Abstract
To study the orientation and dynamics of myosin, we measured fluorescence polarization of single molecules and ensembles of myosin decorating actin filaments. Engineered chicken gizzard regulatory light chain (RLC), labeled with bisiodoacetamidorhodamine at cysteine residues 100 and 108 or 104 and 115, was exchanged for endogenous RLC in rabbit skeletal muscle HMM or S1. AEDANS-labeled actin, fully decorated with labeled myosin fragment or a ratio of ∼1:1000 labeled:unlabeled myosin fragment, was adhered to a quartz slide. Eight polarized fluorescence intensities were combined with the actin orientation from the AEDANS fluorescence to determine the axial angle (relative to actin), the azimuthal angle (around actin), and RLC mobility on the ≪10 ms timescale. Order parameters of the orientation distributions from heavily labeled filaments agree well with comparable measurements in muscle fibers, verifying the technique. Experiments with HMM provide sufficient angular resolution to detect two orientations corresponding to the two heads in rigor. Experiments with S1 show a single orientation intermediate to the two seen for HMM. The angles measured for HMM are consistent with heads bound on adjacent actin monomers of a filament, under strain, similar to predictions based on ensemble measurements made on muscle fibers with electron microscopy and spectroscopic experiments.
INTRODUCTION
Repeated cycles of myosin attachment to actin, tilting and detachment, fueled by hydrolysis of ATP, are thought to translocate interdigitating filaments in muscle (1,2) or various cellular cargoes in nonmuscle cells (3). The “lever arm hypothesis” suggests that the motor domain (MD) of the myosin head binds rigidly to actin, whereas the light chain domain (LCD) produces the motion by rotating about a fulcrum within the MD. Evidence from x-ray crystallography, spectroscopic experiments, electron microscopy, low-angle x-ray diffraction from muscle fibers, and mechanics in vitro have provided strong evidence that the LCD serves as a lever arm, amplifying subnanometer sized motions in the MD associated with ATP hydrolysis and product release into nanometer-scale motions at the head-tail junction (4–6). Some motions or tilting within the MD may also contribute to force production and filament sliding (7,8).
With the processive myosin isoform V, tilting motions of the LCD large enough to explain the step length have been detected in single molecules (9). In muscle fibers, tilting motions of the RLC have mostly been observed during filament sliding after length changes (10–13), leading some to question whether tilting is the result of sliding rather than causing it (14). The orientation distribution of the LCD in an isometrically contracting muscle fiber is very broad due to thermal rotational motions on the nanosecond to microsecond timescales and due to cycling through all of the states of the enzymatic cycle (13,15,16). In addition to these dynamic processes, static disorder, due to the mismatched periodicities of the actin and myosin filaments (17) and/or strain in the LCD (18,19), is expected to broaden the angular distribution among the attached myosin heads. The orientational disorder in rigor (in the absence of ATP) is less than that during active contraction, but the contributions of static and dynamic disorder have not been accurately determined. Spectroscopic evidence suggests broader distributions of the LCD (20,21) than the MD (22,23).
To address these issues directly, we applied the total internal reflection fluorescence polarization technique described in the accompanying article (24) to simultaneously detect orientations and dynamics of single myosin molecules in rigor actomyosin complexes. Orientation of the LCD was determined using actin filaments decorated with labeled subfragment-1 (S1) and heavy meromyosin (HMM). Low and high ratios of labeled to unlabeled myosin subunits enabled measurements from individual molecules and populations, respectively. Data from highly labeled filaments facilitated comparison with measurements on muscle fibers. In data from individual molecules, the average orientation and the extent of motions on the microsecond timescale were determined simultaneously (24). The data suggest that in rigor, the heads of HMM bind at two orientations that differ in both tilt and twist. Microsecond dynamic disorder accounts for ∼35° of orientational dispersion. The two average orientations are consistent with the two heads bound to adjacent actin monomers and strained toward each other. S1 binds at an intermediate orientation. Some of these results have been presented in preliminary form (25,26).
MATERIALS AND METHODS
Protein purification and labeling
Fast skeletal muscle myosin II and actin were isolated, purified, and stored as described in Forkey et al. (24). HMM was made according to Kron et al. (27) and was stored on ice for up to 1 week. S1 was made by papain digestion in the presence of MgCl2 to retain the regulatory light chain (RLC) ((28) as modified by Okamoto and Sekine (29)). It was either stored on ice for up to 1 week or rapidly frozen and stored at −80°C for up to 2 months. Actin was labeled at Cys374 with N-(iodoacetyl)-N′-(5-sulfo-1-napthyl)ethylenediamine (1,5-I-AEDANS, Sigma (St. Louis, MO) No. I8879 (24)).
Chicken gizzard RLC mutants, labeled with bisiodoacetamidorhodamine (BR) at pairs of engineered cysteine residues (100 and 108 or 104 and 115; (13)), were a gift from Dr. John E. T. Corrie, MRC Mill Hill, London. The labeled RLC mutants are termed 100-BR-108 and 104-BR-115. They were exchanged for the endogenous RLC subunits in purified myosin proteolytic fragments, HMM or S1. The myosin fragment was combined with labeled RLC at threefold molar excess (RLC/myosin head) in exchange buffer (12 mM EDTA, 120 mM KCl, 10 mM HEPES, pH 7.0, 2 mM DTT). After a 30 min incubation at 30°C, 12 mM MgC12 was added, and the solution was cooled on ice for 15 min. Myosin was separated from free RLC by centrifugation with actin in the absence of ATP, resuspension of the pellet, and a second centrifugation in the presence of ATP. A typical exchange (starting with 2 μM myosin) yielded 0.5 μM protein (∼100% active) with ∼50% of the heads labeled, as estimated by absorbance of rhodamine (ɛ549 = 89,000 M−1cm−1; (30)) and protein concentration determined by Bradford assay. For HMM, molecules with one and two labeled heads were both present. Exchanged protein was stored on ice for up to 1 week. No significant differences in sliding velocity were detected between labeled and unlabeled myosin or labeled and unlabeled actin with in vitro gliding assays (31,32), using actin stabilized with Bodipy 650/665-conjugated phalloidin for visualization (Molecular Probes (Eugene, OR), B-12382).
Experimental apparatus
The experimental apparatus, signal acquisition, and analysis are described in the accompanying article (24). The rhodamine-labeled myosin was excited with a frequency-doubled Nd:YAG laser (532 nm), cycling between four combinations of excitation paths and polarizations every 40 ms (Figs. 1 and S1 of Forkey et al. (24)). Fluorescence emission was directed either onto an intensified charge-coupled device camera or through a polarizing beamsplitter onto two photon-counting avalanche photodiodes (APDs). The resulting photon counts corresponded to eight raw intensity time courses (see Fig. 2 and Forkey et al. (24)). Two additional laser beams approached the sample at glancing angles to generate evanescent waves. An ultraviolet (355 nm) laser was used to excite AEDANS-actin for colocalization of F-actin with labeled myosin (Fig. 1). A HeNe (633 nm) laser was used to excite Bodipy 650/665-phalloidin in control sliding velocity experiments.
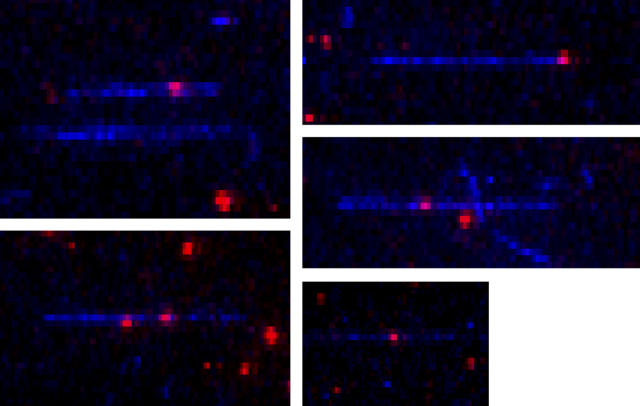
FIGURE 1.
