Abstract
The flow of information through the epidermal growth factor receptor (EGFR) is shaped by molecular interactions in the plasma membrane. The EGFR is associated with lipid rafts, but their role in modulating receptor mobility and subsequent interactions is unclear. To investigate the role of nanoscale rafts in EGFR dynamics, we used single-molecule fluorescence imaging to track individual receptors and their dimerization partner, human epidermal growth factor receptor 2 (HER2), in the membrane of human mammary epithelial cells. We found that the motion of both receptors was interrupted by dwellings within nanodomains. EGFR was significantly less mobile than HER2. This difference was likely due to F-actin because its depolymerization led to similar diffusion patterns between the EGFR and HER2. Manipulations of membrane cholesterol content dramatically altered the diffusion pattern of both receptors. Cholesterol depletion led to almost complete confinement of the receptors, whereas cholesterol enrichment extended the boundaries of the restricted areas. Interestingly, F-actin depolymerization partially restored receptor mobility in cholesterol-depleted membranes. Our observations suggest that membrane cholesterol provides a dynamic environment that facilitates the free motion of EGFR and HER2, possibly by modulating the dynamic state of F-actin. The association of the receptors with lipid rafts could therefore promote their rapid interactions only upon ligand stimulation.
INTRODUCTION
The epidermal growth factor receptor (EGFR) conveys extracellular information to the intracellular compartment. The information transfer is initiated by ligand binding that induces dimerization of the receptor with itself or with other members of the erbB family. Receptor dimerization and other molecular interactions in the plane of the membrane are likely affected by the membrane microenvironment. Both EGFR and its dimerization partner, human EGFR 2 (HER2 or erbB2), have been found associated with lipid rafts (1,2), which are membrane microdomains that are enriched in cholesterol and sphingolipids. It is thought that small and transient “reserved” rafts coalesce into larger and relatively stable rafts upon cell stimulation or receptor oligomerization (3) and serve as intermediate structures in the signaling process (4–6). The involvement of lipid rafts in EGFR signaling has been investigated by manipulating the content of membrane cholesterol, which alters both raft structures and signaling pathways (7–11). However, the mechanism by which lipid rafts affect EGFR signaling is unclear.
Lipid rafts, defined by their isolation as a low-density fraction from cold detergent membrane extracts, are dispersed by depletion of membrane cholesterol. Cholesterol depletion, therefore, might indicate the involvement of lipid rafts with certain cellular functions but cannot exclude the involvement of other structures, such as the actin cytoskeleton, in these processes (12,13). For example, phosphatidylinositol (4,5)-bisphosphate (PIP2), which is a major regulator of the actin cytoskeleton, has been shown to be delocalized from the membrane with cholesterol depletion (14,15). The sequestration of PIP2, like cholesterol depletion, alters the organization of actin and inhibits the lateral diffusion of membrane proteins (16). The motion of membrane proteins is confined by the cortical, membrane-associated F-actin (17–19). The actin strands in the cytoskeleton network are thought to stericly interact with the cytoplasmic tail of the proteins, confining them into microdomains. Single particle tracking (SPT) techniques have supported this view (12,20–24). These studies have suggested that corrals are formed by immobilized membrane-associated proteins that interact with the cytoskeleton, together creating fences and pickets within the plasma membrane (25–28). Membrane proteins were found to dwell, on the order of seconds, within transient confinement zones that could represent lipid rafts (29–35). The EGFR itself has also been shown to be associated with F-actin (36–38) and this interaction is thought to play an important role in receptor signaling (39–42). Interestingly, the EGFR has been shown to evoke cortical actin polymerization and stress fiber breakdown (43–45). The effect of EGFR on F-actin has been linked to PIP2 (46,47), which also accumulates in lipid rafts (48).
To better understand the functional significance of the association of the EGFR and its dimerization partner with lipid rafts, we used time-dependent single-molecule fluorescence imaging to identify and quantify the motion patterns of the receptor and its dimerization partner, HER2. Individual EGFR and HER molecules were followed in human mammary epithelial (HME) cells while manipulating the cholesterol content of the membrane and the dynamic state of F-actin. Our observations suggest that membrane cholesterol plays a potent modulatory role in the lateral mobility of both EGFR and HER2 in the membrane and thereby provides a possible mechanism by which cholesterol modulation affects receptor activation.
MATERIALS AND METHODS
Fluorescence microscopy
A fluorescence laser microscope (Axiovert 200, Zeiss, Jena, Germany) equipped with a 100× oil-immersion objective (Plan-Apochromat, numerical aperture = 1.4, Zeiss) and a 2× relay lens in the emission path to the charge-coupled device (CCD) camera was used for wide-field single-molecule imaging. The overall magnification was 200×, leading to 100 nm per image pixel. A green laser (Nd:YAG Verdi V-10, Coherent, Santa Clara, CA) was used to excite the dye (Alexa Fluor (AF)-546, Molecular Probes, Eugene, OR) at 532 nm wavelength. The laser was coupled by a fiber coupler to the microscope, and the illumination intensity was adjusted to 5 kW/cm2. An ultrafast shutter, controlled by the CCD controller, was set in front of the laser beam to produce 10 ms laser exposures at 7.5 Hz. The dichroic beam-splitter and band-pass emission filters (Chroma Technology, Brattleboro, VT) were set to collect the emission between 550 nm and 600 nm. Single-molecule fluorescence images were acquired by a back-illuminated cooled CCD camera (Spec-10 1340 × 400B, Roper Scientific, Trenton, NJ) with 90% quantum efficiency and single-molecule sensitivity. It is estimated that one intensity count in the CCD image corresponds to two detected photons.
Calculating the position and intensity of individual receptors
The center of individual fluorescent spots was determined by the Gaussian mask algorithm (49) by iterations of the following equation:
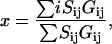 |
where Sij is the photon counts at pixel (i, j), Gij is a two-dimensional (2D) Gaussian function with a peak x and a full width at half maximum (FWHM) that equals 300 nm, which is given by the point-spread function of the microscope. Because Gij depends on x, the calculation of x is iterated until x is converged. The accuracy of the Gaussian mask algorithm was tested both experimentally and by mathematical simulations. Experimentally, individual dye molecules were fixed on a polymer surface and were used as a model system to determine the uncertainty in the position of the fluorescent spot. Simulations were done using single-molecule images of 600 photons per spot, with 30 background photons per pixel, which is at the lower end of the value found in our experiment. Both simulations and experimental results suggest that the accuracy of the fitting is within 20 nm.
Single-molecule fluorescence intensities were calculated by the summation of a 3 × 3 pixels area, which is at the diffraction limit size. For a 2D Gaussian intensity profile with 300 nm at FWHM, 60% of the intensity falls within the 3 × 3 pixels (100 nm/pixel). The intensities of single fluorophores were determined by spin coating glass coverslips with picomolar concentrations of the dye. The individual fluorescent hotspots were regarded as single molecules if their size met the diffraction limit and they photobleached in single steps. Intensity histograms, generated from hundreds of hotspots, indicated the expected fluorescence counts of single fluorophores under our experimental parameters. Intensity histograms of antibodies labeled with the fluorophores were generated as described above and were compared with the histograms generated from the fluorophores alone.
Calculating mean square displacement, diffusion coefficient, and μm scale domains
The mean square displacement (MSD) values of individual receptors are defined by the following equation: MSD = 〈r(t)2〉 where r is the distance between steps in time t. When the change in MSD was nonlinear with time and the curve could be fitted by the equation for confined diffusion (20,32), a restricted motion was assumed where MSD reached maximum at infinity: 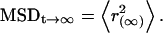 Closer to time 0 where the change in MSD was linear, single-molecule diffusion was considered as Brownian motion and the diffusion coefficient, D, was calculated according to:
Closer to time 0 where the change in MSD was linear, single-molecule diffusion was considered as Brownian motion and the diffusion coefficient, D, was calculated according to: 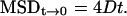
The confinement area was calculated following Kusumi and colleagues (32). Assuming a square shape, the confinement area was calculated according to 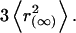
Distinguishing nanoscale confinement episodes from random behavior
To distinguish random behavior from true confinement episodes, the method described by Simson and colleagues (29) was applied. A circle was created around any given point in a trajectory to include an increasing number of following points. The radius of all these circles was used to calculate the probability of staying within the circles by random motion, using the diffusion coefficient we found and the uncertainty in our tracking (0.25 pixel). The smallest probability was then tested against a threshold to identify the confinement episodes that are beyond random motion, within every point in the trajectory. If a point was determined to be within a confinement area, the radius was taken for calculating the average confinement size. The threshold value that we found depicted molecules that were confined within a circle with a radius (R) = 50 nm. When simulated trajectories of a random walk were tested, hardly any episodes of dwellings within R = 50 nm were found.
FRAP measurements
Fluorescence recovery after photobleaching (FRAP) was done by photobleaching a circular area (2.5 μm at FWHM) with 10 ms laser exposures. Using the intensity values that were found before and immediately after photobleaching, K, which describes the fractional depth of photobleaching (50), was calculated. Using K and the value of the recovery time at half maximum derived from our data, the characteristic τ (τc) was calculated and found to be 80 s for EGFR and 50 s for HER2. The diffusion coefficients were then calculated according to D = w2/4τc.
Cell culture and transfection
The HME 184A1 cell line was a kind gift from Martha Stampfer (Lawerence Berkeley National Laboratory, Berkeley, CA). The cell line was used to create the HER2 overexpressing cell line by retroviral transduction, as is described elsewhere (51). Briefly, a retroviral vector containing HER2 was constructed and transfected into the ΨCRIP packaging cell line. The transfectant clones were screened for HER2 expression using mAb 4D5 against HER2, and the supernatants were screened for high virus titers. Individual clones of retrovirus transfected cells were isolated using cloning rings, and the degree of HER2 expression was determined by immunofluorescence, flow cytometry, and equilibrium-binding studies using labeled Fab fragments. The average number of HER2 molecules per cell was 3 × 105. The cells were grown in DFCI-1 medium supplemented with 12.5 ng/ml EGF, and HER2 overexpressing cells were grown in the same medium with the addition of 100 μg/ml G418-sulfate. All cells were grown in 30 mm tissue culture plates with a glass coverslip bottom.
