Abstract
Filopodium, a spike-like actin protrusion at the leading edge of migrating cells, functions as a sensor of the local environment and has a mechanical role in protrusion. We use modeling to examine mechanics and spatial-temporal dynamics of filopodia. We find that >10 actin filaments have to be bundled to overcome the membrane resistance and that the filopodial length is limited by buckling for 10–30 filaments and by G-actin diffusion for >30 filaments. There is an optimal number of bundled filaments, ∼30, at which the filopodial length can reach a few microns. The model explains characteristic interfilopodial distance of a few microns as a balance of initiation, lateral drift, and merging of the filopodia. The theory suggests that F-actin barbed ends have to be focused and protected from capping (the capping rate has to decrease one order of magnitude) once every hundred seconds per micron of the leading edge to initiate the observed number of filopodia. The model generates testable predictions about how filopodial length, rate of growth, and interfilopodial distance should depend on the number of bundled filaments, membrane resistance, lamellipodial protrusion rate, and G-actin diffusion coefficient.
INTRODUCTION
The crawling motion of animal cells over a substrate has been described as the succession of protrusion, attachment, and retraction (1). The first step in this sequence, protrusion, is driven by actin polymerization at the leading edge of the cell (2). A common type of protrusive specialization of the leading edge of the cell is the lamellipodium—a flat, leaf-like extension filled with a dense branched network of short (tenths of micron long) actin filaments (Fig. 1). According to the dendritic nucleation model (3), nascent filaments branch from the sides or tips of existing filaments in a sterically precise way. Filaments' barbed ends are oriented forward at roughly ±35° to the direction of protrusion (4). The barbed ends elongate at tenths of micron per second and are capped within seconds. After capping, the filaments lag behind the leading edge and are replaced by the next generation of filaments.
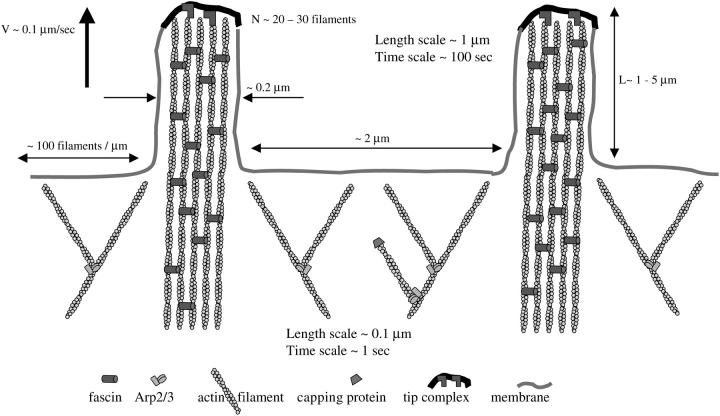
FIGURE 1.
Organization and characteristic scales of filopodia and lamellipodia.
The lamellipodial leading edge is interspersed with filopodia—bundles of actin filaments that are packed tightly together and protrude forward (5,6) (Fig. 1). Similar to the lamellipodial filaments, filopodial filaments are polarized with their barbed ends in the direction of protrusion, but in contrast, they are parallel, long, and turn over very slowly (7) (Fig. 1). Filopodial and lamellipodial protrusions rely on different mechanisms: filament treadmilling (8) and array (9) treadmilling, respectively. They are regulated by different signaling pathways (10,11), yet they are intimately connected, because the filopodial bundles emerge from the lamellipodial network (12,13).
Filopodial protrusions can be “guiding” devices probing space ahead of the lamellipodium. They can also be mechanical devices “penetrating” the environment and serving as a robust scaffold for the lamellipodial protrusion. The role of filopodia as the sensors of the local environment and as sites for adhesion and signaling is well documented (14). In some cells, filopodia are essential for navigation: when filopodia are suppressed, the nerve growth cones can advance but cannot navigate (15). However, fish keratocytes, for example, migrate without filopodia at all (16). It is also worth mentioning that three-dimensional (3D) cell migration through extracellular matrices or engineered scaffolds seems to rely more on the filopodial protrusions in contrast with two-dimensional (2D) cell crawling on flat surfaces (17,18).
There are two major questions about the filopodial protrusion: how is it maintained and how is it initiated? One possibility is that the filopodial filaments are initiated from the cell leading edge by specialized structures (19). Recent evidence, however, points out that the lamellipodial filaments themselves can bend together and “zipper” into parallel bundles of actin filaments (12,20). First, these bundles do not protrude much from the leading edge (such bundles are called Λ-precursors (12) because of their shape). Then, they either mature into the filopodia, or merge with other bundles. To initiate such bundles, the barbed ends have to be locally associated with each other and protected from capping. Protein VASP, elevated in the region of frequent filopodial emergence (12), inhibits capping (21). VASP also transiently binds the barbed ends (22), so association of VASP molecules with each other or some protein cluster could be sufficient to create the tip of the actin bundle. Bundling protein fascin, which is enriched near the tip of the bundle (12), assists VASP and likely other proteins in filopodial initiation. Emerged filopodia have to “outrun” the lamellipodial protrusion, so they have to overcome the membrane resistance, and the G-actin has to be delivered to the filopodial tips.
Available quantitative data (12,23,24) allow theoretical examination of the filopodial mechanics. In the next section, we investigate the restrictions that buckling, membrane resistance, and G-actin diffusion impose on the filopodial dynamics. Then, we find the connection between the spacing between adjacent filopodia and the rates of the filopodial initiation and lateral drift. Finally, we discuss the modeling implications for the biology of filopodial protrusions.
MECHANICS AND MAINTENANCE OF FILOPODIA
Filopodial protrusions are a few tenths of micron in diameter (25), a few microns in length (7,12,13) (see Discussion for exceptions), and contain 10–30 bundled filaments (12,25) (Fig. 1). The distances between neighboring filopodia are in the micron range. In this article, we explain how these characteristic scales emerge from the physics of the actin bundle. Models' parameters and variables are listed in Tables 1 and 2, respectively.
TABLE 1.
Model variables
| Symbol | Meaning | Estimated value |
|---|---|---|
| L | Filopodial length | ∼1–2 μm |
| a(x, t) | G-actin concentration | ∼10 μM |
| λ | No. of Λ-precursors per micron of the leading edge | ∼0.5/μm |
| f | No. of filopodial protrusions per micron of the leading edge | ∼0.5/μm |
| xi(t) | Position of actin bundle in the stochastic model | – |
| vi(t) | Rate of lateral movement in the stochastic model | – |
| mi | Maturity index in the stochastic model | 0,1 |
| t | Time | Seconds |
| x | Distance | Microns |
TABLE 2.
Model parameters
| Symbol | Meaning | Value | References |
|---|---|---|---|
| d | Diameter of filopodium | ∼0.2 μm | (25) |
| N | No. of filaments in filopodial bundle | ∼10–30 | (12,25) |
| F | Membrane resistance force | ∼20 (10–50) pN | (26–28) |
| kBT | Thermal energy | 4.1 pN × nm | (26) |
| Lp | F-actin persistence length | 10 (1–20) μm | (29,30) |
| B | Membrane bending resistance | 50 kBT | References in Peskin et al. (26) |
| δ | Half-size of actin monomer | ≈2.7 nm | References in Peskin et al. (26) |
| D | Effective G-actin diffusion coefficient | ∼5 μm2/s | (55) |
| a0 | G-actin concentration at the leading edge | ∼10 μM | References in Mogilner and Edelstein-Keshet (34) |
| η | Geometric conversion coefficient | ∼20 μM−1μm−1 | This article |
| kon | G-actin assembly rate | ≈10 μM−1s−1 | References in Mogilner and Edelstein-Keshet (34) |
| θc | Critical angle between filaments and direction of protrusion | 60–75–80° | Guessed using data from Maly and Borisy and others (4,12) |
| N0 | No. of filaments to support filopodium | ≈13 | This article |
| Vl | Protrusion rate | 0.05 μm/s | (12) |
| b | Rate of bundling | ∼0.01 μm−1s−1 | This article |
| m | Rate of Λ-precursors maturation | ∼0.01/s | This article |
| Vd | Rate of the lateral drift | ∼0.01 μm/s | This article |
| lc | Distance between fascin cross-links | ∼20–200 nm | Guessed using data from Svitkina et al. (12) |
More than 10 bundled filaments are required for filopodia not to buckle
Membrane bending and tension result in the resistance force at the filopodial tip estimated theoretically as F ∼10–20 pN for a membrane cylinder of radius 50–100 nm (26). More detailed recent modeling results in a similar estimate (S. Sun, Johns Hopkins University, personal communication). Experiments with pulling membrane tethers the width and length of which are similar to characteristic filopodial dimensions also give the forces in similar range of 10–50 pN (27,28). Note that in these experiments both membrane bending force and breaking of membrane-cortex links contribute to the resistance, which is likely similar to the resistance to the filopodial protrusion.
Mechanically, the cross-linked filopodial bundle is an effective elastic rod, to which tip the membrane resistance force is applied. The critical force that buckles such rod is equal to:
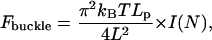 |
(1) |
where kBT ≈ 4.1 pN nm is the thermal energy (26), Lp ∼ 10 μm is the F-actin persistence length (29,30), and L is the length of the filopodial protrusion. Here π2kBTLp/4L2 is the buckling force for one filament (31), and I(N) is the nondimensional factor, which is responsible for the dependence of the bundle stiffness on the number of the bundled filaments, N. There are two limiting cases: if the filaments are bundled weakly (for example, the distance between the cross-links along the filaments is large and bundling protein is very flexible), then the filaments buckle independently, and I(N) = N. If, in the opposite limit, the bundling is so frequent and tight that the filaments are effectively “glued” to each other, then the bundle can be considered as a single thick rod. The cross-section area of such rod is equal to the number of the filaments times one filament's cross-section area, and the rod's effective radius is proportional to the square root of the number of the filaments. The stiffness is proportional to the radius in power four (31), so in this case I(N) ∼ N2. Numerical simulations described below suggest that I(N) ≈ 0.5 × N2.
