Abstract
The Epstein-Barr virus (EBV) immediate-early protein BZLF1 is a transcriptional activator that mediates the switch between the latent and the lytic forms of EBV infection. It was previously reported that BZLF1 inhibits p53 transcriptional function in reporter gene assays. Here we further examined the effects of BZLF1 on p53 function by using a BZLF1-expressing adenovirus vector (AdBZLF1). Infection of cells with the AdBZLF1 vector increased the level of cellular p53 but prevented the induction of p53-dependent cellular target genes, such as p21 and MDM2. BZLF1-expressing cells had increased p53-specific DNA binding activity in electrophoretic mobility shift assays, increased p53 phosphorylation at multiple residues (including serines 6, 9, 15, 33, 46, 315, and 392), and increased acetylation at lysine 320 and lysine 382. Thus, the inhibitory effects of BZLF1 on p53 transcriptional function cannot be explained by its effects on p53 phosphorylation, acetylation, or DNA binding activity. BZLF1 substantially reduced the level of cellular TATA binding protein (TBP) in both normal human fibroblasts and A549 cells, and the inhibitory effects of BZLF1 on p53 transcriptional function could be partially rescued by the overexpression of TBP. Thus, BZLF1 has numerous effects on p53 posttranslational modification but may inhibit p53 transcriptional function in part through an indirect mechanism involving the suppression of TBP expression.
The p53 gene encodes a multifunctional protein that participates in cell cycle control, programmed cell death, genomic stability, and DNA replication, transcription, and repair (42, 53, 54, 71). Under normal conditions, p53 is maintained at low levels by ubiquitin-dependent degradation, but in response to stresses, such as genotoxic insults, viral infection, nucleotide depletion, hypoxia, and oncogenic activation, p53 is modified. These alterations then contribute to p53 stabilization and stimulate the accumulation of p53 in the nucleus, where it acts as a transcriptional factor (5, 71). Posttranscriptional modifications to the p53 protein, which include phosphorylation, acetylation, and sumoylation, also play a role in the activation of p53 as a transcriptional factor (6). p53 binds DNA in a sequence-specific manner and regulates its many downstream gene targets both positively and negatively (34, 70). Transcriptional activation by p53 is mediated through an acidic domain at the N terminus that binds directly to the TATA binding protein (TBP) and to TBP-associated factors (44, 57, 62, 80).
DNA tumor viruses interfere with p53 function through multiple different mechanisms. The papillomavirus E6 protein interacts directly with p53 and promotes its degradation (8, 79). The simian virus 40 large T antigen interacts directly with p53 and prevents its binding to DNA (9, 92). The hepatitis B virus X protein inhibits the nuclear translocation of p53 (84). The adenovirus E1A protein and the papillomavirus E7 protein repress p53 transactivation by binding to and apparently sequestering limiting quantities of cellular TBP (65, 78). Adenovirus E1A also competes with p53 for limiting quantities of the acetyltransferase p300 (78). The adenovirus E1B 55-kDa protein interacts directly with p53 and inhibits its acetylation. (64, 66, 90). Thus, the inhibition of p53 function clearly is advantageous for the replication of many viruses, and numerous and diverse strategies have been developed to inhibit p53 function.
Epstein-Barr virus (EBV) is a human herpesvirus associated with a number of different malignancies. EBV predominantly infects B lymphocytes, where it exists in a latent state, expressing only a small subset of viral genes (51, 72). The EBV lytic program is initiated by the expression of the viral immediate-early (IE) protein BZLF1 (15, 16, 74, 83). The BZLF1 protein is a DNA binding, b-zip transcriptional factor which binds to AP-1-like sequences present in the promoters of early lytic genes (13, 25, 28, 29, 41, 49, 55, 88). The BZLF1-induced cascade of viral gene expression eventually results in viral DNA replication and virion production (26, 27, 51, 72).
As is the case for the smaller DNA viruses, members of the herpesvirus family have also been found to manipulate p53 for their own purposes. For example, the cytomegalovirus IE2 protein (11, 82, 85, 87), the Kaposi's sarcoma-associated herpesvirus open reading frame (ORF) K8 protein (67), and the human herpesvirus 6 ORF 1 protein (20) all stabilize p53, thus increasing overall levels of p53 protein but inhibiting its transactivation ability. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus and latent membrane protein 1 of EBV also interfere with p53 function (30, 31). However, in comparison to the effects of the smaller DNA viruses, considerably less is known regarding the various mechanisms by which herpesviruses inhibit the transcriptional function of p53.
Zhang et al. reported that the EBV IE protein BZLF1 directly binds p53 and inhibits its ability to activate a reporter construct containing p53 binding motifs (95). However, the effects of BZLF1 on p53 function remain somewhat controversial. While other researchers similarly found that BZLF1 increases the level of cellular p53, these groups reported that this effect is accompanied by increased, rather than decreased, p53 transcriptional function (12, 14, 21). The reason for these different results is not clear but could reflect cell type-dependent effects of BZLF1 on p53 function. BZLF1 induces cell growth arrest in certain cell types, and this effect is associated with increased expression of the cyclin-dependent kinase inhibitors p21 and p27 (12, 73). Although p53 activates p21 expression, inhibition of cell cycle progression by BZLF1 has been shown to be independent of p53 (73).
In this report, we further examined the effects of BZLF1 on p53. Using an adenovirus vector to express BZLF1, we demonstrated that BZLF1 expression resulted in an increased p53 level in A549 cells. Although BZLF1 expression induced many posttranslational modifications of p53 that would be anticipated to increase p53 transcriptional function, including phosphorylation at seven different sites and acetylation at two sites, we found that p53 transcriptional function was nevertheless inhibited by BZLF1 in A549, U-2 OS, and normal human fibroblast cells. BZLF1 also enhanced p53-specific DNA binding. However, we showed that BZLF1 significantly reduced the levels of TBP in both normal human fibroblast and A549 cells and that the inhibitory effects of BZLF1 on p53 function were partially overcome by the overexpression of TBP. Thus, BZLF1 expression in cells induces both potentially activating and inhibitory effects on p53 function. We speculate that in certain cell types, the activating effects of BZLF1 on p53 function may predominate, thus explaining the apparently contradictory results in the previous literature.
MATERIALS AND METHODS
Cell cultures.
Normal human fibroblasts (NHF5-neo or NHF) were derived from neonatal foreskin. NHF were maintained in Eagle's minimal essential medium supplemented with 10% fetal bovine serum (FBS) and nonessential amino acids. A549 cells (ATCC CCL-185), derived from a human lung carcinoma which expresses wild-type p53, were maintained in Dulbecco's modified essential medium supplemented with 10% FBS. U-2 OS (ATCC HTB-96), a human osteosarcoma cell line which expresses wild-type 53, was maintained in McCoy's 5a medium with 15% FBS. The AGS-EBV cell line (a generous gift from Lindsey Hutt-Fletcher) was obtained by G418 selection of AGS cells (gastric carcinoma cells) that were infected with recombinant Akata virus in which a neomycin resistance cassette had been inserted into the nonessential BDLF3 ORF. The AGS-EBV cell line was maintained in Ham's F12 medium supplemented with 10% FBS. All media contained penicillin (100 U/ml) and streptomycin (100 μg/ml). The cells were maintained at 37°C in a humidified atmosphere containing 5% CO2.
Adenovirus vectors and infections.
An E1- and E3-deficient adenovirus type 5 vector expressing EBV IE protein BZLF1 cDNA (under the control of a cytomegalovirus promoter) was made by using the recombinant Cre-Lox-mediated recombination system as previously described (AdBZLF1) (91). The control adenovirus vector (AdLacZ) is identical to AdBZLF1, except that it contains the bacterial β-galactosidase gene in place of BZLF1 cDNA. An E1- and E3-deficient adenovirus type 5 vector expressing wild-type p53 also was used (a generous gift from Wendell Yarbrough, University of North Carolina, Chapel Hill).
NHF were plated at a cell density of 107 per 150-mm plates. Cells were infected with no adenovirus (mock infected), AdLacZ, or AdBZLF1 at a multiplicity of infection (MOI) of 250. A549 and U-2 OS cells were plated at a cell density of 2 × 106 per 150-mm plate and infected at an MOI of 50 as described above. Cells were harvested 24 to 72 h postinfection.