Fab fragments, generation, and labeling
mAb 13A9 against EGFR and the Fab fragment of mAb 7C2 against HER2 were gifts from Genentech (San Francisco, CA). It has been shown that both 13A9 and 7C2 antibodies do not activate the receptors (52,53). Fab fragments of mAb 13A9 were generated using agarose bead-immobilized papain (Pierce, Rockford, IL). The immunoglobulins were dialyzed against 20 mM Na2HPO4/10 mM EDTA for 24 h, followed by incubation with immobilized papain in digestion buffer (20 mM cysteine-HCl, 10 mM EDTA, pH 7.0) for 48 h in 37°C. The Fab fragments were then separated from undigested whole IgG and Fc by a protein A column. The fraction containing Fab fragments was dialyzed against phosphate buffer saline (PBS; pH 7.4), and its purity was confirmed by SDS gel electrophoresis. The Fab fragments (50 KD) were labeled with AF-546 using succinimidyl ester derivatives of the dyes in PBS at pH 8.0. The coupling reaction was carried out at room temperature for 60 min with continual stirring. Labeled Fab fragments were separated from the fluorophore using G-25 gel filtration. The degree of labeling (DOL) was calculated according to Lambert-Beer Law and ranged within 0.7–1.5 for all labeled Fab fragments used in this study. These values were verified by spin coating nM concentrations of the labeled Fab fragments on glass coverslips followed by the generation of intensity histograms of individual diffraction-limited hotspots, as described above.
For tagging receptors with labeled Fab fragments, cells were grown to 70% confluence and were brought to quiescence by overnight incubation in minimal growth medium (without EGF, bovine pituitary extract, or fetal bovine serum) containing 0.1% bovine serum albumin (BSA). Cells were incubated for 1 h with 1% BSA before labeling. Receptor labeling was carried out by 10 min incubation at room temperature with AF-546-tagged Fab fragments at 20–30 nM concentration. Cells were washed and imaged in physiological buffer containing NaCl (162 mM), KCl (2.5 mM), CaCl2 (1.0 mM), glucose (10 mM), HEPES (10 mM), pH 7.4.
Drug application
All reagents were purchased from Sigma (St. Louis, MO) unless noted otherwise. To deplete membrane cholesterol, cells were preincubated in the minimal growth medium with no HEPES, serum, or BSA. Cells were then incubated with 10 mM Methyl-β-cyclodextrin (MβCD) in the same medium for 30 min at 37°C. Staining with labeled antibodies was followed using the protocol described above, except no BSA was added to the incubation medium. To enrich the membrane with cholesterol, cells were preincubated in the growth medium containing 1% BSA and 300 μg/ml water-soluble cholesterol, balanced with MβCD (54,55). This gives a 1:6.25 mol ratio of cholesterol to MβCD at a final concentration of 5 mM MβCD and 0.8 mM cholesterol. The incubation was carried out at 37°C for 3 h. Cells were then incubated with the antibody as described above. To depolymerize F-actin, cells were incubated for 3 h in their normal growth medium with 5 μM latrunculin A (Biomol, Plymouth Meeting, PA). Cytochalasin D, which leads to depolymerization of F-actin by capping the (+) end of the filament, was also used. Cells were incubated for 30 min in their normal growth medium with 5 μM cytochalasin D. Cells were then washed and incubated for 10 min with the growth medium containing 1% BSA and were labeled with the antibody as described above.
RESULTS
Single-molecule fluorescence microscopy was used to track individual nonactivated EGF receptors and HER2 molecules in the membrane of HME cells to determine the rate and pattern of the lateral motion of the receptors. Individual receptors, tagged by fluorescent Fab fragments of the corresponding antibodies, were tracked as they appeared in a small membrane area that was photobleached (3 μm2, corresponding to the FWHM of the laser profile).
Identifying single molecules
Single molecules were identified by the following criteria: (A), The size of the fluorescent spot was the size of the diffraction limit, about half of the excitation wavelength with the 1.4 numerical aperture objective we used. A spot containing one AF-546 dye molecule is expected to be ∼270 nm (with 532 nm laser excitation), slightly <3 × 3 pixels (see Fig. 2 A). (B), The fluorescent spot blinked and photobleached in one discrete step. Fig. 1 shows a typical example of antibody-tagged receptors at the membrane adjacent to the culture dish, demonstrating blinking and single-step photobleaching (see also movie-1 and movie-2 in the supplementary material). (C), The intensity of the fluorescent spot fell within the intensity range of single fluorophores, spin coated on glass slides at picomolar concentrations. The expected fluorescence intensity of a single AF-546 molecule under our specific experimental conditions was 251 ± 109 counts per 3 × 3 pixels during 10 ms laser exposure, as determined by imaging hundreds of individual dye molecules (Fig. 2, B and C, solid bars). The distribution of individual fluorescent spots in the cell membrane was slightly shifted to a higher intensity (Fig. 2, B and C, open bars). The peak distributions of hotspots containing antibodies against EGFR and HER2 were 298 ± 78 and 306 ± 100, respectively. The slight increase in the peak distributions could result from the presence of antibodies labeled with more than one dye molecule as the DOL ranged from 0.7 to 1.5. The shift could also potentially result from the presence of a small pool of preformed dimers. However, the spots that met all three criteria described above were treated as individual receptors and were pursued for further analysis.
FIGURE 2.
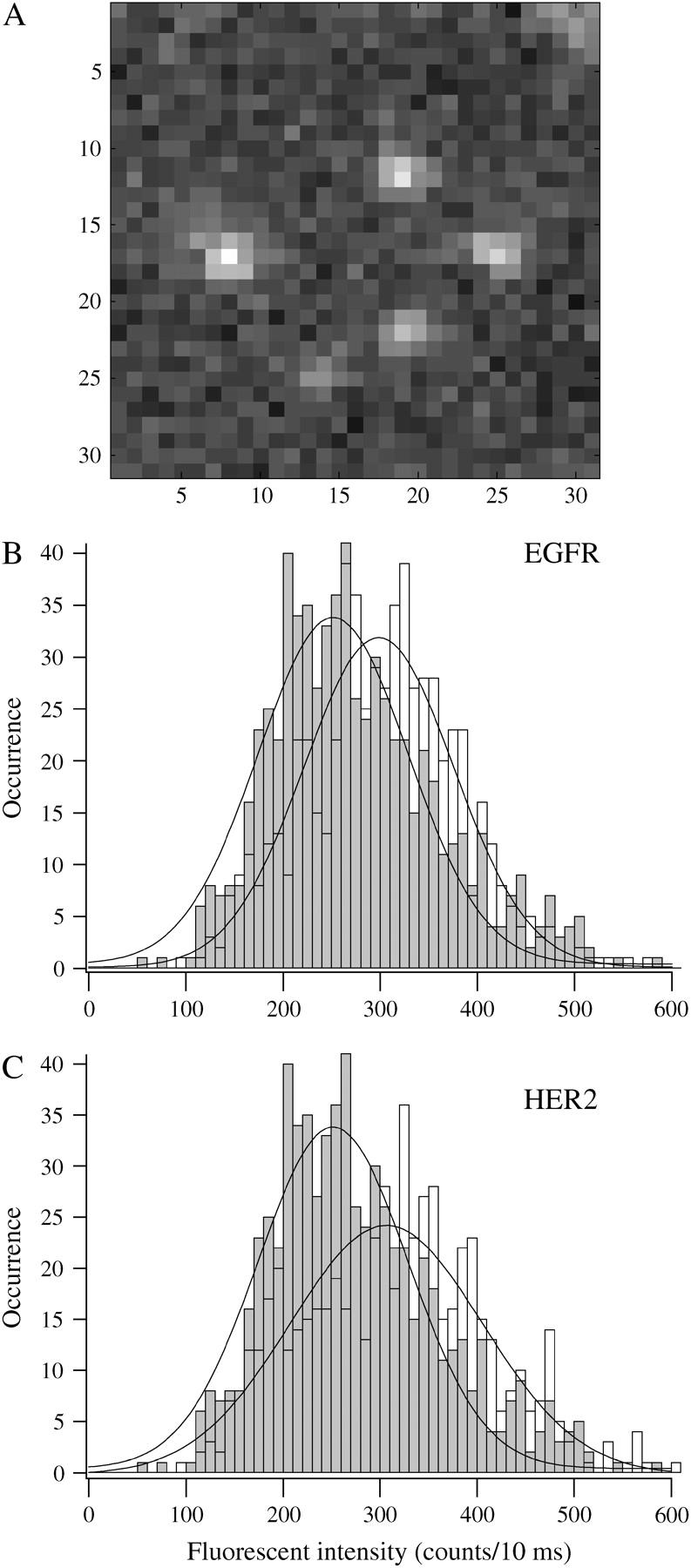
Expected fluorescence intensity of individual molecules was determined from the histogram that was generated by imaging the fluorophore alone, spin coated on glass slides at picomolar concentrations. (A) An example of raw images of diffraction-limited fluorescent spots of <300 nm2 (3 × 3 pixels) that were pursued for further analysis. The spots were fitted by a 2D Gaussian distribution with 300 nm at FWHM. The peak of the distribution was considered the center of the molecule, which was determined with 20 nm resolution. (B) The histogram, generated from 600 fluorescent spots of AF-546 dye molecules spin coated on glass slides at pM concentrations (solid bars), peaked at 252 ± 78 counts per 3 × 3 pixels during a 10 ms laser exposure. This value indicates the expected intensity of individual receptors under our specific experimental conditions. The distribution that was generated from the intensities of spots containing AF-546-tagged EGFR that appeared in the photobleached membrane areas (open bars) peaked at 298 ± 78 counts per 300 nm2 (n = 600). (C) The fluorescence intensity distribution, generated from the fluorescent spots of AF-647-tagged HER2 molecules in the cell membrane (open bars), peaked at 306 ± 100 (n = 600). Because the degree of Fab fragment-labeling was distributed around 1, it is expected that some Fab fragments were labeled with more than one dye molecule, which could explain the shift in the intensity distribution of the spots in the cell membrane. This shift could also reflect the appearance of dimers in the photobleached area.
FIGURE 1.
