Abstract
Numerous mutations are found in subacute sclerosing panencephalitis (SSPE) viruses, and the M gene is the gene most commonly affected. In some SSPE viruses, such as the MF, Osaka-1, Osaka-2, and Yamagata-1 strains, translation of the M protein is complicated by a transcriptional defect that leads to an almost exclusive synthesis of dicistronic P-M mRNA. To understand the molecular mechanisms of this defect, we sequenced the P gene at the P-M gene junction for several virus strains and probed the involvement of several mutations in the readthrough region via their expression in measles virus minigenomes containing different sequences of the P-M gene junction and flanking reporter genes. The deletion of a single U residue in the U tract of the Osaka-1 strain (3′-UAAUAUUUUU-5′) compared with the consensus sequence resulted in a marked reduction of the expression of the downstream reporter gene. In addition, the expression of the downstream gene was markedly decreased by (i) the substitution of a C residue in the U tract of the P gene end of the OSA-2/Fr/B strain of the Osaka-2 virus (3′-UGAUAUUCUU-5′ compared with the sequence 3′-UGAUAUUUUU-5′ from a sibling virus of the same strain, OSA-2/Fr/V), and (ii) the substitution of a G in the sequence of the P gene end of the Yamagata-1 strain at a variable site immediately upstream from the six-U tract (3′-UGAUGUUUUUU-5′ instead of 3′-UGAUUUUUUUU-5′). Mutations at the P gene end can account for the readthrough transcription variation at the P-M gene junction, which directly affects M protein expression.
Measles virus (MV) is a member of Paramyxoviridae, and its genome is a nonsegmented single-stranded RNA of negative polarity. The MV genome contains N, P/C/V, M, F, H, and L genes, with the L gene encoding an RNA-dependent RNA polymerase (8). The viral RNA polymerase synthesizes the antigenome, which serves as a template for a precise copy of the genomic RNA. Also, each gene is transcribed sequentially from the 3′ end of the genome by the same RNA polymerase (24). One of the characteristics of the MV genome is that the conserved transcriptional control regions are at the gene boundaries. Each gene is separated by an intergenic region of three nucleotides that is not copied into mRNA (17). Usually, most mRNAs are monocistronic RNA because the transcription terminates before the intergenic region and only relatively small amounts of dicistronic readthrough products are synthesized. On the other hand, the viral RNA polymerase must read through the intergenic region in order to synthesize antigenomic RNA. Encapsidation of newly synthesized RNA from the 3′ end of the genome is thought to trigger antigenome synthesis (24). The encapsidation proceeds to the 5′ end of the genome and includes each junction. Because mRNA is not encapsidated by the N protein, the mechanisms of readthrough at the gene junction for the replication and transcription modes are apparently different.
Subacute sclerosing panencephalitis (SSPE) is a fatal degenerative disease caused by persistent MV infection of the central nervous system. The molecular mechanisms of cell-to-cell spread of the virus in the patient brain are unclear. Some mutations in the viral envelope protein genes appear to have been selected for virus survival in specific environments. The M gene of SSPE virus seems particularly vulnerable to mutation, and its expression is restricted (2, 3, 13, 14, 15, 16, 35). For instance, the M protein of the Biken strain is extremely unstable and becomes antigenically altered because of the high frequency of mutations that occur throughout the M gene (2). The IP-3-Ca strain also contains a posttranslational defect that affects M protein stability (13, 35). The M gene protein-coding frames of viruses isolated from SSPE case K and from the Niigata-1 strain were found to be interrupted by the mutational creation of an in-frame termination codon (3, 15), and, in the Yamagata-1 strain, another mutation was found to have destroyed the normal codon for translation initiation (40). Furthermore, transcriptional alteration of the M gene is found in some SSPE viruses (13, 15). Readthrough at the P-M gene junction was directly detected in the RNA derived from the brain of SSPE case K (15). A similar transcriptional alteration at the P-M gene junction of the MF strain of SSPE virus also has been reported (12). The P-M dicistronic RNA product has also been detected in cells infected with the Osaka-1 strain or with one of the sibling viruses of the Osaka-2 strain (1, 34). In addition, the transcriptional patterns for the P and M genes differed from lot to lot of the Yamagata-1 strain (our unpublished observation). Since the second cistron of the dicistronic mRNA is not generally translatable in infected cells—a conclusion that has been demonstrated specifically in experiments using P-M dicistronic constructs (39)—readthrough at the P-M gene junction may be the primary explanation for observations of defective M protein expression. However, the reasons for readthrough at the P-M gene junction have not been clarified (12, 15). In other viruses with genome structures similar to that of MV, such as vesicular stomatitis virus, respiratory syncytial virus, simian virus 5, Sendai virus, and human parainfluenza virus type 1, sequence alterations at the gene junction have been found to be responsible for readthrough at the junction (5, 9, 20, 23, 28, 30, 31). In this study, we have identified the mutations occurring in the end of the P gene in three strains of SSPE virus that may have affected readthrough at the P-M gene junction.
The nucleotide sequence of the P gene end varies among strains.