Cellular irradiation.
A549 cells were infected (or mock infected) with adenovirus vectors 24 to 48 h before gamma irradiation. Cells were irradiated with 8 Gy of gamma radiation and then harvested at 1 and 6 h after gamma irradiation to examine the levels of p53 and p21 proteins, respectively. For examination of p53 acetylation, Trichostatin A (Sigma) was added at a final concentration of 5 μM immediately after gamma irradiation. N-acetylleucylleucylnorleucinal (ALLN), a proteosome inhibitor, also was used as a control in acetylation experiments (75)
Immunoblotting.
Whole-cell extracts were harvested as previously described (1). A total of 50 to 100 μg of protein was separated by polyacrylamide gel electrophoresis and blotted onto nitrocellulose membranes. The membranes were incubated first in blocking buffer (phosphate-buffered saline, 0.1% Tween 20, 5% milk) at room temperature for 60 min and then with primary antibody in blocking buffer for 60 min. Primary antibodies included anti-p53 mouse monoclonal antibody (1:500; DO-1 [Santa Cruz]), anti-p21 rabbit polyclonal antibody (1:500; C-19 [Santa Cruz]), anti-MDM2 mouse monoclonal antibody (1:500; SMP14 [Santa Cruz]), anti-TBP mouse monoclonal antibody (1:500; 58C9 [Santa Cruz]), and anti-BZLF1 mouse monoclonal antibody (1:200; AZ-69 [Argene]). The membranes were incubated in wash buffer (phosphate-buffered saline, 0.1% Tween 20) and then with horseradish peroxidase-conjugated secondary antibody (1:10,000; Promega) at room temperature for 60 min. The membranes were further washed, and proteins were detected by enhanced chemiluminescence (Amersham).
EMSA.
Whole-cell extracts were prepared from A549 cells approximately 48 h after adenovirus infection as previously described (4). The p53 electrophoretic mobility shift assay (EMSA) was performed as previously described (17). The radiolabeled double-stranded p53 oligonucleotide probe (5′-GATCCCGGCATGTCCGGGCATGTCCGGGCATGT-3′) contains two tandem repeats of the nearly palindromic sequence GGCATGTCC, known to be bound by p53 with a high affinity. Ten micrograms of extract was added to a mixture containing 4 μl of 5× binding buffer (50 mM Tris-Cl [pH 8], 0.5 mM zinc acetate, 5 mM dithithreitol, 25% glycerol). Specific (p53) and nonspecific (AP-1) cold competitor DNAs were added to some reactions (200-fold excess) for 15 min prior to the addition of the oligonucleotide probe master mixture [2 × 105 cpm and 1 μg of poly(dI-dC)-poly(dI-dC)]. To enhance p53-DNA complex formation, 100 ng of affinity-purified Pab421 (p53; Ab-1 [Oncogene]) was added to the binding reactions (36). The total reaction volume was 20 μl. After the addition of the probe, the reaction mixtures were incubated at room temperature for 30 min and subsequently were electrophoresed at 4°C on a 6% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA (45 mM Tris-borate, 1 mM EDTA) at 400 V. Complexes were visualized by autoradiography.
Phosphorylation- and acetylation-specific p53 antibodies.
Rabbit polyclonal antibodies specific for p53 phosphorylated at Ser6, Ser9, Ser15, or Ser33 or acetylated at Lys320 or Lys382 have been described elsewhere (40, 46, 75, 76). A similar approach was used to generate antibodies specific for p53 phosphorylated at Ser46, Ser315, or Ser392. Briefly, the PAbSer(P)46, PAbSer(P)315, and PAbSer392 antibodies were raised against the human p53 sequences Ac-41-51(46P)C [i.e., Ac-Asp-Asp-Leu-Met-Leu-Ser(P)-Pro-Asp-Asp-Ile-Glu-Cys-NH2], Ac-310-321(315P)C [i.e., Ac-Asn-Asn-Thr-Ser-Ser-Ser(P)-Pro-Gln-Pro-Lys-Lys-Lys-Cys-NH2], and Ac-C-385-393(392P) [i.e., Ac-Cys-Phe-Lys-Thr-Glu-Gly-Pro-Asp-Ser(P)-Asp-NH2], respectively, coupled to keyhole limpet hemocyanin. Phosphorylation-specific antibodies were affinity purified from the resulting serum by use of each phosphorylated peptide coupled with Sulfolink (Pierce). The purified antibodies were then passed through a column coupled with the respective unphosphorylated peptide to deplete antibodies that reacted with unphosphorylated p53. The specificity of each antibody was confirmed by enzyme-linked immunosorbent and immunoblot assays.
Immunoprecipitation and immunoblotting for phosphorylated and acetylated p53.
Cells were solubilized in ice-cold lysis buffer (50 mM Tris-HCl [pH 7.5], 5 mM EDTA, 150 mM NaCl, 1% Triton X-100, 50 mM NaF, 10 mM sodium pyrophosphate, 25 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 mM sodium molybdate, 10 μg of aprotinin/ml, 10 μg of leupeptin/ml, 5 μg of pepstatin/ml, 0.5 mM phenylmethylsulfonyl fluoride). Immunoprecipitation and Western blotting were performed as described previously (40, 76).
Reverse transcription and DNA amplification.
Total RNA was isolated from NHF by using an RNeasy minikit (Qiagen) according to the manufacturer's instructions. The synthesis of single-stranded cDNA from total cellular RNA was carried out by using a reverse transcription system (Promega). One microgram of total cellular RNA was placed at 70°C for 10 min and then placed on ice. Avian myeloblastosis virus reverse transcriptase (RT), random primers, deoxynucleoside triphosphates, RT buffer, MgCl, and recombinant RNasin RNase inhibitor were added according to the manufacturer’s instructions, and the reaction mixture was incubated at room temperature for 10 min and then at 42°C for 1 h. The samples were heated to 99°C for 5 min and then placed on ice for 5 min to inactivate the avian myeloblastosis virus RT.
The primers used for PCR were as follows: human TBP-specific sequences were amplified by using sense-strand primer 1, 5′-GCTGCAGCCGTTCAGCAGTC-3′ (residues 295 to 314), and antisense-strand primer 2, 5′-GCTCTGACTTTAGCACCTGT-3′ (residues 937 to 956), to give a 662-bp product. β2-Microglobulin-specific sequences were amplified by using sense-strand primer 1, 5′-TTCTGGCCTGGAGGGCATCC-3′ (residues 925 to 946) (exon 1), and antisense strand primer 2, 5′-ATCTTCAAACCTCCATGATG-3′ (residues 2244 to 2263) (exon 3).
For PCR, 5 μl of the RT reaction mixture was added to a final volume of 50 μl containing PCR buffer (10 mM Tris-HCl [pH 9.0 at 25°C], 50 mM KCl, 0.1% Triton X-100), 2.5 mM MgCl, 0.2 mM deoxynucleoside triphosphates, sequence-specific primers (each at a concentration of 5 μM), and 2.5 U of thermostable Taq DNA polymerase (Promega). Amplification was carried out with a Perkin-Elmer DNA thermal cycler 480 with an initial denaturing step at 95°C for 10 min, followed by 50 cycles of annealing at 56°C for 1 min, extension at 72°C for 1 min, and denaturing at 95°C for 1 min. PCR products were examined on a 2% agarose gel stained with ethidium bromide.
CAT assay and plasmids.
Construction of the BZLF-1 (pCMV-Z), p53 (pC53-SN3; a generous gift from Bert Vogelstein), and TBP (LTReTBP; a generous gift from Deborah Johnson) expression plasmids has been described elsewhere (7, 10, 95). The p53 reporter construct, pG13-CAT (a generous gift from Bert Vogelstein), contains 13 copies of the consensus p53 binding site upstream of the polyomavirus early promoter linked to the chloramphenical acetyltransferase (CAT) gene (50). Plasmid DNA was purified with a Qiagen maxikit as specified by the manufacturer. DNA was transfected by electroporation with 11 μg of DNA and approximately 7.5 × 106 cells per condition. The cells were shocked at 1,500 V with a Zapper electroporation unit (Medical Electronics Shop, University of Wisconsin, Madison). A549 cells were trypsinized and resuspended in RPMI medium with 10% FBS prior to electroporation. Protein extracts were harvested 72 h after transfection. CAT assays were performed as previously described (33). Briefly, 65 μl of whole-cell extract was incubated with [14C]chloramphenicol in the presence of acetyl coenzyme A at 37°C for 2 h. The percent acetylation of chloramphenicol was quantitated by thin-layer chromatography followed by analysis with a phosphorimager screen.
