Abstract
Some recently studied biological noncovalent bonds have shown increased lifetime when stretched by mechanical force. In each case these counterintuitive “catch-bonds” have transitioned into ordinary “slip-bonds” that become increasingly shorter lived as the tensile force on the bond is further increased. We describe analytically how these results are supported by a physical model whereby the ligand escapes the receptor binding site via two alternative routes, a catch-pathway that is opposed by the applied force and a slip-pathway that is promoted by force. The model predicts under what conditions and at what critical force the catch-to-slip transition would be observed, as well as the degree to which the bond lifetime is enhanced at the critical force. The model is applied to four experimentally studied systems taken from the literature, involving the binding of P- and L-selectins to sialyl LewisX oligosaccharide-containing ligands. Good quantitative fit to the experimental data is obtained, both for experiments with a constant force and for experiments where the force increases linearly with time.
INTRODUCTION
The concept of a catch-bond was first introduced by Dembo et al. (1) in a mathematical description of membrane-to-surface adhesion and detachment. In this work, a catch-bond was defined as a bond that increased in lifetime when the bond was stretched by mechanical force, whereas a slip-bond was defined as one that decreased in lifetime in these conditions. Up until this time it was generally assumed that mechanical force should shorten the lifetime of receptor-ligand pairs as for slip-bonds (2). And, for 16 years after Dembo's mathematical prediction, this appeared to be true, as none of the experimental efforts to establish the catch-bond phenomenon were successful. Recently, however, a mechanism for a real catch-bond was demonstrated when the lectin-like bacterial adhesion protein FimH was shown to undergo a force-induced conformational change that led to stronger binding of bacteria to host cells in flow chambers (3–5). Additional evidence for catch-bonds was offered by single-molecule lifetime studies with an atomic force microscope (AFM), which demonstrated that the blood adhesion proteins P- and L-selectin formed bonds with sialyl LewisX oligosaccharide ligands that last longer as force increases (5–7).
However, these experiments demonstrated that as force further increases beyond a critical value, the selectin catch-bonds transition into slip-bonds whose lifetime decreases with increasing mechanical force. This catch-to-slip transition was observed with the binding of both P-selectin (6) and L-selectin (7) with dimeric P-selectin glycoprotein ligand-1 (PSGL-1) purified from human neutrophils, and with monomeric recombinant PSGL-1 (sPSGL-1). A similar biphasic response to shear-stress-induced force was also observed in the FimH-mediated attachment of bacteria to host cells (3) and of beads to surfaces (8), suggesting that FimH also transitions from a catch-bond to a slip-bond at sufficiently high force. Linearly ramping the force with a constant loading rate results in a bimodal distribution of the rupture forces, with the low force peak dominating at a low loading rate but a switch to the high force peak dominating at higher loading rates or with an initial jump to a high force (9). Evans et al. (9) presented a five-parameter quantitative model to explain this data in which force alters the equilibrium between two rapidly equilibrating low-energy wells in the bound state. Except for the assumption of rapid equilibration, the Evans model is similar to that proposed for FimH, in which the effect of force is to switch the state of the bond, thus favoring a slower unbinding pathway (3) (W. Thomas, M. Forero, O. Yakovenko, L. Nilsson, P. Vicini, E. Sokurenko, and V. Vogel, unpublished data).
We present here an alternative four-parameter physical model where the ligand escapes the receptor binding site via two alternative routes, a catch-pathway that is opposed by the applied force and a slip-pathway that is promoted by force. This model was suggested conceptually by Sarangapani et al. (7) in discussing the L-selectin data. The quantitative model we present here shows good mathematical agreement with the published constant force, constant loading rate, and jump-ramp experiments on selectins (6,7,9). Further mathematical analysis of the present model for the jump-ramp scenario is presented elsewhere (10). It is impossible with the existing published data to distinguish between the two-pathway, two-energy well model presented by Evans et al. and the two-pathway, one-energy well model presented here because the two models give very similar mathematical behavior. However, the interpretation of how force affects the three-dimensional receptor-ligand bond structure is quite different for the two models.
Modeling a catch-bond
A physically intuitive description for the receptor-ligand complex under the influence of the detaching force follows by considering a potential energy profile (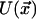 ) for the receptor-ligand interaction (11–15). This potential energy landscape is multidimensional. However, it can be energy weighted and projected upon the direction of an external force to obtain an effective one-dimensional energy profile as shown in Fig. 1. The energy landscape commonly used to study the force dependence of a slip-bond contains a potential energy minimum 1 at coordinate
) for the receptor-ligand interaction (11–15). This potential energy landscape is multidimensional. However, it can be energy weighted and projected upon the direction of an external force to obtain an effective one-dimensional energy profile as shown in Fig. 1. The energy landscape commonly used to study the force dependence of a slip-bond contains a potential energy minimum 1 at coordinate 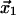 corresponding to the bound state of the ligand. The bound state is separated from the free state 0 by a potential energy barrier 2 at coordinate
corresponding to the bound state of the ligand. The bound state is separated from the free state 0 by a potential energy barrier 2 at coordinate 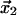 corresponding to the transition state. The barrier on the other side of the minimum is infinitely high. In the absence of the force, the rate of transition from the bound to the free state is determined by the level of thermal fluctuations of the ligand within the binding site relative to the height of the energy barrier that prevent escape of the ligand from the bound state. The rate constant of the ligand escape is proportional to the thermal probability of reaching the barrier top
corresponding to the transition state. The barrier on the other side of the minimum is infinitely high. In the absence of the force, the rate of transition from the bound to the free state is determined by the level of thermal fluctuations of the ligand within the binding site relative to the height of the energy barrier that prevent escape of the ligand from the bound state. The rate constant of the ligand escape is proportional to the thermal probability of reaching the barrier top
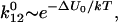 |
(1) |
with 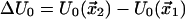 being the barrier height, T the absolute temperature, and k the Boltzmann constant.
being the barrier height, T the absolute temperature, and k the Boltzmann constant.
FIGURE 1.
Potential energy profile projected onto the direction of force for a simple slip-bond. A simple catch-bond would have the opposite energy profile with respect to the direction of force.