Using Eq. 1, we can estimate the critical length at which the membrane resistance force buckles the filopodial bundle:
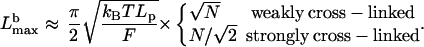 |
(2) |
We plotted 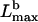 as function of N in two limiting cases (I(N) = N and
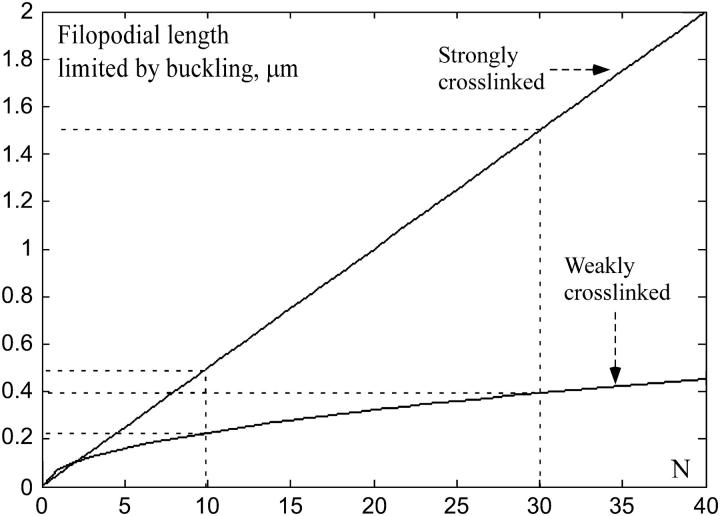
as function of N in two limiting cases (I(N) = N and 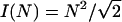 ) in Fig. 2. The plot demonstrates that the weakly cross-linked bundle of 10–30 filaments would buckle at length below 0.5 μm. Strong bundling can support the length in micron range, in agreement with numerous observations.
) in Fig. 2. The plot demonstrates that the weakly cross-linked bundle of 10–30 filaments would buckle at length below 0.5 μm. Strong bundling can support the length in micron range, in agreement with numerous observations.
FIGURE 2.
Dependence of the critical length, at which the filopodium would buckle, on the number of cross-linked and not cross-linked filaments (solid curves), as predicted by Eq. 2. The dotted lines show the predicted length range for the characteristic numbers of the filaments.
We used computer simulations to derive the function I(N) in biologically relevant situation. In the filopodial bundle, the filaments are not packed densely (the cross-section area of the protrusion is ∼0.01 μm2, whereas the total cross-section area of the 25 filopodial filaments is ∼0.001 μm2), so it is likely that each filament is cross-linked with only a few neighbors. We considered variable number (3–15) of elastic rods, 2 μm in length, with the same mechanical characteristics as those of F-actin. The rods were arranged in a parallel stack, and each rod was connected by elastic cross-links to 2–4 nearest neighbors. The bundling protein, fascin, has length between 10 and 15 nm (32), and its stiffness is likely to be similar to that of F-actin (33). We varied both the length of the cross-links from 10 to 50 nm, and their stiffness from 10 times less to 10 times more than that of F-actin. The electron microscopy (EM) data (12) indicate that the interfascin distance along a filament in the filopodia is of the order of tens of nanometers, so we varied the average corresponding distance from 20 to 500 nm. (In the Appendix, we consider a model of fascin distribution along the bundle.)
We used FEMLAB to solve the buckling problem of elasticity theory (the “Structural Mechanics” module solves an eigenvalue problem, such that the lowest eigenvalue corresponds to the buckling force, whereas the corresponding eigenfunction gives the shape of the buckled bundle). The shape of the buckled actin bundle in a sample simulation is shown in Fig. 3 A. (Filaments were arranged in parallel in 2D; general principles of the linear elasticity theory suggest that the 3D bundle would buckle at similar forces.) The simulation results showed that if the average distance between the neighboring cross-links along an actin filament in the bundle is of the order of 1 μm, then the stiffness of the bundle scaled as ≈N (Fig. 3, B and C), in agreement with simple physical arguments above. Also in agreement with these qualitative arguments, at small average distance between the neighboring cross-links ≈0.1 μm, the stiffness of the bundle scaled as ≈0.5 × N2 (Fig. 3, B and C). We conclude that the observed bundling is tight and in the biologically relevant regime the stiffness of the filopodial bundle increases with the number of the bundled filaments approximately as ≈0.5 × N2. The observed number of the filaments can support the bundle of 1–1.5-μm long against buckling (Fig. 2).
FIGURE 3.
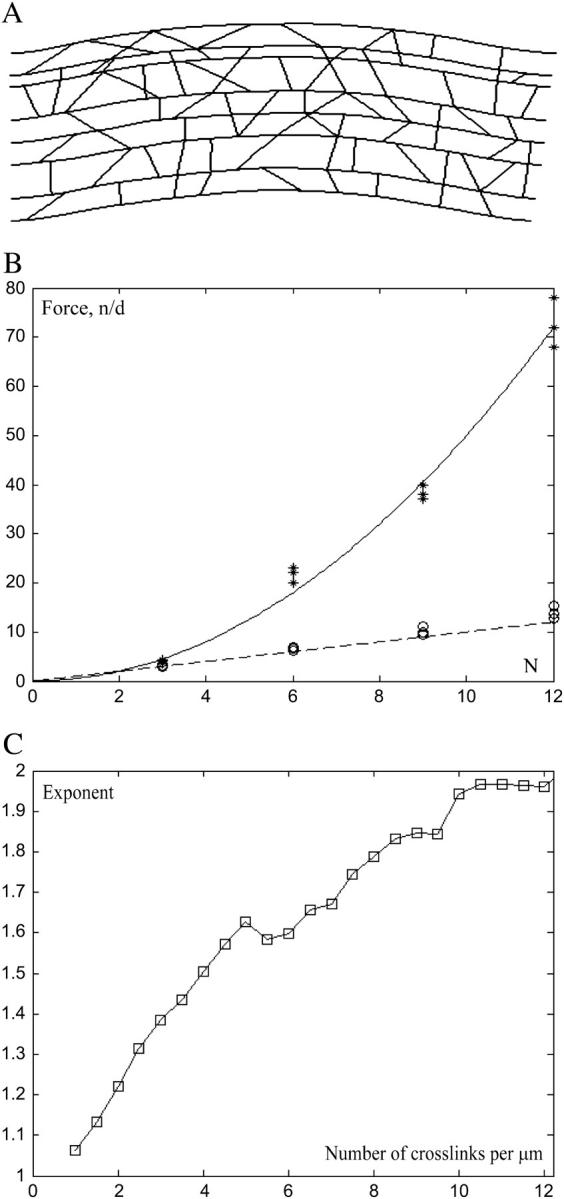
(A) Computed shape of the actin filaments bundled by short elastic links. (B) The computed buckling force (scaled by the buckling force for one 2-μm-long filament) is plotted as the function of the number of bundled filaments N for the average number of cross-links per 1 μm of each filament's length equal to 0.5, 1, 1.5 (circles) and to 10, 11, 12 (stars). At small cross-linking density, function I(N) is approximated well by I(N) = N (dashed line). At large cross-linking density, function I(N) is approximated well by I(N) = 0.5 × N2 (solid curve). (C) The buckling force was computed as the function of the average number of cross-links per 1 μm of each filament's length for the bundles consisting of N = 3, 6, 9, 12 filaments. Then, for each value of the cross-linking density, the computed force dependence on N was fitted with function A × Nx. The exponent x is plotted as the function of the average number of cross-links per 1 μm of each filament's length (squares). The plot confirms that at small cross-linking density I(N) ∼ N, whereas at large cross-linking density I(N) ∼ N2.
D. Mullins (University of California, San Francisco, personal communication) observed recently that a filopodial bundle of 25 filaments has effective persistence length of 14 mm. This agrees with our simulations, according to which 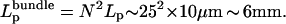 In this observation, bundle grew to be 40-μm long, an order of magnitude greater than predicted by the model. We discuss the difference in the Discussion.
In this observation, bundle grew to be 40-μm long, an order of magnitude greater than predicted by the model. We discuss the difference in the Discussion.
G-actin diffusion limits the length of thick bundles; membrane resistance limits the length of thin bundles
G-actin diffusion, in addition to the membrane resistance, limits the length of the filopodium. In this article, by G-actin we mean a part of the G-actin pool that can assemble onto the barbed ends (GTP-G-actin not sequestered by thymosin; see Mogilner and Edelstein-Keshet (34)). Let L(t) be the time-dependent length of the N-filament bundle, and a(x, t) be the concentration of the G-actin along the filopodial length (Fig. 4 A). The x axis is oriented forward, and its origin is at the base of the filopodium at the leading edge (Fig. 4 A). In the filopodium, G-actin diffuses and drifts with the cytoplasmic fluid. This drift rate is roughly equal to the rate of the filopodial protrusion dL/dt, because on the relevant timescale the membrane is impermeable to water (35), and due to incompressibility, the cytoplasm has to fill the filopodium at the rate of protrusion. Equation for the G-actin concentration has the form:
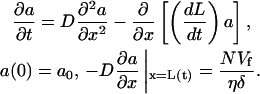 |
(3) |
FIGURE 4.