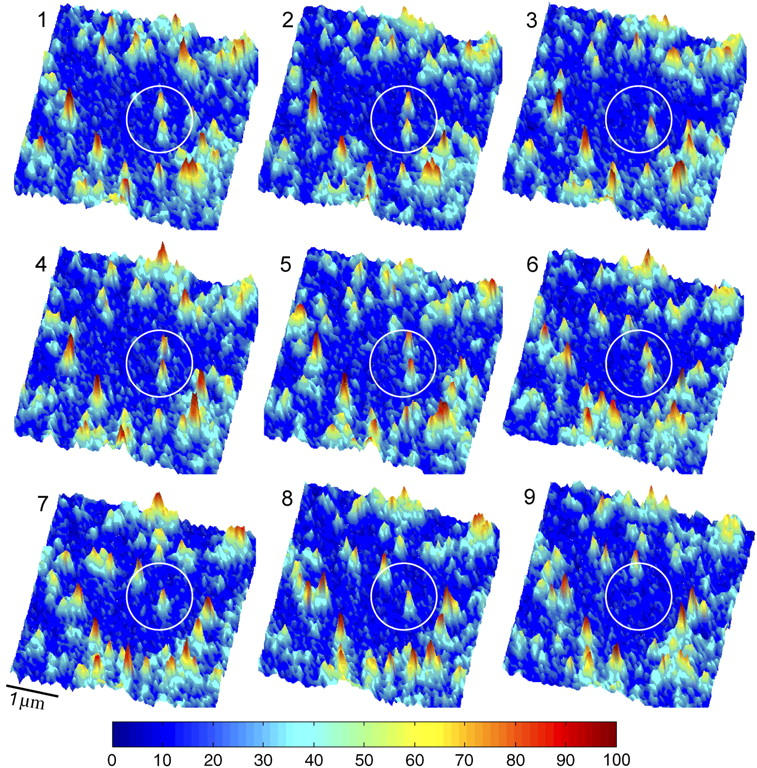
A series of consecutive images, taken at 130 ms intervals, showing individual AF-546-Fab fragment-tagged receptors as they entered a small photobleached area in the membrane of HME cells. Individual molecules were detected by their fluorescence blinking as shown in the third image (upper fluorescent spot within the circle) and by their single-step photobleaching as shown in the seventh image (upper spot) and the ninth image (lower spot). Also see movie-1 and movie-2 in the supplementary material.
Relative mobility of EGFR and HER2 in the plane of the membrane
Individual EGFR and HER2 molecules were tracked at the lower and upper membrane of the cells (movie-2 in the supplementary material). Although the size of the fluorescent spot is limited by the resolution of the microscope, the center of the spot can be determined by fitting a 2D Gaussian curve to the spot (56). Fitting 2D Gaussian curves using iterations of least-squares estimators allowed us to determine the motion of the spot to an ∼20 nm resolution (49). Examples of traces, taken by tracking individual EGFR and HER2 molecules in the lower membrane of the cells, are shown in Fig. 3, A and B, respectively, encased within 1 μm2 squares. The images were taken every 130 ms, which was the time resolution of our experiments. With this time resolution, it was possible to capture short periods, on the order of 1 s, where the trajectories interrupted by receptor dwellings within nanoscale domains (red circles) before diffusing away. To quantify the diffusion pattern and rate of the two receptors, we first calculated the MSD of individual molecules. Fig. 4 A shows the changes in MSD over time, as determined from traces of 180 EGF receptors (open circles) and 120 HER2 molecules (solid circles). The nonlinear change of MSD with time suggests that the motion of the receptors was restricted, and the curves were fitted by the equation for confined diffusion (20,32) (solid line). From the plateau of the fitted curves, and assuming a square shape, we estimated that EGFR and HER2 diffused within microdomains of 1.23 ± 0.06 μm and 1.65 ± 0.16 μm, respectively. The diffusion coefficient (D) that was calculated from the linear part of the curve (closer to time 0) showed 0.023 ± 0.002 μm2/s for EGFR and 0.035 ± 0.004 μm2/s for HER2. The significantly smaller diffusion coefficient of EGFR relative to HER2 could result from the direct interaction of EGFR with F-actin (36,39,40). To test this possibility, the cells were treated with drugs that depolymerize F-actin. Cytochalasin D, which is a fungal alkaloid that caps the (+) end of F-actin, was used. The drug blocks further addition of subunits and leads to F-actin depolymerization over time. By incubating the cells with cytochalasin D, we found that the diffusion coefficient of EGFR but not HER2 was increased (0.027 ± 0.003 μm2/s). Both EGFR and HER2 restricted domains were increased (1.5 ± 0.19 μm and 1.95 ± 0.29 μm, respectively) as well (Fig. 4 B). Using a highly potent drug, latrunculin A, which is secreted by sponges and inhibits the addition of G-actin to the filament end, yielded more profound results. From the traces of the receptors in latrunculin-treated cells, it was found that the diffusion coefficient of EGFR (0.032 ± 0.003 μm2/s, n = 180) and the size of its restricting microdomains (1.68 ± 0.1 μm) became similar to those of HER2 under control conditions (Fig. 4 C, open circles). The diffusion coefficient of HER2 was not changed by latrunculin treatment (0.029 ± 0.003 μm2/s; n = 70; Fig. 4 C, solid circles). However, the boundaries of HER2 restricted domains disappeared, as indicated by the linear change of MSD with time. It is possible that the boundaries of the restricted domains were extended by the toxin, but the molecules photobleached before these boundaries were reached. The difference in the degree of domain extensions between EGFR and HER2 could be explained if the depolymerization of F-actin was incomplete, leading to a greater restriction of EGFR motion via its direct interaction with intact filaments.
FIGURE 3.
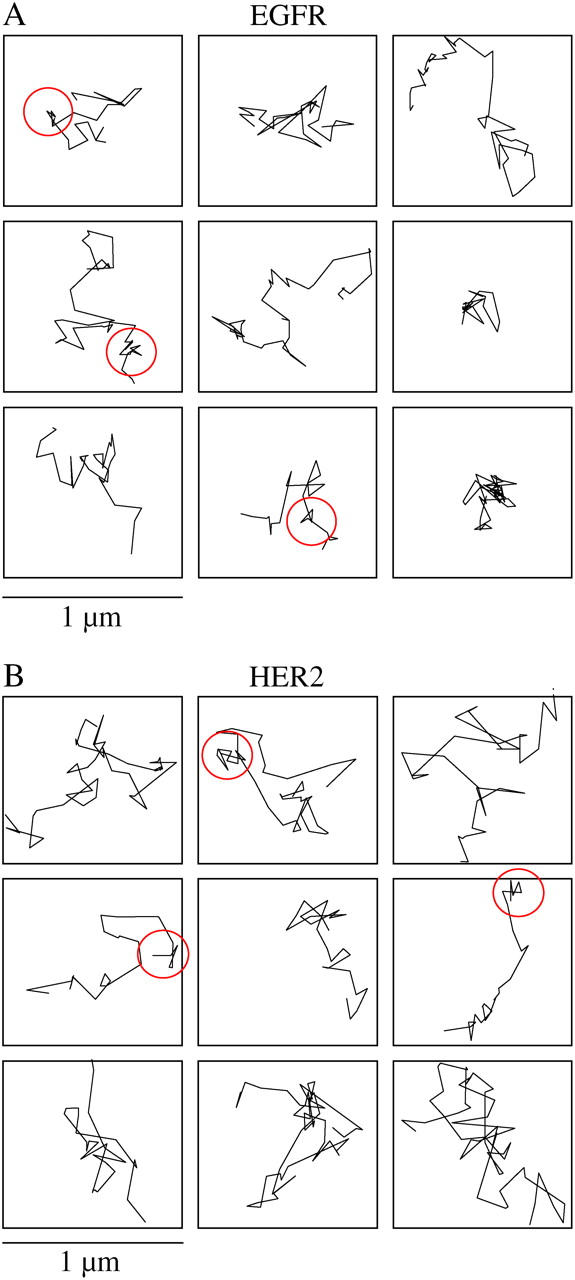
Traces of individual molecules show that the receptors diffuse freely within larger microscale domains, interrupted by short confinement episodes within nanoscale domains. (A) Typical traces of individual EGFR receptors showing their diffusion pattern in the plane of the membrane under normal conditions. The traces were taken in the lower membrane of the cell and are shown within 1 μm2 (10 × 10 pixels) frames. Short dwellings within nanodomains are indicated by the circles. (B) The same pattern is observed by tracking individual HER2 molecules.
FIGURE 4.