Fluorescence-activated cell sorting analysis.
The level of cellular BZLF1 expression was determined by fixing cells in 60% acetone and incubating them with anti-BZLF1 antibody (1:100; Argene) and then with fluorescein isothiocyanate-conjugated anti-mouse antibody (1:100; Sigma).
RESULTS
BZLF1 inhibits p21 induction in gamma-irradiated cells.
One of the best-characterized targets of p53 transcriptional function is p21 (23). The p21 gene encodes a cyclin-dependent kinase inhibitor that stops cell cycle progression in G1 by binding to G1 cyclin-dependent kinases (Cdk2, Cdk3, Cdk4, and Cdk6) (37). Cayrol and Flemington previously reported that BZLF1 induces a G1 cell cycle block by activating p21 expression (12).
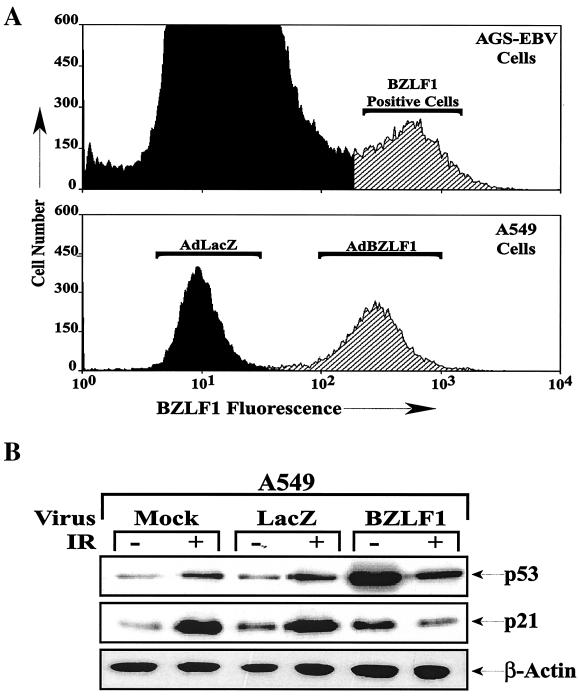
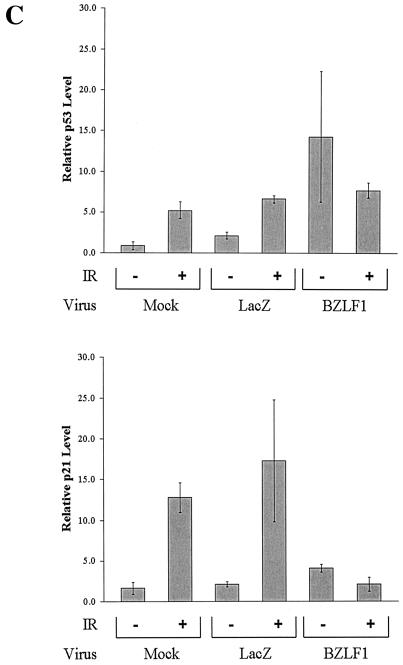
To determine whether BZLF1 expression at a physiologic level alters the transcriptional function of endogenous p53, we examined its effects in gamma-irradiated A549 cells by using adenovirus vectors; gamma irradiation of cells activates p53 transcriptional function through multiple mechanisms (32, 47). The level of BZLF1 expression in A549 cells infected with the AdBZLF1 vector at an MOI of 50 was similar to that in the lytically infected population of AGS gastric carcinoma cells infected with the intact EBV genome (Fig. 1A). As shown in Fig. 1B, when A549 cells were infected with the AdLacZ or AdBZLF1 vector 48 h prior to treatment of the cells with gamma irradiation, BZLF1 expression significantly increased the level of endogenous p53 expression and slightly increased the level of p21 expression in untreated cells but prevented the induction of p21 expression that normally occurs following gamma irradiation. Similar results were observed in normal human fibroblasts (data not shown). Thus, BZLF1 inhibited the ability of endogenous p53 to transcriptionally activate the p21 gene following gamma irradiation.
FIG. 1.
BZLF1 inhibits the induction of p21 in gamma-irradiated cells. A549 cell cultures were mock infected or infected with adenovirus vectors (MOI, 50) expressing the LacZ and BZLF1 genes. (A) The level of cellular BZLF1 expression was determined by fluorescence-activated cell sorting analysis of A549 cells infected with the adenovirus vectors and EBV-positive gastric carcinoma cells (AGS-EBV), of which approximately 5% express BZLF1. (B) At 48 h after adenovirus infection, A549 cells were treated with 8 Gy of gamma radiation (IR). Cells were harvested for immunoblot analysis at 1 h after gamma irradiation to quantitate the level of cellular p53 and at 6 h after gamma irradiation to quantitate p21 and β-actin levels. (C) The results of two separate experiments were quantitated to show the effects of BZLF1 on p53 and p21 expression in A549 cells in the presence and absence of gamma irradiation. The average and the range are shown; the level of p21 or p53 expression in mock-infected cells in the absence of gamma irradiation was set at 1.
BZLF1 prevents the induction of p21 and MDM2 in U-2 OS cells overexpressing p53.
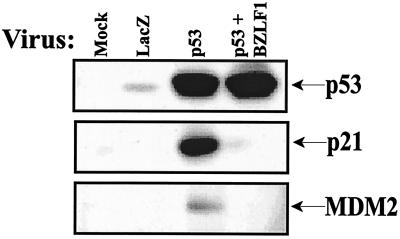
To determine whether the ability of BZLF1 to inhibit p53 transcriptional effects is unique to the p21 gene or affects other p53 target genes as well, U-2OS cells were infected with an adenovirus vector expressing the p53 protein (Adp53), with or without concomitant infection with a BZLF1 adenovirus vector. As previously observed, the overexpression of p53 by itself induced p21 expression, while the expression of BZLF1 and p53 together, while not affecting the level of adenovirus vector-produced p53, inhibited the ability of p53 to activate the expression of p21 (Fig. 2). Infection of cells with the p53 adenovirus vector by itself also induced the expression of another known p53 target gene, MDM2. The MDM2 protein negatively regulates p53 function by inducing its degradation and preventing its interaction with TBP (38, 43, 52, 86). As was the case for p21, coinfection of cells with both the p53 and the BZLF1 adenovirus vectors also inhibited the ability of p53 to induce MDM2 expression. Thus, BZLF1 inhibition of p53 transcriptional function is not limited to the p21 target gene.
FIG. 2.
BZLF1 inhibits p53 transcriptional function. U-2 OS cells were mock infected or infected with equal amounts of AdLacZ, AdBZLF1, Adp53 and AdLacZ, or Adp53 and AdBZLF1. Cells were harvested for immunoblot analysis at 48 h postinfection to quantitate the levels of cellular p53, p21, and MDM2.
BZLF1 does not inhibit p53-specific DNA binding in A549 cells.
To further define the mechanism(s) by which BZLF1 inhibits p53 transcriptional function, we examined its effects on p53-specific DNA binding. A549 cells were infected with the control adenovirus vector (AdLacZ), AdBZLF1, or Adp53, and the amount of p53 binding activity in cell extracts was quantitated by an EMSA with a radiolabeled oligonucleotide probe containing two copies of a high-affinity p53 binding motif. The Pab421 antibody, which has been shown to dramatically enhance p53-specific DNA binding in EMSAs (36), was added to identify the p53 binding complex.