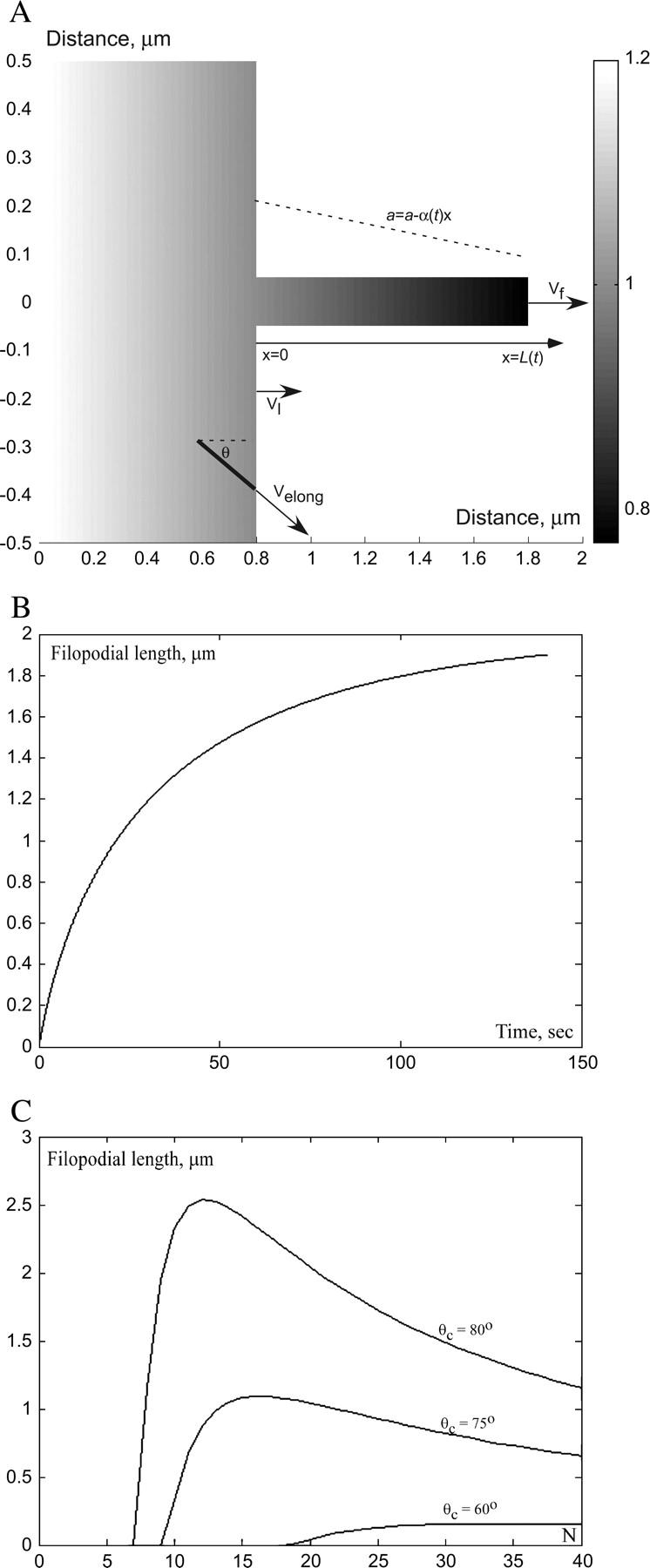
(A) Results of computer simulations of the 2-D G-actin distribution (a(x, t)) in the filopodium and the small adjacent part of the lamellipodium (distance is in microns; G-actin concentration illustrated with shading is in nondimensional units, see Appendix II for details). Approximate analytical one-dimensional solution described in the text coincides with the computer-simulated G-actin concentration in the filopodium. Both in the lamellipodium, and the filopodium, the linear gradient of G-actin develops in the direction of protrusion. One sample lamellipodial filament at the leading edge is shown. (B) Filopodial length as a function of time predicted by the solution of Eqs. 3 and 4. (C) Stationary filopodial length as a function of the number of bundled filaments for three different values of the lamellipodial filaments' critical angle, as predicted by Eq. 8. Higher value of the critical angle corresponds to slower lamellipodial protrusion.
Here D is the effective G-actin diffusion coefficient. The boundary condition at the filopodial base (x = 0) is that the G-actin concentration there is equal to that at the leading lamellipodial edge, a0. (In the Appendix, we examine this assumption by simulating a two-dimensional G-actin distribution (shown in Fig. 4 A) in the filopodium and adjacent part of the lamellipodium.) The boundary condition at the filopodial tip (x = L(t)), which is similar to that at the lamellipodial leading edge (34), is that the G-actin diffusive flux there, −D(∂a/∂x)(L), is equal to the number of monomers assembling per second onto the tips of N filaments. This number is equal to the number of the filaments times the rate of elongation of the filopodial filaments Vf and divided by the half-monomer size δ; η is the geometric coefficient converting the number of monomers into micromolar units (see the Appendix). The drift part of the G-actin flux is not included into the boundary condition, because the filopodial tip and the cytoplasm move together.
The rate of the filopodial extension, dL/dt, is equal to the difference between the rates of the filopodial filaments' elongation Vf and of the lamellipodial filaments' extension in the direction of protrusion, Vl: dL/dt = Vf − Vl. Neglecting small rate of filaments' disassembly, Vf ≈ konδa(L) exp(−Fδ/kBTN) (34). Here V0 = konδa(L) is the free polymerization rate proportional to the G-actin concentration at the filopodial tip (kon is the assembly rate); F/N is the membrane resistance load force per filament. The exponential factor is responsible for slowing the protrusion rate by the membrane resistance (34). It is convenient to introduce the effective number of filaments that can support the filopodial protrusion, N0 = Fδ/kBT ≈ 13, then Vf ≈ konδa(L) exp(−N0/N).
In the lamellipodium, the leading edge filaments are distributed over a wide range of angles (4). Those elongating at acute angles to the direction of protrusion grow slower, than those elongating at greater angles, simply because the filaments' elongation rate is the increasing function of the angle, Velong = Vl/ cos θ, where Vl is the same for all filaments (Fig. 4 A). This means that slower growing filaments at small angles are generating disproportionately large force, whereas the filaments at greater angles are “free-loaders” growing faster against smaller force. This also means that a large “critical angle” θc exists, such that filaments growing at this angle elongate against zero force at free polymerization rate V0 = konδa0, whereas filaments growing at even greater angles simply cannot keep up with the leading edge and lag behind it (36,24). Then, Vl = konδa0 cos θc, and
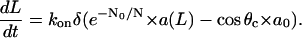 |
(4) |
Equations 3 and 4 together represent a difficult free boundary problem. Fortunately, the timescale separation (G-actin diffusion is much faster than the filopodial growth and cytoplasmic drift) allows approximate analytical solution of the problem, which is sketched in the Appendix. The result is that the G-actin concentration decreases linearly from the base to the tip of the filopodium (Fig. 4 A), so that the concentration gradient is the function of the filopodial length:
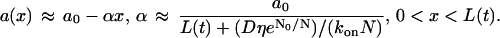 |
(5) |
This gradient induces the G-actin flux, which is “consumed” at the tip and makes it grow, but the longer the filopodium becomes, the smaller is the G-actin concentration at the tip, and the slower is the rate of growth. Solution of Eqs. 3 and 4 (see the Appendix) is plotted in Fig. 4 B for N = 20, θc = 80°. It has the following asymptotics:
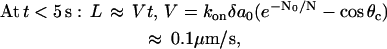 |
(6) |
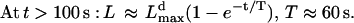 |
(7) |
Thus, for the first few seconds, when the filopodium is short, the G-actin concentration at its tip is almost equal to that at the lamellipodial leading edge, and the filopodial filaments grow in the direction of protrusion much faster than tilted lamellipodial filaments. Then, the G-actin concentration decreases, and the filopodial growth slows down exponentially at great (over micron) lengths. The maximal length of the filopodium, when its elongation slows down to match the lamellipodial expansion rate, is given by the formula:
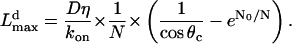 |
(8) |
In Fig. 4 C, we plotted the stationary filopodial length Lmax as a function of N at three different values of the critical angle, θc = 60°, 75°, 80°. The data (4) indicate that the critical angle in rapidly moving cells is likely to be not <60°.
The greater the critical angle is, the slower the lamellipodial protrusion, and the farther the filopodium can extend (Fig. 4 C). This is in agreement with the observation that in the rapidly and steadily moving keratocyte cells the filopodia are absent (16). Another prediction is that the filopodial length is linearly proportional to the G-actin diffusion coefficient.
Dependence of the filopodial length on the number of the bundled filaments is biphasic. At great N, the factor exp(N0/N) ≈ 1 (many filaments easily overcome the membrane resistance), and the length is inversely proportional to the filament number, because many growing filament tips deplete the actin monomeric pool. When N is small, the exponential factor exp(N0/N) increases rapidly, and the filopodial length dramatically decreases. In fact, if the filament number is less than:
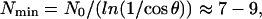 |
(9) |
the bundle cannot protrude at all. These results are similar to effects of the membrane resistance and G-actin diffusion in lamellipodial protrusion (35,34).
Note that we made the estimates assuming that the filaments are not moving relative to the substratum. In fact, the model is also valid in the presence of retrograde flow of actin, which is almost always the case (37). Indeed, let Ve be the elongation rate of the lamellipodial filaments in the direction of protrusion, and Vr be the rate of lamellipodial network's retrograde movement. Then, the lamellipodial extension rate would be Ve = Vl − Vr. The filopodial bundles are embedded into the lamellipodial network and move rearward with the same rate (7,24), so the rate of the filopodial extension is Vf − Vr, where Vf is the rate of growth of the filopodial filaments. Then, the filopodial length changes with the rate (Vl − Vr) − (Vf − Vr), same as in Eq. 4. The G-actin diffusion and drift are unaffected by the retrograde flow. Note also, that our theory is not applicable to the acrosomal protrusion of Thyone (38,39), where the physics and biology is different, and length, rate of extension, and actin concentration are many times greater. Finally, there is a possibility that filopodial tip complex proteins, such as formins, change the polymerization kinetics, in which case the estimates would change.