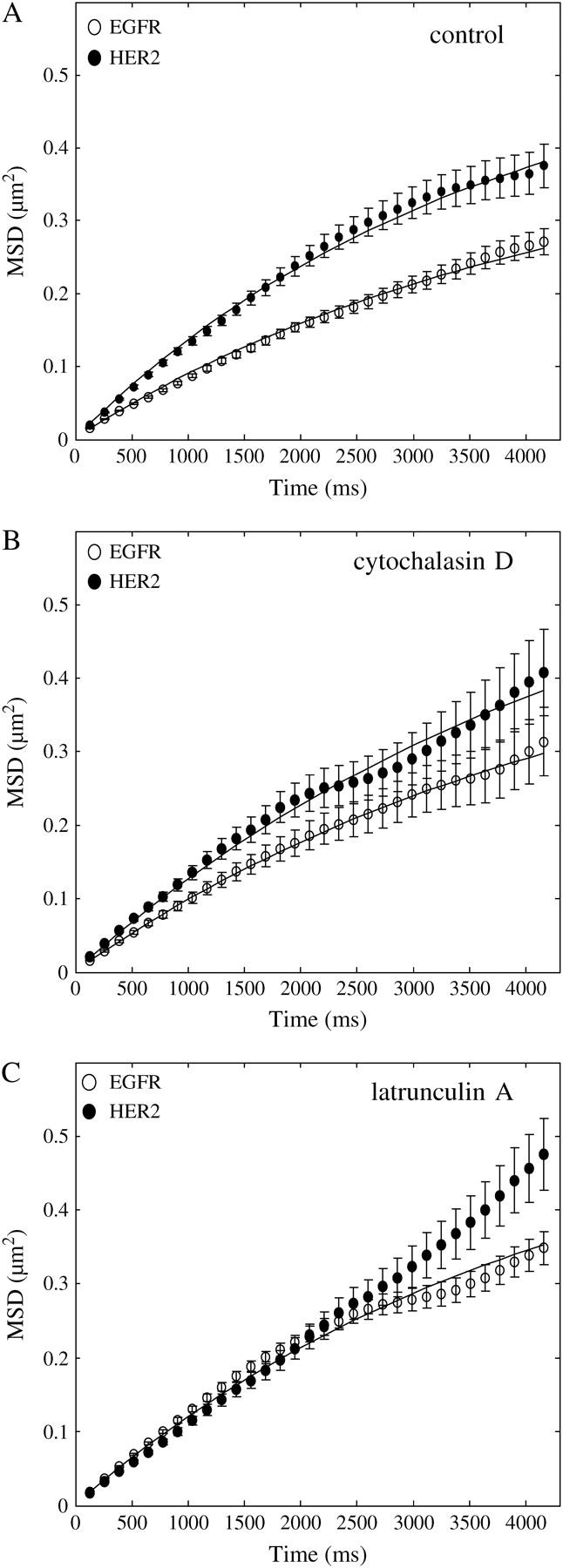
Change in MSD over time, as calculated from traces of individual receptors, identified microdomain confinements within which HER2 diffused faster than EGFR. (A) The nonlinear change of MSD over time, as calculated from 180 traces of individual EGFR (○) and 120 traces of individual HER2 molecules, suggests that the receptors were confined within microscale domains. By fitting the curves with the equation for confined diffusion (solid line) and assuming a square shape, the confinement area was found to be 1.23 ± 0.06 μm for EGFR and 1.65 ± 0.16 μm for HER2. The diffusion coefficient of the receptors was calculated from the linear part of the curve (closer to time 0), showing that HER2 was more mobile (0.035 ± 0.004 μm2/s) than EGFR (0.023 ± 0.002 μm2/s). (B) Treating the cells with cytochalasin D to depolymerize F-actin increased the diffusion coefficient of EGFR (0.027 ± 0.003 μm2/s, n = 40, ○) but did not change the diffusion coefficient of HER2 (0.035 ± 0.004 μm2/s, n = 40, •). The toxin also extended the boundaries of the restricted domains of both receptors (EGFR: 1.5 ± 0.19 μm; HER2: 1.95 ± 0.29 μm). (C) Using a more potent drug to depolymerize F-actin, latrunculin A, led to the increase in the diffusion coefficient of EGFR (0.032 ± 0.003 μm2/s, n = 180, ○) to the same level as that of HER2. The diffusion coefficient of HER2 was not changed by the toxin (0.029 ± 0.003 μm2/s, n = 70, •). As indicated by the linear change of MSD with time, the toxin extended the boundaries of HER2 restricted domains beyond the ability of the experiment to detect them, which was limited by the photobleaching time of the individual molecules. Latrunculin A extended the boundaries of EGFR restricted domains to the same level as that of HER2 under normal conditions (1.65 ± 0.1 μm). These observations suggest that the direct interaction of EGFR with F-actin slows down the motion of the receptor in the plane of the membrane.
Identification of short nanoscale confinement episodes of both HER2 and the EGFR
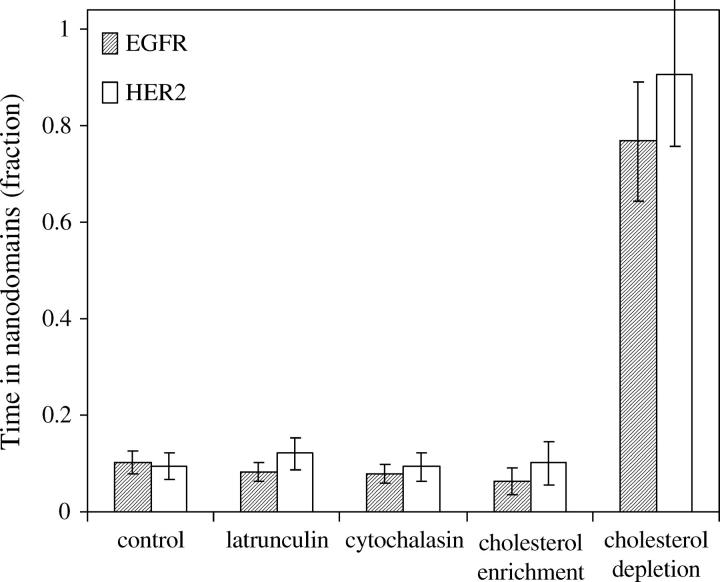
Within the time resolution of our experiment (130 ms), it was not possible to capture the “hop” diffusion between nanodomains that dominated the behavior of other membrane proteins reported earlier (24,57–60). However, it was possible to capture episodes, on the order of 1 s, where the trajectories interrupted by receptor dwellings within nanoscale domains (Fig. 3, red circles). To identify whether these episodes were guided by specific cellular structures or interactions, it was necessary to rule out the possibility that they could occur by a random behavior. We followed the approach for determining the probability threshold above which such confinement episodes would not occur by random walk (29), as described under Materials and Methods. The average radius (R) of all trajectory points that fell above the threshold was 50 nm, for both EGFR and HER2. Using the diffusion coefficient that we found for each receptor type under normal conditions and the experimental tracking uncertainty (25 nm), simulation of random walk showed almost no cases of three consecutive trajectory points within R = 50 nm. However, episodes of three or more consecutive trajectory points within R = 50 nm were identified in EGFR and HER2 traces. The sum of all the points in the confinement episodes showed that EGFR spent 10% ± 2% of the time within these nanodomains, for an average time of 1.4 ± 0.2 s (total of 180 traces) under control conditions (Fig. 5, control). Similarly, HER2 spent 9% ± 3% of the time inside the nanodomains, for an average time of 1.3 ± 0.2 s (total of 120 traces). Interestingly, F-actin depolymerization by latrunculin A or cytochalasin D did not change significantly the fraction of time that EGFR and HER2 spent within the 50 nm radius domains (Fig. 5).
FIGURE 5.
Episodes of dwellings within nanodomains, on the order of 1 s, were identified by their probabilistic deviation from random walk, suggesting the involvement of specific cellular structures. The average radius of all trajectory points that fell above the threshold for nonrandom behavior was 50 nm for both receptors. Whereas simulations of random walk showed almost no episodes of three consecutive points within 50 nm radius domains, both EGFR and HER2 showed a significant number of such confinement episodes. EGFR spent 10% ± 2% of its time for an average time of 1.4 ± 0.2 s, and HER2 spent 9% ± 3% of its time for an average time of 1.3 ± 0.2 s within the nanodomains under normal conditions. Similar values were observed in latrunculin A or cytochalacin D treated cells and in membrane enriched with cholesterol. However, a dramatic increase in the fraction of time that the receptors spent within the nanodomains was observed in cholesterol-depleted membranes. EGFR spent 77% ± 12%, and HER2 spent 91% ± 15% of their time inside the 50 nm radius domains, for an average time of 2.2 s and 2.9 s, respectively.
Modulation of both EGFR and HER2 diffusion by altering membrane cholesterol and F-actin polymerization
To identify the role of membrane cholesterol in governing the pattern of EGFR and HER2 motion in the plane of the membrane, Fab-tagged receptors were tracked in the membrane of cells that were enriched or depleted of cholesterol. A dramatic change was observed with both manipulations. Fig. 6 A shows traces of individual EGFR within 1.2 μm2 squares taken from cholesterol-enriched membranes. On average, the traces covered a larger area of the membrane than traces obtained from control cells (Fig. 3 A). In contrast, cholesterol depletion led to an almost complete confinement of the receptors to nanoscale domains, with short and rare escapes. Examples of traces taken from cholesterol-depleted membranes are shown in Fig. 6 B framed by 0.5 μm2 squares. Similar patterns were observed for HER2. Calculations of MSD changes over time revealed that although cholesterol enrichment did not change the diffusion coefficient of HER2 (0.029 ± 0.003 μm2/s) (Fig. 7 A, solid circles), it increased the diffusion coefficient of EGFR (0.030 ± 0.003 μm2/s) to the same level as that of HER2 (Fig. 7 A, open circles) and extended the boundaries of the domains of both receptors. We inferred that the boundaries were extended from the linear change of MSD over the duration of the experiment but could not measure them directly because of the relatively rapid photobleaching times of the individual fluorophores. Interestingly, the time that the two receptors spent within the nanodomains did not change significantly from control conditions by cholesterol enrichment (Fig. 5, cholesterol enrichment) as determined by the identification of the nanoscale confinement episodes based on their probabilistic deviation from random walk. In contrast, the time that EGFR and HER2 spent within the nanoscale domains increased dramatically in cholesterol-depleted membranes from ∼10% to 77% ± 12% and 91% ± 15%, respectively (Fig. 5, cholesterol depletion). The diffusion coefficient of both receptors was smaller than the resolution of our experiment (0.002 μm2/s; Fig. 7 B) in cholesterol-depleted cells. However, when cholesterol-depleted cells were treated with latrunculin A, the diffusion coefficient of EGFR was partially restored (0.017 ± 0.002 μm2/s; Fig. 7 B, asterisks). These observations suggest that the confinement of the receptors by cholesterol depletion might occur via modulation of the dynamic state of F-actin, possibly via membrane dislocation of (4,5) PIP2 (12–15). Sequestering of PIP2, like cholesterol depletion, has been reported to alter cell actin organization and inhibit the lateral diffusion of membrane proteins (16).
FIGURE 6.