As shown in Fig. 3, extracts from A549 cells infected with the AdBZLF1 vector had much more p53-specific DNA binding activity than those from cells infected with the AdLacZ vector. Cells infected with the Adp53 vector, as expected, had massive quantities of p53-specific DNA binding activity. In a separate experiment, the level of p53-specific DNA binding activity in AdBZLF1-infected A549 cells (without gamma irradiation) was similar to that observed in gamma-irradiated A549 cells (data not shown). These results indicate that BZLF1 actually enhances the amount of cellular p53-specific DNA binding and thus clearly does not interfere with p53 transcriptional function by inhibiting p53-specific DNA binding.
FIG. 3.
BZLF1 enhances p53-specific DNA binding activity. A549 cells were infected with adenovirus vectors expressing LacZ, BZLF1, and p53. Whole-cell extracts were prepared at 48 h postinfection, and the level of p53-specific DNA binding activity was quantitated by EMSA analysis as described in Materials and Methods. The Pab421 antibody (421 Ab) has been shown to enhance p53 sequence-specific DNA binding activity.
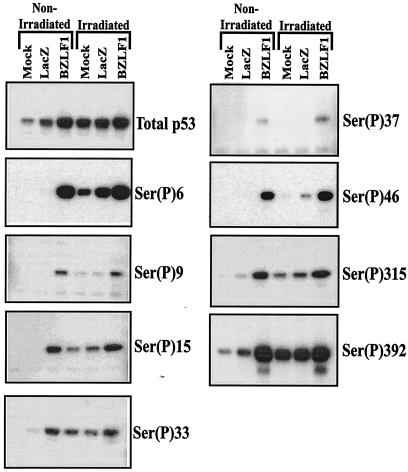
BZLF1 expression is associated with enhanced phosphorylation at multiple serine residues.
The transcriptional function of p53 is enhanced by phosphorylation. The amino terminus of p53, which contains the transcriptional regulatory domain, has eight known phosphorylation sites (serines 6, 9, 15, 20, 33, 37, and 46 and threonine 18) (5). The carboxyl terminus contains the nuclear import and export signals, the tetramerization domain, a non-sequence-specific DNA binding domain, and four phosphorylation sites (serines 315, 376, 378, and 392).
By using antibodies that recognize specific sites when phosphorylated, we examined the effects of BZLF1 on p53 phosphorylation in A549 cells. Treatment of cells with gamma irradiation served as a positive control for p53 phosphorylation in these experiments. As shown in Fig. 4, both BZLF1 expression and gamma irradiation increased the level of total p53 in A549 cells; both also increased the level of p53 phosphorylation at a number of different sites. Most notably, the phosphorylation of p53 serine 15, which is phosphorylated by the ATM and ATR kinases in vivo and which is important for efficient p53 transcriptional function (6), was dramatically enhanced in cells expressing BZLF1 as well as in gamma-irradiated cells. Likewise, BZLF1 expression in A549 cells also induced the phosphorylation of p53 at serine 33 and serine 46, which also are thought to be involved in the activation of p53 transcriptional function (5, 6). BZLF1 also increased phosphorylation at several other serines in p53 for which the functional effects of phosphorylation are unclear, including serine 6, serine 9, serine 315, and serine 392 (5, 6). These results indicate that the ability of BZLF1 to reduce p53 transcriptional function cannot be attributed to decreased phosphorylation of p53 in the presence of BZLF1.
FIG. 4.
BZLF1 enhances p53 phosphorylation at multiple serine residues. A549 cells were either mock infected or infected with AdLacZ or AdBZLF1. Some cells were gamma irradiated with 8 Gy. Cells were harvested at 48 h postinfection (and 1 h postirradiation) for immunoblot analysis with antibodies recognizing total p53 or antibodies specific for various phosphorylated (P) p53 residues.
BZLF1 expression increases p53 acetylation.
Acetylation of the carboxyl terminus of p53 at lysine 320 and lysine 382 enhances p53 transcriptional function (6). BZLF1 interacts directly with the CREB binding protein (CBP), an acetyltransferase (2, 94), and thus could modulate p53 acetylation. Therefore, we examined the effects of BZLF1 expression in A549 cells on the levels of p53 acetylation at lysine 320 and lysine 382 by using antibodies that specifically recognize p53 acetylated at each lysine (Fig. 5). In the experiment shown, the mock-infected cells were also treated with a proteosome inhibitor (ALLN) in an attempt to compensate for the effects of BZLF1 on total p53. As expected, gamma irradiation increased the level of total p53 as well as p53 acetylation at lysine 320 and lysine 382. However, even in nonirradiated cells, BZLF1 expression dramatically increased p53 acetylation, particularly at lysine 382. Thus, inhibition of p53 acetylation cannot explain the absence of p53 transcriptional function in the presence of BZLF1.
FIG. 5.
BZLF1 enhances p53 acetylation. A549 cells were either mock infected or infected with AdLacZ or AdBZLF1. Some cells were also treated with gamma irradiation or treated with the proteosome inhibitor ALLN. Cells were harvested and analyzed as described in the legend to Fig. 4, except that antibodies recognizing specific acetylated (Ac) p53 lysines were used.
BZLF1 decreases protein and RNA levels for TBP.
To further investigate BZLF1 repression of p53 transcriptional function, we examined the effects of BZLF1 on TBP, since it is known that p53 must interact directly with TBP to transcriptionally activate promoters (24, 80) and since certain inhibitors of p53 have been shown to prevent the interaction between p53 and TBP (63, 65, 78). In the course of performing coimmunopreciptation experiments with TBP and p53 in the presence and absence of BZLF1 (data not shown), we discovered that BZLF1 expression substantially reduces the level of cellular TBP. The effects of BZLF1 expression on TBP are shown in Fig. 6. Compared to mock-infected and AdLacZ-infected cells, AdBZLF1-infected NHF and A549 cells had decreased levels of TBP, and these effects were not reversed by a proteosome inhibitor.
FIG. 6.
BZLF1 reduces the level of cellular TBP. NHF or A549 cells were either mock infected or infected with AdLacZ or AdBZLF1. Some cells were also treated with the proteosome inhibitor ALLN. Immunoblot analysis was performed at 48 h postinfection to quantitate the levels of TBP and β-actin in A549 cells and NHF (top and bottom panels) or in a time course experiment with A549 cells (middle panel; time points indicate hours after adenovirus infection).
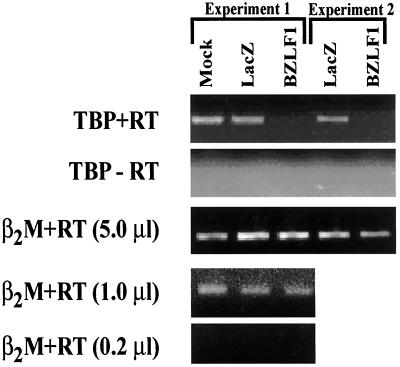
We therefore examined the effects of BZLF1 expression on the level of cellular TBP mRNA. As shown in Fig. 7, BZLF1 decreased the levels of TBP mRNA in NHF in two separate experiments while having no effect on a control transcript (β2-microglobulin). Thus, BZLF1 likely decreases the amount of TBP in cells by decreasing TBP transcription.
FIG. 7.
BZLF1 reduces the level of TBP mRNA. NHF cells were infected with AdLacZ or AdBZLF1. RNA was prepared at 48 h postinfection, and RT PCR analysis was performed (+RT) with primers specific for either the TBP or the β2-microglobulin (β2M) message. PCR also was performed in the absence of reverse transcriptase (−RT). Two separate experiments are shown. In experiment 1, RT PCR analysis with the β2-microglobulin primers was performed with decreasing amounts of the cDNA product.
Overexpression of TBP partially rescues BZLF1 inhibition of p53 transactivation.
