Abstract
To define structural elements critical for RNA replication in human parechovirus 1 (HPeV1), a replicon with chloramphenicol acetyltransferase as a reporter gene and an infectious virus cDNA clone have been used. It was observed that there are cis-acting signals required for HPeV1 replication located within the 5′-terminal 112 nucleotides of the genome and that these include two terminal stem-loops, SL-A and SL-B, together with a pseudoknot element. Significant disruption of any of these structures impaired both RNA replication and virus growth. In view of the similarity in terminal structures to several picornaviruses, such as cardioviruses and hepatoviruses, the insights generated in this work are of wider significance for understanding picornavirus replication.
Picornaviridae is a large family of important human and animal pathogens, including such members as Poliovirus, human rhinoviruses (the major cause of the common cold), Hepatitis A virus (HAV), and Foot-and-mouth disease virus of ungulates. Currently, nine picornavirus genera are recognized: Enterovirus, Rhinovirus, Cardiovirus, Aphthovirus, Hepatovirus, Parechovirus, Erbovirus, Kobuvirus, and Teschovirus (16). Picornaviruses have a single-stranded RNA genome of positive polarity, encoding one open reading frame, which is preceded by a 5′ untranslated region (5′ UTR) and followed by a short 3′ UTR and poly(A) tract (31). Picornavirus 5′ UTRs contain several secondary and tertiary structure elements. Detailed studies have demonstrated that this genomic region is implicated in two key cis-acting functions (3). One of these is initiation of translation of the virus polyprotein, which takes place via a cap-independent process with an internal ribosome entry site (IRES). Extensive regions of the 5′ UTR make up the IRES and allow virus protein synthesis to continue when the virus has shut off cap-dependent, cellular translation (35). In terms of the structure of the IRES, seven of the nine picornavirus genera can be assigned to two groups: enteroviruses and rhinoviruses constitute one group and are said to have a type I IRES, while cardioviruses, aphthoviruses, hepatoviruses, erboviruses, and parechoviruses have a type II IRES, although the hepatovirus structure is somewhat diverse.
Rhinovirus and enterovirus 5′ UTRs have a stable cloverleaf-like domain at their 5′ terminus (3). A requirement for this structure in another cis-acting function, RNA replication, through RNA-protein complex formation with virus and cellular proteins, is well established (2, 8-11, 23, 26, 30). There is, however, relatively little information on the second 5′-UTR group in terms of RNA replication. These presumably need to provide structures to fulfill critical roles analogous to those of rhinovirus-enterovirus cloverleaves. Our previous data on human parechovirus 1 (HPeV1) showed that 5′-proximal deletions were lethal but did not affect in vitro translation (20). Thus, they may not be involved in translation, but in other replication steps, including initiation of synthesis of genomic RNA. The HPeV1 5′-proximal structure consists of a long stem-loop (SL-A), together with a short stem-loop (SL-B) whose loop appears to participate in a pseudoknot structure (pk-C). Downstream, there is a variable AC-rich tract (12). To further understand the role of SL-A, SL-B, and pk-C in RNA replication of HPeV1, mutational analysis (deletion, insertion, and substitution mutations) of the HPeV1 5′ UTR was carried out. This used a complete HPeV1 cDNA clone and a chloramphenicol acetyltransferase (CAT) replicon, and the ability of mutant RNA to produce infectious virus and/or CAT reporter gene expression upon transfection of green monkey kidney (GMK) cells was then assessed. The results identified and mapped cis-acting RNA replication signals to the 5′ terminus of the viral genome. RNA transcripts derived from HPeV1 and its replicon were unable to replicate unless they contained stable forms of each of the 5′-most structures, SL-A, SL-B, and pk-C, indicating that these are critical for HPeV1 RNA replication.
Subgenomic HPeV1 replicon mRNAs.