We have previously documented readthrough at the P-M gene junction in cells infected with the Osaka-1 strain or with one of the sibling viruses (OSA-2/Fr/B) of the Osaka-2 strain (1, 34). We have also noticed differences in the pattern of M gene transcription among stocks of the Yamagata-1 strain. The Yamagata-1 strain passaged in the IMR-32 neuroblastoma cell line had been previously reported to be defective in M gene expression, due in part to readthrough at the P-M gene junction (40). The Yamagata-1 strain passaged in Vero cells, however, was not transcriptionally defective (data not shown). Subsequently, we discovered that there were genetic variants in the original stocks in Vero cells and that the observed transcriptional difference was due to variation among lots of Vero cells rather than to passaging in IMR-32 cells (unpublished observation).
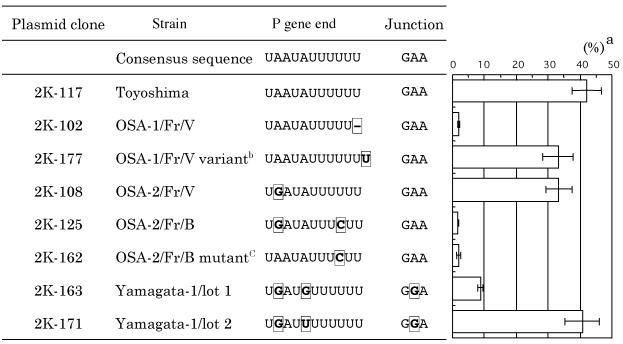
To discover the relationship between nucleotide sequence and readthrough at the P-M gene junction, the region containing the junction was amplified by reverse transcription-PCR (RT-PCR) from the total RNA of cells infected with each virus and the sequences were compared. Methods for total cellular RNA preparation from cells infected with virus strains (19, 26) and reverse transcription primed with a random primer (nonadeoxynucleotide mixture; Takara Biomedicals) have been described previously (4). The region containing the P-M gene junction was amplified by PCR with a primer set (MVP-m1, 5′-CCTGCATCACGCAGTGTAATCC-3′; MVM-g5, 5′-CCAGTTTTTCATTGAGCCCTGC-3′) based on the published sequence for the Edmonston strain of MV. Amplification involved 40 cycles of 1 min at 95°C, 1 min at 50°C, and 3 min at 72°C, with Pfu DNA polymerase used as recommended by the manufacturer (Stratagene). Amplified products were purified as previously described (4), and sequences were determined directly with a Thermosequenase II dye terminator cycle sequencing kit (Amersham Pharmacia Biotech) and a model 373S sequencer (Applied Biosystems). The sequences of the transcriptional start region and the intergenic sequence at the P-M gene junction were well conserved among strains (Table 1). The only exception was the SSPE virus strain Yamagata-1, which contained an A-to-G substitution in the intergenic sequence (Table 1; Yamagata-1 lots 1 and 2). In contrast, the end of the P gene varied among strains. The Toyoshima strain is a laboratory strain of MV containing the P gene end sequence (genome sense) 3′-UAAUAUUUUUU-5′. This sequence is identical to that of the Edmonston and is conserved among MV strains, including recent field isolates. The P gene end of the Osaka-1 strain, however, contained the sequence 3′-UAAUAUUUUU-5′, in which a single U residue in the U tract was deleted. Variation in the Osaka-1 strain was explored further by sequencing 50 randomly selected plasmids following the cloning of the PCR products into the pBluescript II KS(−) plasmid (Table 2). The sequences of the inserts (550 bp excluding the primer regions) of most clones were identical to the sequence determined by direct sequencing except for 12 sporadic substitutions found in nine clones, which may be attributable in part to polymerase error during RT-PCR. For comparison, the region containing the N-P gene junction, which was chosen because it closely resembled the P-M gene junction, was similarly amplified by PCR from the same stock of RNA with a different primer set (MVN-m7, 5′-GAGAGGCCGAGGACCAGAACAA-3′; MVP-g5, 5′-GAGTCAGCATCTTGGATTCC-3′). The products (500 bp) were cloned into the same plasmid vector, and 50 randomly selected plasmids were sequenced. Eleven sporadic nucleotide substitutions were found in 11 clones. This number of substitutions was comparable to the numbers of substitutions in the 50 plasmids containing the P-M gene junction described above. However, no sequence variation in the N gene end of the Osaka-1 strain was found among 50 clones, and all the clones contained the six-U tract (Table 2). In contrast, sequence variation occurring in the P gene end of the Osaka-1 strain was apparently frequent and was considered significant. Three variations were found: 45 clones possessed the major sequence consisting of a five-U tract, two clones contained the six-U tract of the wild-type sequence, and the sequences of three clones possessed a seven-U tract. These variant sequences were not apparent from the direct sequencing of PCR products.
TABLE 1.
Nucleotide sequences of P gene ends of several MV strains
| Strain | Readthroughc | Sequenced of:
|
||
|---|---|---|---|---|
| P gene end | Junction | M gene start | ||
| Toyoshima | +/− | UAAUAUUUUUU | GAA | UCCUCGUUUC |
| OSA-1/Fr/V | +++ | UAAUAUUUUU- | GAA | UCCUCGUUUC |
| OSA-2/Fr/V | +/− | UGAUAUUUUUU | GAA | UCCUCGUUUC |
| OSA-2/Fr/B | +++ | UGAUAUUUCUU | GAA | UCCUCGUUUC |
| OSA-3/Bs/V | +/− | UAAUAUUUUUU | GAA | UCCUCGUUUC |
| Yamagata-1 lot 1 | +++ | UGAUGUUUUUU | GGA | UCCUCGUUUC |
| Yamagata-1 lot 2 | +/− | UGAUUUUUUUU | GGA | UCCUCGUUUC |
| MFa | +++ | UAAUAUUUUU- | GAA | UCCUCGUUUC |
| Case Kb | +++ | UAAUAUUUUU- | GAA | UCCUCGUUUC |
TABLE 2.