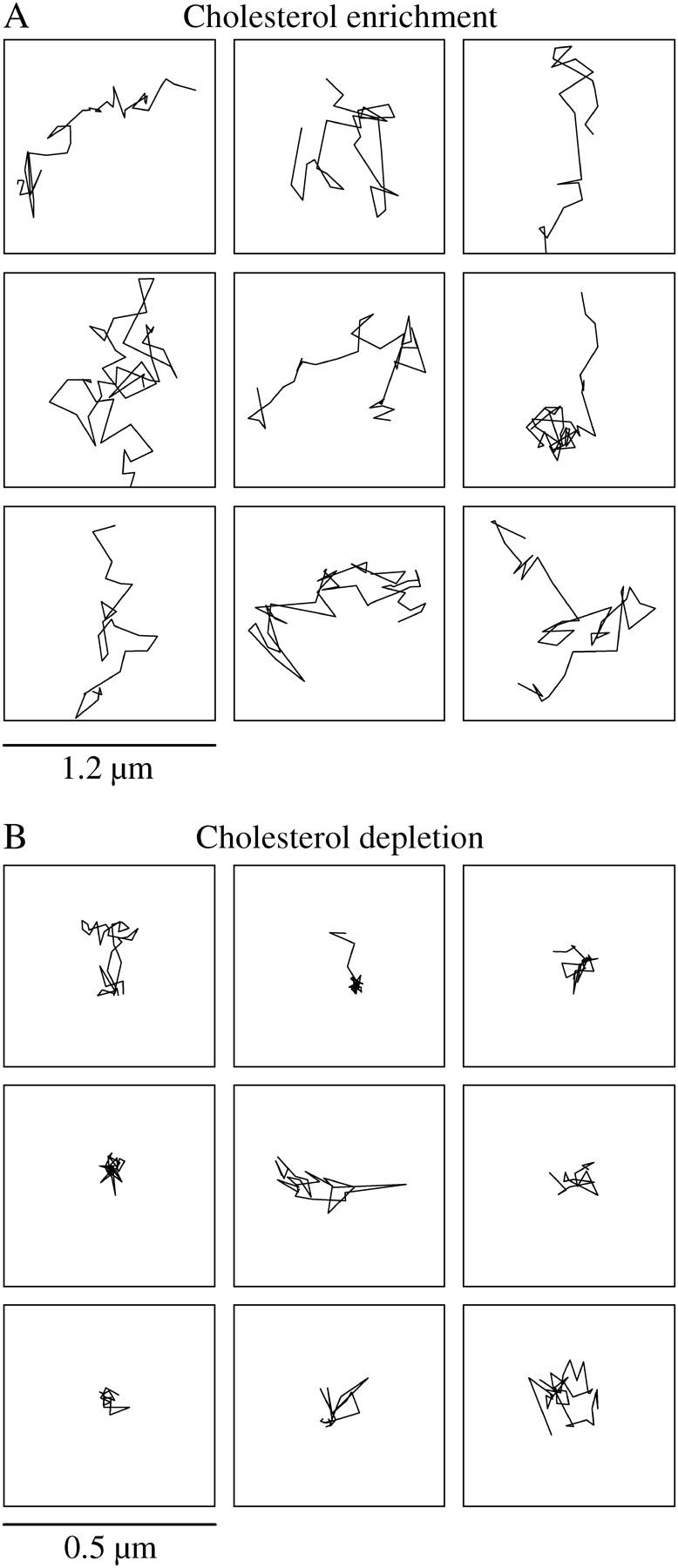
Manipulations of the cholesterol content of the membrane changed the pattern of the lateral motion of the receptors. (A) Examples of EGFR traces in cholesterol-enriched membranes encased within 1.2 μm2 boxes. The receptors covered, on average, larger areas than those covered under normal conditions. (B) Traces of the receptors in cholesterol-depleted membranes, shown within 0.5 μm2 boxes, were highly confined to nanoscale domains, with short and rare escapes.
FIGURE 7.
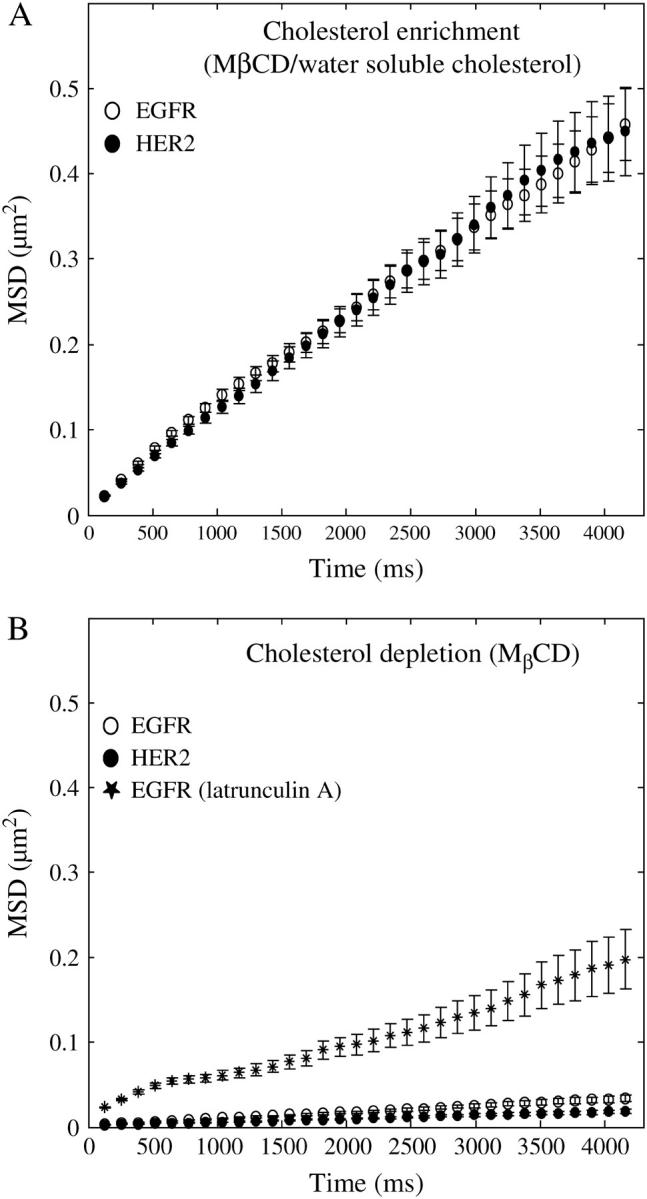
Change of MSD with time, calculated from traces that were taken in cholesterol-enriched or -depleted membranes, shows that cholesterol provides a dynamic environment that supports the free motion of the receptors. (A) Cholesterol enrichment did not change the diffusion coefficient of HER2 (0.029 ± 0.003 μm2/s, n = 50; filled circles) but increased the diffusion coefficient of EGFR to the same level as that of HER2 (0.03 ± 0.003 μm2/s, n = 80, ○). Cholesterol enrichment caused the boundaries of the restricted domains to extend beyond the tracking time window as determined by the linear change of MSD with time for both receptors. (B) Cholesterol depletion led to a dramatic decrease in the diffusion coefficient of EGFR and HER2 (○ and •, respectively) below 0.002 ± 0.0002 μm2/s, which is the resolution limit of our experiments. However, treating cholesterol-depleted cells with latrunculin A led to a partial recovery of the diffusion coefficient (0.017 ± 0.002 μm2/s, n = 30, *) of the receptors, suggesting that the modulation of receptor mobility by membrane cholesterol occurs via modulation of F-actin.
Comparison of single-molecule measurements to whole receptor populations
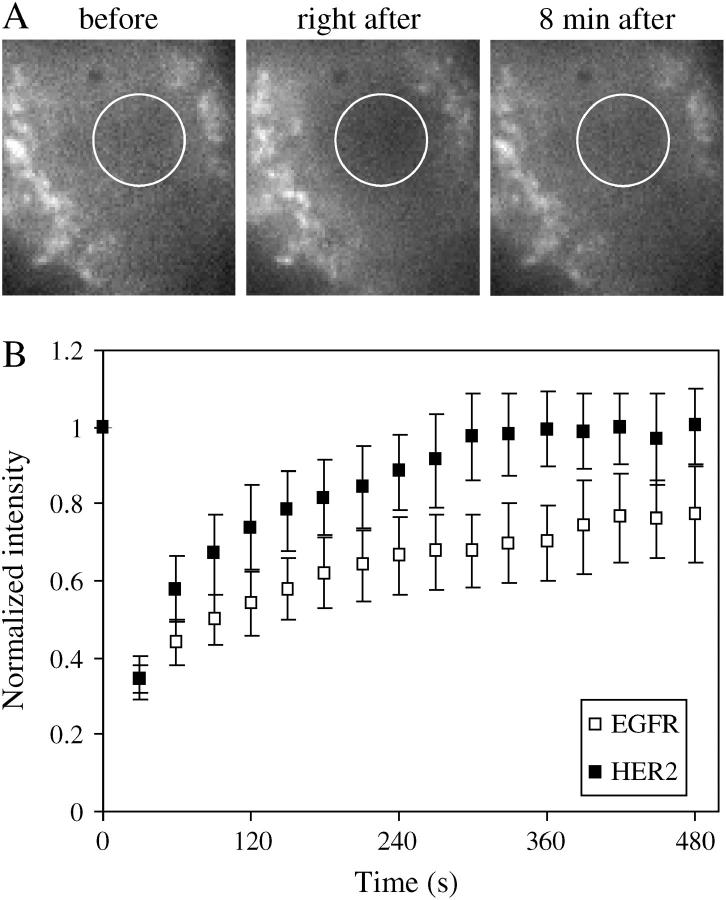
Ensemble FRAP measurements cannot identify diffusion patterns such as restricted diffusion and nanoscale short confinement episodes or identify subtle differences in diffusion coefficients. However, FRAP measurements were used here to verify that the diffusion coefficients, found by single-molecule measurements, reflect the diffusion coefficient of the whole population rather than a selected subpopulation. An example of the fluorescence images that were used for calculating FRAP are shown in Fig. 8 A. The first image was followed by photobleaching with 10 s exposure to the laser beam, indicated by the circle at FWHM. Images were taken every 30 s for the next 8 min, where the fluorescence recovery leveled off (third image). From the fluorescence recovery over time and following the approach of Axelrod and colleagues (50) described in the Materials and Methods section, the diffusion coefficients of EGFR and HER2 were found to be 0.014 ± 0.007 μm2/s and 0.022 ± 0.01 μm2/s, respectively (Fig. 8 B). These values are slightly lower than those we found using single-molecule tracking. In the case of single-molecule tracking, the diffusion coefficients are calculated from the initial part of the MSD curve, reflecting the free diffusion. However, in the case of FRAP, the calculations include both the free and the confined diffusion and therefore are expected to be lower. The values we found by FRAP show the same tendency for a higher diffusion coefficient for HER2 relative to EGFR and suggest that the single-molecule approach describes the mobility pattern of the population at large rather than a subpopulation.
FIGURE 8.
Single-molecule measurements represent the whole population as determined by ensemble FRAP measurements. (A) An example of images that were used for calculating FRAP, where the diameter of the laser beam is indicated by the circle at FWHM (2.5 μm). (B) Normalized fluorescence intensities were plotted against time (images taken every 30 s), and the diffusion coefficient of the receptors was calculated as described in the text. The diffusion coefficient calculated by FRAP for EGFR (0.014 ± 0.007 μm2/s, n = 10 cells, □) and HER2 (0.022 ± 0.01 μm2/s, n = 10 cells, ▪) show the same tendency of a higher diffusion coefficient for HER2 relative to EGFR, as was found by single-molecule tracking.
