Abstract
We have developed a new method for immobilization of single proteins by utilizing streptavidin-biotin and protein L-antibody interactions on glass coverslips coated with polyethylene glycol. The method is particularly well suited for single-molecule fluorescence studies. A monomeric, detergent-solubilized bacterial transport protein, GlpT, and the dimeric cytoplasmic region of a mammalian transporter, cdAE1, were immobilized by our method with a high degree of specificity. The fluorescence from single molecules attached to the immobilized proteins was detected with a high signal/noise ratio. Single-pair fluorescence resonance energy transfer (spFRET) measurements on cdAE1 dimers indicate that the structure of the protein is not compromised and provide evidence that the cdAE1 protein can exist in at least two conformations under physiological conditions.
Although single-molecule fluorescence has rapidly developed into a powerful technique for observing dynamic behavior in biochemical systems (1), methodological challenges remain in achieving suitable immobilization of biological molecules on a surface without compromising their structure and function. Biotinylation at the 3′ or 5′ end has enabled immobilization of nucleic acids and proteins interacting with them on avidin-coated surfaces (2). For studying proteins alone, one method involves trapping labeled proteins within lipid vesicles and tethering them via biotin-avidin chemistry (3). This technique, however, presents difficulties in studying protein-protein interactions or changing buffer conditions. Proteins expressed with (His)6 tags have been immobilized on coverslips derivatized with Ni2+ (4). This method can potentially cause denaturation by bringing the protein close to the charged glass surface. Also, Ni2+ coating of glass surfaces is a complicated three-step process that takes over 24 h (4).
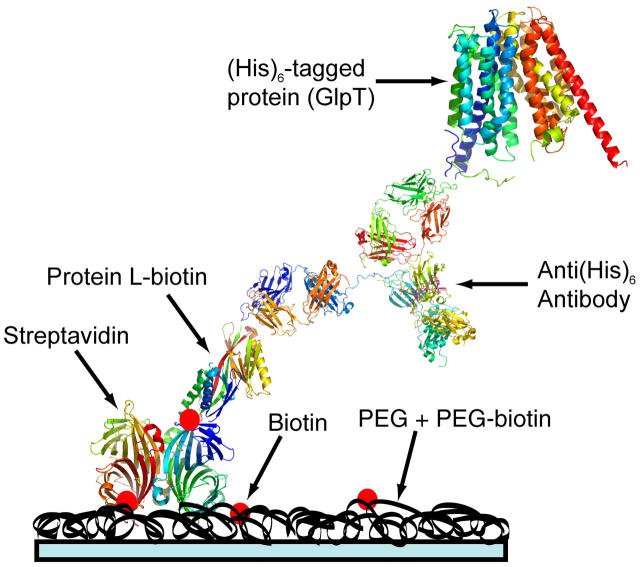
We have developed an alternative technique for protein immobilization on polyethylene glycol (PEG) derivatized glass coverslips, using a “brick-laying” approach. Clean glass coverslips were coated with a layer of PEG + 1% PEG-biotin followed by addition of streptavidin, protein L-biotin, and finally the protein of interest preincubated with an antibody (Fig. 1).
FIGURE 1.
Schematic representation of the immobilization scheme (see Supplemental Materials for protocols).
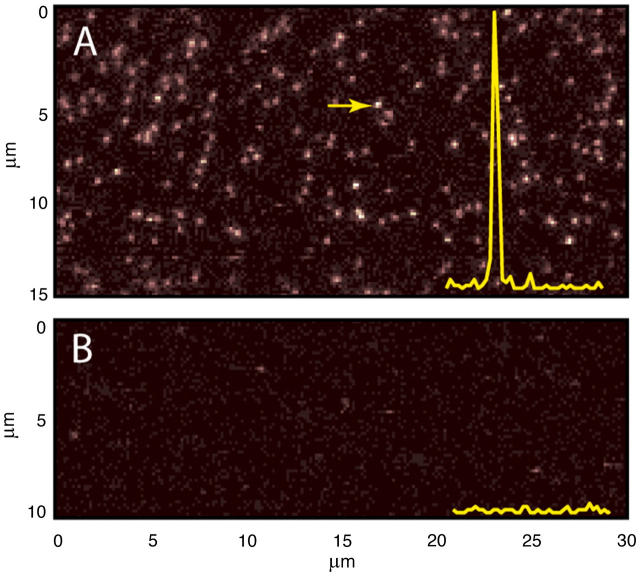
As a test of this method, we selected two proteins: i), a detergent-solubilized monomeric bacterial membrane transporter, GlpT (5), and ii), the cytoplasmic domain of a homodimeric human erythrocyte transporter, cdAE1 (6). Fig. 2 A shows a raster-scanned fluorescence image of detergent-solubilized GlpT (N-terminal (His)6-tagged) protein labeled by Cy3-maleimide and immobilized by the above method via an anti-(His)6 antibody. With a final protein concentration of 250 pM, we observe ∼200 single molecule spots in a 15×30-μm area. Each spot exhibits a high signal/noise ratio as indicated by the inset.