The replicon construct, pSN/REP (Fig. 1a), was based on pHPeV1, which contains a complete HPeV1 cDNA (20). The 659-nucleotide CAT coding region was inserted in frame between the 5′ UTR and a SalI site (position 1001, in the VP0-encoding region). A SalI site was introduced C-terminal to the CAT-encoding region, and the 5-amino-acid sequence Leu-Ser-Asn-Gln-Gly was added. This is a cleavage site for HPeV1 3C and is based on the 2B-2C junction (15). The CAT initiation codon was fused to the HPeV1 5′ UTR so that the Kozak consensus sequence was maintained. MluI or ApaI sites immediately downstream of HPeV1 cDNA allowed linearization prior to T7 polymerase transcription of pSN/REP.
FIG. 1.
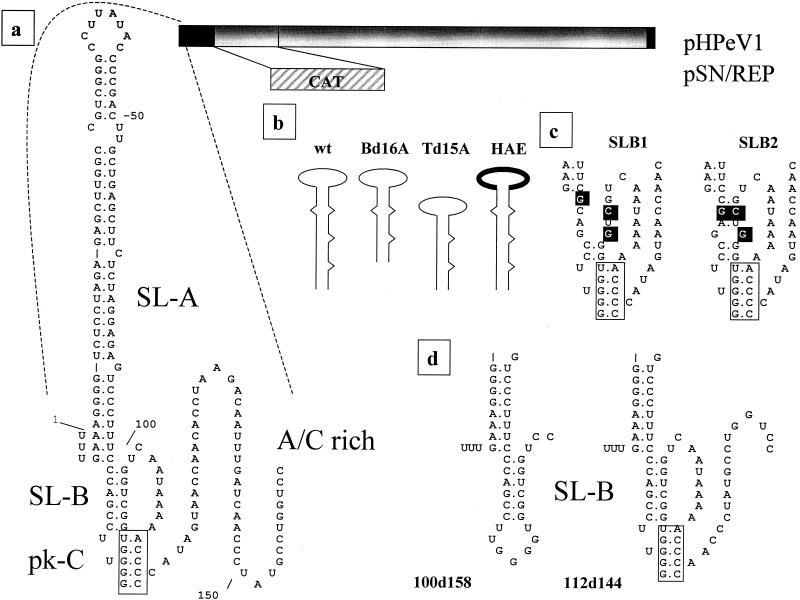
(a) Representation of HPeV1 cDNA contained within the infectious cDNA clone pHPeV1. Noncoding regions are shown in black. Part of the coding region is replaced by an in-frame fusion of the CAT coding sequence in the replicon pSN/REP. An expanded view of the 5′-proximal region is also shown, indicating the domains studied. The position of pk-C is indicated by a box. (b) SL-A mutants where the bottom (Bd16A) and top (Td15A) of the domain have been deleted and the terminal loop replaced by that of EMCV (HAE) compared to the wild-type (wt) structure. (c) Predicted effect of the mutations introduced into SL-B in the mutants SLB1 and SLB2. Mutations are shown in reverse contrast, and pk-C is boxed. (d) Predicted structure of mutants 100d158 and 112d144 which have deletions in the AC-rich region. pk-C is boxed.
Several deletions (mutants named according to the nucleotides deleted) were constructed by digestion with restriction enzymes. These were 1d168 (SstI-BstBI), 87d168 (XcmI-BstBI), 168d298 (BstBI-BstXI), 298d538 (BstXI-SpeI), and 1d538 (SstI-SpeI) (20). Precise deletions, together with insertions and point mutations, were introduced by single or overlap PCR. These involved mutation of the AC-rich tract region (100d158 and 112d144), stem-loop A (Td15A, Bd16A, and HAE), stem-loop B (SLB1 and SLB2), and pk-C region (dpk and Rdpk). PCR-amplified DNA fragments were ligated into pGEMT-Easy (Promega) and sequenced to confirm the presence of only the desired mutation. The following mutagenesis primer pairs were used: OL827(s) (GAGCTCTTTTCTCCTAGAGAGCTTGGC) and OL828(c) (CCACCCCAAGGCTGGCTCTCCTAGAGAAGCTC) for Bd16A, OL825(s) (GAGAGCTTGGCCCTTATACGCTGAGCTTCTCTAGGAG) andOL826(c) (GTATAAGGGCCAAGCTCTCTAGGAGA) for Td15A, OL782(s) (CGTCGGGTCGATATCCCCGACTTGCTGAGCT) and OL783(c) (AGCTCAGCAAGTCGGGGATATCGACCCGACG) for