Colocalization of actin filaments with sparsely labeled myosin. Actin filaments labeled with AEDANS (blue) at Cys374 are decorated with HMM or S1. Labeled and unlabeled myosin fragments are mixed ∼1:1000 so individual rhodamine (red) molecules can be distinguished along the length of the actin filaments. When the decorated filaments are added in the presence of excess free HMM or S1, respectively, to a poly-L-lysine coated flow chamber, the filaments are aligned well, as pictured here, due to a balance between flow forces and attachment frequency.
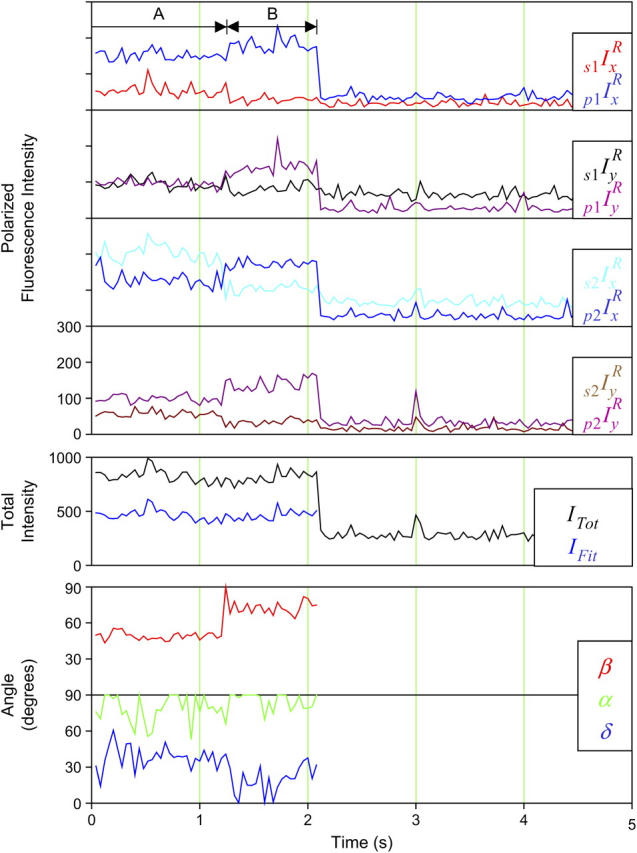
FIGURE 2.
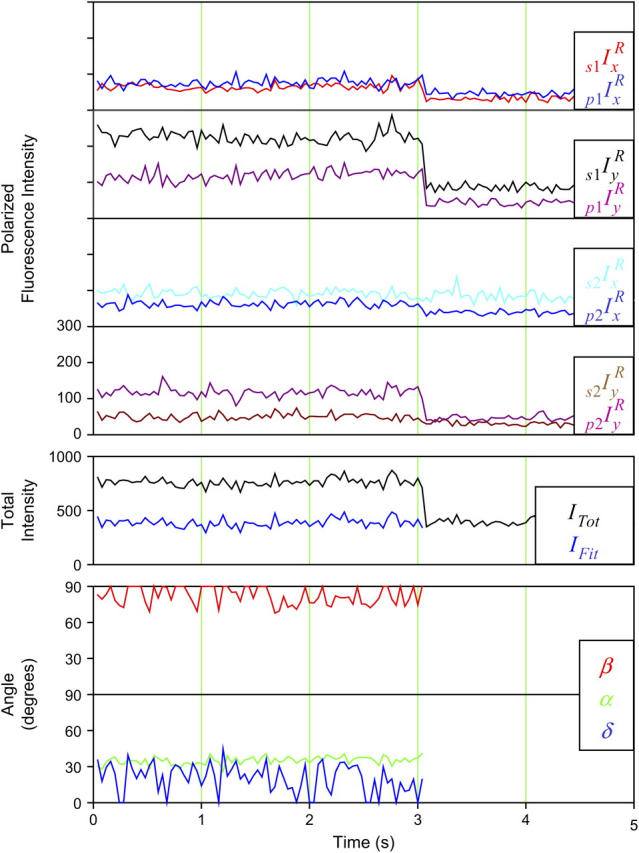
Typical single molecule polarized fluorescence data. Measurements were made from a single HMM(104-BR-115) molecule bound to an actin filament in the presence of apyrase as described in Materials and Methods. Alternation of excitation pathways (1 and 2) and excitation polarization (s and p) with simultaneous collection with two detector polarizations (x and y) produces eight measurements every 40 ms. The numbers of photons detected by the APDs in each 10 ms interval are plotted as eight polarized fluorescence intensities. The weighted sum (ITot (24)) of these intensities is plotted in black. The eight intensities are then fit to determine the orientation, β (red), α (green), slow wobble, δ (blue), and IFit (blue), a value proportional to the total intensity (minus background). The average angles for the molecule shown here are β = 82°, α = 35°, and δ = 20°. The average 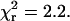 The intensity levels and angles are constant as expected for actomyosin in the absence of ATP. A high β value is common for the highly polarized probe (104-BR-115).
The intensity levels and angles are constant as expected for actomyosin in the absence of ATP. A high β value is common for the highly polarized probe (104-BR-115).
Slide preparation
The surfaces of a ∼10 μl flow chamber (24) were coated with 1 mg/ml poly-L-lysine (Sigma, P-0879) for 1 min, rinsed with water, and then track buffer (TB: 25 mM KCl, 20 mM HEPES, pH 7.4, 4 mM MgCl2, 100 mM DTT) was added. Equimolar concentrations of actin and myosin heads (10 nM actin and 5 nM HMM or 10 nM S1 in TB) were mixed with actin in suspension, and the decorated actin was applied to flow chambers to produce “tracks” of actomyosin (33). An excess of unlabeled HMM or S1 (∼200 nM) was added to the actomyosin suspension <1 min before adding two volumes of the mixture to the flow chamber. The excess unlabeled myosin fragment helped to align the actin filament axis with the flow direction (Fig. 1). The actomyosin complexes were allowed to bind to the surfaces for 2 min and then the chamber was washed with TB or TB plus 0.5 mg/ml bovine serum albumin to block the surfaces, and then TB plus 5 U/ml apyrase (grade VII, Sigma A6535). For ensemble experiments, ∼50% of the myosin heads were labeled, whereas for single molecule experiments a ∼1:1000 ratio of labeled/unlabeled myosin heads was used. Actomyosin tracks were attached to the surface via the myosin fragments as demonstrated by movement of the actin filaments upon addition of ATP.
Experimental protocol for single molecule observations
Actomyosin tracks were added to a flow chamber at typical concentrations of 5 nM actin monomers, 5 nM unlabeled myosin, and 5 pM labeled heads for single molecule observations. A single image of AEDANS-actin fluorescence was acquired with 83 ms integration time. The same field was then illuminated with the time-multiplexed 532 nm laser beams and an image of the rhodamine-labeled myosin was recorded with 149 ms integration time. On an overlay of the two images, a spot was selected containing a rhodamine fluorophore based on its intensity and its colocalization with a well-aligned actin filament. The piezo-electric stage was then moved, under computer control, to shift this spot in the x-y plane into a point conjugate with the APDs. The eight intensity traces described above were collected for 10 s (250 cycles, 40 ms each). If all of the polarized traces from a molecule bleached simultaneously to background levels in a stepwise manner, indicating bleaching of a single molecule, it was analyzed further. For traces showing two stepwise bleaches, only the interval immediately before the last bleaching step was analyzed. The polarized intensity levels after photobleaching were taken as the eight background intensities for each molecule.
Spots giving ITot = 200 − 850 counts per 40 ms recording cycle and a single step bleach were retained for further analysis. Optical correction factors X1, X2, X12, and Cd, defined in Forkey et al. ((24), Materials and Methods and Supplemental Information, Eq. B.1), were measured after each data set was recorded to calculate corrected intensities.
