that the interaction of TCF-4 with Tat may lead to the inactivation of this potent viral protein in the cells. To examine the association of Tat with TCF-4, in the first series of experiments, in vitro-synthesized TCF-4 was incubated with glutathione S-transferase (GST) or GST-Tat fusion protein coupled to glutathione-Sepharose beads. Two variants of the Tat protein, one encompassing amino acids 1 to 86 (GST-Tat 86) and the other encompassing amino acids 1 to 72 (GST-Tat 72), were utilized in this experiment. As shown in Fig. 4A, TCF-4 bound to both the GST-Tat 86 and GST-Tat 72 proteins (lanes 3 and 4, respectively) but not to the GST protein alone (lane 2). In the next series of experiments, we investigated the interaction of TCF-4 with Tat in cells expressing both TCF-4 and β-catenin. Protein extracts from U-87MG cells transfected with plasmids expressing TCF-4, β-catenin, and TCF-4 plus β-catenin were prepared and examined for their binding activity to GST-Tat or GST prior to Western blot analysis for detection of TCF-4. As shown in Fig. 4B, analysis of the protein that bound to GST-Tat showed the detection of a band corresponding to TCF-4 in protein extracts obtained from cells transfected with TCF-4 and TCF-4 plus β-catenin (lanes 8 and 9, respectively). The absence of this protein band in protein fractions from GST showed that the association of TCF-4 with GST-Tat is a rather specific event. Interestingly, the appearance of the protein band in lane 9 indicates that the interaction of TCF-4 and Tat can occur in the presence of its partner, β-catenin.
FIG. 3.
Effect of mutant TCF-4 on LTR transcription. Transfection of U-87MG cells was performed as described for Fig. 1, except that 10 μg of a mutant TCF-4 (mut.TCF-4) lacking the β-catenin binding site (described in reference 23) was utilized. All values were determined as described for Fig. 1.
FIG. 4.
Interaction of Tat with Wnt TCF-4. (A) In vitro-translated, 35S-labeled TCF-4 was incubated with GST (lane 2), GST-Tat 86 (lane 3), and GST-Tat 72 (lane 4). After incubation for 1 h at 4°C, the resins were pelleted and washed with binding buffer as described previously (5). Bound proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by autoradiography. Lane 1 shows an amount equivalent to 10% of the material used for the binding assay. (B) U-87MG cells were transfected with plasmids expressing β-catenin (β-cat.) (lanes 3 and 7), TCF-4 (lanes 4 and 8), or TCF-4 plus β-catenin (lanes 5 and 9), and after 48 h, protein extracts from transfected and untransfected control cells (lanes 2 and 6) were prepared and incubated with GST or GST-Tat 86; the bound proteins were then analyzed by Western blotting by using anti-TCF-4 antibody according to the procedure described in our previous report (5). Lane 1 shows Western blot analysis of 30 μg of protein extract from cells transfected with TCF-4. (C) Approximately 300 μg of protein extract from U-87MG cells transfected with 5 μg of plasmid DNA expressing β-catenin (lane 2), CFP-Tat (lanes 3 and 7), β-catenin plus CFP-Tat (lane 4), or TCF-4 plus CFP-Tat (lane 8) was incubated with anti-β-catenin antibody or anti-TCF-4 antibody as indicated in the figure. The protein complex was analyzed by Western blotting with anti-Tat antibody. Lane 9 shows Western blot analysis of 30 μg of total protein extract from cells transfected with CFP-Tat. The position of CFP-Tat is shown by an arrow.