Sequence variations found in the P gene end of the Osaka-1 strain
| Gene end | Sequence | No. of clones/total no. sequenced |
|---|---|---|
| P | UAAUAUUUUU | 45/50 |
| P | UAAUAUUUUUU | 2/50 |
| P | UAAUAUUUUUUU | 3/50 |
| N | CAAUAUUUUUU | 50/50 |
Both Osaka-2 strains (OSA-2/Fr/V and OSA-2/Fr/B) had an A-to-G substitution in the P gene end, and the latter sibling virus possessed an additional U-to-C substitution in the U tract (Table 1). The sequence of the Osaka-3 strain was identical to that of the Toyoshima strain. Two lots of stock for the Yamagata-1 strain were checked. The sequence of the Yamagata-1 strain lot 1 was identical to that previously reported. The Yamagata-1 lot 1 strain had the same A-to-G substitution as OSA-2/Fr/V plus an additional A-to-G substitution just before the U tract, and this sequence is identical to the sequence of the Yamagata-1 strain described in a previous report (22). The Yamagata-1 lot 2 strain possessed the same substitution shared by OSA-2/Fr/V and the Yamagata-1 lot 1 strain but carried a different substitution (A to U) at the position before the U tract (Table 1). Hypothesizing that some of these substitutions might contribute to the readthrough at the P-M gene junction, we applied the minireplicon system to determine the significance of the individual substitutions in the P gene ends of these strains.
Mutations responsible for reduced expression of the downstream gene at the P-M gene junction.
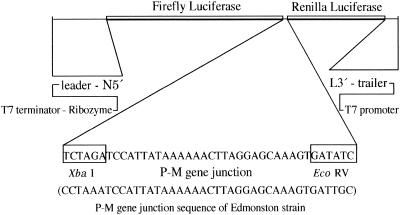
Minigenomes of MV containing different sequences at the P-M gene junction were constructed to investigate the mutations responsible for upstream and downstream gene expression at the junction (Fig. 1). The minigenome was designed essentially as described by Sidhu et al. (36). The basic minigenome cDNA of 1,152 nucleotides was constructed as follows. The cDNA contained the 107 3′-terminal nucleotides corresponding to the leader sequence of the MV genomic RNA and the 109 5′-terminal nucleotides corresponding to its trailer sequence. These MV terminal sequences flanked a Renilla reniformis luciferase open reading frame of 936 nucleotides (Promega) (25). The positions of the T7 RNA polymerase promoter and hepatitis delta virus ribozyme (33) were designed so that the transcribed RNA created the specific 3′ and 5′ termini of the MV genome. Two T7 terminator sequences were aligned downstream from the ribozyme sequence to stop RNA transcription by the T7 RNA polymerase. The constructed cDNA was subcloned into the pUC118 plasmid, and the resultant plasmid was designated pMGKLuc01. Primer pairs for inserting plasmid 2K-177 (Minig-1, 5′-CGATCCTAAGGACCACCATGGTCGACTCTAGATCCATTATAA-3′; Minig-2, 5′-ATCACTTTGCTCCTAAGTTTTTTATAATGGATCTAGAGTCGA-3′) were hybridized and treated with DNA polymerase Klenow fragments to make the insert. The resulting fragment was inserted into the EcoRV site of plasmid pMGKLuc01 to construct the precursor plasmid pMGKLuc01A. The firefly luciferase gene—the NcoI-XbaI fragment of the pGL3-Control Vector plasmid (Promega)—was then inserted into pMGKLuc01A. The two reporter genes (the firefly and the Renilla luciferase genes) were separated by the insertion of the P-M gene junction sequence between the XbaI and EcoRV restriction sites (Fig. 1). Therefore, we could quantitatively analyze the extent of the readthrough by the polymerase complex at the P-M gene junction in the same background by determining the ratio between the levels of firefly and Renilla luciferase activity. Mutation-containing fragments (see Fig. 3) were prepared similarly to the fragment prepared with the Minig-1 and Minig-2 primer pair and inserted between the XbaI and EcoRV sites. The primer pairs prepared for the insertion of these mutant plasmids were as follows: 2K-102, Minig-3 (5′-GACTCTAGATCCATTATAAAAACTTAG-3′) and Minig-4 (5′-ATCACTTTGCTCCTAAGTTTTT-3′); 2K-177, Minig-14 (5′-GACTCTAGACCATTATAAAAAAACTTA-3′) and Minig-15 (5′-ATCACTTTGCTCCTAAGTTTTTTTATA-3′); 2K-108, Minig-7 (5′-GACTCTAGATCCACTATAAAAAACTTA-3′) and Minig-8 (5′-ATCACTTTGCTCCTAAGTTTTTTATAG-3′); 2K-125, Minig-5 (5′-GACTCTAGATCCACTATAAAGAACTTA-3′) and Minig-6 (5′-ATCACTTTGCTCCTAAGTTCTTTATA-3′); 2K-162, Minig-9 (5′-GACTCTAGATCCATTATAAAGAACTTA-3′) and Minig-6 (5′-ATCACTTTGCTCCTAAGTTCTTTATA-3′); 2K-163, Minig-10 (5′-GACTCTAGATCCACTACAAAAAACCTA-3′) and Minig-11 (5′-ATCACTTTGCTCCTAGGTTTTTTGTG-3′); 2K-171, Minig-12 (5′-GACTCTAGATCCACTAAAAAAAACCTA-3′) and Minig-13 (5′-ATCACTTTGCTCCTAGGTTTTTTTTAG-3′). To satisfy the “rule of six” criterion (10), the total length of the minigenomic RNA to be transcribed was adjusted to a multiple of six by treatment with Klenow DNA polymerase after digestion with the restriction enzymes ClaI (for plasmids 2K117, 2K-177, 2K-108, 2K-125, 2K-162, 2K-163, 2K-171) and Bsu36I (for 2K-102). These restriction sites were located upstream of the firefly luciferase gene. To provide the MV