Values for β and α, corresponding to the axial and azimuthal angles of the probe dipoles, respectively, relative to the F-actin axis (see Fig. 2 b of Forkey et al. (24)), were determined from θ and φ and the actin orientation using an Euler rotation matrix (34). Gaussian curves were fitted to the distributions of β by minimizing the negative log of the likelihood that the data are described by a single Gaussian component (−ln(LC1) or the sum of two Gaussians (−ln(LC2); (35)):
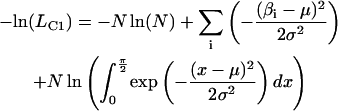 |
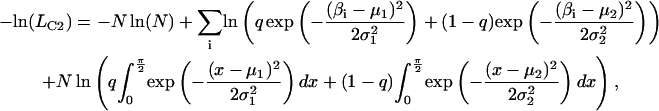 |
where the five free parameters are q, the proportion of area under component 1, μ1, and σ1, the mean and standard deviation of component 1 and μ2 and σ2, the mean and standard deviation of component 2. To evaluate the goodness of each fit, the binned distributions were compared to the predicted values (Fig. 5) by calculating the reduced χ2,
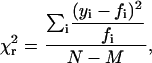 |
where yi is the measured value of each bin and fi is the predicted value for i = 1..N bins and N − M is the degrees of freedom with M equal to the number of free parameters. To determine whether the data for S1 and HMM statistically justified fitting with two Gaussian peaks rather than one, the parameter 2Δln(L) = 2(ln(LC1) − ln(LC2)) was calculated. This metric is distributed as χ2 (36) with three degrees of freedom (the difference of fitting one curve with two free parameters, μ and σ, and two curves with five free parameters, μ1, σ1, μ2, σ2, and q, the fraction of component 1). This procedure was carried out with Gaussian components either weighted by sin β or not. The peak angles were not affected by this weighting, although the partition between the two components was shifted slightly. The curves without sin β weighting fit the distributions slightly better, so those results are reported.
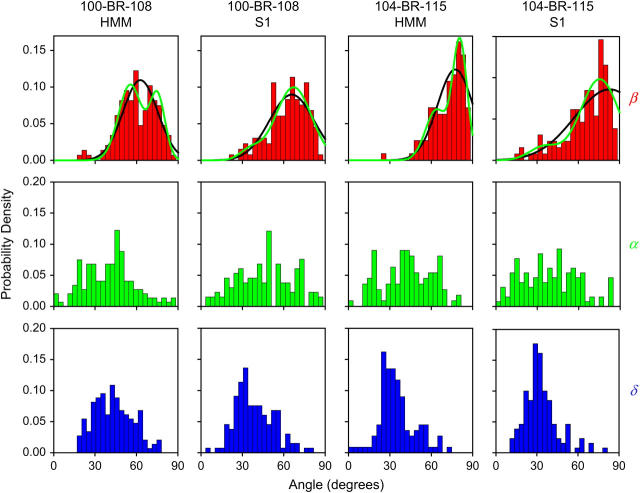
FIGURE 5.
Single molecule orientation distributions. The orientation of each molecule is found by fitting the average values of the eight polarized intensities to find the dipole orientation in the laboratory frame and then transforming those values into the actin frame to give β, α, and δ. Histograms are plotted with 3.4° bins and normalized to the same total area for comparison. Higher average β-values for (104-BR-115) relative to (100-BR-108) are apparent. α is approximately uniformly distributed. The average δ is 35–45° in all cases. The β-distributions were fit with one and two Gaussian components by minimizing −ln(L), as described in the text. Black lines are the best fits of single Gaussian components to the data; green lines are the best fit of two summed components. For both HMM cases, two components are clearly distinguished (see Table 1). For S1, the one and two distribution fits are similar, as reflected in the statistics shown in Table 1. HMM(100-BR-108) n = 147; S1(100-BR-108) n = 132; HMM(104-BR-115) n = 111; and S1(104-BR-115) n = 130.
To compare single molecule and multiple molecule experiments, order parameters were calculated directly from the distributions of single molecules (see Supplementary Information). Alternatively, the sum of intensities from many single molecules on sparsely decorated filaments was assumed to be equal to that of highly decorated filaments. Under that assumption, intensities from all of the single molecule experiments were added and the resulting values analyzed as if they had been measured from one filament.
Experimental protocol for multiple molecule observations
In experiments on actomyosin tracks with higher labeling density, the same procedure was followed to make up the sample except that myosin fragments with ∼50% of the RLC labeled were used directly instead of diluting them into unlabeled myosin fragments as above. The excitation intensity was reduced to ∼3 mW such that photobleaching was negligible for a 2 s illumination. Each of the eight polarized intensities was averaged over the entire recording period. Intensities from a clear region between filaments were subtracted from averaged filament intensities to correct for background light. The analysis is described in Supplementary Information.
RESULTS AND DISCUSSION
The term “myosin fragment” will be used throughout the article for HMM or S1. In all cases, experiments were performed on AEDANS-labeled actin filaments decorated at approximately equimolar ratios of S1 or HMM heads to actin monomers. The degree of labeling refers to the fraction of the myosin fragments containing labeled RLC.
Multiple molecule experiments
Fluorescence polarization was measured from actin filaments decorated with myosin fragments containing ∼50% labeled RLC per myosin head. The filaments selected were aligned either along the x- or y-axis as described in Materials and Methods. Experiments on these samples, similar to those described for labeled actin in the accompanying article (24), showed that the microscope and analytical procedures result in reliable angular measurements. These tests are reported in Supplementary Information. Polarization ratios, as defined in Forkey et al. (24), averaging as high as 0.4 and 0.6 were measured with 100-BR-108 and 104-BR-115, respectively, with both HMM and S1 samples (Fig. S1), indicating that the fluorophores were well ordered in these samples. The single molecule data were compared to the data from more heavily labeled filaments by adding together the intensities of many single molecules, which label actin filaments oriented in the same direction. The polarization ratio values from the “summed singles” (yellow dots in Fig. S1) closely track those from heavily labeled filaments, providing a strong indication that the single molecule data represent the same orientation distributions. Further comparisons of single molecules and muscle fibers are discussed in Supplementary Information (Table S1 and Fig. S3).
Single molecule experiments
Actin filaments were fully decorated with either HMM or S1 as above, but the labeled myosin fragments were diluted ∼1:1000 with the corresponding unlabeled fragment, allowing single rhodamine-labeled heads to be resolved (Fig. 1 and (24)).
A typical measurement from a single HMM(104-BR-115) molecule bound to an actin filament in the presence of apyrase is shown in Fig. 2. Data from HMM(100-BR-108) and both S1 samples are similar. When the fluorescence emission from a single rhodamine-labeled myosin head was projected onto the detectors, the eight polarized intensities were usually constant for 0.5–5 s and then all bleached to the background intensity level at one time (at 3 s in Fig. 2). The relative magnitudes of the eight polarized fluorescence intensities from each individual molecule were variable as expected for molecules oriented in different directions. A weighted, relatively orientation-independent, sum of these intensities (see Forkey et al. (24); Fig. 2, ITot) and the fitted value reflecting the total intensity less background, IFit, were also constant until the fluorophore bleached. Histograms of the average IFit values from individual molecules have one narrow peak (data not shown). The homogeneous intensity distribution and the single-step bleaching indicate that the data from the traces are from single fluorophores. Occasionally (17% of recordings), the total intensity trace decreased to background in two discrete steps, presumably indicating that two rhodamine molecules were located very close to each other. Only the portion of the recording preceding the last bleaching step was considered.
Analytical expressions for the probe angles (β and α) and mobility on the 4 ns ≪ τ ≪ 10 ms timescale (δ), fitted to the traces (see Materials and Methods and (24)), showed that the orientation and extent of motion are unvarying (Fig. 2, lowest two panels). The fitted total intensity, IFit (blue trace in the fifth panel), matches the ITot minus background, and the average 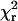 for this trace was 2.2, a typical value, indicating that the fitting procedure predicts the experimental recordings well.
for this trace was 2.2, a typical value, indicating that the fitting procedure predicts the experimental recordings well.