FIGURE 2.
(A) Raster-scanned fluorescence images of immobilized Cy3-GlpT (a 532-nm laser was used for excitation); inset shows fluorescence intensity at the scan region marked by the arrow. (B) Control experiment with no protein L-biotin.
When no protein L-biotin is added (Fig. 2 B) or when the protein is not preincubated with an appropriate antibody, we see less than five spots in the same area, indicating a very high degree of specificity for this method. A similar specificity was found for Cy3-labeled cdAE1 dimers containing a C-terminal (His)6 tag when incubated with the same antibody (not shown).
This functional C-terminal (His)6-tagged mutant of cdAE1, 201C, which contains a single cysteine at position 201, was also labeled with equimolar concentrations of Cy3- and Cy5.5-maleimide and immobilized by this method for single-pair fluorescence resonance energy transfer (spFRET) studies. By using a dual-channel detection system for simultaneous measurement of donor and acceptor emission (see protocols in Supplemental Materials), we are able to detect immobilized dimers that contain a single acceptor and single donor in a heterogeneous population that also includes donor-only and acceptor-only labeled proteins. An automated program then locates each donor-acceptor spot and records a time trace of simultaneous donor and acceptor emission with 1-ms time resolution.
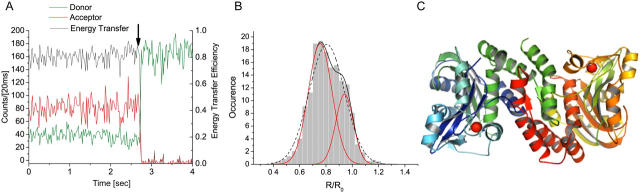
Fig. 3 A shows typical time-series data binned to 20 ms for a single dimer at pH 7.4. The donor (Cy3) fluorescence (green) is initially low and the acceptor (Cy5.5 (red)) is high due to energy transfer; hence the energy transfer efficiency is high, as shown on the same plot in black. After the acceptor Cy5.5 photobleaches (marked by an arrow), the donor fluorescence increases. For a given single dimer, the data from donor and acceptor channels are corrected for background and cross talk and are used up to the bleaching step for calculating the maximum likelihood estimate (7) of the parameter R/R0 for each 20-ms bin. The R/R0 distribution for each dimer is normalized to unity and then the distributions for the single dimers observed in the scanned area are summed to obtain a histogram as shown in Fig. 3 B.
FIGURE 3.
(A) Simultaneous time-series recording of donor and acceptor channels and corresponding energy transfer efficiency. (B) Distribution of R/R0. (C) Crystal structure of cdAE1 dimer showing Cys-201 locations as red dots (image created with PyMol).
The distribution of R/R0 values for 157 single dimers is plotted in Fig. 3 B. A fit to a single Gaussian function (dashed line) exhibits systematic deviations and has a χ2 value of 0.96 and r2 of 0.97. The fit to two Gaussians (solid lines) is much better, with χ2 = 0.27 and r2 = 0.99. The two peaks are centered at 0.756 ± 0.006 and 0.941 ± 0.007. Based on R0 = 47.3 Å calculated for Cy3/Cy5.5 attached to 201C, these correspond to intercysteine distances of 35.7 ± 0.3 and 44.5 ± 0.3 Å.