Length of 10–30-filament bundle is limited by buckling; length of >30-filament bundle is limited by G-actin diffusion
Equations 2 and 8 give the maximal attainable filopodial lengths 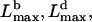 limited by buckling and diffusion, respectively, as functions of the number of the bundled filaments. The resulting observed length is the minimum of these two lengths,
limited by buckling and diffusion, respectively, as functions of the number of the bundled filaments. The resulting observed length is the minimum of these two lengths, 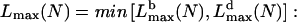 if at given N the filopodium buckles at shorter length than that allowed by diffusion, then the growth would be stopped by buckling, and vice versa. We plotted the function Lmax(N) in Fig. 5 A. Our model predicts that >7–8 bundled filaments can maintain the filopodial protrusion. When the filament number is <10, the membrane resistance limits the filopodial length to submicron range. The length of the bundle of 10–30 filaments is limited by buckling, and is proportional to the filament number. The length of the optimal, 30-filament bundle, reaches 1.5 μm. The length of the thicker bundle decreases inversely proportionally to the filament number, because more filament tips deplete G-actin.
if at given N the filopodium buckles at shorter length than that allowed by diffusion, then the growth would be stopped by buckling, and vice versa. We plotted the function Lmax(N) in Fig. 5 A. Our model predicts that >7–8 bundled filaments can maintain the filopodial protrusion. When the filament number is <10, the membrane resistance limits the filopodial length to submicron range. The length of the bundle of 10–30 filaments is limited by buckling, and is proportional to the filament number. The length of the optimal, 30-filament bundle, reaches 1.5 μm. The length of the thicker bundle decreases inversely proportionally to the filament number, because more filament tips deplete G-actin.
FIGURE 5.
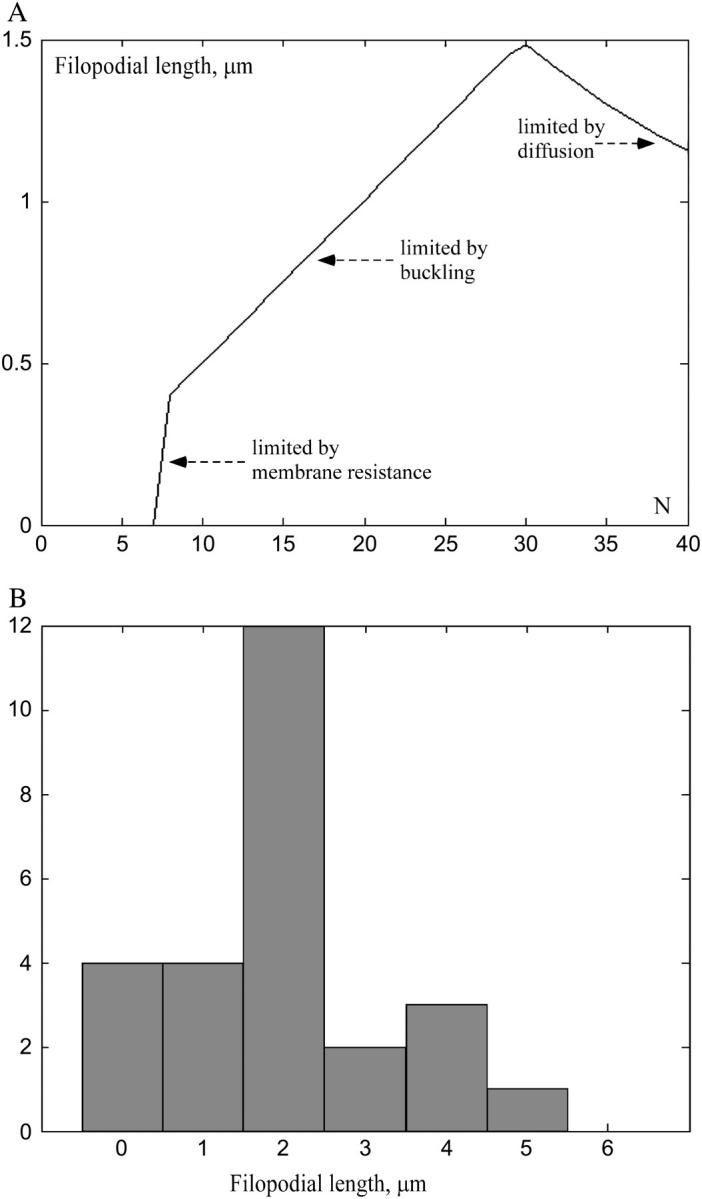
(A) Predicted filopodial length limited by the membrane resistance, buckling, and G-actin diffusion as a function of the number of bundled filaments (θc = 80°). (B) Length distribution for 26 filopodial protrusions gleaned from Fig. 2 of Oldenbourg et al. (24).
Quantitative observations reported in Argiro et al. (23) partially corroborate our theoretical findings. The maximal length of the observed filopodia was 2–10 μm, which is greater, but the same order of magnitude as predicted. In the Discussion, we speculate on the factors that can explain the difference. The observed rate of the filopodial extension, ≈0.12 μm/s, in agreement with Eq. 6, was maximal just after filopodial initiation and declined thereafter, similar to the predicted time series in Fig. 4 B. The growth did not end with asymptotic slowing down, rather, the filopodia collapsed pivoting or buckling when the maximal length was reached. The initial rate of extension directly correlated with the eventual length of the filopodium (23). This is explained by Eq. 6: greater N means faster initial extension rate (more filaments are less affected by the membrane resistance), and also greater final length when the bundle buckles. Interesting model prediction is that for thicker bundles, length of which is limited by the diffusion, the initial extension rate should be correlated negatively with the final filopodial length. Also, we measured the lengths of 26 adjacent filopodial protrusions in Fig. 2 of Oldenbourg et al. (24) (plotted in Fig. 5 B). Most of the filopodia observed have lengths of 2 μm, in agreement with our estimates.
BALANCE OF LATERAL DRIFT AND EMERGENCE RATE OF Λ-PRECURSORS REGULATES INTERFILOPODIAL SPACING
The data on molecular mechanisms are too sketchy to attempt detailed quantitative modeling of the filopodial initiation. Here we address the easier question of spacing of the filopodial protrusions along the lamellipodial leading edge. In the next section we also discuss the implications of the estimates that we derive below for the “convergence-elongation” model (12) of the actin bundle initiation. In the Appendix, we consider a simple model that explains fascin-mediated bundling near the tip of the bundle.
Growing lamellipodial barbed end tilted at angle θ relative to the direction of protrusion drifts with velocity Vl tan θ relative to the leading edge protruding with the rate Vl (8) (Figs. 4 A and 6 A). Convergence, “zippering”, and elongation of a few such lamellipodial filaments would produce an actin bundle, either remaining embedded into the lamellipodium, or making filopodial protrusion (Fig. 6 A). Such bundle would be also tilted at some smaller angle and undergo the lateral drift (24). When two such bundles “collide” at the leading edge, their filaments align with each other and the bundles merge (12). As a result, the number of the bundles decreases. Here we show that the interfilopodial spacing can be explained by the balance between the bundle initiation and merging caused by the lateral drift. We neglect simple disappearance of filipodia, because filaments in a filopodium are stable for >1000 s (11).
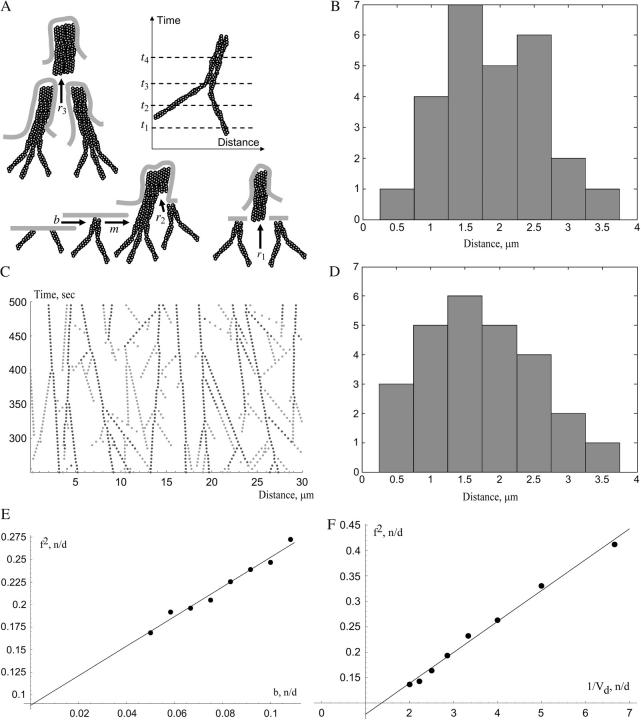
FIGURE 6.