The effect of the external force applied to the bond on the rate constant of Eq. 1 can be included by changing the height of the energy barrier (2,9)
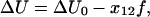 |
(2) |
where 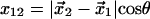 is the transition state distance projected onto the direction of applied force, with θ the angle between the directions of the force and the displacement of the ligand from
is the transition state distance projected onto the direction of applied force, with θ the angle between the directions of the force and the displacement of the ligand from 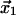 to
to 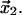 Equations 1 and 2 require several assumptions. First, the height of the potential barrier in the Kramers' Eq. 1 must be sufficiently big compared to temperature,
Equations 1 and 2 require several assumptions. First, the height of the potential barrier in the Kramers' Eq. 1 must be sufficiently big compared to temperature, 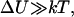 even after the addition of force. Second, the energy barriers must be sharp (
even after the addition of force. Second, the energy barriers must be sharp (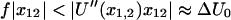 ) so that the location of the barriers and the preexponential coefficient in Eq. 1 do not depend on force. These assumptions are usually made for modeling the effect of force on bonds (9,11,16), and the log-linear relationship between loading rate and rupture force in single-molecule force spectroscopy experiments of both unbinding and unfolding of proteins(11,17,18) validates them. Thus,
) so that the location of the barriers and the preexponential coefficient in Eq. 1 do not depend on force. These assumptions are usually made for modeling the effect of force on bonds (9,11,16), and the log-linear relationship between loading rate and rupture force in single-molecule force spectroscopy experiments of both unbinding and unfolding of proteins(11,17,18) validates them. Thus,
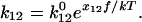 |
(3) |
The ligand-receptor pair forms a slip-bond when the tensile force is applied to the ligand in the direction from the bound state minimum to the transition state maximum so the projected transition state distance is positive (x12 > 0), as in Fig. 1 if force pulls to the right. In this case the force performs positive work on the ligand and promotes bond breaking and the ensuing ligand exit from the binding pocket via a single escape route.
If the geometry of the receptor-ligand pair is such that the projection of tensile force onto the direction from the bound to the transition state is negative (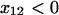 ), force pulls the bond away from the transition state. In this situation, the distance between the anchor points of the receptor-ligand complex is shorter in the transition state than in the bound state. The force applies negative work on the bond during any attempt to overcome the energy barrier, and increases the effective height of the barrier, ΔU. Thus, tensile force applied to the bond decreases rather than increases the unbinding rate according to Eq. 3, making the bond a catch-bond. This assumption that the motion from the bound state to the transition state in the catch-pathway has a negative projection onto the direction of force is analogous to the assumption Dembo et al. (1) make in the Appendix to demonstrate a catch-bond.
), force pulls the bond away from the transition state. In this situation, the distance between the anchor points of the receptor-ligand complex is shorter in the transition state than in the bound state. The force applies negative work on the bond during any attempt to overcome the energy barrier, and increases the effective height of the barrier, ΔU. Thus, tensile force applied to the bond decreases rather than increases the unbinding rate according to Eq. 3, making the bond a catch-bond. This assumption that the motion from the bound state to the transition state in the catch-pathway has a negative projection onto the direction of force is analogous to the assumption Dembo et al. (1) make in the Appendix to demonstrate a catch-bond.
As noted before, in experimental demonstrations of catch-bonds, the catch-bonds invariably transformed into slip-bonds as the force increased over a critical value(6,7,9). It was recently suggested by Sarangapani et al. (7) that this transition could result from a two-pathway model, where the ligand can dissociate from the binding site via two alternative routes. To describe the catch-slip transition within a single model, the potential energy profile of Fig. 1 has to be modified to show two finite energy barriers, in opposite directions from the bound state when projected onto the direction of force. Transformation from a catch-bond at moderate force to a slip-bond at large force can be understood with the potential shown in Fig. 2, where the ligand escapes the bound state via two pathways. A single minimum corresponding to the bound state and located at 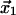 is connected to the free state 0 via two barriers located at
is connected to the free state 0 via two barriers located at 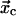 and
and  respectively. The indices “c” and “s” refer to catch and slip barriers that are defined by their orientation with respect to the applied force. The applied force pulls to the right in Fig. 2, preventing the ligand from escaping through the left barrier (
respectively. The indices “c” and “s” refer to catch and slip barriers that are defined by their orientation with respect to the applied force. The applied force pulls to the right in Fig. 2, preventing the ligand from escaping through the left barrier (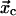 ) and facilitating ligand escape though the right barrier (
) and facilitating ligand escape though the right barrier (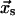 ).
).
FIGURE 2.
Potential energy profile describing the catch-slip transition. The two planes are used to represent a section of the ligand binding pocket that is bent around the catch barrier maximum. For simplicity, the force f (dotted line) is drawn to form the same angle with the two planes. In order for an efficient catch-slip transition to occur from the bound state x1, the catch-barrier “c” opposite to the applied force must be lower than the slip-barrier “s,” and preferably farther from the bound state when projected onto the direction of force as quantified in Eq. 8. Thus, the left barrier grows with force and is responsible for the catch-bond behavior, whereas the right barrier decreases with force, contributing the slip-bond behavior. The effect of force on the energy landscape is illustrated by the dashed line.
Here, we investigate the mean lifetime of the ligand moving in the potential of Fig. 2 under the influence of a constant applied force, and derive the analytic conditions that must be satisfied by the model to show the catch-slip transition The bond survival probability P(t) is the probability that a ligand bound to the receptor at time t = 0 is still bound at a later time t > 0. The probability is one at time 0, (P(0) = 1), and decreases with time according to
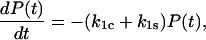 |
(4) |
where 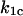 and
and 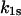 are the rate constants for unbinding through the catch and slip barriers with coordinates
are the rate constants for unbinding through the catch and slip barriers with coordinates 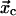 and
and 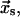 respectively. These rate constants depend exponentially on the applied force according to Eq. 3:
respectively. These rate constants depend exponentially on the applied force according to Eq. 3: 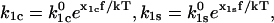 where
where 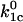 and
and 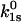 are the rate constants in the absence of force, and as illustrated in Fig. 2,
are the rate constants in the absence of force, and as illustrated in Fig. 2,  and
and 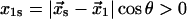 so that they will have opposite responses to force. Eq. 4 omits the terms describing the return of the ligand from the free to the bound state through the catch- and slip-pathways. These terms are small and either do not significantly change (right angle) or even exponentially decrease with force for the acute angle assumed by the bent shape of the interaction potential shown in Fig. 2. A long separation of the free state from the catch and slip barriers stipulated by the softness of the proteins, tethers, and/or cantilevers is sufficient for neglecting the return of the ligand to the bound state, because the long separation requires a large work to overcome even a weak force. The solution of Eq. 4 shows a simple exponential decay in bond survival probability:
so that they will have opposite responses to force. Eq. 4 omits the terms describing the return of the ligand from the free to the bound state through the catch- and slip-pathways. These terms are small and either do not significantly change (right angle) or even exponentially decrease with force for the acute angle assumed by the bent shape of the interaction potential shown in Fig. 2. A long separation of the free state from the catch and slip barriers stipulated by the softness of the proteins, tethers, and/or cantilevers is sufficient for neglecting the return of the ligand to the bound state, because the long separation requires a large work to overcome even a weak force. The solution of Eq. 4 shows a simple exponential decay in bond survival probability:
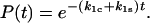 |
(5) |
Indeed, this single exponential decay of bond survival over time is one of the characteristics of the two-pathway catch-bond, which it has in common with the slip-bonds described by Evans and Bell, and with the model of a catch-bond arising from two rapidly equilibrating low-energy wells as suggested by Evans et al. (9). In contrast, the requirement for single exponential decay distinguishes this model from the two-state catch-bond model suggested by Thomas et al. (10), which has two distinct off-rates that permit a double exponential decay in bond survival at a single force.