Transitions
In 44% of the traces recorded from single rhodamine molecules bound to RLC, the polarized fluorescence intensities showed abrupt changes before bleaching. These transitions often occurred in several of the polarized fluorescence traces in opposite directions, so that the total intensity was constant (Fig. 3). This behavior is characteristic of an angular change of the fluorophore and is incompatible with a change in the fluorophore brightness, a translation or laser intensity fluctuation. In Fig. 3, the apparent orientation of the fluorophore shows an abrupt change indicative of a rotation of 24° in β and 18° in δ. The fitted total intensity, IFit, matched the measured sum of intensities, ITot less background, and the average 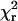 value of the fitted angles was 2.2, indicating that the data are well described by an apparent rotation of the fluorophore. One or more such transitions were observed in 31%–54% of recordings from the four combinations of the two fragments labeled with HMM(100-BR-108) and HMM(104-BR-115). This high frequency of sudden rotational motions was unexpected in experiments in the absence of ATP and possibly indicates detachment of some of the myosin heads from actin, spontaneous rotations while maintaining attachment, or artifactual photochemical events. Grouping the data according to whether a transition was observed or not showed no significant differences in average orientation. Thus, it is unlikely that the molecules undergoing apparent rotations represent a distinct orientation distribution.
value of the fitted angles was 2.2, indicating that the data are well described by an apparent rotation of the fluorophore. One or more such transitions were observed in 31%–54% of recordings from the four combinations of the two fragments labeled with HMM(100-BR-108) and HMM(104-BR-115). This high frequency of sudden rotational motions was unexpected in experiments in the absence of ATP and possibly indicates detachment of some of the myosin heads from actin, spontaneous rotations while maintaining attachment, or artifactual photochemical events. Grouping the data according to whether a transition was observed or not showed no significant differences in average orientation. Thus, it is unlikely that the molecules undergoing apparent rotations represent a distinct orientation distribution.
FIGURE 3.
Large transitions apparent in the polarized fluorescence intensities of a rigor HMM(100-BR-108) molecule. Intervals discussed in the text are indicated by arrows at the top of the plots. The total intensity (ITot) remains constant, indicating that the transitions are not a result of photobleaching or an additional fluorophore passing through the detection region. The intensity transitions indicate an apparent orientation change (Δβ = 24°) and a decrease in slow wobble (Δδ = 18°) but no change in IFit.
Intervals with steady polarized fluorescence intensities ended either with a transition of the polarized intensities (as in Fig. 3, interval A) or when the fluorophore bleached (end of interval B). Assuming both of these processes to be Markovian, the total exit rate from an interval of steady polarized intensities (kdwell, the reciprocal of the dwell time) is the sum of the bleaching and transition rates. Durations of steady intensity intervals and overall recording time before bleach were measured for a series of recordings. The dwell times for steady intensities and the fluorescence durations were both distributed approximately exponentially, giving well-constrained rate constants (Fig. 4, a and b). The rate constant for the transitions, taken as ktrans = kdwell − kbleach, was 0.41 ± 0.06 s−1 for HMM and 0.89 ± 0.05 s−1 for S1 (data from (100-BR-108) and (104-BR-115) combined). Values for the detachment rate of rigor bonds reported in the literature range from 0.01 s−1 to 0.1 s−1 (37,38). Thus the high rate of transitions observed is not consistent with the normal detachment rate of S1 or HMM.
FIGURE 4.
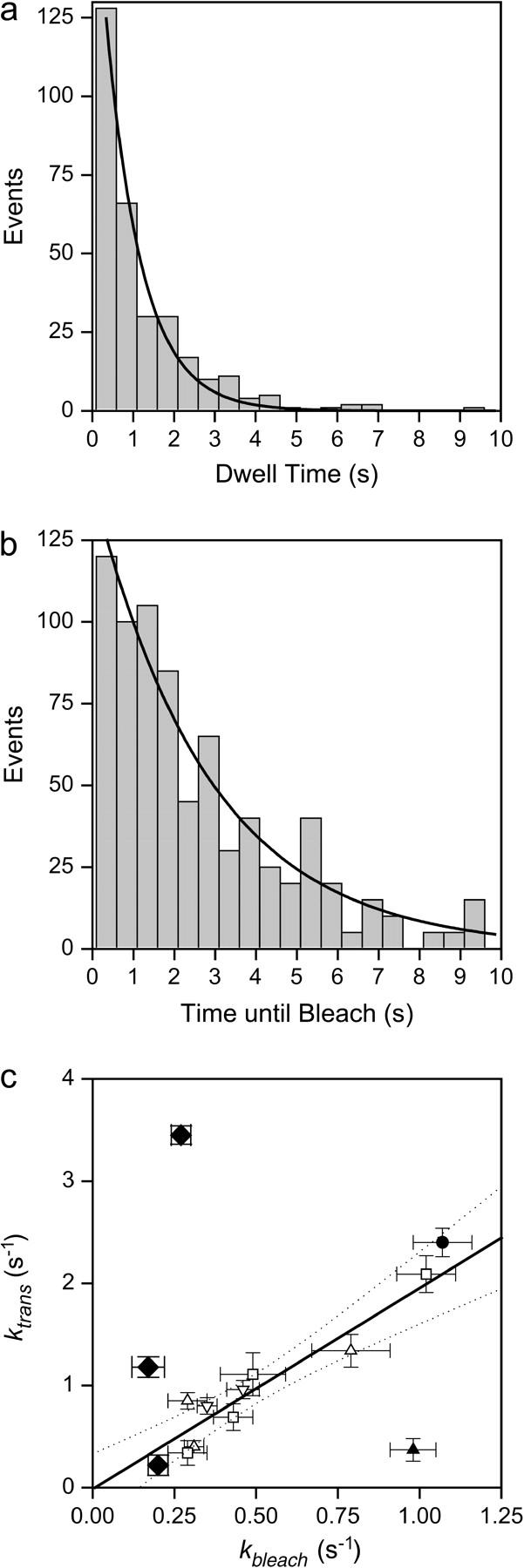
Kinetic analysis of transitions. (a) The interval durations (demonstrated by the arrows in Fig. 3) were plotted as histograms and fit with a single exponential. Here the fit to the histogram of S1(104-BR-115) is shown. The fitted curve indicates a rate constant (kdwell) of 1.14 s−1 with r2 = 0.98. (B) The durations of the traces before photobleaching (the sum of intervals A and B in Fig. 3) were analyzed as in A. Here data for S1(104-BR-115) are shown that have kbleach = 0.35 s−1, r2 = 0.93. (C) ktrans is the difference between kdwell and kbleach as described in the text. kbleach and ktrans are highly correlated for many samples, including HMM, S1, RLC alone, and labeled actin. A linear relationship (solid line) is obtained with slope = 1.97 ± 0.35, and y-intercept = −0.01 ± 0.22, r2 = 0.87. The linear fit was made excluding the apparent outlier, HMM(100-BR-108), in the lower right-hand corner. (□), samples without myosin; (Δ), HMM;, and (∇), S1. The solid circle is HMM(100-BR-108) in the presence of 1 μM ATP. Dotted lines indicate the 95% confidence intervals. (♦), myosin V labeled with BR-calmodulin light chain. The diamonds correspond to 0, 1, and 5 μM ATP in ascending order.
Similar transitions were also observed in several other samples not expected to rotate (see Fig. 4 c legend). A strong correlation (r2 = 0.97) was found between the photobleaching rates, kbleach, and transition rates, ktrans, of these different samples (Fig. 4 c, squares). kbleach was markedly different between the various experiments due either to varied laser power level or to varying resistance to photobleaching. ktrans was close to 2 × kbleach and the intercept of the regression line was near zero (Fig. 4 c). This result strongly suggests that the sudden transitions represent a photochemical artifact.