To further investigate the in vivo interaction of Tat with TCF-4, cells were transfected with various plasmids expressing β-catenin, TCF-4, and Tat in fusion with cyan fluorescent protein (CFP). After 48 h, protein extracts were prepared and reacted with either anti-β-catenin or anti-TCF-4 antibodies. The immunocomplexes were subsequently analyzed by Western blotting with anti-Tat antibody to detect CFP-Tat. As shown in Fig. 4C, no signal corresponding to CFP-Tat was detected in immunocomplexes obtained upon incubation of protein extracts from cells expressing β-catenin (lane 2), CFP-Tat (lane 3), and β-catenin plus CFP-Tat (lane 4) with anti-β-catenin antibody. Further, immunocomplexes obtained by incubation of anti-TCF-4 antibody with cell extracts transfected with TCF-4 and CFP-Tat showed no evidence for the presence of CFP-Tat in the complex (lanes 6 and 7). However, the immunocomplex which was pulled down by anti-TCF-4 antibody from extracts derived from cells expressing TCF-4 plus CFP-Tat showed a strong band corresponding to CFP-Tat, pointing to the interaction of TCF-4 with CFP-Tat in these cells (lane 8). This interaction is specific to Tat, as CFP showed no ability to bind to TCF-4 under similar conditions and the normal control serum failed to pull down any specific complexes that contained CFP-Tat (data not shown). Also, our recent preliminary observation from analysis of protein extracts from HIV-1-infected cells indicated that the immunocomplex that was pulled down by the anti-TCF-4 antibody contains the HIV-1 Tat protein, again suggesting the association of Tat and TCF-4 during the infection cycle (B. Sawaya, data not shown). All together, these studies established the interaction of Tat with TCF-4, both in vitro and in protein extracts from human astrocytic cells.
As stated earlier, TCF-4 is a transcription factor which enters the nuclei, where it exerts its regulatory action on the transcription of responsive genes. Ectopic expression of TCF-4 led to rapid nuclear accumulation of this protein in transfected cells. To investigate the subcellular localization of TCF-4 in cells expressing Tat, a plasmid expressing yellow fluorescent protein (YFP)-Tat was introduced with a plasmid expressing TCF-4 in fusion with CFP, and after 24 h, subcellular locations of both proteins were assessed. As shown in Fig. 5A and B, transfection of cells with CFP-TCF-4 or YFP-Tat alone led to the nuclear appearance of both proteins. However, in cells transfected with both proteins, some levels of CFP-TCF-4 (Fig. 5C) and YFP-Tat (Fig. 5D) were detected in the cytoplasm of the cells as well. These observations suggest that the interaction of TCF-4 and Tat may affect their subcellular localization and that both proteins are retained in the cytoplasm of cells. The changes in the compartmentalization of Tat by TCF-4 may contribute to the ability of Tat to stimulate LTR transcription. Also, the association of Tat with TCF-4 in nuclei may prevent full activation of the viral promoter by Tat.
FIG. 5.
Subcellular detection of Tat and TCF-4 in human astrocytic cells. U-87MG cells were transfected with plasmids expressing CFP-TCF-4 (A), YFP-Tat (B), or both (C and D). After 24 h, expression of CFP-TCF-4 and YFP-Tat was observed by fluorescence microscopy using specific filters set to allow detection of the fusion proteins. The IP Lab program (Scanalytics, Inc.) was utilized to overlay and pseudocolor CFP-TCF-4 (blue) and YFP-Tat (green). Magnification, ×400.
TCF-4 is a member of the high-mobility group (HMG) box-containing transcription factor family, whose members were originally identified by their ability to bind the T-cell receptor enhancer (3). Expression of TCF-4 is not restricted to lymphoid tissue, and several studies suggested its involvement in a variety of developmental processes, including neurogenesis (11). Here, we focused our attention on regulation of HIV-1 LTR transcription by Wnt factors, including β-catenin and TCF-4, in human astrocytic cells. Our results demonstrate that ectopic expression of TCF-4 in astrocytes suppresses the basal transcriptional activity of the HIV-1 LTR promoter. This finding is in contrast to earlier reports on the activation of the LTR by LEF-1, another member of the HMG protein family, in T cells (19). The differences may be attributed to diverse functions of the various members of HMG proteins and/or the cell types which were utilized in this study. As pointed out earlier, TCF-4 was able to decrease transcription of the LTR in primary astrocytes as well as human astrocytic cells transformed with simian virus 40 T antigen, verifying that the observed event is not restricted to U-87MG human astrocytic cells. Furthermore, we demonstrated that TCF-4 has the capacity to diminish Tat activation of LTR transcription independently from the TCF-4 binding sites within the LTR, most likely by associating with Tat protein. In support of this notion, biochemical examination of the interaction between TCF-4 and Tat protein verified their ability to form a stable complex, which was detected by gel electrophoresis. Another interesting observation which allowed us to understand the mechanism of the TCF-4 inhibitory effect emerged from subcellular localization of TCF-4 and Tat in human astrocytes which were ectopically transfected with plasmids expressing TCF-4 and YFP-Tat. It was evident that while TCF-4 and Tat alone are localized to the nuclei of cells, once they are coexpressed, both proteins can also be found in the cytoplasm, suggesting that the TCF-4:Tat complex is transported from the nucleus to the cytoplasm and/or that formation of the complex in the cytoplasm after its synthesis affects its nuclear import. Our future studies are aimed at unraveling the mechanism of LTR suppression through TCF-4:Tat association and the implication of this event in the poor expression of the HIV-1 genome and in viral replication in astrocytes.