It follows from this that the inverse mean lifetime of the bound state at a constant force is given by the sum of the two rate constants and can be written in a more explicit form as
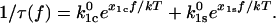 |
(6) |
This equation for the inverse mean lifetime of the population (Eq. 6) also follows from a more general definition of the mean lifetime 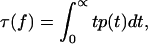 where
where
 |
(7) |
is the probability density function. Equation 6 contains four parameters 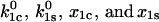 that can be estimated from the experimental data. However, first we consider how these parameter values affect the model behavior.
that can be estimated from the experimental data. However, first we consider how these parameter values affect the model behavior.
Provided that the parameters satisfy the relationship
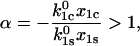 |
(8) |
the inverse lifetime 1/τ(f) reaches a minimum at the critical force, fcr:
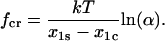 |
(9) |
(Note that 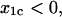 in order for this unbinding pathway to be a catch-bond barrier, so that α is always positive given an energy barrier such as that in Fig. 2.) This value of fcr is determined from the relationship
in order for this unbinding pathway to be a catch-bond barrier, so that α is always positive given an energy barrier such as that in Fig. 2.) This value of fcr is determined from the relationship 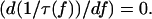 Equation 8 specifies the condition that must be satisfied for a potential energy profile with a single minimum and two maxima as in Fig. 2, to produce a catch-bond that transitions into a slip-bond with increasing force. If, instead, Eq. 8 is not satisfied and α ≤ 1 the two-pathway potential of Fig. 2 describes a slip-bond even if one of the pathways is a catch-pathway.
Equation 8 specifies the condition that must be satisfied for a potential energy profile with a single minimum and two maxima as in Fig. 2, to produce a catch-bond that transitions into a slip-bond with increasing force. If, instead, Eq. 8 is not satisfied and α ≤ 1 the two-pathway potential of Fig. 2 describes a slip-bond even if one of the pathways is a catch-pathway.
The unique nature of catch-bonds is their ability to have a longer lifetime at higher force than at low force, and we will call a quantitative measurement of this ability the “efficiency”. The efficiency of the catch-bond is best characterized by the dimensionless ratio of the bond lifetime at the critical force 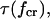 where the lifetime is maximal, to the lifetime in the absence of the force τ(0). It follows from Eqs. 6 and 8 that this ratio equals
where the lifetime is maximal, to the lifetime in the absence of the force τ(0). It follows from Eqs. 6 and 8 that this ratio equals
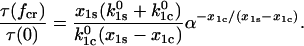 |
(10) |
The parameter conditions under which a catch-bond is most efficient can be understood from the following analysis of Eq. 10. As the α parameter of Eq. 8 decreases and approaches one (α → 1 +), the critical force maximizing the bond lifetime becomes smaller approaching zero (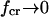 ). Then
). Then 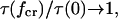 and the catch-bond disappears, replaced by a slip-bond. Biologically, we expect the velocity of cells rolling on catch-bonds as a function of shear stress to be affected by the efficiency. High efficiency may be necessary to cause a shear threshold effect by preventing adhesion at low shear.
and the catch-bond disappears, replaced by a slip-bond. Biologically, we expect the velocity of cells rolling on catch-bonds as a function of shear stress to be affected by the efficiency. High efficiency may be necessary to cause a shear threshold effect by preventing adhesion at low shear.
To understand the relative importance of the barrier heights and the projections of the transition state distances, three conditions can be considered. First, when the catch barrier is significantly farther from the bound state in the direction of force than is the slip barrier ( ), then
), then
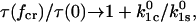 |
(11) |
as long as Eq. 8 holds. In this case, the efficiency is determined by the ratio of the two unstressed off-rates, and can become very high if the unstressed rate constant for the catch barrier is higher than that of the slip barrier as sketched in Fig. 2 (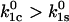 ). In the opposite situation
). In the opposite situation 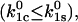 a catch-bond can still be observed but the maximum efficiency is two. Thus, even when inequality 10 holds, an efficient catch-bond (
a catch-bond can still be observed but the maximum efficiency is two. Thus, even when inequality 10 holds, an efficient catch-bond (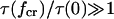 ) requires that the rate constant for escape through the catch-barrier is significantly larger than the rate constant for the escape through the slip-barrier:
) requires that the rate constant for escape through the catch-barrier is significantly larger than the rate constant for the escape through the slip-barrier:
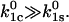 |
(12) |
In the second condition, if the two barriers are equidistant from the bound state in the direction of force (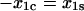 ), then Eq. 10 for the catch-bond efficiency simplifies to
), then Eq. 10 for the catch-bond efficiency simplifies to
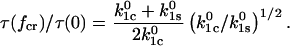 |
(13) |
Thus, this condition also requires that inequality (12) be true for a high efficiency, in which case
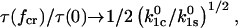 |
(14) |
and the efficiency scales more slowly with the ratio of unstressed unbinding rates than in the first condition.
In the third condition, the projection of the catch barrier along the direction of force is much closer to the bound state than is that for the slip barrier (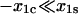 ), thus the catch-pathway is much less force sensitive. In this case, condition 12 must hold even to have
), thus the catch-pathway is much less force sensitive. In this case, condition 12 must hold even to have 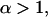 and even in that case,
and even in that case,
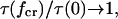 |
(15) |
and there is no catch-bond. Therefore, the catch-bond is particularly efficient only when the catch barrier is significantly lower than the slip barrier, as illustrated in Fig. 2. It is also important that, when projected onto the direction of force, the catch-barrier transition state is similar in distance or farther away from the bound state than the slip barrier transition state, as this condition modulates the effect of the relative barrier heights.
Fitting the model to published experimental data
The two-pathway model described and analyzed above can be applied directly to the published experimental data (6,7,9) on binding and force-induced detachment of P- and L-selectins with the PSGL-1 and sPSGL-1 ligands under constant force. We first address the data for L-selectin, because it appears to function as a monomer, binding to both the soluble monomeric sPSGL-1 ligand and the native dimeric PSGL-1 ligand in a similar fashion via single bonds between the receptor and the ligand. Thus, the derived Eq. 6 for the inverse of bond lifetime 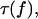 with four unknown parameters
with four unknown parameters 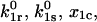 and
and 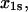 can be fit directly to both sets of data published as Fig. 4, a and b, of Sarangapani et al. (7). The unbinding rates for both L-selectin systems (7) are shown on the same plot in Fig. 3 as a function of force. Since three methods of calculating the unbinding rates were used in the original publication, all three are included in Fig. 3 for both ligands.
can be fit directly to both sets of data published as Fig. 4, a and b, of Sarangapani et al. (7). The unbinding rates for both L-selectin systems (7) are shown on the same plot in Fig. 3 as a function of force. Since three methods of calculating the unbinding rates were used in the original publication, all three are included in Fig. 3 for both ligands.