N, P, and L proteins to cells under control of the T7 polymerase, plasmids expressing these proteins were constructed. The N, P, and L cDNAs derived from the Edmonston strain were subcloned into the pCITE-K plasmid, which was placed downstream of the T7 promoter. The plasmids carrying the genes originating from the Edmonston strain were designated pCIN001 (N gene), pCIP001 (P gene), and pCIL001 (L gene). The negative-sense minigenome RNA was synthesized from each of the HindIII-digested plasmids by in vitro transcription with Ampliscribe T7 transcription kits (Epicenter Technologies). The synthesized RNA was treated with DNase I to remove any template DNA and extracted with phenol-chloroform, followed by chloroform treatment and precipitation with ethanol. RNA was resuspended in RNase-free water, and the concentration was estimated. A subconfluent HeLa cell monolayer prepared in 12-well plates (about 1.5 × 105 cells/well) was infected with T7 RNA polymerase-expressing vaccinia virus vTF7-3 at a multiplicity of infection of 2 to 3 (18). After adsorption for 1 h, the cells were washed with 0.5 ml of Opti-MEM I (GIBCO BRL) and then transfected with 0.5 μg of synthesized negative-sense minigenome RNA and 0.5 μg of pCIN001, 0.25 μg of pCIP001, and 0.1 μg of pCIL001 by using DMRIE-C reagent (GIBCO BRL). After incubation at 32.5°C for 3 h in a 5% CO2 incubator, the medium containing the transfection reagent, minigenome RNA, and helper plasmids was replaced with fresh medium containing 5% fetal calf serum and cytosine arabinofuranoside (40 μg/ml). After a further incubation at 32.5°C for 40 h in 5% CO2, the transfected cells were harvested and used for RNA analysis and luciferase assay.
FIG. 1.
Structure of the minigenome. The P-M gene junction sequence of the Edmonston MV was mutagenized to reproduce the sequences of OSA-1/Fr/V, OSA-2/Fr/V, OSA-2/Fr/B, OSA-3/Bs/V, and the two Yamagata-1 strains.
FIG. 3.
Transcriptional termination efficiency of different P-M gene junction sequences inserted between the firefly and Renilla luciferase genes. Efficiency (superscript a) is expressed as the percentage (mean of four experiments [plusmn] standard deviation) of Renilla luciferase activity relative to that of firefly luciferase. The plasmid clone and the corresponding sequences of the P gene end and P-M gene junction are presented at the left of each bar. Superscript b, clone of a very rare variant found in the OSA-1/Fr/V virus; superscript C, clone containing a single nucleotide back-mutation from clone 2K-125. Nucleotides different from the consensus sequence are boxed.
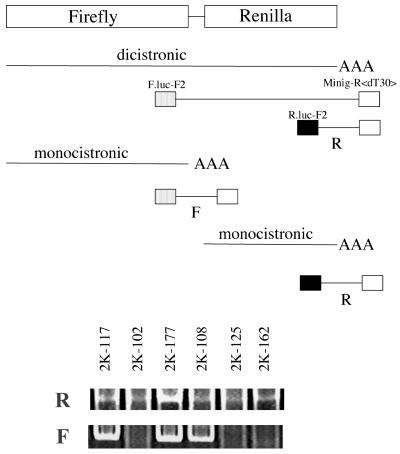
To show monocistronic and dicistronic RNA products expressed by our minigenome system, we first attempted to detect these RNA products by Northern blot analysis of total RNA obtained from cells expressing each construct. However, we could not obtain results satisfactory for distinguishing specific bands. In a previous report of Reutter et al. in which the MV minigenome system was used, mRNA synthesis of a reporter gene was not detected by Northern blot and primer extension analyses (32), and this might be a general problem for this MV minigenome system (36). Then we applied PCR technology to detect the differences among the RNA species expressed from different minigenome constructs. Polyadenylated RNA was selected by the Oligotex-dT30 <super> mRNA purification kit (Takara Biomedicals). The resulting RNA was converted to cDNA by primer Minig-R<T30> (5′-CTGCAGTGAATTCAGGATCCTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT-3′). The 3′-end regions of the firefly and Renilla luciferase genes were amplified by PCR with the same primer and the gene-specific primers F.luc-F2, 5′-ACGAAGTACCGAAAGGTCTT-3′, and R.luc-F2, 5′-AGATGCACCTGATGAAATGG-3′, respectively (Fig. 2, scheme). Amplification involved 50 cycles of 15 s at 95°C, 15 s at 60°C (or 55°C for Renilla), and 15 s at 68°C, with KOD Plus DNA polymerase used as recommended by the manufacturer (Toyobo). PCR products from all the constructs tested corresponding to the expected size of DNA from the 3′-end region of the Renilla luciferase gene were detected (Fig. 2, R). On the other hand, only three constructs (2K-117, 2K-177, and 2K-108) produced a specific band from the 3′-end region of the firefly luciferase gene (Fig. 2, F). Therefore, cDNA derived from minigenome constructs with six-U or seven-U tracts at their P-M gene junctions produced detectable amounts of DNA. In other words, detectable polyadenylated, monocistronic RNA was transcribed from the upstream firefly luciferase genes from minigenomes containing six-U or seven-U tracts at the P gene ends but not from those containing five-U tracts or OSA-2/Fr/B-type sequences at the P gene ends. This was explained by the readthrough at the P-M gene junction and supported the results from the luciferase assay used in this study.