HAE, OL796(s) [GTCCCTTTCC(C,G)A(C,G)CCTTGGGGTGGGTCGTCAATA] and OL797(c) [GGGGTTTTTATTGACGACCCCAAGG(G,C)T(G,C)GGAAAG] for SLB1 and SLB2, OL565(c) (CCAGGCATAGGGTTGGGGTTTTTATTGACC) and OL566(s) (CAACCCTATGCCTGGTC) for 112d144, OL567(c) (TTCGAATAGTGGGGACCAGCCACCCCAAGGC) for 100d158, OL780(s) (GGTCAATAAAAGTCCCCATAGTAACCA) and OL781(c) (TGGTTACTATGGGGACTTTTATTG) for dpk, OL821(s) [(G,C)ACGGCTGGTCAATAAAAGTCCCCATAG] and OL822(c)[GACTTTTATTGACCAGCCGT(G,C)CCAAGGCTG] forRdpk, and OL823(s) (TCCCAGCCTTGGGGTAAGACCCCGGCTGGTCAATA) and OL824(c) (GTCTTACCCCAAGGCTGGGAAAG) for Lpk, where the genomic-sense sequence is shown by “(s)” and the complementary-sense sequence is shown by “(c).” Nucleotide substitutions are indicated in boldface, and nucleotides in parentheses indicate degenerate nucleotides.
In the case of deletions generated by restriction enzyme digestion, a mutated KpnI subclone was used to replace the wild-type fragment of either pHPeV1 or pSN/REP, following KpnI digestion and phosphatase treatment as previously described (20). For the other mutants, it was more convenient to ligate the SstI/BstBI fragment (positions 1 to 168) from pGEMT-Easy mutants into SstI/BstBI-cut pHPeV1 and pSN/REP cDNAs. All constructs were confirmed by restriction enzyme digestion and nucleotide sequencing.
GMK cells were grown in Dulbecco modified Eagle medium (Gibco BRL) with 10% fetal calf serum. T7-mediated, in vitro transcription of linearized clones, RNA transfection into GMK cells with Lipofectin (Gibco BRL), and CAT assay were performed as previously described (14, 26).
The 5′ end of the 5′ UTR functions as a genome replication signal.
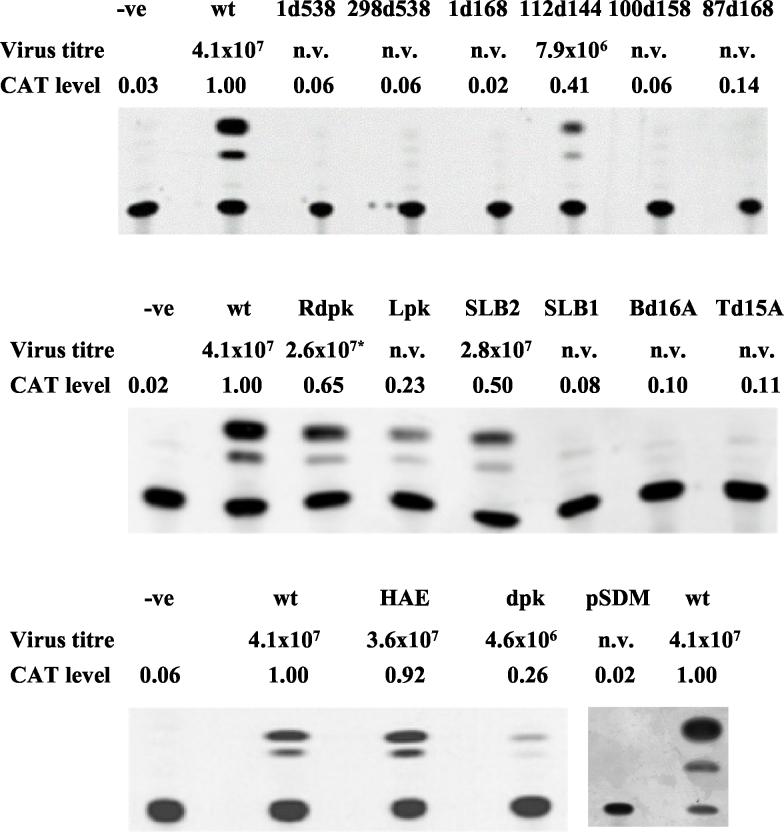
We have found previously that several HPeV1 mutants with 5′-proximal deletions, produced in the context of mapping the HPeV1 IRES, were unable to replicate, although the mutations had little effect on translation in vitro (20). Here, these and several additional mutations were analyzed, by using an infectious cDNA clone (pHPeV1) and a CAT replicon (pSN/REP), to find whether they gave an RNA-replication-defective phenotype (Fig. 1). In order to verify that the CAT replicon was measuring RNA synthesis, pSDM, a negative control construct, was produced. This lacked sequences downstream of position 5479 but had an intact 5′ UTR, and so RNA should have been translated efficiently. RNA transcribed from pSDM did not give any CAT activity after transfection (Fig. 2), presumably because proteins needed for replication were absent. This