FIGURE 4.
The bond lifetime as a function of the applied force for bonds of dimeric P-selectin with monomeric sPSGL-1 (solid symbols) and dimeric PSGL-1 (open symbols). The lifetimes are shown as determined by Marshall et al. (2003) by the mean lifetimes (squares), inverse negative slopes for the off-rate (circles), and the standard deviation of the lifetime (triangles) The left panel shows the data as previously published (6). The thin solid line shows the theoretical fit to Eq. 6 using the parameter estimates reported for P-selectin in Table 1, and this shows good fit to the monomeric sPSGL-1 data. The thick solid line shows the predicted behavior of the dimeric PSGL-1 bond, assuming the relationship of Eqs. A7 and 18, as described in the Appendix, and demonstrates good fit to the data. The thick dotted line shows the predicted behavior of the dimeric bond by the alternative model that is described by Eq. A6 and demonstrates poor fit to the data. The right panel shows the experimental data for PSGL-1 scaled according to Eq. 18, because this was shown to fit the model better in the left panel. The solid line (right) shows the theoretical fit to Eq. 6 using the parameters estimated from all six data sets (right) using SAAM II software (19) as described in the text and reported in Table 1.
FIGURE 3.
The inverse lifetime as a function of the applied force for bonds of L-selectin with sPSGL-1 (open symbols) and PSGL-1 (solid symbols). The inverse lifetimes are shown as determined by Sarangapani et al. (7) by the reciprocal mean lifetimes (squares), negative slopes for the off-rate (circles), and the reciprocal standard deviation of the lifetime (triangles). The data form a single trend supporting the expectation that both systems form single catch-bonds because L-selectin functions as a monomer. The solid line shows the theoretical fit to Eq. 6 using the parameters estimated from all six data sets using SAAM II software (19) as described in the text and reported in Table 1.
The combined six sets of data shown in Fig. 3 are fit with Eq. 6 as follows. The SAAM II software (SAAM Institute, Seattle, WA) (19) was used to obtain maximum likelihood estimates of the model parameters (20). Weighted least squares (model-based, relative weighting scheme in SAAM II) were utilized; the measurement error in the lifetimes was assumed to be zero mean and have constant standard deviation. This method allows estimation not only of the optimal parameter values, but of the asymptotic standard errors in the parameters as well, a common measure of parameter precision (21). The resulting fit to Eq. 6 is shown as a solid line in Fig. 3, and the estimated parameters for this fit are displayed in Table 1.
TABLE 1.
Parameters of the two-pathway model needed to fit published experimental data
| Catch
|
Slip
|
Transition point
|
||||
|---|---|---|---|---|---|---|
| Selectin | 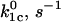 |
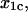 Å Å |
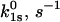 |
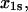 Å Å |
Efficiency | fcr |
| L-selectin (cf) | 34.9 ± 6.3 | −2.8 ± 0.6 | 7.2 ± 1.4 | 0.34 ± 0.09 | 3.5 ± 0.6 | 48.5 ± 4.3 |
| P-selectin (cf) | 120 ± 55 | −21.7 ± 2.4 | 0.25 ± 0.05 | 5.1 ± 0.5 | 91 ± 42 | 11.8 ± 0.2 |
| P-selectin (vf) | 20 ± 10 | −3.8 ± 1.1 | 0.34 ± 0.09 | 2.1 ± 0.1 | – | – |
Lines 1 and 2 give the parameters for fitting the constant force (cf) data for L-selectin (7) and P-selectin (6) to the combined PSGL-1 and sPSGL-1 ligands as described in the text. The fourth line gives the parameters needed to fit the variable force (vf) experiments of P-selectin binding to sPSGL-1 (9).
Fitting Eq. 6 to P-selectin is more complicated, because both the native PSGL-1 ligand and P-selectin are dimeric, and so bind to each other through two identical bonds in Fig. 3 b of Marshall et al. (6). Each of these bonds is equivalent to the bond between PSGL-1 and the monomeric sPSGL-1 monomer, with force dependence reported in Fig. 3 a of Marshall et al. (6). Thus, the dimeric PSGL-1 and monomeric sPSGL-1 experiments with P-selectin should fit with a single model and parameter set. However, this requires a model of how force affects the lifetime of two identical bonds under force as compared to one bond.
In the AFM experiments on P-selectin, the AFM tip was held at a constant distance from the surface instead of being moved away at a constant speed as in most previous studies on single-molecule mechanics (6). Then, the deflection of the cantilever on the tip was used to detect the force on the bond, f, which was assumed to remain constant except for thermal fluctuations (6). In the case of a dimeric bond, this force would be distributed over two identical bonds, so that the force on each monomeric bond would be 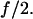 If the spring constant of the remaining monomer is half that of the original dimeric bond, the two bonds act independently and if the cantilever is much stiffer than the dimer, then when the first monomeric bond of the dimer breaks, the force on the remaining monomer should remain at
If the spring constant of the remaining monomer is half that of the original dimeric bond, the two bonds act independently and if the cantilever is much stiffer than the dimer, then when the first monomeric bond of the dimer breaks, the force on the remaining monomer should remain at 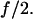 This assumption is a close approximation of the actual situation in these experiments, which applies when the two monomeric bonds are mechanically decoupled, and
This assumption is a close approximation of the actual situation in these experiments, which applies when the two monomeric bonds are mechanically decoupled, and 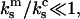 where
where 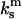 and
and 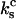 are the spring constants of the monomer bond and the cantilever, respectively. This is discussed in the Appendix of this article, which addresses the various elastic contributions to the system. Using these assumptions, it is appropriate to model the dimeric bond by asking when the second bond breaks.
are the spring constants of the monomer bond and the cantilever, respectively. This is discussed in the Appendix of this article, which addresses the various elastic contributions to the system. Using these assumptions, it is appropriate to model the dimeric bond by asking when the second bond breaks.
The probability 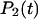 that a system with two equivalent bonds exists in the bound state at time t is related to the corresponding probability
that a system with two equivalent bonds exists in the bound state at time t is related to the corresponding probability  (Eq. 5) for a single-bond system by
(Eq. 5) for a single-bond system by
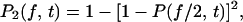 |
(16) |
according to the following arguments. The two-bond system does not dissociate until both individual bonds are broken. The probability that a single bond has already broken at time t is 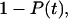 and because the two bonds can be considered independent, the probability that both bonds have already broken is
and because the two bonds can be considered independent, the probability that both bonds have already broken is 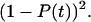 Thus, the probability that one or more bonds are still intact is
Thus, the probability that one or more bonds are still intact is 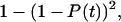 resulting in Eq. 16. Again, this analysis is for the approximation that the force remains at
resulting in Eq. 16. Again, this analysis is for the approximation that the force remains at 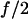 and thus that the two bonds act independently.
and thus that the two bonds act independently.