DISCUSSION
The principal new finding to emerge from this work is the role of membrane cholesterol in providing a dynamic environment that supports the lateral movement of EGFR and HER2 in the plane of the membrane. We propose a possible mechanism whereby the modulation of receptor mobility could arise from F-actin polymerization underneath cholesterol-enriched lipid rafts. We quantified the confined motion pattern of the two receptors and identified, by deviations from random walk, dwellings within 50 nm domains that could indicate specific cellular interactions. This work also demonstrates that HER2 is more mobile in the plane of the membrane than EGFR, a difference that could result from the ability of EGFR to directly interact with F-actin (36–38). This idea is supported by our observation that F-actin depolymerization increased the diffusion coefficient of EGFR, raising it to the same level as that of HER2. The idea is also supported by several studies showing that deletion mutations of the intracellular domains significantly affected the diffusion rate and pattern of certain membrane proteins. These experiments were done using FRAP (22) or single-particle tracking and optical tweezers (23,24). Using FRAP, it was also reported that cytoplasmic deletion mutations did not affect the diffusion coefficient of EGFR, which was tagged by fluorescent ligands (60). The use of fluorescent ligands might have partially masked the effects of the mutations by stabilizing preexisting or inducing new associations at the cell membrane. Our study was done with endogenous EGFR receptors naturally expressed by HME cells. Using heterologous cells that do not express endogenous EGFR, it will be possible to directly evaluate the contribution of EGFR and F-actin interaction by transfecting with either a wild-type or a mutated receptor lacking the actin binding site (39,42).
Whereas the diffusion coefficient of EGFR has been reported earlier using FRAP (60–64) or SPT (32), the diffusion coefficient of HER2 has not previously been measured. The reported values for EGFR ranged around 0.02 μm2/s, which agrees with the value we found here by following individual receptors (0.023 ± 0.002 μm2/s). The diffusion coefficient of HER2 (0.035 ± 0.003 μm2/s) was significantly higher than that of EGFR, a difference that disappeared with F-actin depolymerization (Fig. 4, B and C) or cholesterol enrichment (Fig. 7 A). Both F-actin depolymerization and cholesterol enrichment extended the boundaries of the restricted areas but did not change the degree of receptor dwellings within the nanodomains that we could capture intermittently along the trajectories (Fig. 5). These observations suggest a common mechanism for both treatments, both acting on the dynamic state of F-actin either directly or indirectly. However, this mechanism must be different from the mechanism underlying the infrequent episodes of nanodomain dwellings, on the order of 1 s, that we observed within the larger microdomains.
The time resolution of our experiment (130 ms) could not allow the capturing of the “hop” diffusion between milliseconds dwellings in nanoscale domains, which are thought to be formed by F-actin (24,57–59). The longer-lived nanodomains that we observed were not affected by F-actin manipulations and therefore could not reflect a subpopulation of the short-lived nanodomains mentioned above. The nanodomains that we observed could represent the transient confinement zones reported earlier (29–31,35). However, these zones were suggested to represent lipid rafts and therefore are expected to be affected by cholesterol manipulations. Our results are not entirely consistent with this idea because the time of confinement within nanodomains was affected only by cholesterol depletion but not by enrichment. We therefore believe that different mechanisms underlie the nanodomain dwellings that we observed under normal conditions (or cholesterol enrichment) and those under cholesterol depletion. It is conceivable that the dwellings in nanodomains that we observed intermittently within the microdomains under normal conditions are caused by episodes of spontaneous dimerization (65–67) or molecular interactions with scaffolding and downstream molecules and possibly the consequent formation of more “stable” rafts (68). In contrast, the dramatic increase in nanodomain dwellings that was caused by cholesterol depletion could be explained by the relationship of lipid-rafts with F-actin as described below.
The restriction of the lateral mobility of transmembrane proteins by cholesterol depletion has been observed before (16,69,70) but not in all cases (71,72). Using laser trapping of HLA molecules labeled with antibody-coated beads, Kwik and colleagues (16) found that with cholesterol depletion, the molecules became significantly more confined by elastic elements of the cytoskeleton as determined by the snapping of the molecules back to the point of origin once they encountered an obstacle and escaped from the trap. Using optical laser tweezers, Suzuki and colleagues (73,74) identified two types of cytoskeleton-dependent barriers: elastic barriers involving weak but specific bonds to the actin cytoskeleton and small nonelastic barriers that depended on membrane cholesterol. Together, these observations suggest that cholesterol depletion might lead to receptor confinement by acting on F-actin (12,13). The intimate relationships between lipid rafts and the actin cytoskeleton have been observed before (75–77).
Cholesterol depletion has been shown to result in the reorganization and stabilization of membrane-associated actin (14), and this reorganization could occur, at least in part, as a result of the membrane dislocation of PIP2 that is a consequence of cholesterol depletion (16). PIP2 enhances F-actin polymerization by stimulating the activity of WASP family proteins that activate the nucleation and filament branching of actin, among other mechanisms (15). PIP2 accumulates in rafts by binding to myristoylated alanine-rich PKC-substrate and related proteins (78). It has been suggested that rafts could serve as platforms for the integration of PIP2 action and actin polymerization, creating specialized membrane microdomains that are associated with specific cytoskeletal structures (15). Dislocating PIP2 from the membrane could create a submembrane area that is less dynamic by suppressing F-actin polymerization, increasing the constraints on receptor mobility. In support of this view, we observed the partial recovery of receptor motion by treating cholesterol-depleted cells with latrunculin A (Fig. 7 B), where F-actin depolymerization led to an increase in the diffusion coefficient and a decrease in the confinement that cholesterol depletion caused. Cholesterol enrichment, on the other hand, could create submembrane areas that are more dynamic by enhancing F-actin polymerization. In support of this interpretation is our observation that latrunculin A treatment and cholesterol enrichment had similar effects on the diffusion coefficient of the receptors and the same tendency to increase the size of the restricted areas.
Together, our observations could be explained by a model in which cholesterol-rich membrane areas, or lipid rafts, provide active structures for the polymerization of F-actin, possibly by the recruitment of PIP2, that in turn create a dynamic environment in the submembrane area that allows transmembrane proteins to freely diffuse within the cholesterol-rich areas. Cholesterol enrichment increases the size of the rafts and enhances their ability to form a reticular network at the cell surface. In so doing, cholesterol enrichment creates larger platforms for the active polymerization of F-actin, leading to the significant increase that we observed in the size of the microdomains. This increase can also be achieved by direct manipulation of F-actin, as we observed. Cholesterol depletion disperses or decreases the size of the rafts and dissociates their networks. By doing that, cholesterol depletion arrests F-actin polymerization, possibly via PIP2 dislocation, leading to the observed dramatic increase in the confinement of the receptors within nanodomains. Direct depolymerization of F-actin should counteract the effect of cholesterol depletion, as we observed. It is possible that under normal conditions, the network and the individual rafts determine the size of the microdomains and the nanodomains (observed with high time resolution by others), respectively. Interestingly, F-actin polymerization has been reported to localize to the activated EGFR (43,45), which could be explained by EGFR activation of PLCγ, leading to PIP2 hydrolysis and an increase in F-actin turnover (46), all in association with lipid rafts where these molecules accumulate.
Cholesterol depletion has been shown to increase the binding of EGF to the EGFR (1,7,8) and to increase receptor dimerization, autophosphorylation, and tyrosine kinase activity (1,8,10,11). In contrast, cholesterol enrichment has been reported to decrease EGF binding and EGFR activation in cells (1,7) and increase the affinity of reconstituted receptors in vesicles (9). However, the mechanisms underlying the effects of cholesterol manipulations on the receptor are unclear. Based on our observations, the effects of cholesterol manipulations on EGFR activation and ligand binding could be explained in the context of receptor mobility in the plane of the membrane. It is possible that with cholesterol depletion, receptors that become almost immobile and highly confined tend to persist as activated dimers, which have a higher affinity for EGF (79). In contrast, cholesterol enrichment increases the mobility and freedom of the receptors in the plane of the membrane, which could decrease their likelihood of dimerization and activation. Because HER2 activation depends on its ability to form heterodimers with the EGFR, the presence of both receptors in cholesterol-enriched membrane domains might be an important aspect that regulates their activation upon ligand stimulation. Ligand stimulation is thought to induce formation of larger and more stable rafts that, in turn, could preserve the spatial information conveyed by the receptor through the membrane-associated signal transduction (58). The activation of EGFR has been shown to provide spatial and positional information during normal development (80–82) or tumor invasion (83). The association of EGFR and HER2 with the lipid raft network could therefore increase the reliability of the spatial information transduced by the receptors.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
We thank Gary Holtom for his help with the lasers and optics, Chii-Shiarng Chen for his help with the labeling of the antibodies, and David Panther for his help with the FRAP experiments. We also thank Genentech for the generous donation of the antibodies.
This work was supported by the Pacific Northwest National Laboratory (PNNL)—Directed Research and Development funds. The work was performed in the W. R. Wiley Environmental Molecular Sciences Laboratory, Richland, WA. PNNL is operated by Battelle for the U.S. Department of Energy.
Galya Orr and Dehong Hu contributed equally to this work.