suggests that significant CAT levels are achieved only when the RNA is replicated, not by translation of the input RNA. As pSDM lacks a 3′ UTR and poly(A) tract, it is possible that its translation efficiency is reduced. However, the fact that the constructs 1d168 and 87d168 (Fig. 2), previously shown to translate with wild-type efficiency, also gave no CAT activity supports the conclusion that the CAT assay is indeed measuring RNA replication (20).
FIG. 2.
Results of CAT assays following transfection of cells with RNA transcribed from the mutant replicon constructs indicated. The level of CAT was determined by densitometry and normalized to that obtained from the wild-type construct pSN/REP (labeled wt) assayed with each batch of mutants, to ensure comparability. Each assay was performed twice and gave similar results in each case. −ve is a negative (mock-transfected) control included to show that there is no endogenous acetylating activity, while pSDM is a deletion construct lacking replication ability. The titer of virus obtained when the corresponding mutation was built into the infectious clone pHPeV1 is also indicated in each case. n.v. indicates a nonviable virus. *, only a pseudorevertant virus with an additional mutation was obtained from Rdpk.
RNAs transcribed from cDNA in the wild-type and deletion mutant constructs (1d538, 298d538, 1d168, and 87d168, where the numbers represent nucleotides at the boundaries of the deletion) were initially transfected. No virus plaques were observed after 6 days from the deletion mutants, while transfected RNA transcribed from wild-type cDNA gave plaques after 3 days. There was no detectable CAT expression at ∼30 h posttransfection in any of these mutants (Fig. 2). In contrast, transfection with RNA from the wild-type construct, pSN/REP, gave substantial CAT activity, indicating that the RNA was undergoing replication. The absence of CAT activity from the 1d538 and 298d538 deletions was expected, as these constructs were defective in translation (20). Even if potentially capable of RNA replication, expression of CAT activity requires translation to occur. However, as discussed above, deletions of nucleotides 1 to 168 and 87 to 168 also prevented CAT expression (Fig. 2) but were previously shown to have no effect on in vitro translation (20). Thus, elements within the region of nucleotides 1 to 168 function in RNA synthesis without a substantial role in internal translation initiation.
Disruption of the domain A structure prevents HPeV1 RNA synthesis.