The lifetime 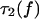 of the two-bond system is determined from the general definition
of the two-bond system is determined from the general definition
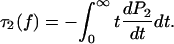 |
(17) |
Using Eq. 16, integration by parts relates 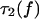 to the lifetime of the monomeric bond at half force
to the lifetime of the monomeric bond at half force 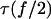
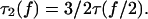 |
(18) |
Thus, the mean lifetime  of the double bond is maximized with a force that is twice as large as the force maximizing the monomeric bond lifetime
of the double bond is maximized with a force that is twice as large as the force maximizing the monomeric bond lifetime 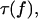 and the maximum achievable lifetime of the dimeric system is two-thirds larger than that of the monomeric system. Equation 18 is used below for the analysis of the P-selectin PSGL-1 dimeric bond and P-selectin sPSGL-1 monomeric bond data (6).
and the maximum achievable lifetime of the dimeric system is two-thirds larger than that of the monomeric system. Equation 18 is used below for the analysis of the P-selectin PSGL-1 dimeric bond and P-selectin sPSGL-1 monomeric bond data (6).
The derived relationship in Eq. 18 relating the lifetimes versus forces for the monomeric and dimeric bonds shows excellent agreement with the experimental data (6) in Fig. 4 for the two-bond P-selectin-PSGL-1 system and the one-bond P-selectin-sPSGL-1 system. The analysis of the experimental data is carried out using Eqs. 6 and 18 in the following way. The original data reported in Marshall et al. (6) for the 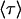 regime is plotted in Fig. 4 (left) for sPSGL-1 using open symbols and for PSGL-1 using solid symbols. The data for the dimeric P-selectin-PSGL-1 system are rescaled according to Eq. 18 to correspond to a single bond. These rescaled PSGL-1 data are shown in Fig. 4 (right) together with the original single-bond sPSGL-1 data and the close overlap of the data suggests the validity of the assumptions used to derive Eq. 18. The monomeric lifetime calculations and the rescaled dimeric lifetime calculations are used to estimate the parameters of Eq. 6 in the same manner as described above for L-selectin. The obtained values of these parameters are shown in the second line of Table 1. The fit is reported in Fig. 4 (left) by a continuous line.
regime is plotted in Fig. 4 (left) for sPSGL-1 using open symbols and for PSGL-1 using solid symbols. The data for the dimeric P-selectin-PSGL-1 system are rescaled according to Eq. 18 to correspond to a single bond. These rescaled PSGL-1 data are shown in Fig. 4 (right) together with the original single-bond sPSGL-1 data and the close overlap of the data suggests the validity of the assumptions used to derive Eq. 18. The monomeric lifetime calculations and the rescaled dimeric lifetime calculations are used to estimate the parameters of Eq. 6 in the same manner as described above for L-selectin. The obtained values of these parameters are shown in the second line of Table 1. The fit is reported in Fig. 4 (left) by a continuous line.
Thus, the two-pathway model presented here describes the published constant force data well, even explaining the relationship between the data for PSGL-1 and sPSGL-1. This model can also explain the experiments where the force changes with time 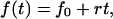 starting from an initial jump value
starting from an initial jump value 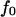 and increasing linearly in time with ramp rate r (9).
and increasing linearly in time with ramp rate r (9).
The open squares in Fig. 5 show the experimental data published as Fig. 2 in Evans et al. (9) for the binding of P-selectin to the monomeric sPSGL-1 in biomembrane force probe experiments. In the constant loading rate experiments, the force was increased linearly (Fig. 5, A–C), whereas the force was jumped to a force of 28–35 pN and then increased linearly in the jump-ramp experiments (Fig. 5, D–F) (9). The probability density 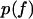 for remaining in the bound state in these jump/ramp experiments can be obtained by solving Eq. 4 with the time-dependent coefficients
for remaining in the bound state in these jump/ramp experiments can be obtained by solving Eq. 4 with the time-dependent coefficients 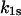 and
and 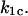
FIGURE 5.
The distribution of rupture forces in variable force experiments for binding of P-selectin to sPSGL-1. The squares show the “jump-ramp” experimental data from Evans et al. (9) where force was jumped to the indicated value and then ramped up at the indicated loading rate. The best fits are included for comparison for two models. The solid line shows the model fit for the two-pathway model described in this article with the parameters in the third line of Table 1. The heavy dotted line shows the fit of two-well two-pathway model used to describe the data by Evans et al. (9) (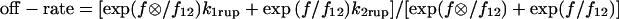 ) using the parameters k1rup = 10, k2rup = 0.36, fb = 19.6, f12 = 9.0,
) using the parameters k1rup = 10, k2rup = 0.36, fb = 19.6, f12 = 9.0,  = 14.5.
= 14.5.
The solid lines in Fig. 5 show the predicted behavior of the two-pathway model we present here with the parameters in line 3 of Table 1. These parameters were determined using the SAAM II software to fit the model as described before, except that the force was linearly dependent on time, and the absolute standard deviations shown in the original figure were used in the fitting. The model fits the data well, showing that the two-pathway model predicts that a jump to ∼30 pN would switch the bond from the low-impedance to the high-impedance pathway.
The maximum of the probability density 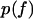 as a function the applied force occurs at large forces in the slip-bond regime, where the contribution of the catch-pathway becomes negligible, as clearly seeing in Fig. 5, B, C, E, and F, with the ramp rates
as a function the applied force occurs at large forces in the slip-bond regime, where the contribution of the catch-pathway becomes negligible, as clearly seeing in Fig. 5, B, C, E, and F, with the ramp rates 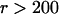 pN/s. In this case, the peak force value
pN/s. In this case, the peak force value 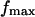 equals
equals
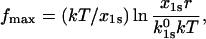 |
(19) |
and depends on the ramp rate r. Notably, the jump force does not affect the value of 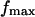 although it does affect the relative height of the high impedance pathway peak as less bonds escape through the low impedance pathway.
although it does affect the relative height of the high impedance pathway peak as less bonds escape through the low impedance pathway.
To compare the predictions of our model to that proposed by Evans et al. (9) to explain the data, the best fit for the two-well, two-pathway model was also included in Fig. 5 as a heavy dotted line. To properly compare the two models, and because the exact parameters were not identified in the original work on the two-well two-pathway model, these parameters were also determined by fitting the model data using SAAM II. However, we found that the five-parameter model overfits the data; a wide range of values of two parameters, “k1rup” and “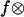 ” fit the model well, while fixing either of these allowed the other four parameters to be determined. We thus fixed k1rup, the fast unbinding rate, at 10 s−1, the value chosen by the authors, and estimated the remaining parameters as indicated in Fig. 5. It can be seen that the two models fit the data nearly identically, with the only significant difference seen at low force in panel C, where a different prefixed value of k1rup could make the two models overlap. This is probably because the mathematical behavior of the models is nearly identical; both show single exponential decay in bond survival at a constant force, and a bimodal response of lifetime as a function of force. The only difference between the two models is greater flexibility of the five-parameter model at low force, a regime that is difficult to measure. Thus, the published data are insufficient to distinguish mathematically between the four-parameter two-pathway model and the five-parameter two-state model.