(A) Illustration of the lateral drift. Dashed lines represent the lamellipodial leading edge at four consecutive moments of time. Barbed ends of the individual filaments and the Λ-precursor change their position along the leading edge as the edge protrudes. The rest of the figure illustrates the dynamics of the actin bundles: bundling of the individual filaments (rate b), maturation of the precursors into the filopodium (rate m), merging of two precursors into the filopodium (rate r1), merging of the precursor with the filopodium (rate r2), and merging of two filopodia into one (rate r3). (B) Distance distribution between neighboring filopodia for 26 protrusions gleaned from Fig. 2 of Oldenbourg et al. (24). (C) Results of Monte Carlo simulations of the initiation, lateral drift, maturation, and merging of Λ-precursors (light gray) and filopodia (dark gray). (Horizontal axis) Distance along the lamellipodial leading edge in microns; the vertical axis is time in seconds. (D) Distance distribution between neighboring filopodia based on the stochastic model simulations. (E) Results of Monte Carlo simulations (dots) confirm the analytical prediction that the density of filopodia along the leading edge is proportional to the square root of the rate of initiation of Λ-precursors. We used the value Vd = 0.06 μm/s; b is plotted in units of 0.25 μm−1s−1. (F) Results of Monte Carlo simulations (dots) confirm the analytical prediction that the density of filopodia along the leading edge is inversely proportional to the square root of the drift rate. We used the value b = 0.02 μm−1s−1; Vd is plotted in units of 0.2 μm/s.
We investigate the spacing between filopodia using first a continuous deterministic model that reveals important biological scales, and then performing realistic stochastic simulations. In the continuous model, we introduce densities (numbers per micron) of Λ-precursors, λ(x, t), and of filopodial protrusions, f(x, t), along the lamellipodial leading edge. These densities change according to the following dynamics:
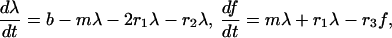 |
(Fig. 6 A). Here b is the rate of initiation of Λ-precursors (bundling), and m is the rate of “maturation” of the Λ-precursors into the filopodial protrusions; r1 is the effective rate of “collision” of the Λ-precursors, as a result of which two colliding Λ-precursors merge into one filopodial protrusion due to the increase of the number of filaments in the merged bundle; r2 is the effective rate of “collision” of a Λ-precursor with a filopodial protrusion, as a result of which the Λ-precursor disappears merging with the filopodial protrusion. Finally, r3 is the effective rate of “collision” of two filopodia that merge into one. This simple model can be made more sophisticated by making the number of filaments in actin bundles the independent model variable and assuming some rules of when merging of Λ-precursors results in a thicker Λ-precursor, and when it results in a filopodial protrusion depending on the numbers of bundled filaments. However, this does not change qualitatively simple results derived below.
Order of magnitude of the rate r1 can be estimated as average inverse time before collision of two Λ-precursors, which is equal to the average distance between the precursors, 1/λ, divided by the average rate of the lateral drift, vd: r1 ≈ vdλ. Similarly, r2 ≈ vdf, r3 ≈ vdf. Thus, we obtain the system of equations (in the mean field approximation neglecting correlations between filopodia) for the actin bundle densities:
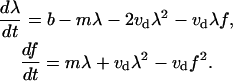 |
(10) |
These equations can be nondimensionalized by scaling the densities using the balance between the bundling rate and merging rate: vdλ2 ∼ b, so the density scale is 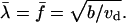 Timescale is equal to the characteristic life time of an individual λ-precursor before it merges with another,
Timescale is equal to the characteristic life time of an individual λ-precursor before it merges with another, 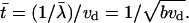 Equations for nondimensional variables
Equations for nondimensional variables 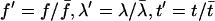 have the form:
have the form:
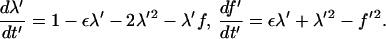 |
(11) |
Here 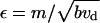 is the dimensionless ratio of the maturation rate to the effective rate of merging of actin bundles.
is the dimensionless ratio of the maturation rate to the effective rate of merging of actin bundles.
Phase plane analysis of nonlinear Eq. 11 shows that there is a unique stable biologically relevant stationary solution. It can be found analytically in two limits. First, when the maturation rate is very slow, 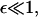 the average precursor and filopodial densities are almost equal:
the average precursor and filopodial densities are almost equal: 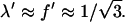 On the other hand, when the λ-precursors mature fast,
On the other hand, when the λ-precursors mature fast, 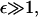 the λ-precursors density is low, λ′ ≈ 1/ε, while f′ ≈ 1. For intermediate values of ε, f′ ∼ 1. The important conclusion is that the order of magnitude of the stationary filopodial density is
the λ-precursors density is low, λ′ ≈ 1/ε, while f′ ≈ 1. For intermediate values of ε, f′ ∼ 1. The important conclusion is that the order of magnitude of the stationary filopodial density is 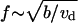 (in dimensional variables).
(in dimensional variables).
We measured the distances between 26 adjacent filopodia in Fig. 2 of Oldenbourg et al. (24) and plotted the results in Fig. 6 B. The average interfilopodial distance is ∼2 μm, and f ∼ 0.5/μm. The average angle at which the actin bundles are tilted relative to the direction of protrusion is <30°, and the average lateral drift rate is a few fold less than the rate of protrusion, vd ∼ 0.01μm/s. We can estimate the rate of emergence of the Λ-precursors as b ∼ vdf2 ∼ 0.001–0.01 μm−1s−1. Thus, a new Λ-precursor has to appear once every few hundreds of seconds per micron of the leading edge. There are no measurements of this rate available, but this estimate seems to compare well with observations reported in Svitkina et al. (12).
Also, the micrographs in Svitkina et al. (12) indicate that the densities of Λ-precursors and filopodial protrusions are comparable, so, according to our analysis, the rate of precursors' maturation cannot be faster than 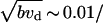 s. In other words, on the average, individual Λ-precursors can be observed for ∼100 s before merging or maturing into a filopodium.
s. In other words, on the average, individual Λ-precursors can be observed for ∼100 s before merging or maturing into a filopodium.
This analysis is supported by the following stochastic simulations, which are essential because of the dispersal of the actin bundles' orientations, correlations between the bundles and large fluctuations of the bundles' number. Each actin bundle (either Λ-precursor, or filopodium) is characterized by its position along the lamellipodial leading edge, xi(t), rate of lateral movement, vi(t), and maturity index, mi, equal to zero for a precursor and to unity for a filopodium. We consider a 30-μm-long segment of the leading edge (Fig. 6 C) and generate new precursors on it at random location with the rate b = 0.01 μm−1s−1. Each nascent precursor is tilted to the protrusion direction at a random angle uniformly distributed in the interval −30° < θi < 30°, and vi(t) = v × tan(θi), where v = 0.05 μm/s is the protrusion rate. The precursors mature (mi switches from 0 to 1) into the filopodia with the constant rate m = 0.01/s. The trajectories of the precursors (light gray) and filopodia (dark gray) from a sample simulation are shown in Fig. 6 C, where time in seconds is shown on the y axis. In the simulations, we update the positions of the tips of the precursors and bases of the filopodia along the leading edge each time step (5 s). Actin bundles that run into the edges of the segment “disappear”.
We consider “collision events” of pairs of the actin bundle, when the distance between them is smaller than 30 nm. Each collision results in the merger of the pair. When two precursors collide, the resulting actin bundle becomes another precursor, or changes into a filopodium with equal probability. (Numerical experiments show that assigning weighed probabilities does not change the results qualitatively.) When either a precursor collides with a filopodium, or two filopodia collide, a single filopodium results. The lateral movement rate of the resulting bundle is equal to that of one of the colliding pair having minimal absolute value, if either two precursors, or two filopodia collide. If a precursor and a filopodium collide, then the lateral movement rates of “mother” and “daughter” filopodia are the same.
Repeating simulations like those shown in Fig. 6 C, we plotted the histogram of the interfilopodial distances (Fig. 6 D). We chose the rate of initiation of Λ-precursors so that the observed and calculated mean interfilopodial distances are the same order of magnitude. Simulations demonstrate that the observed and calculated variances of these distances are also similar (Fig. 6, B and D). We also tested numerically the predicted dependence 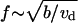 of the analytical model. The stochastic simulations confirm that the density of filopodia along the leading edge is proportional to the square root of the rate of initiation of Λ-precursors and inversely proportional to the square root of the drift rate (Fig. 6, E and F).
of the analytical model. The stochastic simulations confirm that the density of filopodia along the leading edge is proportional to the square root of the rate of initiation of Λ-precursors and inversely proportional to the square root of the drift rate (Fig. 6, E and F).
DISCUSSION
Filopodial length
Estimates in this article show that to overcome the membrane resistance, >10 actin filaments have to be bundled in filopodia. The length of the filopodial bundle of 10–25 filaments is limited to 1–2 μm due to buckling of the bundle by the membrane resistance force. Thicker bundles are stronger, but growth of >30 filaments bundled together is limited by G-actin diffusion: more barbed ends consume so many monomers that diffusion cannot maintain bundles longer than 2 μm. This analysis explains the observed number (tens) of actin filaments in filopodia and their length (microns): in fibroblasts, macrophages, and nerve growth cones the filopodial length rarely exceeds 10 μm. Our findings can also explain the experimental observations (23) of the rate of the filopodial growth of the order of 0.1 μm/s and its correlation with the final filopodial length.
However, filopodia sometimes grow longer: in sea urchin embryo, where filopodia were first seen live in 1961 (40), they were 5–35-μm long. R. D. Mullins (University of California, San Francisco, personal communication cited above) observed recently the filopodial bundle 40-μm long. Also, some observations indicate that the rate of filopodial elongation does not slow down with filopodial length as fast as predicted by our theory (7). These discrepancies point out that simple G-actin diffusion and linear elastic stability of the cross-linked filament bundle cannot fully explain the observed filopodial behavior. Thus, perhaps the most valuable lesson from our model is that additional mechanisms have to be at work in filopodia.