FIG. 2.
Analysis of mRNA of reporter genes. After conversion to cDNA from oligo(dT)-selected RNA with an oligo(dT)-containing primer (minig-R<dT30>), the 3′-end regions of the firefly (F) and Renilla (R) luciferase genes were amplified by PCR with the same reverse primer and the gene-specific primer (scheme). For a detail of each plasmid clone, see Fig. 3.
The firefly and the Renilla luciferase activities were assayed with a dual-luciferase assay system (Promega). The luciferase activity was measured for 2 s by TopCount (Packard Instruments). The value of the luciferase activity was high but was different in different experiments. For example, the firefly and Renilla luciferase activities of clone 2K-117 in four experiments ranged from 4.8 × 105 to 3.5 × 106 relative light units (RLU) and from 1.7 × 105 to 1.6 × 106 RLU, respectively. However, this was not a problem because we designed this system by using a dual-luciferase assay to overcome this fluctuation. To compare the effects on upstream and downstream gene expression of different P-M gene junction sequences, the activity of the Renilla luciferase for each construct was compared to that of the firefly luciferase (Fig. 3). The relative activity of clone 2K-117 (representing the consensus P-M gene junction), which is presented in Fig. 3 as Renilla luciferase activity as a percentage of firefly luciferase activity, is 42%. For clone 2K-102 (constituting the majority of the OSA-1/Fr/V strain), however, the activity of the Renilla luciferase was greatly reduced; thus, the deletion of a single U in the U tract at the end of the P gene can account for the readthrough observed at the P-M gene junction of the Osaka-1 strain. A single U insertion, on the other hand, had no significant effect on the transcriptional termination of the upstream gene (Fig. 3, 2K-177). Likewise, the single mutation found in the OSA-2/Fr/V virus had little effect on transcriptional-termination efficiency (Fig. 3, 2K-108). However, an additional U-to-C mutation in the U tract of the sibling virus OSA-2/Fr/B markedly reduced the transcription of the downstream Renilla luciferase gene (Fig. 3, 2K-125). Back-mutation of clone 2K-125 from G to A (clone 2K-162) did not improve the relative transcription of Renilla luciferase (Fig. 3), indicating that the U-to-C mutation in the U tract was solely responsible for readthrough at the P-M gene junction of virus OSA-2/Fr/B. Renilla luciferase activity relative to firefly luciferase activity for clone 2K-171 (corresponding to the P-M gene junction of the Yamagata-1 lot 2 strain) was 41%, a value for transcription termination efficiency equivalent to that for consensus clone 2K-117 (Fig. 3). However, the A-to-G mutation upstream from the U tract designed into clone 2K-163 (corresponding to the P-M gene junction of the Yamagata-1 lot 1 strain) reduced transcription of the downstream Renilla luciferase gene to 9% (Fig. 3, 2K-163). Since the two Yamagata-1 strains share the other two substitutions at the P-M gene junction, the single nucleotide difference at the position immediately upstream from the U tract was responsible for the readthrough at the P-M gene junction of the Yamagata-1 strain.
We have determined the nucleotide sequences at the P-M gene junctions of several SSPE viruses and have discovered variation among the strains at the end of the P gene. The Osaka-1 strain sequence, 3′-UAAUAUUUUU-5′, has one less U residue in the U tract than the sequence reported for the Edmonston strain (6, 7, 29, 39). This sequence is identical to that of the SSPE virus MF strain and to that isolated from SSPE patient case K (12, 15). The number of U residues in the U tract of the P gene previously had been reported to be variable, either five, six, or seven residues (12, 15, 17). Indeed, our sequencing of individual plasmid clones of the Osaka-1 strain also revealed that, although the majority of clones had five U residues in the P gene end, the U tracts of some clones were six or seven residues in length. In addition, whereas the sequence for the P gene end of the Nagahata strain has been reported to contain five U residues (DDBJ accession no. D63927), our results with the same strain yield a stretch of six U residues at the end of the P gene. If one considers that a stretch of five to seven U residues constitutes the consensus sequence for the end of the P gene end, then the occurrence of five U residues in the U tract of the Osaka-1 strain cannot be regarded as a deviation from the consensus. Cattaneo et al. previously concluded that there were no changes in the P-M gene junction sequence that were related to the readthrough observed at the P-M gene junction in the MF strain and in case K (12, 15). However, our experience based on using standard sequencing methods with a regular polyacrylamide gel and the Applied Biosystems 373S automated DNA sequencer has been that sequencing the end of the P gene is complicated by compression problems. However, compression difficulties were easily overcome either by using an improved gel matrix such as Super Reading DNA sequence solution (Toyobo) with the same DNA sequencer or by using the Applied Biosystems 310 capillary sequencer. Based on observations following resolution of this problem and the sequences of many MV field isolates obtained by us and other investigators (27, 37, 38), we conclude that the actual number of U residues at the end of the P gene end is six, with the consensus sequence being 3′-UAAUAUUUUUU-5′. As a consequence, the sequences of the P gene end of the Osaka-1 strain and of the MF strain and case K can be inferred to be alternates. Another type of variation occurred at the P gene end of a sibling virus of the Osaka-2 strain. The sequence of the OSA-2/Fr/B sibling virus (readthrough type) was 3′-UGAUAUUUCUU-5′, whereas that of the OSA-2/Fr/V sibling virus (nonreadthrough type) was 3′-UGAUAUUUUUU-5′, a difference of a single nucleotide in the U tract between the two sibling viruses. A third type of variation was discovered in the Yamagata-1 strain, in which a single nucleotide substitution immediately upstream from the U tract greatly affected the transcriptional-termination efficiency.