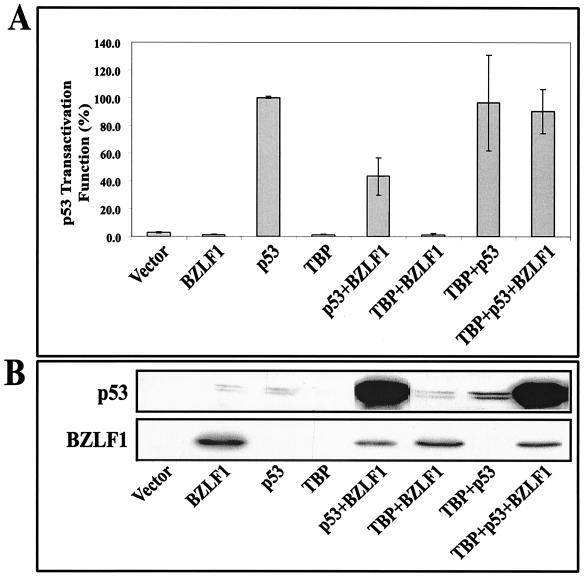
Inhibition of TBP expression in cells suggests a mechanism by which BZLF1 could repress p53 transcriptional function in spite of also inducing multiple posttranslational modifications of p53. If the inhibition of p53 transcriptional function by BZLF1 were due to the relative lack of TBP, then we would expect that the overexpression of TBP would reverse the BZLF1 inhibitory effects. The effects of TBP overexpression were examined in transient transfection experiments by using a p53-responsive promoter construct (pG13-CAT, which contains 13 copies of the p53 binding motif) in A549 cells. As shown in Fig. 8, a p53 expression vector by itself activated the expression of the pG13-CAT construct. BZLF1, while dramatically increasing the level of transfected p53, nevertheless decreased the transcriptional function of p53. TBP by itself did not activate the pG13-CAT construct and did not enhance the ability of transfected p53 to activate this construct, implying that the level of endogenous TBP in cells is sufficient for maximal activation. Most notably, however, the overexpression of TBP reduced the inhibitory effects of BZLF1 on p53 function while not affecting the levels of p53 or BZLF1. In contrast, the overexpression of CBP, which is also required for both BZLF1 and p53 transcriptional function and which is thought to be limiting in cells, did not significantly affect the inhibitory effects of BZLF1 on p53 function (data not shown). Therefore, it appears that BZLF1 at least partially inhibits p53 transcriptional function by reducing the level of cellular TBP.
FIG. 8.
BZLF1 inhibition of p53 transactivation is partially reversed by TBP overexpression. (A) The p53-responsive pG13-CAT construct was cotransfected into A549 cells with vector DNA, the BZLF1 expression plasmid alone, the p53 expression plasmid alone, the TBP expression plasmid alone, or various combinations of these plasmids (with total DNA kept constant). The level of CAT activity produced under each condition at 72 h after transfection was quantitated. Results are expressed as the relative amount of p53 transactivator function for each condition (average and range are shown), with the activity of the p53 expression vector alone set at 100%. (B) Immunoblot analyses of the extracts used in the CAT assays were performed to quantitate the expression of BZLF1 and p53 under each condition.
DISCUSSION
Virus infection may activate p53-mediated responses, leading to cell cycle arrest or apoptosis; thus, many viruses encode proteins that interfere with p53 function to ensure an optimal environment for viral replication, either by releasing cells from cell cycle checkpoints or by protecting cells from p53-dependent apoptosis (23, 89, 93). Virus-encoded proteins that modulate p53 function include simian virus 40 large T antigen, adenovirus E1A and E1B, hepatitis B virus X protein, and human papillomavirus E6 and E7. These interfere with p53 function in a variety of ways. Large T antigen and E1B bind to p53 and increase its stability but inhibit its function (64, 66). Human papillomavirus E6, on the other hand, induces the degradation of p53 in a ubiquitin-dependent manner (66, 79). Moreover, the hepatitis B virus X protein binds p53 and inhibits its translocation to the nucleus (84, 90). Here we show that an IE protein of EBV, BZLF1, has both potentially activating and inhibitory effects on p53. We speculate that the balance between the activating and the repressive effects of BZLF1 on p53 function in EBV-infected host cells has a profound impact on the efficiency of viral replication and the fate of the cells.
Herpesviruses can infect cells in either a latent or a lytic form. For EBV, the switch between the latent and the lytic forms of infection is mediated by BZLF1. While the latent type of infection allows herpesviruses to persist in the host for life, protected from the immune system, the lytic form of infection is required for the transmission of these viruses from cell to cell and from host to host. Inhibition of p53 function appears to be important for efficient lytic herpesvirus infection, given that a number of these viruses encode lytic proteins that inhibit p53 function (11, 20, 67, 82, 85, 87). Decreased p53 activity during lytic herpesvirus infection likely serves to protect the virus from cellular apoptosis, thereby allowing sufficient time for virus assembly and release.
Although BZLF1 expression in A549 cells was associated with decreased p53 transcriptional function, it nonetheless induced a variety of posttranslational p53 modifications that would normally serve to activate p53 function. The ability of BZLF1 expression in A549 cells to enhance the level of p53 phosphorylation at multiple different serine residues was unexpected and, to our knowledge, unprecedented. The cellular kinases responsible for phosphorylating p53 include ATM and ATR kinases (serine 15), casein kinase II (serine 392), c-Jun N-terminal kinase (serine 33), p38 kinase (serine 33 and serine 46), Cdk kinases (serine 315), and unknown kinases (serines 6 and 9) (5). Interestingly, Adamson et al. have shown that BZLF1 expression upregulates at least two of these kinases, p38 kinase and c-Jun N-terminal kinase (1). The dramatic activation of p53 phosphorylation (as well as acetylation) by BZLF1 expression in cells suggests the possibility that BZLF1 induces DNA damage. Consistent with this notion, early lytic EBV infection has been shown to induce DNA breakage in host cells (48). Alternatively, the ability of BZLF1 to increase the level of p53 phosphorylation at most phosphorylation sites suggests that it may instead inhibit the function of one or more cellular protein phosphatases.
BZLF1 expression in A549 cells also dramatically increased the level of p53 acetylation. Recent studies have shown that p53 is acetylated at two different lysine residues in the carboxyl region by p300/CBP and p300/CBP-associated factor (PCAF) and that acetylation increases DNA binding affinity and transcriptional function (56, 75). p300/CBP and PCAF exhibit specificity for different lysine residues of p53; while lysine 320 is the preferential target for PCAF, lysine 382 is the target for p300/CBP (56, 75). Adenovirus has been shown to alter p53 acetylation at two different levels: E1B interacts with p53 and inhibits its acetylation induced by PCAF (58), whereas E1A interacts with p300/CBP and PCAF and represses their acetylation activity for p53 (60, 78). Like E1A, BZLF1 also interacts directly with cellular CBP and requires this interaction for efficient transcriptional function (2, 94). Although we anticipated that BZLF1 would inhibit the acetylation of p53 by competing for limiting quantities of cellular CBP, the opposite proved to be the case, especially at lysine 382. Since it is known that BZLF1 binds to the carboxyl-terminal end of p53 (95), it is possible that it enhances p53 acetylation by bringing CBP to p53. Alternatively, BZLF1 may indirectly enhance p53 acetylation by inducing cellular DNA damage (48).
As we observed here with BZLF1, both the human papillomavirus E7 and the adenovirus E1A proteins were shown to perturb p53 function through their interactions with TBP (65, 78). This inhibition is thought to be due to competition for limiting quantities of cellular TBP, and the overexpression of TBP is able to release the E1A-mediated repression of p53 (65, 78). We show here that BZLF1 reduces the expression of TBP in normal human fibroblasts and A549 cells and that the inhibitory effects of BZLF1 on p53 transcriptional function are partially ameliorated by the overexpression of TBP. Thus, BZLF1 likely inhibits p53 transcriptional function at least partially by reducing the level of cellular TBP. Since BZFL1 can also interact directly with TBP (18), this interaction could also play a role in inhibiting p53 transcriptional activity. However, since we have not found that BZLF1 expression reduces the activity of a variety of other cellular promoters in A549 cells, the decreased TBP level in BZLF1-positive cells is presumably still capable of supporting transcription by some transcription factors.