To identify crucial elements of the terminal domain SL-A, a deletion of 16 nucleotides (designated Bd16A) was constructed to remove the complementary nucleotides 4 to 11 and 74 to 81 and thus truncate the bottom of the domain A stem (Fig. 1b). A second mutant (Td15A), with nucleotides 30 to 36 and 45 to 54 deleted, had the top part of the stem truncated. To characterize the importance of the loop structure, mutant HAE was made. Here the HPeV1 loop sequence was replaced with that of a picornavirus with a related 5′ UTR, encephalomyocarditis virus (EMCV), by changing the sequence from CCUUAUAC to UCGAUAUC. Mutant HPeV1 and replicon RNAs were transfected onto monolayers of GMK cells. Three days posttransfection, HPeV1-HAE viral plaques were observed and were similar to those from wild-type RNA. The transfection efficiency was also similar to that of wild-type RNA (ca. 104 plaques/μg of RNA), indicating that the virus probably did not result from reversion or pseudoreversion events. The sequence of the first 170 nucleotides of the virus recovered showed that it had retained the EMCV loop A sequence and had no other changes. The virus obtained gave a similar titer as wild-type HPeV1 (Fig. 2). In contrast, HPeV1 derivatives Bd16A and Td15A did not give plaques or evident cytopathic effect even after 6 days. To confirm that this resulted from lesions in RNA synthesis, CAT activities of the corresponding replicons at 24 h posttransfection were assayed. Bd16A and Td15A had not undergone RNA synthesis, as the level of CAT was minimal, while the HAE mutant replicated with near-wild-type efficiency (Fig. 2). Thus, virus viability and the CAT assay indicated that the stem of SL-A has an essential role in RNA synthesis, while the loop can be exchanged, at least between HPeV1 and EMCV.
Structural stability of SL-B is of functional significance in HPeV1 RNA synthesis.
The failure of 87d168 to replicate suggests that SL-B may be of functional significance in HPeV1 RNA synthesis and that this domain is highly conserved in parechoviruses, indicating a critical role in the virus life cycle (12, 15, 21, 32). Further analysis was carried out by using point mutations to disrupt SL-B. One mutant, SLB1, contained a G-to-C substitution at position 98, a C-to-G change at position 96, and a nondesigned C-to-G mutation at position 81. These disrupted potential base pairing considerably (Fig. 1c). A second construct, SLB2, had mutations of G to C at position 98, C to G at position 96, and C to G at position 82. The mutation of C to G at position 82 maintains the position 82-98 interaction, and so this mutant had only one mismatch (84-96 interaction) (Fig. 1c).
Transfection of HPeV1-SLB1 RNA gave no infectious virus, and the substitutions were clearly lethal for the virus. The HPeV1-SLB2 transcript gave a virus which replicated almost as well as wild-type HPeV1 in terms of titer. CAT assays showed similar effects, as the SLB2, but not the SLB1, replicon gave CAT activity, although reduced by 50% from wild-type levels (Fig. 2). The results obtained indicate that the structure, but not the sequence, of the SL-B stem is important for RNA replication and virus viability.
Deletion mutations involving the AC-rich tract.
Parechoviruses have a variable sequence rich in A and C bases between nucleotides 100 and 156, predicted to be unstructured and corresponding in genomic location to the poly(C) tract of cardioviruses-aphthoviruses and the polypyrimidine tract of hepatoviruses (12, 15, 21). To attempt to define the role of this region in the replication of HPeV1, two precise deletion mutations were created (Fig. 1d). No virus growth was observed in flasks transfected with the 100d158 mutant, even after 6 days of incubation, while plaque formation of the mutant 112d144 took longer (4 days) than that for wild-type HPeV1 (3 days). Partial sequencing of the virus recovered showed that it had the deletion of nucleotides 112 to 144, while plaque titration showed that it grew to a lower titer than did the wild type (Fig. 2). HPeV1 can therefore be recovered with a deletion of nucleotides 112 to 144, including much of the AC-rich region, although its growth is impaired. The longer deletion, through the whole AC-rich region, was lethal. However, this had also removed sequences involved within the predicted pseudoknot (pk-C). The effects of these deletions on CAT expression are shown in Fig. 2. 112d144 gave significant CAT activity, although reduced by nearly 60% below that from the wild-type replicon. The deletion extending to nucleotides 100 to 158 abolished CAT activity. Thus, nucleotides 112 to 144 of HPeV1 RNA are dispensable for RNA synthesis and virus growth in GMK cells but affect both to some extent.
The pk-C pseudoknot functions in HPeV1 RNA synthesis.