References
- 1.Ringerike, T., F. D. Blystad, F. O. Levy, I. H. Madshus, and E. Stang. 2002. Cholesterol is important in control of EGF receptor kinase activity but EGF receptors are not concentrated in caveolae. J. Cell Sci. 115:1331–1340. [DOI] [PubMed] [Google Scholar]
- 2.Nagy, P., G. Vereb, Z. Sebestyen, G. Horvath, S. J. Lockett, S. Damjanovich, J. W. Park, T. M. Jovin, and J. Szollosi. 2002. Lipid rafts and the local density of ErbB proteins influence the biological role of homo- and heteroassociations of ErbB2. J. Cell Sci. 115:4251–4262. [DOI] [PubMed] [Google Scholar]
- 3.Kusumi, A., I. Koyama-Honda, and K. Suzuki. 2004. Molecular dynamics and interactions for creation of stimulation-induced stabilized rafts from small unstable steady-state rafts. Traffic. 5:213–230. [DOI] [PubMed] [Google Scholar]
- 4.Simons, K., and E. Ikonen. 1997. Functional rafts in cell membranes. Nature. 387:569–572. [DOI] [PubMed] [Google Scholar]
- 5.Simons, K., and E. Ikonen. 2000. How cells handle cholesterol. Science. 290:1721–1726. [DOI] [PubMed] [Google Scholar]
- 6.Ikonen, E. 2001. Roles of lipid rafts in membrane transport. Curr. Opin. Cell Biol. 13:470–477. [DOI] [PubMed] [Google Scholar]
- 7.Roepstorff, K., P. Thomsen, K. Sandvig, and B. van Deurs. 2002. Sequestration of epidermal growth factor receptors in noncaveolar lipid rafts inhibits ligand binding. J. Biol. Chem. 277:18954–18960. [DOI] [PubMed] [Google Scholar]
- 8.Pike, L. J., and L. Casey. 2002. Cholesterol levels modulate EGF receptor-mediated signaling by altering receptor function and trafficking. Biochemistry. 41:10315–10322. [DOI] [PubMed] [Google Scholar]
- 9.den Hartigh, J. C., P. M. van Bergen en Henegouwen, J. Boonstra, and A. J. Verkleij. 1993. Cholesterol and phosphoinositides increase affinity of the epidermal growth factor receptor. Biochim. Biophys. Acta. 1148:249–256. [DOI] [PubMed] [Google Scholar]
- 10.Chen, X., and M. D. Resh. 2002. Cholesterol depletion from the plasma membrane triggers ligand-independent activation of the epidermal growth factor receptor. J. Biol. Chem. 277:49631–49637. [DOI] [PubMed] [Google Scholar]
- 11.Westover, E. J., D. F. Covey, H. L. Brockman, R. E. Brown, and L. J. Pike. 2003. Cholesterol depletion results in site-specific increases in epidermal growth factor receptor phosphorylation due to membrane level effects. Studies with cholesterol enantiomers. J. Biol. Chem. 278:51125–51133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edidin, M. 2001. Shrinking patches and slippery rafts: scales of domains in the plasma membrane. Trends Cell Biol. 11:492–496. [DOI] [PubMed] [Google Scholar]
- 13.Edidin, M. 2003. The state of lipid rafts: from model membranes to cells. Annu. Rev. Biophys. Biomol. Struct. 32:257–283. [DOI] [PubMed] [Google Scholar]
- 14.Pike, L. J., and J. M. Miller. 1998. Cholesterol depletion delocalizes phosphatidylinositol bisphosphate and inhibits hormone-stimulated phosphatidylinositol turnover. J. Biol. Chem. 273:22298–22304. [DOI] [PubMed] [Google Scholar]
- 15.Yin, H. L., and P. A. Janmey. 2003. Phosphoinositide regulation of the actin cytoskeleton. Annu. Rev. Physiol. 65:761–789. [DOI] [PubMed] [Google Scholar]
- 16.Kwik, J., S. Boyle, D. Fooksman, L. Margolis, M. P. Sheetz, and M. Edidin. 2003. Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin. Proc. Natl. Acad. Sci. USA. 100:13964–13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsuji, A., and S. Ohnishi. 1986. Restriction of the lateral motion of band 3 in the erythrocyte membrane by the cytoskeletal network: dependence on spectrin association state. Biochemistry. 25:6133–6139. [DOI] [PubMed] [Google Scholar]
- 18.Yechiel, E., and M. Edidin. 1987. Micrometer-scale domains in fibroblast plasma membranes. J. Cell Biol. 105:755–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edidin, M., and I. Stroynowski. 1991. Differences between the lateral organization of conventional and inositol phospholipid-anchored membrane proteins. A further definition of micrometer scale membrane domains. J. Cell Biol. 112:1143–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saxton, M. J., and K. Jacobson. 1997. Single-particle tracking: applications to membrane dynamics. Annu. Rev. Biophys. Biomol. Struct. 26:373–399. [DOI] [PubMed] [Google Scholar]
- 21.Mirchev, R., and D. E. Golan. 2001. Single-particle tracking and laser optical tweezers studies of the dynamics of individual protein molecules in membranes of intact human and mouse red cells. Blood Cells Mol. Dis. 27:143–147. [DOI] [PubMed] [Google Scholar]
- 22.Edidin, M., M. C. Zuniga, and M. P. Sheetz. 1994. Truncation mutants define and locate cytoplasmic barriers to lateral mobility of membrane glycoproteins. Proc. Natl. Acad. Sci. USA. 91:3378–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sako, Y., A. Nagafuchi, S. Tsukita, M. Takeichi, and A. Kusumi. 1998. Cytoplasmic regulation of the movement of E-cadherin on the free cell surface as studied by optical tweezers and single particle tracking: corralling and tethering by the membrane skeleton. J. Cell Biol. 140:1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomishige, M., Y. Sako, and A. Kusumi. 1998. Regulation mechanism of the lateral diffusion of band 3 in erythrocyte membranes by the membrane skeleton. J. Cell Biol. 142:989–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujiwara, T., K. Ritchie, H. Murakoshi, K. Jacobson, and A. Kusumi. 2002. Phospholipids undergo hop diffusion in compartmentalized cell membrane. J. Cell Biol. 157:1071–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang, Q., and M. Edidin. 2003. Lowering the barriers to random walks on the cell surface. Biophys. J. 84:400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakada, C., K. Ritchie, Y. Oba, M. Nakamura, Y. Hotta, R. Iino, R. S. Kasai, K. Yamaguchi, T. Fujiwara, and A. Kusumi. 2003. Accumulation of anchored proteins forms membrane diffusion barriers during neuronal polarization. Nat. Cell Biol. 5:626–632. [DOI] [PubMed] [Google Scholar]
- 28.Ritchie, K., R. Iino, T. Fujiwara, K. Murase, and A. Kusumi. 2003. The fence and picket structure of the plasma membrane of live cells as revealed by single molecule techniques (Review). Mol. Membr. Biol. 20:13–18. [DOI] [PubMed] [Google Scholar]
- 29.Simson, R., E. D. Sheets, and K. Jacobson. 1995. Detection of temporary lateral confinement of membrane proteins using single-particle tracking analysis. Biophys. J. 69:989–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simson, R., B. Yang, S. E. Moore, P. Doherty, F. S. Walsh, and K. A. Jacobson. 1998. Structural mosaicism on the submicron scale in the plasma membrane. Biophys. J. 74:297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dietrich, C., B. Yang, T. Fujiwara, A. Kusumi, and K. Jacobson. 2002. Relationship of lipid rafts to transient confinement zones detected by single particle tracking. Biophys. J. 82:274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kusumi, A., Y. Sako, and M. Yamamoto. 1993. Confined lateral diffusion of membrane receptors as studied by single particle tracking (nanovid microscopy). Effects of calcium-induced differentiation in cultured epithelial cells. Biophys. J. 65:2021–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sako, Y., and A. Kusumi. 1994. Compartmentalized structure of the plasma membrane for receptor movements as revealed by a nanometer-level motion analysis. J. Cell Biol. 125:1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saxton, M. J. 1995. Single-particle tracking: effects of corrals. Biophys. J. 69:389–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sheets, E. D., G. M. Lee, R. Simson, and K. Jacobson. 1997. Transient confinement of a glycosylphosphatidylinositol-anchored protein in the plasma membrane. Biochemistry. 36:12449–12458. [DOI] [PubMed] [Google Scholar]
- 36.den Hartigh, J. C., P. M. van Bergen en Henegouwen, A. J. Verkleij, and J. Boonstra. 1992. The EGF receptor is an actin-binding protein. J. Cell Biol. 119:349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Bergen en Henegouwen, P. M., J. C. den Hartigh, P. Romeyn, A. J. Verkleij, and J. Boonstra. 1992. The epidermal growth factor receptor is associated with actin filaments. Exp. Cell Res. 199:90–97. [DOI] [PubMed] [Google Scholar]
- 38.Gronowski, A. M., and P. J. Bertics. 1995. Modulation of epidermal growth factor receptor interaction with the detergent-insoluble cytoskeleton and its effects on receptor tyrosine kinase activity. Endocrinology. 136:2198–2205. [DOI] [PubMed] [Google Scholar]
- 39.van der Heyden, M. A., P. M. Van Bergen en Henegouwen, N. de Ruiter, M. A. Verdaasdonk, J. G. van den Tweel, G. Rijksen, J. Boonstra, and P. Joling. 1997. The actin binding domain of the epidermal growth factor receptor is required for EGF-stimulated tissue invasion. Exp. Cell Res. 234:521–526. [DOI] [PubMed] [Google Scholar]
- 40.Tang, J., and D. J. Gross. 2003. Regulated EGF receptor binding to F-actin modulates receptor phosphorylation. Biochem. Biophys. Res. Commun. 312:930–936. [DOI] [PubMed] [Google Scholar]
- 41.Gronowski, A. M., and P. J. Bertics. 1993. Evidence for the potentiation of epidermal growth factor receptor tyrosine kinase activity by association with the detergent-insoluble cellular cytoskeleton: analysis of intact and carboxy-terminally truncated receptor. Endocrinology. 133:2838–2846. [DOI] [PubMed] [Google Scholar]
- 42.Stoorvogel, W., S. Kerstens, I. Fritzsche, J. C. den Hartigh, R. Oud, M. A. van der Heyden, J. Voortman, and P. M. van Bergen en Henegouwen. 2004. Sorting of ligand-activated epidermal growth factor receptor to lysosomes requires its actin-binding domain. J. Biol. Chem. 279:11562–11569. [DOI] [PubMed] [Google Scholar]
- 43.Peppelenbosch, M. P., L. G. Tertoolen, W. J. Hage, and S. W. de Laat. 1993. Epidermal growth factor-induced actin remodeling is regulated by 5-lipoxygenase and cyclooxygenase products. Cell. 74:565–575. [DOI] [PubMed] [Google Scholar]