Although our results suggest that its effects on TBP are an important mechanism by which BZLF1 inhibits p53 transcriptional function, it is likely that other mechanisms also contribute to its effects. For example, it recently was shown that p53 transport to promyelocytic leukemia (PML) (ND10) nuclear bodies is important for its transcriptional function (35, 69). PML bodies are also known to bind to CBP, and it has been shown that PML bodies, p53, and CBP can form a stable complex (68). Since it was recently shown that BZLF1 disperses PML bodies (3), we speculate that this effect also contributes to decreased p53 function in BZLF1-expressing cells. In addition, it was shown that BZLF1 is strongly SUMO-1 modified and can compete for limiting quantities of SUMO-1 in host cells (3). Given that sumoylation was reported to be an activating modulator of p53 (6), BZLF1 may also be expected to potentially decrease p53 function through its inhibition of p53 sumoylation, similar to its ability to inhibit SUMO-1 modification of the PML body protein.
Zhang et al. reported that the EBV IE protein BZLF1 directly interacts with p53 (95). At this point, we are uncertain which, if any, of the BZLF1 effects on p53 require a direct interaction between BZLF1 and p53. Using BZLF1-specific antibodies, we have been unable to show that the p53-specific DNA binding complex in AdBZLF1-infected cells contains BZLF1. Furthermore, it is unlikely that the ability of BZLF1 to decrease the level of cellular TBP, which we show here is a major mechanism for the inhibition of p53 function, requires a direct interaction between BZLF1 and p53. Therefore, if the direct interaction between BZLF1 and p53 is functionally important in vivo, it more likely serves to enhance p53 function. For example, it is possible that a larger complex containing CBP, p53, and BZLF1 promotes the acetylation of p53.
Although our results here clearly document that BZLF1 inhibits p53 transcriptional function in at least some cell types, Cayrol and Flemington reported that BZLF1 induces a G1 block in HeLa cells and suggested that this effect is mediated through the activation of p21 (12). Although we have found that BZLF1 inhibits p53 function in HeLa cells (unpublished data), our results reported here do not exclude the possibility that in some situations, the activating effects of BZLF1 on p53 function predominate, resulting in a G1 block through the p53-dependent activation of p21. In contrast to the situation for the smaller DNA viruses, which often activate cell cycle progression, there is growing evidence that it may be advantageous for herpesviruses to induce a G1 block during the lytic form of infection, thus avoiding competition with host cells for limiting nucleotide pools (19, 22, 45, 59, 77, 81). The BZLF1-induced activation of p53 function may therefore enhance viral replication in certain situations, particularly since BZLF1 activates the expression of other lytic EBV proteins with known antiapoptotic functions (39, 61). The development of cell lines that can efficiently support primary lytic EBV infection in vitro will be necessary to fully explore the interaction between BZLF1 and p53 in the context of the intact virus.
In summary, we show here that BZLF1 expression in host cells has dramatic effects on p53 posttranslational modifications as well as p53 transcriptional function. While many of the effects of BZLF1 expression in cells would be anticipated to enhance p53 function, including increased p53 protein stability, increased p53-specific DNA binding activity, increased p53 phosphorylation, and increased p53 acetylation, in many cell types these effects are apparently outweighed by the ability of BZLF1 to reduce TBP expression. We speculate that this complex regulation of p53 function by BZLF1 serves to enhance the efficiency of lytic EBV replication.
Acknowledgments
This work was supported by grants NCI CA64852 and RO1-CA58853 from the National Institutes of Health. C.W.A. and S.S. were supported in part by a laboratory directed research and development award at Brookhaven National Laboratory under contract with the U.S. Department of Energy.
REFERENCES
- 1.Adamson, A. L., D. Darr, E. Holley-Guthrie, R. A. Johnson, A. Mauser, J. Swenson, and S. Kenney. 2000. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 activate the ATF2 transcription factor by increasing the levels of phosphorylated p38 and c-Jun N-terminal kinases. J. Virol. 74:1224-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamson, A. L., and S. Kenney. 1999. The Epstein-Barr virus BZLF1 protein interacts physically and functionally with the histone acetylase CREB-binding protein. J. Virol. 73:6551-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adamson, A. L., and S. Kenney. 2001. Epstein-Barr virus immediate-early protein BZLF1 is SUMO-1 modified and disrupts promyelocytic leukemia bodies. J. Virol. 75:2388-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews, N. C., and D. V. Faller. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Appella, E., and C. W. Anderson. 2000. Signaling to p53: breaking the posttranslational modification code. Pathol. Biol. (Paris) 48:227-245. [PubMed] [Google Scholar]
- 6.Appella, E., and C. W. Anderson. 2001. Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268:2764-2772. [DOI] [PubMed] [Google Scholar]
- 7.Baker, S. J., S. Markowitz, E. R. Fearon, J. K. Willson, and B. Vogelstein. 1990. Suppression of human colorectal carcinoma cell growth by wild-type p53. Science 249:912-915. [DOI] [PubMed] [Google Scholar]
- 8.Band, V., S. Dalal, L. Delmolino, and E. J. Androphy. 1993. Enhanced degradation of p53 protein in HPV-6 and BPV-1 E6-immortalized human mammary epithelial cells. EMBO J. 12:1847-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bargonetti, J., P. N. Friedman, S. E. Kern, B. Vogelstein, and C. Prives. 1991. Wild-type but not mutant p53 immunopurified proteins bind to sequences adjacent to the SV40 origin of replication. Cell 65:1083-1091. [DOI] [PubMed] [Google Scholar]
- 10.Bryant, G. O., L. S. Martel, S. K. Burley, and A. J. Berk. 1996. Radical mutations reveal TATA-box binding protein surfaces required for activated transcription in vivo. Genes Dev. 10:2491-2504. [DOI] [PubMed] [Google Scholar]
- 11.Castillo, J. P., A. D. Yurochko, and T. F. Kowalik. 2000. Role of human cytomegalovirus immediate-early proteins in cell growth control. J. Virol. 74:8028-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cayrol, C., and E. K. Flemington. 1996. The Epstein-Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J. 15:2748-2759. [PMC free article] [PubMed] [Google Scholar]
- 13.Chang, Y. N., D. L. Dong, G. S. Hayward, and S. D. Hayward. 1990. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J. Virol. 64:3358-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen, H., J. M. Lee, Y. Wang, D. P. Huang, R. F. Ambinder, and S. D. Hayward. 1999. The Epstein-Barr virus latency BamHI-Q promoter is positively regulated by STATs and Zta interference with JAK/STAT activation leads to loss of BamHI-Q promoter activity. Proc. Natl. Acad. Sci. USA 96:9339-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chevallier-Greco, A., E. Manet, P. Chavrier, C. Mosnier, J. Daillie, and A. Sergeant. 1986. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 5:3243-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Countryman, J., and G. Miller. 1985. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl. Acad. Sci. USA 82:4085-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuddihy, A. R., S. Li, N. W. Tam, A. H. Wong, Y. Taya, N. Abraham, J. C. Bell, and A. E. Koromilas. 1999. Double-stranded-RNA-activated protein kinase PKR enhances transcriptional activation by tumor suppressor p53. Mol. Cell. Biol. 19:2475-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng, Z., C. J. Chen, D. Zerby, H. J. Delecluse, and P. M. Lieberman. 2001. Identification of acidic and aromatic residues in the Zta activation domain essential for Epstein-Barr virus reactivation. J. Virol. 75:10334-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dittmer, D., and E. S. Mocarski. 1997. Human cytomegalovirus infection inhibits G1/S transition. J. Virol. 71:1629-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doniger, J., S. Muralidhar, and L. J. Rosenthal. 1999. Human cytomegalovirus and human herpesvirus 6 genes that transform and transactivate. Clin. Microbiol. Rev. 12:367-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dreyfus, D. H., M. Nagasawa, C. A. Kelleher, and E. W. Gelfand. 2000. Stable expression of Epstein-Barr virus BZLF-1-encoded ZEBRA protein activates p53-dependent transcription in human Jurkat T-lymphoblastoid cells. Blood 96:625-634. [PubMed] [Google Scholar]