There are few possible explanations for these discrepancies between theory and experiment. First, decreasing membrane resistance by two orders of magnitude increases the filopodial length limited by buckling by one order of magnitude, from a few microns to a few tens of microns (buckling length is proportional to the square root of the force). This can be accomplished by regulation of the membrane tension (41). Second, adhesion of the filopodia to the substratum, which we did not consider, can strengthen the filopodia significantly: long filopodia adheres to the surface, whereas filopodia without adhesions bends laterally (13). Third, as far as the diffusion-limited growth is concerned, our estimates were made for the steadily protruding lamellipodial leading edge. In fact, this protrusion in most cells consists of irregular cycles of protrusion and retraction (21). Filopodial growth can continue past the micron range, with slowing speed, if the lamellipodial leading edge is stalled. Finally, other means of transport not considered here, for example those mediated by unconventional myosin motors (42), can contribute to filopodial elongation. Indeed, there are indications that unconventional myosin motors are responsible for transport of adhesion molecules (14) and of Mena/VASP proteins (43).
Note about protrusion force generation
The polymerization ratchet mechanism of force generation (44) requires frequent bending of either filament tips, or membrane, or both, so that the transient gap between the filaments' tips and membrane is >δ ≈ 2.7 nm. Unlike the tilted lamellipodial filaments, the filopodial filaments are perpendicular to the resisting membrane, and the transient gap due to their thermal bending is smaller. Its magnitude can be estimated as the shortening of the end-to-end distance for elastic rod of length lc and persistence length Lp due to the thermal bending: 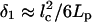 (45). For actin, Lp ∼ 10 μm, and lc ∼ 20–30 nm is of the order of the average distance between the fascin cross-links. The value of δ1 is <1 nm at these parameters, so filament bending is not sufficient. However, the membrane bending is sufficient: in Mogilner and Oster (44) we derived the formula:
(45). For actin, Lp ∼ 10 μm, and lc ∼ 20–30 nm is of the order of the average distance between the fascin cross-links. The value of δ1 is <1 nm at these parameters, so filament bending is not sufficient. However, the membrane bending is sufficient: in Mogilner and Oster (44) we derived the formula: 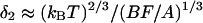 for the corresponding gap. Substituting the values of the membrane bending modulus, B ∼ 50 kBT, the membrane resistance force, F ∼ 20 pN, and the area of the filopodial tip, A ∼ 0.01 μm2 we estimate the corresponding gap as ∼10 nm.
for the corresponding gap. Substituting the values of the membrane bending modulus, B ∼ 50 kBT, the membrane resistance force, F ∼ 20 pN, and the area of the filopodial tip, A ∼ 0.01 μm2 we estimate the corresponding gap as ∼10 nm.
More thorough stochastic simulations taking into account detailed membrane dynamics and polymerization kinetics confirm this conclusion (S. Sun, Johns Hopkins University, personal communication). However, future modeling is needed because the filopodial tip is loaded with proteins and its mechanical properties are unknown. Also, abundance of VASP at the tip could lead to frequent attachment of the filaments to the membrane (46). It is possible that other models of force generation based on complex mechanochemical cycles of barbed ends associated with auxiliary proteins are relevant for the filopodial protrusion (47,48). If this is the case, then the exact values for the generated polymerization force and G-actin kinetics rates at the filopodial tip would change, but their orders of magnitude would not, so the order of magnitude estimates in this article would remain valid.
Model implications for molecular mechanisms of filopodia initiation
The model explains the characteristic distance between adjacent filopodia in micron range as the balance of initiation and lateral drift and merging of the actin bundles. The theory suggests that F-actin barbed ends have to be locally focused and protected from capping approximately once every hundred seconds per micron of the lamellipodial leading edge to initiate the observed number of filopodia.
From the EM data reported in Svitkina et al. (12) we can glean ∼100 barbed ends per micron of the leading edge (this estimate compares well with 250 ends per micron reported in (49)). At protrusion rate v = 0.05 μm/s and average angle between filament growth and leading edge protrusion θ = 35°, the average lateral drift rate is v × tan(θ) ≈ 0.035 μm/s, so barbed ends tilted to the right/left converge at the rate 0.07 μm/s. Total height of the lamellipod is ∼0.2 μm (49), so each filament (which is ∼0.005 μm in diameter) would, on the average, “collide” due to the lateral drift with another filament at the rate (∼50 filaments/μm) × (0.07 μm/s) × (0.005 / 0.2 μm) ∼ 0.1/s. Filaments are tenths of microns long, so the capping rate at the leading edge is of the order of 0.1/s, so there is a significant probability that any growing filament would “collide” with an oppositely tilted filament. If the barbed ends of such pair of filaments are kept together either by a dynamic cross-linker that stays close to the growing filament tips, or by a “processive capper”, such as formin (50), which in turn is associated with a nascent “filopodial tip complex” (reviewed in Small et al. (51)), then the filaments would bend into parallel configuration and start to grow almost in the direction of protrusion. The corresponding bending force is in subpiconewton range and can be easily generated by the polymerization ratchet mechanism (44,50).
Other filament tips would collide with the pair and could be trapped in the growing bundle creating a nascent Λ-precursor. In order for this precursor to assemble a bundle of ∼10 filaments, the effective capping rate in the vicinity of the precursor tip has to decrease to ∼0.01/s. Indeed, the average stationary number of the growing tips in the bundle can be estimated as the ratio of the rate of collisions of the bundle with lamellipodial barbed ends, ∼0.1/s, to the capping rate, so the latter can be estimated as ∼(0.1/s) / 10 = 0.01/s. It would take ∼10 / (0.1/s) = 100 s to assemble the actin bundle.
Such a low capping rate cannot be maintained along the whole leading edge, because the filaments would grow a few microns long and buckle (21). Therefore, our estimates suggest that once every hundred seconds per micron of the lamellipodial leading edge, a nascent filopodial tip complex (or part of it) self-assembles, such that its components both associate with the filament tips physically, and protect them from capping. Then, in hundred seconds, a Λ-precursor develops and matures into a filopodium or merges with other actin bundles. Likely, some positive feedbacks are involved in this process. For example, transient changes in membrane curvature have been shown to cause filopodia perhaps by activating pathways that trigger actin polymerization (52), and in turn concentration of filaments in actin bundles curves the membrane locally. It is premature to speculate about specific pathways of the filopodial initiation. The value of our model is that it predicts the rate of filopodial precursors initiation and the local capping rate posing quantitative constraints for future models.
Model predictions
The model generates the following testable predictions:
There is an “optimal” filament number at which the maximal filopodial length is achieved.
Decreasing membrane stiffness would lead to increasing of the filopodial lengths for actin bundles with small filament numbers (the length of which is limited by the membrane resistance), whereas the length of thicker filament bundles (the length of which is limited by the G-actin diffusion) would not change.
Faster lamellipodial protrusion correlates with shorter average filopodial lengths.
Faster lamellipodial protrusion correlates with greater average distance between adjacent filopodia.
Initial growth rate of thin (thick) filopodial bundles is an increasing (decreasing) function of the filament number and correlates positively (negatively) with the final filopodial length.
From the physical point of view, it is tempting to compare the filopodial and lamellipodial protrusions. In terms of G-actin “consumption”, the lamellipodial filaments (hundreds per micron of the leading edge (49)) deplete G-actin pool equally with the filopodial bundle (tens of filaments per one-tenth of micron of the leading edge). On smooth surfaces, lamellipodial organization of actin filaments is optimal for the elastic polymerization ratchet mechanism of protrusion force generation, because in the lamellipodium the filaments are cross-linked neither too heavily, nor too lightly, so they do not buckle, yet are flexible enough (46). However, filopodial protrusions would be more efficient for crawling through extracellular matrix and on surfaces of other cells. Another possible role of relatively rigid actin bundles embedded into the lamellipodial actin sheet is to strengthen the lamellipodium against buckling, by analogy with engineered macroscopic structures (53). Future modeling efforts can help to elucidate other filopodial important functions, such as being guides for microtubules (54).
Acknowledgments
We are grateful to G. G. Borisy, T. Schaus, R. Cheney, T. Switkina, and K. Tosney for fruitful discussions, and to R. D. Mullins, G. G. Borisy, and S. Sun for sharing unpublished data.
The work is supported by National Institutes of Health GLUE grant “Cell Migration Consortium” (NIGMS U54 GM64346) and National Science Foundation grant DMS-0315782.
APPENDIX I: ANALYSIS OF THE G-ACTIN DIFFUSION AND THE GROWTH OF THE FILOPODIUM
The factor η [μM−1μm−1] converts μM concentration units into the number of molecules per unit length of the filopodium, given that the filopodial radius is ∼0.1 μm. As noted above, most of the volume inside the filopodium is free for the monomers to diffuse in. A concentration of 1 μM corresponds to 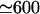 molecules per μm3, and this figure corresponds to π × (0.1 μm)2 × 600/μm
molecules per μm3, and this figure corresponds to π × (0.1 μm)2 × 600/μm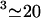 molecules per 1 μm of the filopodium. Thus,
molecules per 1 μm of the filopodium. Thus, 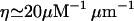 .
.