The GGGGU sequence (positions 89 to 93), in the loop of SL-B, and nucleotides 109 to 113 (ACCCC) are complementary and potentially form a pseudoknot (pk-C) (Fig. 1a). An analogous structure is predicted for the other human parechoviruses, and similar pseudoknots can also form in HAV and EMCV and may have a role in viral RNA amplification in infected cells (22). The lethal phenotype of 100d158 may be due to disruption of pk-C, and to study this structure further, an insertion of eight nucleotides was introduced downstream of nucleotide 93. This had nucleotides complementary to the SL-B loop pseudoknot residues and might therefore interfere with it (Fig. 3). This mutation was designated Lpk. No virus was generated from the mutant RNA, and CAT synthesis was reduced by 77%, suggesting that interference with the pseudoknot has a marked effect on replication (Fig. 2).
FIG. 3.
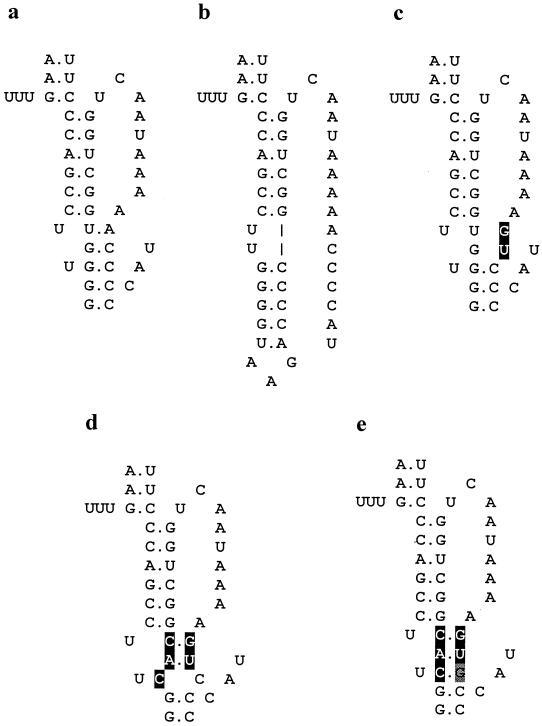
Predicted structure of pk-C mutants. (a) Wild-type structure. (b) Lpk, an insertion of 8 nucleotides designed to complement SL-B loop nucleotides and thus prevent pseudoknot formation. (c) dpk, a substitution of two nucleotides (shown in reverse contrast) which disrupt pk-C. (d) Rdpk, three additional changes compared to dpk which partially restore pk-C. (e) A revertant, arising from passaging Rdpk, which has a fully repaired structure due to an additional substitution (gray highlight).
To further study the effect of disruption of the pseudoknot, a double mutation was introduced at positions 109-111 (AC→GU, designated dpk) to weaken the structure by removing 2 bp (Fig. 3). Weakening pk-C in this way gave a virus which grew to a titer 1 log less than that of the wild type. Reverse transcription-PCR confirmed that the virus recovered had retained the mutations. The dpk replicon showed CAT activity reduced by 74% (Fig. 2). This further implies that the pk-C structure is of functional importance in HPeV1 RNA synthesis.
A mutant (Rdpk) with three additional mutations compared to dpk was then produced. In this, two of the mutations (GU→CA, positions 92 and 93) are complementary to those introduced into dpk. The third change (position 91) was introduced to give a single mismatch in the center of pk-C. CAT synthesis from pSN/REP-Rdpk was reduced by 35% compared to wild type (Fig. 2). Plaques were obtained from pHPeV1-Rdpk, and the virus from one of these was passaged and sequenced. The virus recovered had near-wild-type growth properties and had acquired a C→G change at position 111, complementary to the manipulated C base at position 91, thus giving a perfectly restored pk-C of 5 bp (Fig. 3).
5′-terminal sequences in picornavirus replication.