When kbleach was purposefully varied by illuminating HMM(100-BR-108) with different laser power levels (Fig. 4 c, open triangles), the data were again highly correlated with approximately the same relationship as the other samples. One set of HMM(100-BR-108) (solid triangle in Fig. 4 c) is very different, having an unexpectedly low rate of angular transitions. When this outlier is excluded, the slope and intercept are 1.97 ± 0.35 and −0.01 ± 0.22, respectively (mean ± SE, r2 = 0.87). When this point is included in the regression, the slope is 1.03 ± 0.56 and the y-intercept is 0.34 ± 0.34 (r2 = 0.30). The similar relationship between bleaching rate and transition rate for these different protein samples indicates that the predominant mechanism for the apparent rotation is an artifactual photochemical phenomenon in which the transitions are proportional to the number of excitation-emission cycles.
In another study, data from myosin V labeled with BR on a calmodulin light chain (9) were acquired at several ATP concentrations (Fig. 4 c, black diamonds). In the absence of ATP, ktrans versus kbleach fell on the same regression line shown in Fig. 4 c. In the presence of 1 and 5 μM ATP, however, ktrans increased with increasing [ATP], whereas kbleach remained unchanged. Under these conditions, the rotations far outnumbered the photochemical transitions making dynamic orientation measurements reliable (9).
The single molecule data plotted in Fig. S1 (yellow dots) and all of the data presented below are taken from the initial portion of each single molecule recording before the first photochemically induced transition has occurred (e.g., interval A in Fig. 3). By accepting only the initial portion of each recording, the major effects of this artifact are removed from the orientation data in the absence of ATP. However, the photochemical phenomena present under these conditions prevented us from studying active rotational motions of myosin II in the presence of ATP.
Two additional apparent photochemical effects were detected in the single molecule data. In ∼5% of the single molecule traces, the total fluorescence intensity suddenly increased for 40–120 ms just before the bleach. Wazawa et al. (39) detected similar intensity spikes in single molecule fluorescence recordings from tetramethylrhodamine probes bound to Cys707 of S1, and they found a simultaneous blue shift in the fluorophore emission spectrum. Presumably some processes that lead to photobleaching briefly alter the spectral properties and orientations.
When measurements were made in the presence of antifade enzymes (glucose oxidase and catalase as per Harada et al. (40)) and high concentrations of DTT (>50 mM), apparent transient darkening and recovery (“blinking”) was observed. A high concentration of DTT was more effective than enzymes alone or enzymes with low DTT concentrations in delaying bleaching without introducing blinking. Therefore, deoxygenating enzymes were not included in the experiments. We also observed blinking in rhodamine-labeled actin (41). This behavior may indicate reversible production of a nonfluorescent chemical species of the chromophore. These phenomena would be difficult to detect in ensemble experiments, but their presence should be considered in studies of protein rotational motions using single molecule fluorescence polarization.
Angular distributions of individual molecules
Orientation distributions of the probe dipoles relative to the actin axis (β) in the absence of ATP, and before any apparent photochemically induced rotational motions, are plotted in Fig. 5. Fitting the same equations to the time-course data successively for each 40 ms time point and then calculating the average of the fitted orientations gave similar results. Fitted values for β, α, or δ >89.5° or <0.5° were not reliable, due to the shallow χ2 surfaces in these angular regions, and were discarded (24). The average 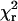 was 1.3 ± 0.1, indicating that the equations fit the intensity data well.
was 1.3 ± 0.1, indicating that the equations fit the intensity data well.
The azimuthal angle of the probes, α, is expected to be uniformly distributed around the actin axis. In all four experimental samples, the mean value of α is near 45° (Fig. 5), and the spread of values is broad (SD ∼ 20°). Values of α close to 0° and 90°, however, are populated less than expected for a perfectly uniform distribution. This behavior also occurs in simulated data (42) and thus accounts for the observed distribution of α. There is no correlation (r < 0.1) for any of the samples between α and β or δ, further suggesting that the probes are randomly distributed around the actin axis.
The distributions of axial angle, β, have mean angles of 61° (HMM(100-BR-108)), 63° (S1(100-BR-108)), 73° (HMM(104-BR-115)), and 67° (S1(104-BR-115)). The higher values of β (more perpendicular to the actin axis) for S1(104-BR-115) and HMM(104-BR-115), relative to those for S1(100-BR-108) and HMM(100-BR-108), are consistent with data on these probes within muscle fibers (13,21).
The distributions of β were more narrow (SD 11°–17°) than those for α. The distributions of individual β-values for both S1 samples are broader than the corresponding distributions from HMM (Fig. 5). The β-distributions for HMM(100-BR-108) and HMM(104-BR-115) each show two clear peaks (Fig. 5, Table 1).
TABLE 1.
Gaussian fits to single molecule distributions
| Peak 1
|
Peak 2
|
||||||
|---|---|---|---|---|---|---|---|
| βo | σ | βo | σ | q |
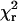 (df) (df) |
p2 | |
| HMM 100-BR-108 | 62 | 13 | – | – | – | 0.65 (21) | <0.01 |
| 55 | 9 | 75 | 5 | 54 | 0.39 (18) | ||
| S1 100-BR-108 | 65 | 16 | – | – | – | 0.84 (24) | 0.38 |
| 35 | 8 | 67 | 13 | 12 | 0.81 (21) | ||
| HMM 104-BR-115 | 75 | 13 | – | – | – | 0.45 (23) | 0.04 |
| 63 | 8 | 80 | 5 | 30 | 0.18 (20) | ||
| S1 104-BR-115 | 82 | 26 | – | – | – | 1.14 (24) | 0.13 |
| 35 | 11 | 75 | 14 | 14 | 0.89 (21) | ||
One or two Gaussian distributions were fit to the β-distributions of single molecule measurements (red histograms with black and green curves in Fig. 5). The peak (βo) and spread (σ) of the best fits are reported. In the case of two Gaussians, “q” indicates the fraction of molecules that fall into the lower angle component. The goodness of fit of each model is indicated by 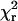 (df = degrees of freedom). p1 is the probability that the improvement of the fit, when adding a second Gaussian component, is due to chance. In both HMM cases, fitting with two Gaussians is statistically justified.
(df = degrees of freedom). p1 is the probability that the improvement of the fit, when adding a second Gaussian component, is due to chance. In both HMM cases, fitting with two Gaussians is statistically justified.
Model angular distributions with one or two Gaussian components were fit to the β-distributions of all of the samples (Fig. 5), by minimizing the negative logarithm of the likelihood (−ln(L)) that the model is the best fit (see Materials and Methods). For HMM(100-BR-108) and HMM(104-BR-115), including the few molecules having β-values <30° increased the 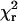 values of the fitted orientation distributions 3- to 10-fold and were thus discarded as outliers. No outliers were discarded for S1(100-BR-108) or S1(104-BR-115).
values of the fitted orientation distributions 3- to 10-fold and were thus discarded as outliers. No outliers were discarded for S1(100-BR-108) or S1(104-BR-115).
Values of 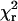 were ≤1.1 for all of the fitted curves, indicating that the Gaussian curves describe the data well (Fig. 5 and Table 1). The
were ≤1.1 for all of the fitted curves, indicating that the Gaussian curves describe the data well (Fig. 5 and Table 1). The 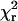 values for both HMM species were decreased substantially by using two components rather that one, whereas minimal reduction of
values for both HMM species were decreased substantially by using two components rather that one, whereas minimal reduction of 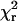 was obtained by fitting S1 data with two components (see Materials and Methods and Table 1). For both HMM samples, the likelihood (p1 in Table 1) that the improvement of fit obtained by adding the second Gaussian component is due to chance is <0.05, indicating that the data justify fitting with two components.
was obtained by fitting S1 data with two components (see Materials and Methods and Table 1). For both HMM samples, the likelihood (p1 in Table 1) that the improvement of fit obtained by adding the second Gaussian component is due to chance is <0.05, indicating that the data justify fitting with two components.