- 44.Toral, C., M. C. Solano-Agama, J. Luna, M. C. Romano, and M. E. Mendoza-Garrido. 2003. Epidermal growth factor induces an increase in cell adhesion and an arrangement of actin skeleton in stress fibres in pituitary cultured cells from infantile rats but not adult rats. J. Cell. Physiol. 195:80–91. [DOI] [PubMed] [Google Scholar]
- 45.Rijken, P. J., S. M. Post W. J. Hage, P. M. van Bergen en Henegouwen, A. J. Verkleij, and J. Boonstra. 1995. Actin polymerization localizes to the activated epidermal growth factor receptor in the plasma membrane, independent of the cytosolic free calcium transient. Exp. Cell Res. 218:223–232. [DOI] [PubMed] [Google Scholar]
- 46.Chen, P., J. E. Murphy-Ullrich, and A. Wells. 1996. A role for gelsolin in actuating epidermal growth factor receptor-mediated cell motility. J. Cell Biol. 134:689–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brandt, B. H., A. Roetger, T. Dittmar, G. Nikolai, M. Seeling, A. Merschjann, J. R. Nofer, G. Dehmer-Moller, R. Junker, G. Assmann, and K. S. Zaenker. 1999. c-erbB-2/EGFR as dominant heterodimerization partners determine a motogenic phenotype in human breast cancer cells. FASEB J. 13:1939–1949. [DOI] [PubMed] [Google Scholar]
- 48.Caroni, P. 2001. New EMBO members' review: actin cytoskeleton regulation through modulation of PI(4,5)P(2) rafts. EMBO J. 20:4332–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson, R. E., D. R. Larson, and W. W. Webb. 2002. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 82:2775–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Axelrod, D., D. E. Koppel, J. Schlessinger, E. Elson, and W. W. Webb. 1976. Mobility measurement by analysis of fluorescence photobleaching recovery kinetics. Biophys. J. 16:1055–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hendriks, B. S., L. K. Opresko, and H. S. Wiley. 2003. Lauffenburger D. coregulation of epidermal growth factor receptor/human epidermal growth factor receptor 2 (HER2) levels and locations: quantitative analysis of HER2 overexpression effects. Cancer Res. 63:1130–1137. [PubMed] [Google Scholar]
- 52.Burke, P., K. Schooler, and H. S. Wiley. 2001. Regulation of epidermal growth factor receptor signaling by endocytosis and intracellular trafficking. Mol. Biol. Cell. 12:1898–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fendly, B. M., M. Winget, R. M. Hudziak, M. T. Lipari, M. A. Napier, and A. Ullrich. 1990. Characterization of murine monoclonal antibodies reactive to either the human epidermal growth factor receptor or HER2/neu gene product. Cancer Res. 50:1550–1558. [PubMed] [Google Scholar]
- 54.Christian, A. E., M. P. Haynes, M. C. Phillips, and G. H. Rothblat. 1997. Use of cyclodextrins for manipulating cellular cholesterol content. J. Lipid Res. 38:2264–2272. [PubMed] [Google Scholar]
- 55.Rodal, S. K., G. Skretting, O. Garred, F. Vilhardt, B. van Deurs, and K. Sandvig. 1999. Extraction of cholesterol with methyl-beta-cyclodextrin perturbs formation of clathrin-coated endocytic vesicles. Mol. Biol. Cell. 10:961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schmidt, T., G. J. Schutz, W. Baumgartner, H. J. Gruber, and H. Schindler. 1995. Characterization of photophysics and mobility of single molecules in a fluid lipid membrane. J. Phys. Chem. 99:17662–17668. [Google Scholar]
- 57.Kusumi, A., K. Suzuki, and K. Koyasako. 1999. Mobility and cytoskeletal interactions of cell adhesion receptors. Curr. Opin. Cell Biol. 11:582–590. [DOI] [PubMed] [Google Scholar]
- 58.Murase, K., T. Fujiwara, Y. Umemura, K. Suzuki, R. Iino, H. Yamashita, M. Saito, H. Murakoshi, K. Ritchie, and A. Kusumi. 2004. Ultrafine membrane compartments for molecular diffusion as revealed by single molecule techniques. Biophys. J. 86:4075–4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ritchie, K., X. Y. Shan, J. Kondo, K. Iwasawa, T. Fujiwara, and A. Kusumi. 2005. Detection of non-Brownian diffusion in the cell membrane in single molecule tracking. Biophys. J. 88:2266–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Livneh, E., M. Benveniste, R. Prywes, S. Felder, Z. Kam, and J. Schlessinger. 1986. Large deletions in the cytoplasmic kinase domain of the epidermal growth factor receptor do not affect its lateral mobility. J. Cell Biol. 103:327–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yarden, Y., M. Gabbay, and J. Schlessinger. 1981. Primary amines do not prevent the endocytosis of epidermal growth factor into 3T3 fibroblasts. Biochim. Biophys. Acta. 674:188–203. [DOI] [PubMed] [Google Scholar]
- 62.Hillman, G. M., and J. Schlessinger. 1982. Lateral diffusion of epidermal growth factor complexed to its surface receptors does not account for the thermal sensitivity of patch formation and endocytosis. Biochemistry. 21:1667–1672. [DOI] [PubMed] [Google Scholar]
- 63.Cuatrecasas, P. 1982. Epidermal growth factor: uptake and fate. Ciba Found. Symp. 92:96–108. [DOI] [PubMed] [Google Scholar]
- 64.Rees, A. R., M. Gregoriou, P. Johnson, and P. B. Garland. 1984. High affinity epidermal growth factor receptors on the surface of A431 cells have restricted lateral diffusion. EMBO J. 3:1843–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gadella, T. W. J., and T. M. Jovin. 1995. Oligomerization of epidermal growth factor receptors on A431 cells studied by time-resolved fluorescence imaging microscopy. A stereochemical model for tyrosine kinase receptor activation. J. Cell Biol. 129:1543–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nagy, P., L. Bene, M. Balazs, W. C. Hyun, S. J. Lockett, N. Y. Chiang, F. Waldman, B. G. Feuerstein, S. Damjanovich, and J. Szollosi. 1998. EGF-induced redistribution of erbB2 on breast tumor cells: flow and image cytometric energy transfer measurements. Cytometry. 32:120–131. [DOI] [PubMed] [Google Scholar]
- 67.Moriki, T., H. Maruyama, and I. N. Maruyama. 2001. Activation of preformed EGF receptor dimers by ligand-induced rotation of the transmembrane domain. J. Mol. Biol. 311:1011–1026. [DOI] [PubMed] [Google Scholar]
- 68.Kusumi, A., H. Ike, C. Nakada, K. Murase, and T. Fujiwara. 2005. Single-molecule tracking of membrane molecules: plasma membrane compartmentalization and dynamic assembly of raft-philic signaling molecules. Semin. Immunol. 17:3–21. [DOI] [PubMed] [Google Scholar]
- 69.Kenworthy, A. K., B. J. Nichols, C. L. Remmert, G. M. Hendrix, M. Kumar, J. Zimmerberg, and J. Lippincott-Schwartz. 2004. Dynamics of putative raft-associated proteins at the cell surface. J. Cell Biol. 165:735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vrljic, M., S. Y. Nishimura, W. E. Moerner, and H. M. McConnell. 2005. Cholesterol depletion suppresses the translational diffusion of Class II major histocompatibility complex proteins in the plasma membrane. Biophys. J. 88:334–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pralle, A., P. Keller, E. L. Florin, K. Simons, and J. K. Horber. 2000. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. J. Cell Biol. 148:997–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ike, H., A. Kosugi, A. Kato, R. Iino, H. Hirano, T. Fujiwara, K. Ritchie, and A. Kusumi. 2003. Mechanism of Lck recruitment to the T-cell receptor cluster as studied by single-molecule-fluorescence video imaging. ChemPhysChem. 4:620–626. [DOI] [PubMed] [Google Scholar]
- 73.Suzuki, K., R. E. Sterba, and M. P. Sheetz. 2000. Outer membrane monolayer domains from two-dimensional surface scanning resistance measurements. Biophys. J. 9:448–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Suzuki, K., and M. P. Sheetz. 2001. Binding of cross-linked glycosylphosphatidylinositol-anchored proteins to discrete actin-associated sites and cholesterol-dependent domains. Biophys. J. 81:2181–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rodgers, W., and J. Zavzavadjian. 2001. Glycolipid-enriched membrane domains are assembled into membrane patches by associating with the actin cytoskeleton. Exp. Cell Res. 267:173–183. [DOI] [PubMed] [Google Scholar]
- 76.Deckert, M., M. Ticchioni, and A. Bernard. 1996. Endocytosis of GPI-anchored proteins in human lymphocytes: role of glycolipid-based domains, actin cytoskeleton, and protein kinases. J. Cell Biol. 133:791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Harder, T., and K. Simons. 1999. Clusters of glycolipid and glycosylphosphatidylinositol-anchored proteins in lymphoid cells: accumulation of actin regulated by local tyrosine phosphorylation. Eur. J. Immunol. 29:556–562. [DOI] [PubMed] [Google Scholar]
- 78.Laux, T., K. Fukami, M. Thelen, T. Golub, D. Frey, and P. Caroni. 2000. GAP43, MARCKS, and CAP23 modulate PI(4,5)P(2) at plasmalemmal rafts, and regulate cell cortex actin dynamics through a common mechanism. J. Cell Biol. 149:1455–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burgess, A. W., H. S. Cho, C. Eigenbrot, K. M. Ferguson, T. P. Garrett, D. J. Leahy, M. A. Lemmon, M. X. Sliwkowski, C. W. Ward, and S. Yokoyama. 2003. An open-and-shut case? Recent insights into the activation of EGF/ErbB receptors. Mol. Cell. 12:541–552. [DOI] [PubMed] [Google Scholar]
- 80.Zhou, B., A. Bagri, and S. K. Beckendorf. 2001. Salivary gland determination in Drosophila: a salivary-specific, fork head enhancer integrates spatial pattern and allows fork head autoregulation. Dev. Biol. 237:54–67. [DOI] [PubMed] [Google Scholar]
- 81.Shvartsman, S. Y., C. B. Muratov, and D. A. Lauffenburger. 2002. Modeling and computational analysis of EGF receptor-mediated cell communication in Drosophila oogenesis. Development. 129:2577–2589. [DOI] [PubMed] [Google Scholar]
- 82.Maly, I. V., H. S. Wiley, and D. A. Lauffenburger. 2004. Self-organization of polarized cell signaling via autocrine circuits: computational model analysis. Biophys. J. 86:10–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kearsley, J. H., K. L. Furlong, R. A. Cooke, and M. J. Waters. 1990. An immunohistochemical assessment of cellular proliferation markers in head and neck squamous cell cancers. Br. J. Cancer. 61:821–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.