- 22.Ehmann, G. L., T. I. McLean, and S. L. Bachenheimer. 2000. Herpes simplex virus type 1 infection imposes a G(1)/S block in asynchronously growing cells and prevents G(1) entry in quiescent cells. Virology 267:335-349. [DOI] [PubMed] [Google Scholar]
- 23.el Deiry, W. S., T. Tokino, V. E. Velculescu, D. B. Levy, R. Parsons, J. M. Trent, D. Lin, W. E. Mercer, K. W. Kinzler, and B. Vogelstein. 1993. WAF1, a potential mediator of p53 tumor suppression. Cell 75:817-825. [DOI] [PubMed] [Google Scholar]
- 24.Farmer, G., J. Colgan, Y. Nakatani, J. L. Manley, and C. Prives. 1996. Functional interaction between p53, the TATA-binding protein (TBP), andTBP-associated factors in vivo. Mol. Cell. Biol. 16:4295-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farrell, P. J., D. T. Rowe, C. M. Rooney, and T. Kouzarides. 1989. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 8:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feederle, R., M. Kost, M. Baumann, A. Janz, E. Drouet, W. Hammerschmidt, and H. J. Delecluse. 2000. The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19:3080-3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fixman, E. D., G. S. Hayward, and S. D. Hayward. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998-3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Flemington, E., and S. H. Speck. 1990. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. J. Virol. 64:1227-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flemington, E. K., A. M. Borras, J. P. Lytle, and S. H. Speck. 1992. Characterization of the Epstein-Barr virus BZLF1 protein transactivation domain. J. Virol. 66:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 31.Fries, K. L., W. E. Miller, and N. Raab-Traub. 1996. Epstein-Barr virus latent membrane protein 1 blocks p53-mediated apoptosis through the induction of the A20 gene. J. Virol. 70:8653-8659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giaccia, A. J., and M. B. Kastan. 1998. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 12:2973-2983. [DOI] [PubMed] [Google Scholar]
- 33.Gorman, C. M., L. F. Moffat, and B. H. Howard. 1982. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol. Cell. Biol. 2:1044-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 35.Guo, A., P. Salomoni, J. Luo, A. Shih, S. Zhong, W. Gu, and P. P. Paolo. 2000. The function of PML in p53-dependent apoptosis. Nat. Cell Biol. 2:730-736. [DOI] [PubMed] [Google Scholar]
- 36.Halazonetis, T. D., L. J. Davis, and A. N. Kandil. 1993. Wild-type p53 adopts a ′mutant'-like conformation when bound to DNA. EMBO J. 12:1021-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harper, J. W., S. J. Elledge, K. Keyomarsi, B. Dynlacht, L. H. Tsai, P. Zhang, S. Dobrowolski, C. Bai, L. Connell-Crowley, and E. Swindell. 1995. Inhibition of cyclin-dependent kinases by p21. Mol. Biol. Cell 6:387-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haupt, Y., R. Maya, A. Kazaz, and M. Oren. 1997. Mdm2 promotes the rapid degradation of p53. Nature 387:296-299. [DOI] [PubMed] [Google Scholar]
- 39.Henderson, S., D. Huen, M. Rowe, C. Dawson, G. Johnson, and A. Rickinson. 1993. Epstein-Barr virus-coded BHRF1 protein, a viral homologue of Bcl-2, protects human B cells from programmed cell death. Proc. Natl. Acad. Sci. USA 90:8479-8483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Higashimoto, Y., S. Saito, X. H. Tong, A. Hong, K. Sakaguchi, E. Appella, and C. W. Anderson. 2000. Human p53 is phosphorylated on serines 6 and 9 in response to DNA damage-inducing agents. J. Biol. Chem. 275:23199-23203. [DOI] [PubMed] [Google Scholar]
- 41.Holley-Guthrie, E. A., E. B. Quinlivan, E. C. Mar, and S. Kenney. 1990. The Epstein-Barr virus (EBV) BMRF1 promoter for early antigen (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell-specific manner. J. Virol. 64:3753-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hollstein, M., D. Sidransky, B. Vogelstein, and C. C. Harris. 1991. p53 mutations in human cancers. Science 253:49-53. [DOI] [PubMed] [Google Scholar]
- 43.Honda, R., H. Tanaka, and H. Yasuda. 1997. Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420:25-27. [DOI] [PubMed] [Google Scholar]
- 44.Horikoshi, N., A. Usheva, J. Chen, A. J. Levine, R. Weinmann, and T. Shenk. 1995. Two domains of p53 interact with the TATA-binding protein, and the adenovirus 13S E1A protein disrupts the association, relieving p53-mediated transcriptional repression. Mol. Cell. Biol. 15:227-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jault, F. M., J. M. Jault, F. Ruchti, E. A. Fortunato, C. Clark, J. Corbeil, D. D. Richman, and D. H. Spector. 1995. Cytomegalovirus infection induces high levels of cyclins, phosphorylated Rb, and p53, leading to cell cycle arrest. J. Virol. 69:6697-6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jimenez, G. S., F. Bryntesson, M. I. Torres-Arzayus, A. Priestley, M. Beeche, S. Saito, K. Sakaguchi, E. Appella, P. A. Jeggo, G. E. Taccioli, G. M. Wahl, and M. Hubank. 1999. DNA-dependent protein kinase is not required for the p53-dependent response to DNA damage. Nature 400:81-83. [DOI] [PubMed] [Google Scholar]
- 47.Kastan, M. B., O. Onyekwere, D. Sidransky, B. Vogelstein, and R. W. Craig. 1991. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 51:6304-6311. [PubMed] [Google Scholar]
- 48.Kawanishi, M. 1993. Epstein-Barr virus induces fragmentation of chromosomal DNA during lytic infection. J. Virol. 67:7654-7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kenney, S., E. Holley-Guthrie, E. C. Mar, and M. Smith. 1989. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J. Virol. 63:3878-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kern, S. E., J. A. Pietenpol, S. Thiagalingam, A. Seymour, K. W. Kinzler, and B. Vogelstein. 1992. Oncogenic forms of p53 inhibit p53-regulated gene expression. Science 256:827-830. [DOI] [PubMed] [Google Scholar]
- 51.Kieff, E. 1996. Epstein-Barr virus and its replication, p. 2343-2395. In B. N. Fields, D. N. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 52.Kubbutat, M. H., S. N. Jones, and K. H. Vousden. 1997. Regulation of p53 stability by Mdm2. Nature 387:299-303. [DOI] [PubMed] [Google Scholar]
- 53.Levine, A. J. 1997. p53, the cellular gatekeeper for growth and division. Cell 88:323-331. [DOI] [PubMed] [Google Scholar]
- 54.Levine, A. J., J. Momand, and C. A. Finlay. 1991. The p53 tumour suppressor gene. Nature 351:453-456. [DOI] [PubMed] [Google Scholar]
- 55.Lieberman, P. M., J. M. Hardwick, and S. D. Hayward. 1989. Responsiveness of the Epstein-Barr virus NotI repeat promoter to the Z transactivator is mediated in a cell-type-specific manner by two independent signal regions. J. Virol. 63:3040-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu, L., D. M. Scolnick, R. C. Trievel, H. B. Zhang, R. Marmorstein, T. D. Halazonetis, and S. L. Berger. 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19:1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu, X., C. W. Miller, P. H. Koeffler, and A. J. Berk. 1993. The p53 activation domain binds the TATA box-binding polypeptide in Holo-TFIID, and a neighboring p53 domain inhibits transcription. Mol. Cell. Biol. 13:3291-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu, Y., A. L. Colosimo, X. J. Yang, and D. Liao. 2000. Adenovirus E1B 55-kilodalton oncoprotein inhibits p53 acetylation by PCAF. Mol. Cell. Biol. 20:5540-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu, M., and T. Shenk. 1996. Human cytomegalovirus infection inhibits cell cycle progression at multiple points, including the transition from G1 to S. J. Virol. 70:8850-8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lundblad, J. R., R. P. Kwok, M. E. Laurance, M. L. Harter, and R. H. Goodman. 1995. Adenoviral E1A-associated protein p300 as a functional homologue of the transcriptional co-activator CBP. Nature 374:85-88. [DOI] [PubMed] [Google Scholar]