For HMM(104-BR-115), the two peaks of the β-distribution are nearly equally populated (54%/46%), as indicated by the fitted Gaussian components (Fig. 5, Table 1). These peaks (55° and 75°) straddle the peak of S1(104-BR-115) (62°). The most straightforward explanation of this result is that the two heads of HMM bind at two different angles to adjacent actin monomers in the long pitch (13,43). For HMM(100-BR-108), two statistically justified populations which straddle the peak of S1(100-BR-108) are also evident (Fig. 5, Table 1), but the partition between the two components is less equal (30%/70%). Some of the HMM molecules may have only one head bound, although the angular spread of the major component (σ = 9°) is comparable to that of HMM(104-BR-115), suggesting that single-headed binding does not make a major contribution.
The orientation distributions of the two labeled S1 species both showed a main peak and a tail extending toward low angles (Fig. 5). One Gaussian component fit these data and adding a second component did not cause substantial improvement of the fit, as measured by 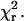 For both S1(100-BR-108) and S1(104-BR-115), the peak of the distribution representing the majority of S1 heads is positioned between the two angles found for HMM. This behavior again suggests that both heads of actin-bound HMM are bent away from the strain-free S1 orientation. The small tails of the S1 distributions at small angles may represent heads bound to actin in nonrigor conformations or bound to the poly-L-lysine surface instead of actin.
For both S1(100-BR-108) and S1(104-BR-115), the peak of the distribution representing the majority of S1 heads is positioned between the two angles found for HMM. This behavior again suggests that both heads of actin-bound HMM are bent away from the strain-free S1 orientation. The small tails of the S1 distributions at small angles may represent heads bound to actin in nonrigor conformations or bound to the poly-L-lysine surface instead of actin.
Microsecond wobble
The 40 ms time resolution enabled measurement of “slow” wobble on the 4 ns ≪ τ ≪ 10 ms timescale (δ). The average amplitude of δ ranged from 33° to 43° for all samples, with a distribution as tight as that for β (SD = 12°–14°). No difference in δ between HMM and S1 or the two probes was detected. This amplitude of wobbling suggests a significant degree of protein motion on the microsecond timescale. If the stiffness of S1 bound to actin is assumed to be 1 pN per nm of translation at its C-terminus (44,45) and the 9.5 nm lever arms of myosin fragments undergo rigid thermal motions, the value of δ would be expected to be only ∼17°. The larger value of δ measured in our experiments suggests that internal motions within the lever arm, such as cantilever bending (19,46) or twisting (13,21,47), also contribute to the μs wobble.
With ensemble data, the static and mobile contributions to disorder of the distributions cannot be differentiated using conventional fluorescence polarization techniques. Transient methods, such as polarized fluorescence depletion (16), can access dynamic rotational information. The amplitude of microsecond motion estimated by polarized fluorescence depletion was 31° in rigor muscle fibers (δp in Table 2 of Bell et al. (16)), quite similar to the amplitude of microsecond motions found here (δ = 33°–43° for the various samples). Although the probe labeling the RLC in Bell et al. (16) was a monofunctional rhodamine, which adopts a different local orientation, the microsecond motions are expected to reflect protein wobble.
Several groups have measured microsecond motions in the MD and LCD of muscle fibers and myosin fragments (48–50). None of these measurements were simultaneously sensitive to orientation. Brown et al. (50) interpreted the motion as wobble in a cone and can, therefore, be directly compared with our measurements. They found that the LCD in thick filaments wobbles with a half-cone angle of 29°, a value somewhat smaller than that reported here. Spectroscopic measurements (saturation transfer electron paramagnetic resonance and phosphorescence anisotropy) show that the MD and LCD rotate independently of one another, though they find that the degree of microsecond motion in these two domains is indistinguishable in rigor. It is possible that myosin is rigid in rigor, causing the MD and LCD to lose their independence. In the accompanying article (24), we report microsecond motions of actin (δ = 37°) on the same scale as that measured for the LCD. This result is consistent with a rigid link between actin and myosin. We have not measured δ for the MD but, as mentioned above, others have found the same degree of microsecond motion in the MD and LCD.
Protein orientations calculated by combining data from the two probe sites
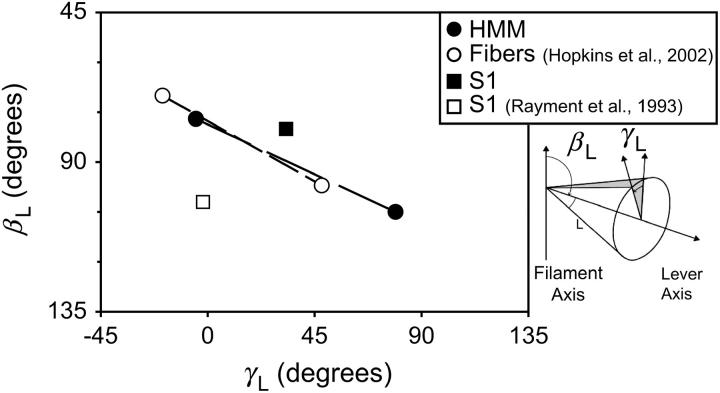
The known orientations of the two probes relative to the myosin crystal structure allow estimation of the orientation distribution of the protein subunits in three dimensions (21). The orientation of the myosin neck can be defined by a “lever axis” along a line from Cys707 to Lys843 (residue numbers from chicken skeletal muscle myosin) near the two probable pivot points in myosin straddling the light chains. The axial angle between the actin axis and the lever is termed βL, and the rotation of the lever about its axis is γL (Fig. 6, S2; see also Hopkins et al. (13)). Orientation data from the two separate probe sites ((100-BR-108) and (104-BR-115)) can be combined with their known local orientations in the RLC (13) to calculate the angular position of the LCD (βL, γL). Along with each β-value from the two peaks in Fig. 5 for each HMM probe site, its reflection across the equatorial plane at (180° − β) has to be considered, leading to 16 sets of possible βL, γL angles for the lever axis. Among these possible orientations for the lever arm, one pair at (βL, γL) = (77°, −5°) and (105°, 79°) (plotted as solid circles in Fig. 6) is similar to the set of orientations found for endogenous heads in rigor muscle fibers ((13); open circles in Fig. 6 here). βL of each head is similar in single molecule HMM measurements and muscle fiber experiments (∼5° greater in both cases); γL measured here is smaller than that measured in fibers (twisted clockwise) by ∼25°. The differences may represent the effects of mismatched periodicities in thick and thin filaments in muscle fibers that are presumably relieved for HMM. The orientations measured in HMM lead to an axial separation (Δz) of 4.6 nm between the two catalytic domains if the head-rod junctions meet (Δz = LL (cos (βL1) − cos (βL2)), where LL is the length of the lever arm, 9.5 nm if taken from Cys707 to Lys843). Thus these data are compatible with two heads of an HMM molecule binding to adjacent actin monomers with their head-rod junctions very close.
FIGURE 6.
Three-dimensional orientation of myosin LCD. The polar angles βL and γL, measured for HMM (•) and S1 (▪), are plotted. For comparison, polar angles measured in muscle fibers (○) with the same fluorescent probes by Hopkins et al. (13) are shown. Dashed lines connect pairs of heads from HMM or muscle fibers. Orientations of HMM and muscle fibers are in good agreement. Polar angles determined for S1 docked into cryoelectron micrographs (□) by Rayment et al. (4) are also shown. The orientation of S1 measured with the same technique is intermediate to the two-headed structures and S1 from cryoelectron microscopy is in the same angular vicinity.