” fit the model well, while fixing either of these allowed the other four parameters to be determined. We thus fixed k1rup, the fast unbinding rate, at 10 s−1, the value chosen by the authors, and estimated the remaining parameters as indicated in Fig. 5. It can be seen that the two models fit the data nearly identically, with the only significant difference seen at low force in panel C, where a different prefixed value of k1rup could make the two models overlap. This is probably because the mathematical behavior of the models is nearly identical; both show single exponential decay in bond survival at a constant force, and a bimodal response of lifetime as a function of force. The only difference between the two models is greater flexibility of the five-parameter model at low force, a regime that is difficult to measure. Thus, the published data are insufficient to distinguish mathematically between the four-parameter two-pathway model and the five-parameter two-state model.
With either model, slightly different parameters are needed to fit the constant force data of Marshall et al. (6) from those needed to fit the variable force data of Evans et al. (9); compare lines two and three of Table 1. This might be expected because the two sets of experiments were performed with completely different equipment and biological constructs. In addition, Eq. 3 for the bond dissociation rate is used both for constant and time-dependent forces. This equation is rigorously valid for the constant force case and may require further analysis and modification for the time-dependent force case (22).
It is instructive to compute the efficiencies of the catch-bonds in the P- and L-selectin systems, as defined by the ratio of the maximum bond lifetime to the bond lifetime in the absence of the force (Eq. 10). The catch-bond efficiencies computed using the parameters of Table 1 are 91 +/− 42 for P-selectin and 3.5 +/− 0.6 for L-selectin, indicating that both systems exhibit strong catch-bonds, with P-selectin producing over an order of magnitude better catch binding than L-selectin. Table 1 and Eq. 10 show that the improved efficiency of P- over L-selectin is primarily due to a much bigger ratio of unstressed rate constants for P-selectin than for L-selectin (∼500-fold difference versus approximately fivefold). For P-selectin, this efficiency is even greater than the sevenfold increase that was observed in the published AFM data, because the lifetime at low force could not be accurately measured in those experiments. In each system, the distance to the transition state in the direction of force is four to eight times farther in the catch-pathway than in the slip-pathway, as predicted for an efficient catch-bond. The distances to the transition state in the L-selectin fit are much shorter than the corresponding distances in P-selectin, causing the response to force to be much lower in L-selectin than in P-selectin.
DISCUSSION
The two-pathway model we present here has been conceptually suggested as an explanation for the biphasic behavior of L-selectin (7). It is distinct from another model proposed by Evans et al. (9) because it has only one minimum, with the catch-behavior arising from a backward unbinding pathway rather than a force-dependent equilibration between two states. Here, quantification of the two-pathway model shows that it can explain all the available P- and L-selectin data. It predicts the biphasic response in constant force experiments (Figs. 3 and 4). Even more important as it is not as intuitively obvious, this model explains the double peaks in the ramped force experiments (Fig. 5) and the effect of an initial jump in force to switch to the high-impedance pathway.
The mathematical model also allows estimation of the parameters and a comparison between different sets of data. The parameters for the slip-pathway are comparable to those already measured in earlier experiments that only detected the higher-force slip behavior. For L-selectin, 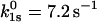 (compare to 3–8.6 s−1 and
(compare to 3–8.6 s−1 and 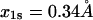 (compare to 0.16–4 Å) (17,23). For P-selectin,
(compare to 0.16–4 Å) (17,23). For P-selectin, 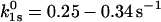 (compare to 0.2 s−1) and
(compare to 0.2 s−1) and 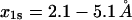 (compare to 1.4 Å) (24). In contrast,
(compare to 1.4 Å) (24). In contrast,  and
and 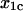 are new parameters that have not previously been measured for either system. The distance parameter
are new parameters that have not previously been measured for either system. The distance parameter  is of special interest because it sets a minimum limit on the scale of conformational change in the bond required during unbinding in the catch-pathway. However, as seen in Table 1, the estimated value of this parameter varies over fivefold between the two sets of data for P-selectin from the Marshall et al. (6) and Evans et al. (9) articles. This difference is not crucial because the experiments are done in different laboratories and with different experimental techniques, which could introduce calibration errors that are consistent within a set of experiments but not between methods. However, it precludes an interpretation about the magnitude of this structural change.
is of special interest because it sets a minimum limit on the scale of conformational change in the bond required during unbinding in the catch-pathway. However, as seen in Table 1, the estimated value of this parameter varies over fivefold between the two sets of data for P-selectin from the Marshall et al. (6) and Evans et al. (9) articles. This difference is not crucial because the experiments are done in different laboratories and with different experimental techniques, which could introduce calibration errors that are consistent within a set of experiments but not between methods. However, it precludes an interpretation about the magnitude of this structural change.
Articulating our four-parameter single-bound-state two-pathway model mathematically allows it to be compared with the alternative models, including the five-parameter two-bound-state, two-pathway models posited by Evans et al. (9) and the seven-parameter model recently proposed by Barsegov and Thirumalai (16).
There are at least two different structural interpretations of the catch-pathway. First, there could be a hook structure that must be shortened to disengage, as pointed out by Isberg and Barnes (4). An alternative interpretation of the two-pathway model is provided by studies on the bacterial adhesive protein FimH (3) (W. Thomas, M. Forero, O. Yakovenko, L. Nilsson, P. Vicini, E. Sokurenko, and V. Vogel, unpublished data). We have shown that FimH undergoes a force-induced conformational change to a higher affinity state (3). This high-affinity state can unbind via two pathways: an unbinding slip-pathway and a catch-pathway to a low-affinity state that unbinds rapidly. It is possible that the P- and L-selectins work via a similar mechanism, with the published data reflecting the lifetime of only the high-affinity state because the low affinity state is too short-lived to be detected. The model proposed by Evans et al. (9) is also similar to the previously proposed two-state mechanism for FimH (3) except that Evans et al. assumes the two states are in rapid equilibrium. Thus, the two-state mechanism that we proposed earlier can reduce to the two-pathway mathematical model we describe here with one set of assumptions (very rapid unbinding from the weak state), but can also reduce to the mathematical model described by Evans et al. (9) with another set of assumptions (rapid equilibrium between states). In either form, the two-state model is an attractive model because it is a natural outcome of the commonly observed phenomenon of conformational states with different strengths of binding. These reducing assumptions are necessary in applying the two-state model to the selectins, to explain why a single exponential decay in bond survival is observed despite the existence of two states.