- 61.Marshall, W. L., C. Yim, E. Gustafson, T. Graf, D. R. Sage, K. Hanify, L. Williams, J. Fingeroth, and R. W. Finberg. 1999. Epstein-Barr virus encodes a novel homolog of the bcl-2 oncogene that inhibits apoptosis and associates with Bax and Bak. J. Virol. 73:5181-5185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martin, D. W., R. M. Munoz, M. A. Subler, and S. Deb. 1993. p53 binds to the TATA-binding protein-TATA complex. J. Biol. Chem. 268:13062-13067. [PubMed] [Google Scholar]
- 63.Martin, D. W., M. A. Subler, R. M. Munoz, D. R. Brown, S. P. Deb, and S. Deb. 1993. p53 and SV40 T antigen bind to the same region overlapping the conserved domain of the TATA-binding protein. Biochem. Biophys. Res. Commun. 195:428-434. [DOI] [PubMed] [Google Scholar]
- 64.Martin, M. E., and A. J. Berk. 1998. Adenovirus E1B 55K represses p53 activation in vitro. J. Virol. 72:3146-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Massimi, P., and L. Banks. 1997. Repression of p53 transcriptional activity by the HPV E7 proteins. Virology 227:255-259. [DOI] [PubMed] [Google Scholar]
- 66.Mietz, J. A., T. Unger, J. M. Huibregtse, and P. M. Howley. 1992. The transcriptional transactivation function of wild-type p53 is inhibited by SV40 large T-antigen and by HPV-16 E6 oncoprotein. EMBO J. 11:5013-5020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park, J., T. Seo, S. Hwang, D. Lee, Y. Gwack, and J. Choe. 2000. The K-bZIP protein from Kaposi's sarcoma-associated herpesvirus interacts with p53 and represses its transcriptional activity. J. Virol. 74:11977-11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pearson, M., R. Carbone, C. Sebastiani, M. Cioce, M. Fagioli, S. Saito, Y. Higashimoto, E. Appella, S. Minucci, P. P. Pandolfi, and P. G. Pelicci. 2000. PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406:207-210. [DOI] [PubMed] [Google Scholar]
- 69.Pearson, M., and P. G. Pelicci. 2001. PML interaction with p53 and its role in apoptosis and replicative senescence. Oncogene 20:7250-7256. [DOI] [PubMed] [Google Scholar]
- 70.Pietenpol, J. A., T. Tokino, S. Thiagalingam, W. S. el Deiry, K. W. Kinzler, and B. Vogelstein. 1994. Sequence-specific transcriptional activation is essential for growth suppression by p53. Proc. Natl. Acad. Sci. USA 91:1998-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prives, C., and P. A. Hall. 1999. The p53 pathway. J. Pathol. 187:112-126. [DOI] [PubMed] [Google Scholar]
- 72.Rickinson, A. B., and E. Kieff. 1996. Epstein-Barr virus, p. 2397-2446. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 73.Rodriguez, A., M. Armstrong, D. Dwyer, and E. Flemington. 1999. Genetic dissection of cell growth arrest functions mediated by the Epstein-Barr virus lytic gene product, Zta. J. Virol. 73:9029-9038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rooney, C. M., D. T. Rowe, T. Ragot, and P. J. Farrell. 1989. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J. Virol. 63:3109-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sakaguchi, K., J. E. Herrera, S. Saito, T. Miki, M. Bustin, A. Vassilev, C. W. Anderson, and E. Appella. 1998. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12:2831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sakaguchi, K., S. Saito, Y. Higashimoto, S. Roy, C. W. Anderson, and E. Appella. 2000. Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding. J. Biol. Chem. 275:9278-9283. [DOI] [PubMed] [Google Scholar]
- 77.Salvant, B. S., E. A. Fortunato, and D. H. Spector. 1998. Cell cycle dysregulation by human cytomegalovirus: influence of the cell cycle phase at the time of infection and effects on cyclin transcription. J. Virol. 72:3729-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sang, N., M. L. Avantaggiati, and A. Giordano. 1997. Roles of p300, pocket proteins, and hTBP in E1A-mediated transcriptional regulation and inhibition of p53 transactivation activity. J. Cell. Biochem. 66:277-285. [DOI] [PubMed] [Google Scholar]
- 79.Scheffner, M., B. A. Werness, J. M. Huibregtse, A. J. Levine, and P. M. Howley. 1990. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63:1129-1136. [DOI] [PubMed] [Google Scholar]
- 80.Seto, E., A. Usheva, G. P. Zambetti, J. Momand, N. Horikoshi, R. Weinmann, A. J. Levine, and T. Shenk. 1992. Wild-type p53 binds to the TATA-binding protein and represses transcription. Proc. Natl. Acad. Sci. USA 89:12028-12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Song, B., J. J. Liu, K. C. Yeh, and D. M. Knipe. 2000. Herpes simplex virus infection blocks events in the G1 phase of the cell cycle. Virology 267:326-334. [DOI] [PubMed] [Google Scholar]
- 82.Speir, E., R. Modali, E. S. Huang, M. B. Leon, F. Shawl, T. Finkel, and S. E. Epstein. 1994. Potential role of human cytomegalovirus and p53 interaction in coronary restenosis. Science 265:391-394. [DOI] [PubMed] [Google Scholar]
- 83.Takada, K., N. Shimizu, S. Sakuma, and Y. Ono. 1986. trans-Activation of the latent Epstein-Barr virus (EBV) genome after transfection of the EBV DNA fragment. J. Virol. 57:1016-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takada, S., N. Kaneniwa, N. Tsuchida, and K. Koike. 1997. Cytoplasmic retention of the p53 tumor suppressor gene product is observed in the hepatitis B virus X gene-transfected cells. Oncogene 15:1895-1901. [DOI] [PubMed] [Google Scholar]
- 85.Tanaka, K., J. P. Zou, K. Takeda, V. J. Ferrans, G. R. Sandford, T. M. Johnson, T. Finkel, and S. E. Epstein. 1999. Effects of human cytomegalovirus immediate-early proteins on p53-mediated apoptosis in coronary artery smooth muscle cells. Circulation 99:1656-1659. [DOI] [PubMed] [Google Scholar]
- 86.Thut, C. J., J. A. Goodrich, and R. Tjian. 1997. Repression of p53-mediated transcription by MDM2: a dual mechanism. Genes Dev. 11:1974-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tsai, H. L., G. H. Kou, S. C. Chen, C. W. Wu, and Y. S. Lin. 1996. Human cytomegalovirus immediate-early protein IE2 tethers a transcriptional repression domain to p53. J. Biol. Chem. 271:3534-3540. [PubMed] [Google Scholar]
- 88.Urier, G., M. Buisson, P. Chambard, and A. Sergeant. 1989. The Epstein-Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J. 8:1447-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Waldman, T., K. W. Kinzler, and B. Vogelstein. 1995. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Res. 55:5187-5190. [PubMed] [Google Scholar]
- 90.Wang, X. W., K. Forrester, H. Yeh, M. A. Feitelson, J. R. Gu, and C. C. Harris. 1994. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc. Natl. Acad. Sci. USA 91:2230-2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Westphal, E. M., A. Mauser, J. Swenson, M. G. Davis, C. L. Talarico, and S. C. Kenney. 1999. Induction of lytic Epstein-Barr virus (EBV) infection in EBV-associated malignancies using adenovirus vectors in vitro and in vivo. Cancer Res. 59:1485-1491. [PubMed] [Google Scholar]
- 92.Xing, J., H. M. Sheppard, S. I. Corneillie, and X. Liu. 2001. p53 stimulates TFIID-TFIIA-promoter complex assembly, and p53-T antigen complex inhibits TATA binding protein-TATA interaction. Mol. Cell. Biol. 21:3652-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yin, C., C. M. Knudson, S. J. Korsmeyer, and T. Van Dyke. 1997. Bax suppresses tumorigenesis and stimulates apoptosis in vivo. Nature 385:637-640. [DOI] [PubMed] [Google Scholar]
- 94.Zerby, D., C. J. Chen, E. Poon, D. Lee, R. Shiekhattar, and P. M. Lieberman. 1999. The amino-terminal C/H1 domain of CREB binding protein mediates zta transcriptional activation of latent Epstein-Barr virus. Mol. Cell. Biol. 19:1617-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang, Q., D. Gutsch, and S. Kenney. 1994. Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol. Cell. Biol. 14:1929-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]