When the same kind of analysis is applied to the Gaussian peaks fitted to the S1 data and equatorial reflections of these peaks are taken into account, four possible orientations emerge (Fig. S3). One orientation, (βL, γL) = (80°, 33°), is located between the two most likely peaks for HMM (Fig. 6, solid square). Chicken skeletal muscle S1 heads docked into cryoelectron micrographs of decorated actin (4) are also oriented in the same angular vicinity (βL, γL) = (102°, −2°) (Fig. 6, open square). It is worth noting that to make the Rayment S1 structure agree with data from muscle fibers (13) and HMM (18,19), the S1 structure must be rotated (or bent) about the axis of the actin filament (αL) at least 20°. In addition, we can compare our measured βL to those predicted for HMM using fluorescence resonance energy transfer and luminescence resonance energy transfer. Chakrabarty et al. (18) rotated Rayment's docked S1 structure to correspond to the short distances measured between RLCs of HMM. One possible solution was to rotate one head of HMM in the axial direction, resulting in heads with βL = 102° and 77°, which agrees well with our HMM measurements. This analysis supports the interpretation that both heads of HMM are bent toward each other and nearly meet at the head-rod junction—implying that the α helix of S2 does not have to unwind—and that the strain-free orientation of S1 is intermediate. A number of other EM and spectroscopic studies have reached the same conclusions (13,18,19,43,51–53).
We applied maximum entropy analysis (54) to the HMM and S1 data to obtain possible distributions of (βL, γL) (Supplemental Information). The outcome was similar to that described here using the Gaussian peaks. Given that the current HMM measurements compare well with those using the same fluorescent probes in muscle fibers (13), as do EPR measurements (19,51), strain and motion in the LCD seem responsible for most of the RLC disorder—not mismatching filament periodicities in muscle. The shift between the current measured HMM orientations and those in fibers (13) could reflect the filament periodicities.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
We thank Drs. Annemarie Weber and Earl Homsher for biochemistry protocols, Eric Gallo, Joby Geevarghese, Ilya Gertsman, and Lisa Prentiss for technical assistance, Dr. Martin Pring for help on the fitting and statistical analysis, and Drs. Robert E. Dale, E. Michael Ostap, and Henry Shuman for useful discussion regarding the results presented here.
Joseph N. Forkey's present address is Precision Optics Corp., Gardner, MA 01440.
Margot E. Quinlan's present address is Dept. of Cellular and Molecular Pharmacology, University of California, San Francisco, CA 94143-2200.
References
- 1.Huxley, A. F. 1957. Muscle structure and theories of contraction. Prog. Biophys. Biophys. Chem. 7:255–318. [PubMed] [Google Scholar]
- 2.Huxley, H. E. 1969. The mechanism of muscular contraction. Science. 164:1356–1365. [PubMed] [Google Scholar]
- 3.Sellers, J. R. 2000. Myosins: a diverse superfamily. Biochim. Biophys. Acta. 1496:3–22. [DOI] [PubMed] [Google Scholar]
- 4.Rayment, I., H. M. Holden, M. Whittaker, C. B. Yohn, M. Lorenz, K. C. Holmes, and R. A. Milligan. 1993. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 261:58–65. [DOI] [PubMed] [Google Scholar]
- 5.Goldman, Y. E. 1998. Wag the tail: structural dynamics of actomyosin. Cell. 93:1–4. [DOI] [PubMed] [Google Scholar]
- 6.Geeves, M. A., and K. C. Holmes. 1999. Structural mechanism of muscle contraction. Annu. Rev. Biochem. 68:687–728. [DOI] [PubMed] [Google Scholar]
- 7.Taylor, K. A., H. Schmitz, M. C. Reedy, Y. E. Goldman, C. Franzini-Armstrong, H. Sasaki, R. T. Tregear, K. Poole, C. Lucaveche, R. J. Edwards, L. F. Chen, H. Winkler, and M. K. Reedy. 1999. Tomographic 3D reconstruction of quick-frozen, Ca2+-activated contracting insect flight muscle. Cell. 99:421–431. [DOI] [PubMed] [Google Scholar]
- 8.Tsaturyan, A. K., S. Y. Bershitsky, R. Burns, and M. A. Ferenczi. 1999. Structural changes in the actin-myosin cross-bridges associated with force generation induced by temperature jump in permeabilized frog muscle fibers. Biophys. J. 77:354–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forkey, J. N., M. E. Quinlan, M. A. Shaw, J. E. T. Corrie, and Y. E. Goldman. 2003. Three-dimensional structural dynamics of myosin V by single-molecule fluorescence polarization. Nature. 422:399–404. [DOI] [PubMed] [Google Scholar]
- 10.Irving, M., T. St Claire Allen, C. Sabido-David, J. S. Craik, B. Brandmeier, J. Kendrick-Jones, J. E. T. Corrie, D. R. Trentham, and Y. E. Goldman. 1995. Tilting of the light-chain region of myosin during step length changes and active force generation in skeletal muscle. Nature. 375:688–691. [DOI] [PubMed] [Google Scholar]
- 11.Hopkins, S. C., C. Sabido-David, J. E. T. Corrie, M. Irving, and Y. E. Goldman. 1998. Fluorescence polarization transients from rhodamine isomers on the myosin regulatory light chain in skeletal muscle fibers. Biophys. J. 74:3093–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piazzesi, G., M. Reconditi, M. Linari, L. Lucii, Y.-B. Sun, T. Narayanan, P. Boesecke, V. Lombardi, and M. Irving. 2002. Mechanism of force generation by myosin heads in skeletal muscle. Nature. 415:659–662. [DOI] [PubMed] [Google Scholar]
- 13.Hopkins, S. C., C. Sabido-David, U. A. van der Heide, R. E. Ferguson, B. D. Brandmeier, R. E. Dale, J. Kendrick-Jones, J. E. T. Corrie, D. R. Trentham, M. Irving, and Y. E. Goldman. 2002. Orientation changes of the myosin light chain domain during filament sliding in active and rigor muscle. J. Mol. Biol. 318:1275–1291. [DOI] [PubMed] [Google Scholar]
- 14.Ishii, Y., A. Ishijima, and T. Yanagida. 2001. Single molecule nanomanipulation of biomolecules. Trends Biotechnol. 19:211–216. [DOI] [PubMed] [Google Scholar]
- 15.Baker, J. E., I. Brust-Mascher, S. Ramachandran, L. E. W. LaConte, and D. D. Thomas. 1998. A large and distinct rotation of the myosin light chain domain occurs upon muscle contraction. Proc. Natl. Acad. Sci. USA. 95:2944–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell, M. G., R. E. Dale, U. A. van der Heide, and Y. E. Goldman. 2002. Polarized fluorescence depletion reports orientation distribution and rotational dynamics of muscle cross-bridges. Biophys. J. 83:1050–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huxley, H. E., and W. Brown. 1967. The low-angle x-ray diagram of vertebrate striated muscle and its behaviour during contraction and rigor. J. Mol. Biol. 30:383–434. [DOI] [PubMed] [Google Scholar]
- 18.Chakrabarty, T., M. Xiao, R. Cooke, and P. R. Selvin. 2002. Holding two heads together: stability of the myosin II rod measured by resonance energy transfer between the heads. Proc. Natl. Acad. Sci. USA. 99:6011–6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baumann, B. A. J., H. Liang, K. Sale, B. D. Hambly, and P. G. Fajer. 2004. Myosin regulatory domain orientation in skeletal muscle fibers: application of novel electron paramagnetic resonance spectral decomposition and molecular modeling methods. Biophys. J. 86:3030–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hambly, B., K. Franks, and R. Cooke. 1991. Orientation of spin-labeled light chain-2 exchanged onto myosin cross-bridges in glycerinated muscle fibers. Biophys. J. 59:127–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Corrie, J. E. T., B. D. Brandmeier, R. E. Ferguson, D. R. Trentham, J. Kendrick-Jones, S. C. Hopkins, U. A. van der Heide, Y. E. Goldman, C. Sabido-David, R. E. Dale, S. Criddle, and M. Irving. 1999. Dynamic measurement of myosin light-chain-domain tilt and twist in muscle contraction. Nature. 400:425–430. [DOI] [PubMed] [Google Scholar]