Finally, the mathematical analysis of the two-pathway model determines what properties are required for a bond to be a two-pathway catch-bond. The analysis indicates that in order for the catch-slip transition to exist, three basic conditions must be satisfied regarding the form of the binding potential and the direction of the detaching force: 1), the ligand must be capable of escaping the binding site via two or more routes involving distinct transition states; 2), the detaching force must be directed such that its projection is positive along some escape routes and negative along other routes; and 3), the product of the unstressed off-rate and the projected transition state distance for the catch-pathway are greater than the corresponding product for the slip-pathways as given by Eq. 8. Similar criteria could be established for other catch-bond models.
Acknowledgments
We greatly appreciate the comments of Dr. Viola Vogel. O.V.P. is grateful to Prof. Daniel Borgis at Laboratoire de Physique Théorique des Liquides, Université Pierre et Marie Curie, Paris, France, for hospitality during manuscript preparation.
The research was supported by National Science Foundation (CAREER Award CHE-0094012 to O.V.P.) and National Institutes of Health Bioengineering Research Partnership (grant No. R01 AI050940 to E.V.S.). We gratefully acknowledge the assistance of Dr. Paolo Vicini and the Resource Facility for Population Kinetics (NIH grant No. EB-001975) in data analysis and parameter estimation. O.V.P. is an Alfred P. Sloan Fellow.
APPENDIX
Consider the case of the system with two parallel monomer bonds with the pulling force f equally distributed between the monomers, such that when both monomers are bound, each experiences the 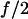 force. To find the lifetime of the dimeric bond under the influence of the pulling force f it is first necessary to find the probability for breaking one of the two bonds. The probability that the first of the two bonds breaks is described by the random variable
force. To find the lifetime of the dimeric bond under the influence of the pulling force f it is first necessary to find the probability for breaking one of the two bonds. The probability that the first of the two bonds breaks is described by the random variable 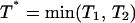 defined in terms of the independent random variables
defined in terms of the independent random variables 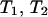 describing the breaking of the individual bonds. The probability density
describing the breaking of the individual bonds. The probability density  of the random variable
of the random variable  is related (25) to the probability density
is related (25) to the probability density  (see Eq. 7) of the random variables
(see Eq. 7) of the random variables 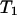 and
and  by
by
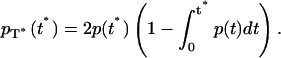 |
(A1) |
The probability density of breaking the remaining bond depends on the random variable 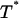 and is equal to:
and is equal to:
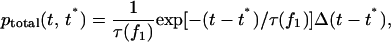 |
(A2) |
where  is the step function and
is the step function and 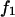 is the value of the force acting on the remaining bond, considered in detail below. After convolving Eq. A2 with
is the value of the force acting on the remaining bond, considered in detail below. After convolving Eq. A2 with
 |
(A3) |
we find
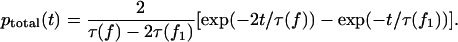 |
(A4) |
It follows from Eq. A4 that the lifetime of the bound state of the dimer is
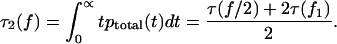 |
(A5) |
The magnitude of the force 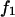 acting on the remaining bond depends on the experimental setup and falls within the two limiting cases: 1), the force f acting on the dimer in the beginning of the experiment is immediately transferred on the remaining bond after the first bond breaks, i.e.,
acting on the remaining bond depends on the experimental setup and falls within the two limiting cases: 1), the force f acting on the dimer in the beginning of the experiment is immediately transferred on the remaining bond after the first bond breaks, i.e.,
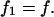 |
(A6) |
This is the case when the experiment is set up to support a constant force (2). The force 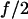 acting on each monomer in the dimer is independent on whether one of the monomer bonds is broken, i.e.,
acting on each monomer in the dimer is independent on whether one of the monomer bonds is broken, i.e.,
 |
(A7) |
This is the case when the experiment is set up to support a constant displacement d of the bonds as shown by the following analysis. The experiments of Marshall et al. (6) can be described by a system of three springs, whose extensions are determined by Hooke's law. The first two springs, corresponding to the monomeric bonds, act in parallel and have the overall spring constant 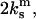 where
where 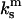 is the spring constant of a single monomer. The third spring corresponds to the cantilever pulling on the dimeric bond. The cantilever with the spring constant
is the spring constant of a single monomer. The third spring corresponds to the cantilever pulling on the dimeric bond. The cantilever with the spring constant 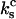 acts sequentially with the dimeric bond, such that the overall spring constant of the system of the dimeric bond plus the cantilever is
acts sequentially with the dimeric bond, such that the overall spring constant of the system of the dimeric bond plus the cantilever is 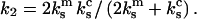 The applied force f is related to the displacement d by Hooke's law
The applied force f is related to the displacement d by Hooke's law 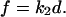 After one of the monomer bonds is broken, the spring constant of the system of the remaining monomer bond plus the cantilever equals
After one of the monomer bonds is broken, the spring constant of the system of the remaining monomer bond plus the cantilever equals 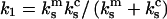 and the force
and the force  that supports the displacement d equals
that supports the displacement d equals 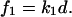 Combining these two relationships for the experiment supporting a constant displacement d, we express the value of the force
Combining these two relationships for the experiment supporting a constant displacement d, we express the value of the force 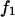 after one bond is broken in terms of the force acting on the double bond:
after one bond is broken in terms of the force acting on the double bond:
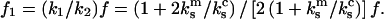 |
(A8) |
In the case where the cantilever is much stiffer than the monomeric bond 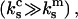 then the
then the 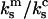 terms in Eq. A8 can be neglected and Eq. A8 reduces to Eq. A7. In this limit Eq. A5 reduces to Eq. 18 of the main text.
terms in Eq. A8 can be neglected and Eq. A8 reduces to Eq. A7. In this limit Eq. A5 reduces to Eq. 18 of the main text.
Two observations justify the simplifying use of Eqs. A7 and 18. First, the spring constants reported for the P-selectin are 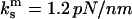 (26), and
(26), and 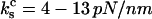 (6), so that
(6), so that 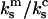 ∼ 0.09–0.3. Second, this assumption accurately describes behavior of the dimeric bond, particularly regarding the 3/2-fold increase in peak lifetime and the twofold increase in force at this peak lifetime (solid lines in Fig. 4, left). In contrast, the other extreme—that the force experienced by the cantilever in the experiments somehow remained constant in each pull as in Eq. A6—does not describe the behavior of the dimeric bond well at all, as shown in the dotted line of Fig. 4, left. This model predicts a jump in force from f to f/2 when the first bond breaks, which could contribute to the instabilities mentioned in Marshall et al. (6). Thus, the assumptions in this Appendix are derived from the described experimental setup and are consistent with the published experimental results.