The alterations discovered in these three strains of SSPE virus are related to the readthrough at the P-M gene junction and affect the downstream transcription of the M gene. At the N-P junction of vesicular stomatitis virus, an intact U tract consisting of seven U residues has been demonstrated to be required for polymerase termination of polyadenylation (5, 20). Shortening the seven U residues at the N gene end of the wild-type vesicular stomatitis virus to either five or six U residues resulted in exclusive synthesis of the dicistronic readthrough product of the N and P genes (20). In our present minigenome expression system, shortening the consensus six U residues to five in the U tract at the end of the MV P gene drastically affects the efficiency of transcriptional termination at the P-M gene junction. Another point mutation in the U tract found in the OSA-2/Fr/B sibling virus was equally effective in generating a readthrough product. The last four U residues in the U tract notably are conserved in the ends of all MV genes.
The position immediately upstream of the U tract also affects the transcriptional termination of the P gene. The U residue found at this position in a variant of the Yamagata-1 strain also occurs commonly at the ends of the M, F, H, and L genes. In contrast, a mutation found in the intergenic region of the Yamagata-1 strain was not implicated in readthrough at the P-M gene junction. These results are consistent with those obtained with similar minigenome systems using other viruses, such as vesicular stomatitis virus and respiratory syncytial virus (5, 20, 23). Since the consensus sequences for MV M, F, H, and L gene ends are 3′-UGUUUU-5′, 3′-AAUUUU-5′, 3′-CUUUUU-5′, and 3′-UCUUUU-5′, respectively, it is possible that the first two U residues in the U tract of the P gene are less important. The mutant G residue four bases upstream from the U tract shared by both sibling viruses of the Osaka-2 strain and the Yamagata-1 strain also occurs in the consensus M gene end and has little apparent effect on readthrough, whereas the mutant G residue immediately upstream from the U tract in a variant of the Yamagata-1 strain caused increased readthrough at the junction. The mutant U residue at the same position in the other variant of the same strain, which is common at the corresponding positions of M, F, H, and L genes, was fully functional.
The reason for the frequent appearance of mutations at the P-M gene junction region in SSPE cases is unclear. Other intergenic boundary regions of SSPE virus strains Osaka-1, -2, and -3 are conserved (unpublished observations). One possible explanation is that the mutated genes are selected in the course of persistent infection in the brain. Determining whether mutation at the P-M gene junction is advantageous for survival or for spread of the virus in the central nervous system is important for our understanding of the pathogenesis of SSPE. Because the M protein plays a central role in virus assembly, defective expression of the M protein would have a great impact on SSPE virus infection. Generally, the M gene of the SSPE virus appears to be highly mutable, and the advantage of such mutability seems to lie not in the creation of new functions for the M protein but rather in possibly furthering the spread of the virus in the brain (11). Readthrough transcription at the P-M gene junction directly affects M gene expression and effectively inactivates M gene function. In addition, readthrough transcription may affect downstream gene expression. Parks et al. (28) described an important role for the gene junction in adjusting the level of polymerase for optimal virus growth. Readthrough transcription across the simian virus 5 M-F junction up-regulated the level of polymerase, which affected overall virus growth. In the present study, we have analyzed only naturally occurring mutations in SSPE viruses. In our previous report, both sibling viruses of the Osaka-2 strain, OSA-2/Fr/B and OSA-2/Fr/V, exhibited neurovirulence in hamsters. However, several small differences between the two sibling viruses with respect to incubation period and the duration of survival also were noted (21). These observations may be related to differences between the two sibling viruses that affect virus growth in the hamster brain. The significance of the junction differences and the possible minor effects of such variation could not be distinguished with the minigenome system but potentially could be evaluated in recombinant MV that contained altered P-M gene junction sequences.
Acknowledgments
We thank K. Tatsuta, E. Uenaka, Y. Deng, and N. Hikita for technical assistance. We acknowledge the generous gift of recombinant vaccinia virus vTF7-3 from B. Moss of the National Institutes of Health.
This work was supported by Grants-in-Aid for Scientific Research (no. 10670290 and 11670779) from the Ministry of Education, Science, Sports, and Culture of Japan, a grant from the Osaka Medical Research Foundation for Incurable Diseases, and by a grant from the Osaka City University Medical Research Foundation Fund.