Acknowledgments
We thank Osamu Tetsu, F. McGormick, B. Kinzler, and A. Levine for providing various plasmids. We also thank past and present members of the Center for Neurovirology and Cancer Biology for their insightful discussion and sharing of ideas and reagents. We thank C. Schriver for editorial assistance and preparation of the manuscript.
This work was made possible by grants awarded by the NIH to K.K., B.E.S., and S.A.
REFERENCES
- 1.Brault, V., R. Moore, S. Kutsch, M. Ishibashi, D. H. Rowitch, A. P. McMahon, L. Sommer, O. Boussadia, and R. Kemler. 2001. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 128:1253-1264. [DOI] [PubMed] [Google Scholar]
- 2.Cho, E. A., and G. R. Dressler. 1998. TCF-4 binds beta-catenin and is expressed in distinct regions of the embryonic brain and limbs. Mech. Dev. 77:9-18. [DOI] [PubMed] [Google Scholar]
- 3.Clevers, H., and M. van de Wetering. 1997. TCF/LEF factor earn their wings. Trends Genet. 13:485-489. [DOI] [PubMed] [Google Scholar]
- 4.Cujec, T. P., H. Okamoto, K. Fujinaga, J. Meyer, H. Chamberlin, D. O. Morgan, and B. M. Peterlin. 1997. The HIV transactivator TAT binds to the CDK-activating kinase and activates the phosphorylation of the carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:2645-2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gallia, G. L., M. Safak, and K. Khalili. 1998. Interaction of the single-stranded DNA-binding protein Purα with the human polyomavirus JC virus early protein T-antigen. J. Biol. Chem. 273:32662-32669. [DOI] [PubMed] [Google Scholar]
- 6.Gallia, G. L., N. Darbinian, A. Tretiakova, S. Ansari, J. Rappaport, J. Brady, M. J. Wortman, E. M. Johnson, and K. Khalili. 1999. Association of HIV-1 Tat with the cellular protein, Purα, is mediated by RNA. Proc. Natl. Acad. Sci. USA 96:11572-11577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorman, C. M., L. F. Moffat, and B. H. Howard. 1982. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol. Cell. Biol. 2:1044-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hovanes, K., T. W. Li, J. E. Munguia, T. Truong, T. Milovanovic, T. Lawrence, J. Marsh, R. F. Holcombe, and M. L. Waterman. 2001. Beta-catenin-sensitive isoforms of lymphoid enhancer factor-1 are selectively expressed in colon cancer. Nat. Genet. 28:53-57. [DOI] [PubMed] [Google Scholar]
- 9.Jacotot, E., K. F. Ferri, C. El Hamel, C. Brenner, S. Druillennec, J. Hoebeke, P. Rustin, D. Metivier, C. Lenoir, M. Geuskens, H. L. Vieira, M. Loeffler, A. S. Belzacq, J. P. Briand, N. Zamzami, L. Edelman, Z. H. Xie, J. C. Reed, B. P. Roques, and G. Kroemer. 2001. Control of mitochondrial membrane permeabilization by adenine nucleotide translocator interacting with HIV-1 viral protein rR and Bcl-2. J. Exp. Med. 193:509-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar, S., M. J. Orsini, J. C. Lee, P. C. McDonnell, C. Debouck, and P. R. Young. 1996. Activation of the HIV-1 long terminal repeat by cytokines and environmental stress requires an active CSBP/p38 MAP kinase. J. Biol. Chem. 271:30864-30869. [DOI] [PubMed] [Google Scholar]