Ensemble measurements on 201C with lanthanide resonance energy transfer (LRET), using a Tb chelate as donor and fluorescein maleimide as acceptor, show a large increase in the intercysteine distance as pH is raised from 5 to 10 (8), consistent with other data indicating a pH-dependent conformational change (6). The LRET data for 201C fit to a two-state model, with an intercysteine distance for the low-pH form of 34.1 Å and for the high-pH form of 44.3 Å. The fact that the LRET distances are in good agreement with the distances for the two spFRET peaks in Fig. 3 B indicates that binding of cdAE1 to the antibody and immobilization have no apparent effect on its structural integrity, as measured by the intermonomeric Cys-201 distance.
The spFRET data suggest that both forms of cdAE1 exist at pH 7.4. Moreover, when spFRET is performed at pH 7.0, the fraction of cdAE1 in the more extended (high-pH) form decreases, as is expected in view of the metastable equilibrium between the two forms.
The cdAE1 protein is attached to the membrane of erythrocytes where it serves as a center for organization of cytosolic proteins (6), including ankyrin (which connects the membrane to the cytoskeleton), glycolytic enzymes, and hemoglobin. The binding of these cytosolic proteins depends on cdAE1 conformation and, conversely, cytosolic protein binding can also affect the conformation of cdAE1. The dynamic equilibrium between two different conformational states of the cdAE1 dimer under physiological conditions may facilitate interactions among these proteins to alter erythrocyte membrane properties.
In summary, we are able to immobilize both a detergent-solubilized membrane protein and a cytoplasmic protein with a high degree of specificity for single-molecule fluorescence detection and biochemical studies. This method provides a general approach for protein immobilization with minimal biological perturbation for single-molecule fluorescence studies, thus enabling future studies focused on transitions between different discrete conformations of the proteins, something that is not possible with ensemble measurements.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
The authors thank Dr. Da-Neng Wang, New York University, for the purified GlpT protein; Dr. Philip Low, Purdue University, for the 201C plasmid; and Brian Holmberg for technical assistance.
This work was funded by National Institutes of Health DK 27495 and National Science Foundation ECS-0210752. M.A.L. acknowledges partial support through a fellowship for prospective researchers by the Swiss National Foundation.
Prithwish Pal's present address is Dept. of Pharmacology, University of North Carolina, Chapel Hill, NC 27599.
M. Andreas Lieb's present address is National Center of Competence in Nanoscale Science, Institute of Physics, University Basel, 4056 Basel, Switzerland.
References
- 1.Weiss, S. 2000. Measuring conformational dynamics of biomolecules by single molecule fluorescence spectroscopy. Nat. Struct. Biol. 7:724–729. [DOI] [PubMed] [Google Scholar]
- 2.Joo, C., S. A. McKinney, D. M. J. Lilley, and T. Ha. 2004. Exploring rare conformational species and ionic effects in DNA Holliday junction using single-molecule spectroscopy. J. Mol. Biol. 341:739–751. [DOI] [PubMed] [Google Scholar]
- 3.Rhoades, E., E. Gussakovsky, and G. Haran. 2003. Watching proteins fold one molecule at a time. Proc. Natl. Acad. Sci. USA. 100:3197–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adachi, K., R. Yasuda, H. Noji, H. Itoh, Y. Harada, M. Yoshida, and K. Kinosita, Jr. 2000. Stepping rotation of F1-ATPase visualized through angle-resolved single-fluorophore imaging. Proc. Natl. Acad. Sci. USA. 97:7243–7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang, Y., M. J. Lemieux, J. Song, M. Auer, and D. N. Wang. 2003. Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science. 301:616–620. [DOI] [PubMed] [Google Scholar]
- 6.Low, P. S. 1986. Structure and function of the cytoplasmic domain of band 3: center of erythrocyte membrane-peripheral protein interactions. Biochim. Biophys. Acta. 864:145–167. [DOI] [PubMed] [Google Scholar]
- 7.Watkins, L. P., and H. Yang. 2004. Information bounds and optimal analysis of dynamic single molecule measurements. Biophys. J. 86:4015–4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pal, P., B. E. Holmberg, and P. A. Knauf. 2005. Conformational changes in the cytoplasmic domain of human anion exchanger 1(AE1) revealed by luminescence resonance energy transfer. Biochemistry. In press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.