The following scales are characteristic for the filopodial protrusion: a0 ≈ 10 μM for the G-actin concentration, 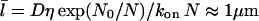 for the lamellipodial length, and
for the lamellipodial length, and 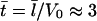 s for time. Rescaling Eqs. 3 and 4, we obtain the nondimensionalized equations for variables
s for time. Rescaling Eqs. 3 and 4, we obtain the nondimensionalized equations for variables 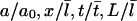 (we keep the same notations for the rescaled variables):
(we keep the same notations for the rescaled variables):
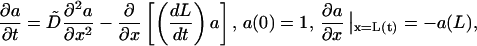 |
(12) |
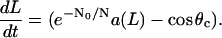 |
(13) |
Here 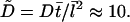 On the relevant scale, the G-actin diffusion is much faster than the cytoplasmic drift and filopodial growth, and the left-hand side and the second term on the right-hand side in Eq. 12 can be neglected: over seconds, the diffusion establishes a quasistationary gradient of the G-actin concentration, which slowly follows changes of the filopodial length over tens of seconds. So,
On the relevant scale, the G-actin diffusion is much faster than the cytoplasmic drift and filopodial growth, and the left-hand side and the second term on the right-hand side in Eq. 12 can be neglected: over seconds, the diffusion establishes a quasistationary gradient of the G-actin concentration, which slowly follows changes of the filopodial length over tens of seconds. So, 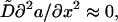 and using the boundary conditions, we obtain: a(x, t) ≈ 1 − α(L(t))x, where α ≈ 1/(1 + L(t)). Corresponding dimensional formula is Eq. 5. Substituting these expressions into Eq. 13 gives:
and using the boundary conditions, we obtain: a(x, t) ≈ 1 − α(L(t))x, where α ≈ 1/(1 + L(t)). Corresponding dimensional formula is Eq. 5. Substituting these expressions into Eq. 13 gives:
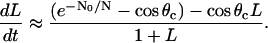 |
Integrating this first order ordinary differential equation (L(0) = 0), we find the solution implicitly:
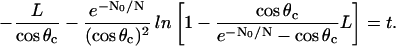 |
This formula can be used to plot the solution numerically (Fig. 4 B) and to find the asymptotic behavior of the filopodial length:
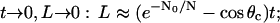 |
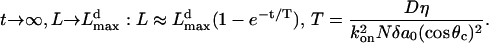 |
APPENDIX II: G-ACTIN GRADIENT IN THE FILOPODIUM “SEAMLESSLY” MATCHES THAT IN THE LAMELLIPODIUM
The boundary condition a(0) = a0 for the G-actin concentration at the base of the filopodium (Fig. 4 A), where a0 is the concentration at the lamellipodial leading edge, is nontrivial, because the “consumption” of the G-actin at the filopodial tip and corresponding diffusive flux can, in principle, locally deplete the G-actin concentration at the leading edge. To examine this boundary condition, we used FEMLAB to solve the following 2D (the lamellipodium is flat) diffusion problem. We considered the 0.1-μm-wide and 1-μm-long filopodium and 1-μm-wide and 0.8-μm-long adjacent part of the lamellipodium (Fig. 4 A). We solved the G-actin diffusion equation on this combined domain using the parameters described above and the following boundary conditions: i), the G-actin concentration at the “back” of the lamellipodial part of the domain is 1.2 (in the units of a0); ii), the G-actin concentration at the “front” of the lamellipodial part of the domain is 1; iii), the G-actin flux at the tip of the filopodial part of the domain is given by Eq. 12; iv), the G-actin flux at the sides of both lamellipodial and filopodial parts of the domain is zero. Conditions i and ii are chosen so that the G-actin flux at the lamellipodial leading edge matches the “consumption” of the G-actin at the edge at characteristic protrusion rate and F-actin density at the edge (34). The stationary solution of this diffusion problem illustrated with shading in Fig. 4 A shows that the G-actin gradients at the lamellipodium and filopodium “seamlessly” match each other, and that the G-actin concentration at the base of the filopodium is, indeed, a0. This is the consequence of the fact that the characteristic number of the filopodial filaments per filopodial size, ∼20/0.1 μm, is the same as the characteristic number of the lamellipodial filaments per leading edge length, ∼200/1 μm (49), so the filopodium “consumes” proportional share of G-actin and does not deplete the lamellipodial G-actin pool.
APPENDIX III: DYNAMIC MODEL OF FASCIN DISTRIBUTION IN THE FILOPODIUM
The bundling protein fascin is turned on and off by regulated phosphorylation (32). It is likely that this regulation takes place at the filopodial tip complex (G. G. Borisy, Northwestern University, personal communication). This suggests the following model that explains the observed increased fascin presence near the tips of actin bundles. Let us consider the (L = 2 μm)-long filopodium, place the x axis directed backward with the origin at the tip, and consider the linear densities of fascin bound to the actin filaments, fb(x, t), diffusing “inactive” fascin dissociated from the actin filaments, fi(x, t), and diffusing “active” fascin dissociated from the actin filaments, fa(x, t). The dynamics of fascin is described by the following system of equations:
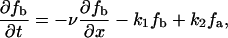 |
(14) |
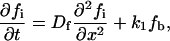 |
(15) |
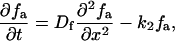 |
(16) |
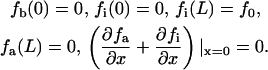 |
(17) |
The first term in the right-hand side of Eq. 14 is responsible for the kinematic drift of the bound fascin with the protrusion rate v, due to treadmilling together with F-actin, away from the filopodial tip. The first terms in the right-hand side of Eqs. 15 and 16 describes the fascin diffusion. We use the value of the diffusion coefficient Df = 2 μm2/s, scaled proportionally to length from the known value of the G-actin diffusivity. The second terms in the right-hand side of Eqs. 14 and 15 describe the inactivation and dissociation of fascin from F-actin with the rate k1. The second term in the right-hand side of Eq. 16 and the third term in the right-hand side of Eq. 14 are responsible for association of activated fascin with F-actin with the rate k2. We use the values k1 = k2 = 1/s, which are characteristic for the kinetics of actin-binding proteins (33). Equation 17 gives the boundary conditions: due to the drift, there is no bound fascin at the tip, fb(0) = 0. All inactivated fascin is activated at the tip, so fi(0) = 0, and the fluxes of the inactivated and activated fascin balance (last formula in Eq. 17). We assume that at the base of the filopodium there is no activated fascin (like in the lamellipodium), and the concentration of the inactivated fascin is equal to that in the lamellipodium, f0.
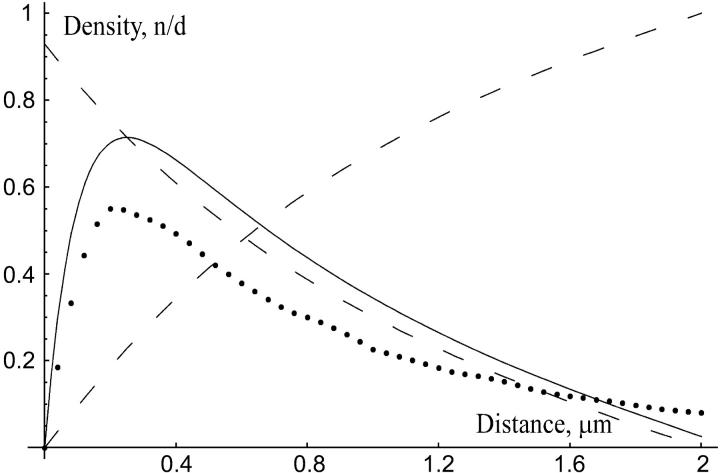
We used FEMLAB to solve Eqs. 14–17. The solutions are plotted in Fig. 7. The model predicts that the bundling fascin concentration is maximal at approximately 300 nm from the filopodial tip, much closer to the tip, than to the base of the filopodium. Near the base, the cross-linking density decreases. This is unlikely to affect the buckling force. Finally, we repeated the simulations for the Λ-precursor (allowing the diffusion to be two-dimensional). The corresponding bound fascin density is shown with the dotted curve in Fig. 7. Again, the fascin density is maximal near the bundle's tip.
FIGURE 7.
Nondimensionalized linear densities of fascin along the filopodial actin bundle. The horizontal axis shows distance in microns; the origin corresponds to the filopodial tip. (Solid line) Fascin associated with F-actin; (dashed line) “activated” (decreasing) and “inactivated” (increasing) fascin; (dotted line) fascin associated with F-actin along the Λ-precursor's bundle.