- 22.Thomas, D. D., and R. Cooke. 1980. Orientation of spin-labeled myosin heads in glycerinated muscle fibers. Biophys. J. 32:891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berger, C. L., J. S. Craik, D. R. Trentham, J. E. T. Corrie, and Y. E. Goldman. 1996. Fluorescence polarization of skeletal muscle fibers labeled with rhodamine isomers on the myosin heavy chain. Biophys. J. 71:3330–3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Forkey, J. N., M. E. Quinlan, and Y. E. Goldman. 2005. Measurement of single macromolecule orientation by total internal reflection fluorescence polarization microscopy. Biophys. J. 89:1261–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quinlan, M. E., J. N. Forkey, J. E. T. Corrie, and Y. E. Goldman. 2002. Orientation of the myosin light chain region by single- and multi-molecule total internal reflection fluorescence polarization microscopy. Biophys. J. 82:404a. (Abstr.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quinlan, M. E., J. N. Forkey, J. E. T. Corrie, and Y. E. Goldman. 1999. Tilting of the light chain region in single myosin molecules using total internal reflection fluorescence polarization microscopy. Biophys. J. 76:A165. (Abstr.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kron, S. J., Y. Y. Toyoshima, T. Q. P. Uyeda, and J. A. Spudich. 1991. Assays for actin sliding movement over myosin-coated surfaces. Methods Enzymol. 196:399–416. [DOI] [PubMed] [Google Scholar]
- 28.Margossian, S. S., and S. Lowey. 1982. Preparation of myosin and its subfragments from rabbit skeletal muscle. Methods Enzymol. 85(Pt. B):55–71. [DOI] [PubMed] [Google Scholar]
- 29.Okamoto, Y., and T. Sekine. 1985. A streamlined method of subfragment one preparation from myosin. J. Biochem. (Tokyo). 98:1143–1145. [DOI] [PubMed] [Google Scholar]
- 30.Corrie, J. E. T., J. S. Craik, and V. R. N. Munasinghe. 1998. A homobifunctional rhodamine for labeling proteins with defined orientations of a fluorophore. Bioconjug. Chem. 9:160–167. [DOI] [PubMed] [Google Scholar]
- 31.Kron, S. J., and J. A. Spudich. 1986. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc. Natl. Acad. Sci. USA. 83:6272–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sase, I., H. Miyata, S. Ishiwata, and K. Kinosita Jr. 1997. Axial rotation of sliding actin filaments revealed by single-fluorophore imaging. Proc. Natl. Acad. Sci. USA. 94:5646–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toyoshima, Y. Y., C. Toyoshima, and J. A. Spudich. 1989. Bidirectional movement of actin filaments along tracks of myosin heads. Nature. 341:154–156. [DOI] [PubMed] [Google Scholar]
- 34.Zare, R. N. 1988. Angular Momentum: Understanding Spatial Aspects in Chemistry and Physics. Wiley-Interscience, New York. 77–81.
- 35.Press, W. H., S. A. Teukolsky, W. T. Vetterling, and B. P. Flannery. 1992. Numerical Recipes in C: The Art of Scientific Computing, 2nd ed. Cambridge University Press, New York.
- 36.Mood, A. M., F. A. Graybill, and D. C. Boes. 1974. Tests of hypotheses. In Introduction to the Theory of Statistics, 3rd ed. McGraw-Hill, New York.
- 37.Marston, S., and A. Weber. 1975. The dissociation constant of the actin-heavy meromyosin subfragment-1 complex. Biochemistry. 14:3868–3873. [DOI] [PubMed] [Google Scholar]
- 38.Nishizaka, T., R. Seo, H. Tadakuma, K. Kinosita Jr., and S. Ishiwata. 2000. Characterization of single actomyosin rigor bonds: load dependence of lifetime and mechanical properties. Biophys. J. 79:962–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wazawa, T., Y. Ishii, T. Funatsu, and T. Yanagida. 2000. Spectral fluctuation of a single fluorophore conjugated to a protein molecule. Biophys. J. 78:1561–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harada, Y., K. Sakurada, T. Aoki, D. D. Thomas, and T. Yanagida. 1990. Mechanochemical coupling in actomyosin energy transduction studied by in vitro movement assay. J. Mol. Biol. 216:49–68. [DOI] [PubMed] [Google Scholar]
- 41.Rosenberg, S. A., M. E. Quinlan, J. N. Forkey, and Y. E. Goldman. 2005. Rotational motions of macromolecules by single-molecule fluorescence microscopy. Acc. Chem. Res. In press. [DOI] [PubMed]
- 42.Quinlan, M. E. 2002. Single molecule fluorescence polarization studies of the myosin light chain domain. PhD thesis. University of Pennsylvania, Philadelphia.
- 43.Schmitz, H., M. C. Reedy, M. K. Reedy, R. T. Tregear, H. Winkler, and K. A. Taylor. 1996. Electron tomography of insect flight muscle in rigor and AMPPNP at 23 degrees C. J. Mol. Biol. 264:279–301. [DOI] [PubMed] [Google Scholar]
- 44.Veigel, C., M. L. Bartoo, D. C. S. White, J. C. Sparrow, and J. E. Molloy. 1998. The stiffness of rabbit skeletal actomyosin cross-bridges determined with an optical tweezers transducer. Biophys. J. 75:1424–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linari, M., I. Dobbie, M. Reconditi, N. Koubassova, M. Irving, G. Piazzesi, and V. Lombardi. 1998. The stiffness of skeletal muscle in isometric contraction and rigor: the fraction of myosin heads bound to actin. Biophys. J. 74:2459–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dobbie, I., M. Linari, G. Piazzesi, M. Reconditi, N. Koubassova, M. A. Ferenczi, V. Lombardi, and M. Irving. 1998. Elastic bending and active tilting of myosin heads during muscle contraction. Nature. 396:383–387. [DOI] [PubMed] [Google Scholar]
- 47.Volkmann, N., D. Hanein, G. Ouyang, K. M. Trybus, D. J. DeRosier, and S. Lowey. 2000. Evidence for cleft closure in actomyosin upon ADP release. Nat. Struct. Biol. 7:1147–1155. [DOI] [PubMed] [Google Scholar]
- 48.Adhikari, B., K. Hideg, and P. G. Fajer. 1997. Independent mobility of catalytic and regulatory domains of myosin heads. Proc. Natl. Acad. Sci. USA. 94:9643–9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roopnarine, O., A. G. Szent-Gyorgyi, and D. D. Thomas. 1998. Microsecond rotational dynamics of spin-labeled myosin regulatory light chain induced by relaxation and contraction of scallop muscle. Biochemistry. 37:14428–14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown, L. J., N. Klonis, W. H. Sawyer, P. G. Fajer, and B. D. Hambly. 2001. Independent movement of the regulatory and catalytic domains of myosin heads revealed by phosphorescence anisotropy. Biochemistry. 40:8283–8291. [DOI] [PubMed] [Google Scholar]
- 51.Zhao, L., J. Gollub, and R. Cooke. 1996. Orientation of paramagnetic probes attached to gizzard regulatory light chain bound to myosin heads in rabbit skeletal muscle. Biochemistry. 35:10158–10165. [DOI] [PubMed] [Google Scholar]
- 52.Chakrabarty, T., C. Yengo, C. Baldacchino, L.-Q. Chen, H. L. Sweeney, and P. R. Selvin. 2003. Does the S2 rod of myosin II uncoil upon two-headed binding to actin? A leucine-zippered HMM study. Biochemistry. 42:12886–12892. [DOI] [PubMed] [Google Scholar]
- 53.Liu, J., M. C. Reedy, Y. E. Goldman, C. Franzini-Armstrong, H. Sasaki, R. T. Tregear, C. Lucaveche, H. Winkler, B. A. J. Baumann, J. M. Squire, T. C. Irving, M. K. Reedy, and K. A. Taylor. 2004. Electron tomography of fast frozen, stretched rigor fibers reveals elastic distortions in the myosin crossbridges. J. Struct. Biol. 147:268–282. [DOI] [PubMed] [Google Scholar]
- 54.Van der Heide, U. A., S. C. Hopkins, and Y. E. Goldman. 2000. A maximum entropy analysis of protein orientations using fluorescence polarization data from multiple probes. Biophys. J. 78:2138–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.