REFERENCES
- 1.Ayata, M., K. Hayashi, T. Seto, R. Murata, and H. Ogura. 1998. The matrix gene expression of subacute sclerosing panencephalitis (SSPE) virus (Osaka-1 strain): a comparison of two sibling viruses isolated from different lobes of an SSPE brain. Microbiol. Immunol. 42:773-780. [DOI] [PubMed] [Google Scholar]
- 2.Ayata, M., A. Hirano, and T. C. Wong. 1989. Structural defect linked to nonrandom mutations in the matrix gene of Biken strain subacute sclerosing panencephalitis virus defined by cDNA cloning and expression of chimeric genes. J. Virol. 63:1162-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayata, M., A. Hirano, and T. C. Wong. 1991. Altered translation of the matrix genes in Niigata and Yamagata neurovirulent measles virus strains. Virology 180:166-174. [DOI] [PubMed] [Google Scholar]
- 4.Ayata, M., T. Kimoto, K. Hayashi, T. Seto, R. Murata, and H. Ogura. 1998. Nucleotide sequences of the matrix protein gene of subacute sclerosing panencephalitis viruses compared with local contemporary isolates from patients with acute measles. Virus Res. 54:107-115. [DOI] [PubMed] [Google Scholar]
- 5.Barr, J. N., S. P. J. Whelan, and G. W. Wertz. 1997. cis-Acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J. Virol. 71:8718-8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellini, W. J., G. Englund, C. D. Richardson, S. Rozenblatt, and R. A. Lazzarini. 1986. Matrix genes of measles virus and canine distemper virus: cloning, nucleotide sequences, and deduced amino acid sequences. J. Virol. 58:408-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bellini, W. J., G. Englund, S. Rozenblatt, H. Arnheiter, and C. D. Richardson. 1985. Measles virus P gene codes for two proteins. J. Virol. 53:908-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blumberg, B. M., J. C. Crowley, J. I. Silverman, J. Menonna, S. D. Cook, and P. C. Dowling. 1988. Measles virus L protein evidences elements of ancestral RNA polymerase. Virology 164:487-497. [DOI] [PubMed] [Google Scholar]
- 9.Bousse, T., T. Takimoto, K. G. Murti, and A. Portner. 1997. Elevated expression of the human parainfluenza virus type 1 F gene downregulates HN expression. Virology 232:44-52. [DOI] [PubMed] [Google Scholar]
- 10.Calain, P., and L. Roux. 1993. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J. Virol. 67:4822-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cathomen, T., B. Mrkic, D. Spehner, R. Drillien, R. Naef, J. Pavlovic, A. Aguzzi, M. A. Billeter, and R. Cattaneo. 1998. A matrix-less measles virus is infectious and elicits extensive cell fusion: consequences for propagation in the brain. EMBO J. 17:3899-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cattaneo, R., G. Rebmann, A. Schmid, K. Baczko, V. ter Meulen, and M. A. Billeter. 1987. Altered transcription of a defective measles virus genome derived from a diseased human brain. EMBO J. 6:681-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cattaneo, R., A. Schmid, M. A. Billeter, R. D. Sheppard, and S. A. Udem. 1988. Multiple viral mutations rather than host factors cause defective measles virus gene expression in a subacute sclerosing panencephalitis cell line. J. Virol. 62:1388-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cattaneo, R., A. Schmid, D. Eschle, K. Baczko, V. ter Meulen, and M. A. Billeter. 1988. Biased hypermutation and other genetic changes in defective measles viruses in human brain infections. Cell 55:255-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cattaneo, R., A. Schmid, G. Rebmann, K. Baczko, V. ter Meulen, W. J. Bellini, S. Rozenblatt, and M. A. Billeter. 1986. Accumulated measles virus mutations in a case of subacute sclerosing panencephalitis: interrupted matrix protein reading frame and transcription alteration. Virology 154:97-107. [DOI] [PubMed] [Google Scholar]
- 16.Cattaneo, R., A. Schmid, P. Spielhofer, K. Kaelin, K. Baczko, V. ter Meulen, J. Pardowits, S. Flanagan, B. K. Rima, S. A. Udem, and M. A. Billeter. 1989. Mutated and hypermutated genes of persistent measles viruses which caused lethal human brain diseases. Virology 173:415-425. [DOI] [PubMed] [Google Scholar]
- 17.Crowley, J. C., P. C. Dowling, J. Menonna, J. I. Silverman, D. Schuback, S. D. Cook, and B. M. Blumberg. 1988. Sequence variability and function of measles virus 3′ and 5′ ends and intercistronic regions. Virology 164:498-506. [DOI] [PubMed] [Google Scholar]
- 18.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Homma, M., M. Tashiro, H. Konno, Y. Ohara, M. Hino, and S. Takase. 1982. Isolation and characterization of subacute sclerosing panencephalitis virus (Yamagata-1 strain) from a brain autopsy. Microbiol. Immunol. 26:1195-1202. [DOI] [PubMed] [Google Scholar]
- 20.Hwang, L. N., N. Englund, and A. K. Pattnaik. 1998. Polyadenylation of vesicular stomatitis virus mRNA dictates efficient transcription termination at the intercistronic gene junctions. J. Virol. 72:1805-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito, N., M. Ayata, M. Shingai, K. Furukawa, T. Seto, I. Matsunaga, M. Muraoka, and H. Ogura. 2002. Comparison of the neuropathogenicity of two SSPE sibling viruses of the Osaka-2 strain isolated with Vero and B95a cells. J. Neurovirol. 8:6-13. [DOI] [PubMed] [Google Scholar]