- 11.Lee, Y. J., B. Swencki, S. Shoichet, and R. A. Shivdasani. 1999. A possible role for the high mobility group box transcription factor Tcf-4 in vertebrate gut epithelial cell differentiation. J. Biol. Chem. 274:1566-1572. [DOI] [PubMed] [Google Scholar]
- 12.Mansky, L. M., S. Preveral, E. Le Rouzic, L. C. Bernard, L. Selig, C. Depienne, R. Benarous, and S. Benichou. 2001. Interaction of human immunodeficiency virus type 1 Vpr with the HHR23A DNA repair protein does not correlate with multiple biological functions of Vpr. Virology 282:176-185. [DOI] [PubMed] [Google Scholar]
- 13.Miller, J. R., and R. T. Moon. 1996. Signal transduction through beta-catenin and specification of cell fate during embryogenesis. Genes Dev. 10:2527-2539. [DOI] [PubMed] [Google Scholar]
- 14.Novak, A., and S. Dedhar. 1999. Signaling through beta-catenin and Lef/Tcf. Cell. Mol. Life Sci. 56:523-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pereira, L. A., K. Bentley, A. Peeters, M. J. Churchill, and N. J. Deacon. 2000. A compilation of cellular transcription factor interactions with the HIV-1 LTR promoter. Nucleic Acids Res. 28:663-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Re, M. C., D. Gibellini, F. Vitone, and M. La Placa. 2001. Antibody to HIV-1 Tat protein, a key molecule in HIV-1 pathogenesis. New Microbiol. 24:197-205. [PubMed] [Google Scholar]
- 17.Rimm, D. L., K. Caca, G. Hu, F. B. Harrison, and E. R. Fearon. 1999. Frequent nuclear/cytoplasmic localization of beta-catenin without exon 3 mutations in malignant melanoma. Am. J. Pathol. 154:325-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sawaya, B. E., K. Khalili, J. Gordon, R. Taube, and S. Amini. 2000. Cooperative interaction between HIV-1 regulatory proteins Tat and Vpr modulates transcription of the viral genome. J. Biol. Chem. 275:35209-35214. [DOI] [PubMed] [Google Scholar]
- 19.Sheridan, P. L., C. T. Sheline, K. Cannon, M. L. Voz, M. J. Pazin, J. T. Kadonaga, and K. A. Jones. 1995. Activation of the HIV-1 enhancer by the LEF-1 HMG protein on nucleosome-assembled DNA in vitro. Genes Dev. 9:2090-2104. [DOI] [PubMed] [Google Scholar]
- 20.Taylor, J. P., R. Pomerantz, O. Bagasra, M. Chowdhury, J. Rappaport, K. Khalili, and S. Amini. 1992. TAR-independent transactivation by Tat in cells derived from the CNS: a novel mechanism of HIV-1 gene regulation. EMBO J. 11:3395-3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor, J. P., R. J. Pomerantz, G. V. Raj, F. Kashanchi, J. N. Brady, S. Amini, and K. Khalili. 1994. Central nervous system-derived cells express a κB-binding activity that enhances human immunodeficiency virus type 1 transcription in vitro and facilitates TAR-independent transactivation by Tat. J. Virol. 68:3971-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao, H., Y. Tao, J. Greenblatt, and R. G. Roeder. 1998. A cofactor, TIP30, specifically enhances HIV-1 Tat-activated transcription. Proc. Natl. Acad. Sci. USA 95:2146-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu, L., R. B. Corcoran, J. W. Welsh, D. Pennica, and A. J. Levine. 2000. WISP-1 is a Wnt-1- and beta-catenin-responsive oncogene. Genes Dev. 14:585-595. [PMC free article] [PubMed] [Google Scholar]