∼ 0.09–0.3. Second, this assumption accurately describes behavior of the dimeric bond, particularly regarding the 3/2-fold increase in peak lifetime and the twofold increase in force at this peak lifetime (solid lines in Fig. 4, left). In contrast, the other extreme—that the force experienced by the cantilever in the experiments somehow remained constant in each pull as in Eq. A6—does not describe the behavior of the dimeric bond well at all, as shown in the dotted line of Fig. 4, left. This model predicts a jump in force from f to f/2 when the first bond breaks, which could contribute to the instabilities mentioned in Marshall et al. (6). Thus, the assumptions in this Appendix are derived from the described experimental setup and are consistent with the published experimental results.
References
- 1.Dembo, M., D. C. Torney, K. Saxman, and D. Hammer. 1988. The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc. R. Soc. Lond. B Biol. Sci. 234:55–83. [DOI] [PubMed] [Google Scholar]
- 2.Bell, G. I. 1978. Models for the specific adhesion of cells to cells. Science. 200:618–627. [DOI] [PubMed] [Google Scholar]
- 3.Thomas, W. E., E. Trintchina, M. Forero, V. Vogel, and E. V. Sokurenko. 2002. Bacterial adhesion to target cells enhanced by shear force. Cell. 109:913–923. [DOI] [PubMed] [Google Scholar]
- 4.Isberg, R. R., and P. Barnes. 2002. Dancing with the host; flow-dependent bacterial adhesion. Cell. 110:1–4. [DOI] [PubMed] [Google Scholar]
- 5.Konstantopoulos, K., W. D. Hanley, and D. Wirtz. 2003. Receptor-ligand binding: “catch” bonds finally caught. Curr. Biol. 13:R611–R613. [DOI] [PubMed] [Google Scholar]
- 6.Marshall, B. T., M. Long, J. W. Piper, T. Yago, R. P. McEver, and C. Zhu. 2003. Direct observation of catch bonds involving cell-adhesion molecules. Nature. 423:190–193. [DOI] [PubMed] [Google Scholar]
- 7.Sarangapani, K. K., T. Yago, A. G. Klopocki, M. B. Lawrence, C. B. Fieger, S. D. Rosen, R. P. McEver, and C. Zhu. 2004. Low force decelerates L-selectin dissociation from P-selectin glycoprotein ligand-1 and endoglycan. J. Biol. Chem. 279:2291–2298. [DOI] [PubMed] [Google Scholar]
- 8.Forero, M., W. E. Thomas, C. Bland, L. Nilsson, E. V. Sokurenko, and V. Vogel. 2004. A catch-bond based smart nano-adhesive sensitive to shear stress. Nano. Lett. 4:1593–1597. [Google Scholar]
- 9.Evans, E., A. Leung, V. Heinrich, and C. Zhu. 2004. Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc. Natl. Acad. Sci. USA. 101:11281–11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pereverzev, Y. V., O. V. Prezhdo, W. E. Thomas, and E. V. Sokurenko. 2005. Distinctive features of the biological catch bond in the jump-ramp force regime predicted by the two-pathway model. Phys. Rev. E. 72:010903. [DOI] [PubMed] [Google Scholar]
- 11.Evans, E. 2001. Probing the relation between force–lifetime–and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 30:105–128. [DOI] [PubMed] [Google Scholar]
- 12.Dudko, O. K., A. E. Filippov, J. Klafter, and M. Urbakh. 2003. Beyond the conventional description of dynamic force spectroscopy of adhesion bonds. Proc. Natl. Acad. Sci. USA. 100:11378–11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derenyi, I., D. Bartolo, and A. Ajdari. 2004. Effects of intermediate bound states in dynamic force spectroscopy. Biophys. J. 86:1263–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garrivier, D., E. Decave, Y. Brechet, F. Bruckert, and B. Fourcade. 2002. Peeling model for cell detachment. Eur. Phys. J. E. Soft Matter. 8:79–97. [DOI] [PubMed] [Google Scholar]
- 15.Izrailev, S., S. Stepaniants, M. Balsera, Y. Oono, and K. Schulten. 1997. Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys. J. 72:1568–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barsegov, V., and D. Thirumalai. 2005. Dynamics of unbinding of cell adhesion molecules: transition from catch to slip bonds. Proc. Natl. Acad. Sci. USA. 102:1835–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans, E., A. Leung, D. Hammer, and S. Simon. 2001. Chemically distinct transition states govern rapid dissociation of single L-selectin bonds under force. Proc. Natl. Acad. Sci. USA. 98:3784–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yuan, C., A. Chen, P. Kolb, and V. T. Moy. 2000. Energy landscape of streptavidin-biotin complexes measured by atomic force microscopy. Biochemistry. 39:10219–10223. [DOI] [PubMed] [Google Scholar]
- 19.Barrett, P. H., B. M. Bell, C. Cobelli, H. Golde, A. Schumitzky, P. Vicini, and D. M. Foster. 1998. SAAM II: simulation, analysis, and modeling software for tracer and pharmacokinetic studies. Metabolism. 47:484–492. [DOI] [PubMed] [Google Scholar]
- 20.Sheiner, L. B., and S. L. Beal. 1985. Pharmacokinetic parameter estimates from several least squares procedures: superiority of extended least squares. J. Pharmacokinet. Biopharm. 13:185–201. [DOI] [PubMed] [Google Scholar]
- 21.Landaw, E. M., and J. J. DiStefano, III. 1984. Multiexponential, multicompartmental, and noncompartmental modeling. II. Data analysis and statistical considerations. Am. J. Physiol. 246:R665–R677. [DOI] [PubMed] [Google Scholar]
- 22.Gardiner, C. E. 1983. Handbook of Stochastic Methods for Physics, Chemistry, and the Natural Sciences. Springer-Verlag, New York, NY.
- 23.Ramachandran, V., M. U. Nollert, H. Qiu, W. J. Liu, R. D. Cummings, C. Zhu, and R. P. McEver. 1999. Tyrosine replacement in P-selectin glycoprotein ligand-1 affects distinct kinetic and mechanical properties of bonds with P- and L-selectin. Proc. Natl. Acad. Sci. USA. 96:13771–13776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanley, W., O. McCarty, S. Jadhav, Y. Tseng, D. Wirtz, and K. Konstantopoulos. 2003. Single molecule characterization of P-selectin/ligand binding. J. Biol. Chem. 278:10556–10561. [DOI] [PubMed] [Google Scholar]
- 25.Feller. 1968. An Introduction to Probability Theory and its Applications. John Wiley & Sons, New York, NY.
- 26.Marshall, B. T., R. P. McEver, and C. Zhu. 2002. Mechanical Properties of the P-selectin/PSGL-1 Interaction. IEEE, Houston, TX.