- 22.Komase, K., T. Haga, Y. Yoshikawa, and K. Yamanouchi. 1992. Complete nucleotide sequence of the phosphoprotein of the Yamagata-1 strain of a defective subacute sclerosing panencephalitis (SSPE) virus. Biochim. Biophys. Acta 1129:342-344. [DOI] [PubMed] [Google Scholar]
- 23.Kuo, L., R. Fearns, and P. L. Collins. 1996. The structurally diverse intergenic regions of respiratory syncytial virus do not modulate sequential transcription by a dicistronic minigenome. J. Virol. 70:6143-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 25.Lorenz, W. W., R. O. McCann, M. Longiaru, and M. J. Cormier. 1991. Isolation and expression of a cDNA encoding Renilla reniformis luciferase. Proc. Natl. Acad. Sci. USA 88:4438-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogura, H., M. Ayata, K. Hayashi, T. Seto, O. Matsuoka, H. Hattori, K. Tanaka, K. Tanaka, Y. Takano, and R. Murata. 1997. Efficient isolation of subacute sclerosing panencephalitis virus from patient brains by reference to magnetic resonance and computed tomographic images. J. Neurovirol. 3:304-309. [DOI] [PubMed] [Google Scholar]
- 27.Parks, C. L., R. A. Lerch, P. Walpita, H.-P. Wang, M. S. Sidhu, and S. A. Udem. 2001. Analysis of the noncoding regions of measles virus strains in the Edmonston vaccine lineage. J. Virol. 75:921-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parks, G. D., K. R. Ward, and J. C. Rassa. 2001. Increased readthrough transcription across the simian virus 5 M-F gene junction leads to growth defects and a global inhibition of viral mRNA synthesis. J. Virol. 75:2213-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radecke, F., and M. A. Billeter. 1995. Measles virus antigenome and protein consensus sequences. Curr. Top. Microbiol. Immunol. 191:181-192. [DOI] [PubMed] [Google Scholar]
- 30.Rassa, J. C., and G. D. Parks. 1998. Molecular basis for naturally occurring elevated readthrough transcription across the M-F junction of the paramyxovirus SV5. Virology 247:274-286. [DOI] [PubMed] [Google Scholar]
- 31.Rassa, J. C., G. M. Wilson, G. A. Brewer, G. D. Parks. 2000. Spacing constraints on reinitiation of paramyxovirus transcription: the gene end U tract acts as a spacer to separate gene end from gene start sites. Virology 274:438-449. [DOI] [PubMed] [Google Scholar]
- 32.Reutter, G. L., C. Cortese-Grogan, J. Wilson, and S. A. Moyer. 2001. Mutations in the measles virus C protein that up regulate viral RNA synthesis. Virology 285:100-109. [DOI] [PubMed] [Google Scholar]
- 33.Rosenstein, S. P., and M. D. Been. 1991. Evidence that genomic and antigenomic RNA self-cleaving elements from hepatitis delta virus have similar secondary structures. Nucleic Acids Res. 19:5409-5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seto, T., M. Ayata, K. Hayashi, K. Furukawa, R. Murata, and H. Ogura. 1999. Different transcriptional expression of the matrix gene of the two sibling viruses of the subacute sclerosing panencephalitis virus (Osaka-2 strain) isolated from a biopsy specimen of patient brain. J. Neurovirol. 5:151-160. [DOI] [PubMed] [Google Scholar]
- 35.Sheppard, R. D., C. S. Raine, M. B. Bornstein, and S. A. Udem. 1986. Rapid degradation restricts measles virus matrix protein expression in a subacute sclerosing panencephalitis cell line. Proc. Natl. Acad. Sci. USA 83:7913-7917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sidhu, M. S., J. Chan, K. Kaelin, P. Spielhofer, F. Radecke, H. Schneider, M. Masurekar, P. C. Dowling, M. A. Billeter, and S. A. Udem. 1995. Rescue of synthetic measles virus minireplicons: measles genomic termini direct efficient expression and propagation of a reporter gene. Virology 208:800-807. [DOI] [PubMed] [Google Scholar]
- 37.Takeda, M., T. Sakaguchi, Y. Li, F. Kobune, A. Kato, and Y. Nagai. 1999. The genome nucleotide sequence of a contemporary wild strain of measles virus and its comparison with the classical Edmonston strain genome. Virology 256:340-350. [DOI] [PubMed] [Google Scholar]
- 38.Takeuchi, K., N. Miyajima, F. Kobune, and M. Tashiro. 2000. Comparative nucleotide sequence analyses of the entire genomes of B95a cell-isolated and Vero cell-isolated measles viruses from the same patient. Virus Genes 20:253-257. [DOI] [PubMed] [Google Scholar]
- 39.Wong, T. C., and A. Hirano. 1987. Structure and function of bicistronic RNA encoding the phosphoprotein and matrix protein of measles virus. J. Virol. 61:584-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yoshikawa, Y., H. Tsuruoka, M. Matsumoto, T. Haga, T. Shioda, H. Shibuta, T. A. Sato, and K. Yamanouchi. 1990. Molecular analysis of structural protein genes of the Yamagata-1 strain of defective subacute sclerosing panencephalitis virus. II. Nucleotide sequence of a cDNA corresponding to the P plus M dicistronic mRNA. Virus Genes 4:151-161. [DOI] [PubMed] [Google Scholar]