References
- 1.Abercrombie, M. 1980. The crawling movement of metazoan cells. Proc. R. Soc. Lond. Biol. Sci. 207:129–147. [Google Scholar]
- 2.Pollard, T. D., and G. G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. [DOI] [PubMed] [Google Scholar]
- 3.Pollard, T. D., L. Blanchoin, and R. D. Mullins. 2000. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29:545–576. [DOI] [PubMed] [Google Scholar]
- 4.Maly, I. V., and G. G. Borisy. 2001. Self-organization of a propulsive actin network as an evolutionary process. Proc. Natl. Acad. Sci. USA. 98:11324–11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis, A. K., and P. C. Bridgman. 1992. Nerve growth cone lamellipodia contain two populations of actin filaments that differ in organization and polarity. J. Cell Biol. 119:1219–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Small, J. V., G. Isenberg, and J. E. Celis. 1978. Polarity of actin at the leading edge of cultured cells. Nature. 272:638–639. [DOI] [PubMed] [Google Scholar]
- 7.Mallavarapu, A., and T. Mitchison. 1999. Regulated actin cytoskeleton assembly at filopodium tips controls their extension and retraction. J. Cell Biol. 146:1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Small, J. V. 1994. Lamellipodia architecture: actin filament turnover and the lateral flow of actin filaments during motility. Semin. Cell Biol. 5:157–163. [DOI] [PubMed] [Google Scholar]
- 9.Borisy, G. G., and T. M. Svitkina. 2000. Actin machinery: pushing the envelope. Curr. Opin. Cell Biol. 12:104–112. [DOI] [PubMed] [Google Scholar]
- 10.Nobes, C. D., and A. Hall. 1995. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 81:53–62. [DOI] [PubMed] [Google Scholar]
- 11.Welch, M. D., and R. D. Mullins. 2002. Cellular control of actin nucleation. Annu. Rev. Cell Dev. Biol. 18:247–288. [DOI] [PubMed] [Google Scholar]
- 12.Svitkina, T. M., E. A. Bulanova, O. Y. Chaga, D. M. Vignjevic, S. Kojima, J. M. Vasiliev, and G. G. Borisy. 2003. Mechanism of filopodia initiation by reorganization of a dendritic network. J. Cell Biol. 160:409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steketee, M. B., and K. W. Tosney. 2002. Three functionally distinct adhesions in filopodia: shaft adhesions control lamellar extension. J. Neurosci. 22:8071–8083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang, H., J. S. Berg, Z. Li, Y. Wang, P. Lang, A. D. Sousa, A. Bhaskar, R. E. Cheney, and S. Stromblad. 2004. Myosin-X provides a motor-based link between integrins and the cytoskeleton. Nat. Cell Biol. 6:523–531. [DOI] [PubMed] [Google Scholar]
- 15.Bentley, D., and A. Toroian-Raymond. 1986. Disoriented pathfinding by pioneer neurone growth cones deprived of filopodia by cytochalasin treatment. Nature. 323:712–715. [DOI] [PubMed] [Google Scholar]
- 16.Lee, J., A. Ishiara, and K. Jacobson. 1993. The fish epidermal keratocyte as a model system for the study of cell locomotion. In Cell Behavior: Adhesion and Motility. G. Jones, C. Wigley, and R. Warn, editors. The Company of Biologists, Cambridge, UK. 73–89. [PubMed]
- 17.Soll, D. R., E. Voss, O. Johnson, and D. Wessels. 2000. Three-dimensional reconstruction and motion analysis of living, crawling cells. Scanning. 22:249–257. [DOI] [PubMed] [Google Scholar]
- 18.Heath, J. P., and L. D. Peachey. 1989. Morphology of fibroblasts in collagen gels: a study using 400 keV electron microscopy and computer graphics. Cell Motil. Cytoskeleton. 14:382–392. [DOI] [PubMed] [Google Scholar]
- 19.Steketee, M., K. Balazovich, and K. W. Tosney. 2001. Filopodial initiation and a novel filament-organizing center, the focal ring. Mol. Biol. Cell. 12:2378–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vignjevic, D., D. Yarar, M. D. Welch, J. Peloquin, T. M. Svitkina, and G. G. Borisy. 2003. Formation of filopodia-like bundles in vitro from a dendritic network. J. Cell Biol. 160:951–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bear, J. E., T. M. Svitkina, M. Krause, D. A. Schafer, J. J. Loureiro, G. A. Strasser, I. V. Maly, O. Y. Chaga, J. A. Cooper, G. G. Borisy, and F. B. Gertler. 2002. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 109:509–521. [DOI] [PubMed] [Google Scholar]
- 22.Samarin, S., S. Romero, C. Kocks, D. Didry, D. Pantaloni, and M. F. Carlier. 2003. How VASP enhances actin-based motility. J. Cell Biol. 163:131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Argiro, V., M. B. Bunge, and M. I. Johnson. 1985. A quantitative study of growth cone filopodial extension. J. Neurosci. Res. 13:149–162. [DOI] [PubMed] [Google Scholar]
- 24.Oldenbourg, R., K. Katoh, and G. Danuser. 2000. Mechanism of lateral movement of filopodia and radial actin bundles across neuronal growth Cones. Biophys. J. 78:1176–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheetz, M. P., D. B. Wayne, and A. L. Pearlman. 1992. Extension of filopodia by motor-dependent actin assembly. Cell Motil. Cytoskeleton. 22:160–169. [DOI] [PubMed] [Google Scholar]
- 26.Peskin, C., G. Odell, and G. Oster. 1993. Cellular motions and thermal fluctuations: the Brownian ratchet. Biophys. J. 65:316–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao, J. Y., and F. M. Hochmuth. 1996. Micropipette suction for measuring piconewton forces of adhesion and tether formation from neutrophil membranes. Biophys. J. 71:2892–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hochmuth, F. M., J. Y. Shao, J. Dai, and M. P. Sheetz. 1996. Deformation and flow of membrane into tethers extracted from neuronal growth cones. Biophys. J. 70:358–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kas, J., H. Strey, M. Barmann, and E. Sackmann. 1993. Direct measurement of the wave-vector-dependent bending stiffness of freely flickering actin filaments. Europhys. Lett. 21:865–870. [Google Scholar]
- 30.Isambert, H., P. Venier, A. Maggs, A. Fattoum, R. Kassab, D. Pantaloni, and M. F. Carlier. 1995. Flexibility of actin filaments derived from thermal fluctuations. Effect of bound nucleotide, phalloidin, and muscle regulatory proteins. J. Biol. Chem. 270:11437–11444. [DOI] [PubMed] [Google Scholar]
- 31.Landau, L., and E. Lifshitz. 1995. The Theory of Elasticity. Butterworth-Heinemann, Boston, MA.
- 32.Adams, J. C. 2004. Roles of fascin in cell adhesion and motility. Curr. Opin. Cell Biol. 16:590–596. [DOI] [PubMed] [Google Scholar]
- 33.Howard, J. 2001. Mechanics of Motor Proteins and the Cytoskeleton. Sinauer, Sunderland, MA.
- 34.Mogilner, A., and L. Edelstein-Keshet. 2002 2002. Regulation of actin dynamics in rapidly moving cells: a quantitative analysis. Biophys. J. 83:1237–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rubinstein, B., K. Jacobson, and A. Mogilner. 2005. Multiscale two-dimensional modeling of a motile simple-shaped cell. SIAM J. Appl. Math. 3:413–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mogilner, A., E. Marland, and D. Bottino. 2001. A minimal model of locomotion applied to the steady “gliding” movement of fish keratocyte cells. In Pattern Formation and Morphogenesis: Basic Processes. H. Othmer and P. Maini, editors. Springer, New York, NY. 269–294.
- 37.Mitchison, T. J., and L. P. Cramer. 1996. Actin-based cell motility and cell locomotion. Cell. 84:371–379. [DOI] [PubMed] [Google Scholar]
- 38.Tilney, L. G., and S. Inoue. 1982. Acrosomal reaction of Thyone sperm. II. The kinetics and possible mechanism of acrosomal process elongation. J. Cell Biol. 93:820–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oster, G., A. Perelson, and L. Tilney. 1982. A mechanical model for elongation of the acrosomal process in Thyone sperm. J. Math. Biol. 15:259–265. [Google Scholar]
- 40.Gustafson, T., and L. Wolpert. 1961. Studies on the cellular basis of morphogenesis in the sea urchin embryo: directed movements of primary mesenchyme cells in normal and vegetalized larvae. Exp. Cell Res. 24:64–79. [DOI] [PubMed] [Google Scholar]
- 41.Sheetz, M. P., and J. Dai. 1996. Modulation of membrane dynamics and cell motility by membrane tension. Trends Cell Biol. 6:85–89. [DOI] [PubMed] [Google Scholar]
- 42.Wang, F. S., J. S. Wolenski, R. E. Cheney, M. S. Mooseker, and D. G. Jay. 1996. Function of myosin-V in filopodial extension of neuronal growth cones. Science. 273:660–663. [DOI] [PubMed] [Google Scholar]
- 43.Tokuo, H., and M. Ikebe. 2004. Myosin X transports Mena/VASP to the tip of filopodia. Biochem. Biophys. Res. Commun. 319:214–220. [DOI] [PubMed] [Google Scholar]
- 44.Mogilner, A., and G. Oster. 1996. The physics of lamellipodial protrusion. Eur. Biophys. J. 25:47–53. [Google Scholar]
- 45.Landau, L., E. Lifshitz, and L. Pitaevskii. 1980. Statistical Physics. Pergamon Press, Oxford, UK.
- 46.Mogilner, A., and G. Oster. 2003. Polymer motors: pushing out the front and pulling up the back. Curr. Biol. 13:R721–R733. [DOI] [PubMed] [Google Scholar]
- 47.Laurent, V., T. P. Loisel, B. Harbeck, A. Wehman, L. Grobe, B. M. Jockusch, J. Wehland, F. B. Gertler, and M. F. Carlier. 1999. Role of proteins of the Ena/VASP family in actin-based motility of Listeria monocytogenes. J. Cell Biol. 144:1245–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dickinson, R. B., L. Caro, and D. L. Purich. 2004. Force generation by cytoskeletal filament end-tracking proteins. Biophys. J. 87:2838–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abraham, V. C., V. Krishnamurthi, D. L. Taylor, and F. Lanni. 1999. The actin-based nanomachine at the leading edge of migrating cells. Biophys. J. 77:1721–1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kovar, D. R., and T. D. Pollard. 2004. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc. Natl. Acad. Sci. USA. 101:14725–14730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Small, J. V., T. Stradal, E. Vignal, and K. Rottner. 2002. The lamellipodium: where motility begins. Trends Cell Biol. 12:112–120. [DOI] [PubMed] [Google Scholar]
- 52.Bettache, N., L. Baisamy, S. Baghdiguian, B. Payrastre, P. Mangeat, and A. Bienvenue. 2003. Mechanical constraint imposed on plasma membrane through transverse phospholipid imbalance induces reversible actin polymerization via phosphoinositide 3-kinase activation. J. Cell Sci. 116:2277–2284. [DOI] [PubMed] [Google Scholar]
- 53.Daniel, I. M., and O. Ishai. 1994. Engineering Mechanics of Composite Materials. Oxford University Press, Oxford, UK.
- 54.Schaefer, A. W., N. Kabir, and P. Forscher. 2002. Filopodia and actin arcs guide the assembly and transport of two populations of microtubules with unique dynamic parameters in neuronal growth cones. J. Cell Biol. 158:139–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McGrath, J. L., Y. Tardy, C. F. Dewey, J. J. Meister, and J. H. Hartwig. 1998. Simultaneous measurements of actin filament turnover, filament fraction, and monomer diffusion in endothelial cells. Biophys. J. 75:2070–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]