Picornavirus RNA replication requires the synthesis of complementary negative-strand RNA to serve as a template for the production of nascent, genomic (positive)-sense RNA. It is expected that the termini of the RNA, by definition being close to the site of initiation, are important in RNA replication, although it has also been shown elsewhere that structures within the RNA (cis-acting replication elements) are also critical in replication (24). Studies of the enterovirus poliovirus have confirmed the requirement for 5′-terminal structures, as disruption of the highly conserved 5′ cloverleaf prevents RNA replication (1, 26). Cardioviruses, aphthoviruses, erboviruses, parechoviruses, and, to a lesser extent, hepatoviruses share a common type of IRES, distinct from that of enteroviruses-rhinoviruses, and several of these have a long, terminal stem-loop structure, rather than a cloverleaf, together with other downstream stem-loops and pseudoknots. The type of 5′ UTR in the other genera, Kobuvirus and Teschovirus, is not yet clear, but a similar terminal stem-loop has been confirmed for kobuviruses recently (27). Although some of these features have been studied for cardioviruses, hepatoviruses, and kobuviruses (7, 17, 27, 28, 29), there has been no comprehensive work defining the contribution of 5′-terminal structures to RNA replication in this non-enterovirus-rhinovirus group. Thus, we analyzed the predicted structural motifs (Fig. 1) in the 5′-proximal region of HPeV1, a frequent human pathogen (33, 34).
Three different parts of SL-A were analyzed. By constructing HPeV1 harboring the EMCV sequence in the top loop of SL-A, a chimeric virus (HAE), with wild-type growth properties, was produced. The aim was to determine whether this is a critical genetic element, possibly involved in forming a ribonucleoprotein complex. Replacement of one of the type I cloverleaf loops has been found elsewhere to be highly virus species restricted, and this was useful in defining a direct interaction with 3CD, as pseudorevertant viruses with mutations in 3C were selected (1). The HAE results imply that this terminal loop is not involved in binding between the HPeV1 5′ UTR and virus protein 3CD, as it is unlikely that the same loop sequence could bind in a specific way to both the EMCV and HPeV1 3CD proteins, since these are highly diverse.
Deletion of either the bottom-most (nucleotides 4 to 11 and 74 to 81, Bd16A) or the topmost (nucleotides 30 to 36 and 45 to 54, Td15A) portions of SL-A resulted in no virus growth and no CAT synthesis (Fig. 1b and 2). The latter finding is not surprising, as a single nucleotide deletion (position 5) in HPeV1 gives highly impaired virus growth (data not shown). Mutational analysis suggested that nucleotides 3 to 8 (in stem “a”) of the negative strand of the poliovirus cloverleaf facilitate the interaction of virus protein 2C with the 3′ terminus (4). Similar results have been obtained for human rhinoviruses and HAV, a virus with a type II-like SL-A (5). The corresponding region deleted in Bd16A may have a similar function, hence explaining the inability of mutant RNA to replicate. However, the Td15A mutation, where the top of the stem-loop is deleted, was also lethal, suggesting that all of the SL-A stem may be critical.
The need for the integrity of the predicted SL-B structure was also studied (Fig. 1c). Extensive disruption of SL-B (mutant SLB1) gave no infectious virus and no CAT activity. This result suggests that the predicted structure exists and is important for RNA synthesis. In contrast, disruption of a single base pair (mutant SLB2) gave a virus which grew with near-wild-type efficiency, with only a minor effect on CAT activity (Fig. 2). The recovery of virus from SLB2, which has three mutations but only one disrupted base pair, suggested that its significance lies in the stem structure rather than its primary sequence.
Deletion of nucleotides 112 to 144, part of the variable, AC-rich region downstream of pk-C, gave a construct with reduced CAT activity in the replicon and a recombinant virus with somewhat impaired growth properties (Fig. 2). Nonetheless, this region is clearly not essential for virus replication, at least in tissue culture. This region is spatially analogous to the hepatovirus polypyrimidine tract and cardiovirus poly(C) tract, both of which can be deleted with little effect on virus replication in tissue culture (28, 29). However, truncation of the cardiovirus poly(C) tract is strongly attenuating, suggesting an important role in natural infections (7), and growth in some murine cell cultures is also affected (18). Analogous deletions in the aphthovirus poly(C) tract are only partially attenuating (25). The fact that HPeVs retain this variable sequence, despite this being dispensable for growth in tissue culture cells (as 112d144 is viable), suggests that it, too, may be important in natural infections.
5′-proximal pseudoknots have been shown to be functionally significant in some picornaviruses. For instance, deletion of a pseudoknot, PKB, located close to the cardiovirus poly(C) tract is lethal, while less radical manipulation of the adjacent PKC markedly impairs RNA synthesis (17). In this work, it was shown that constructs which deleted HPeV1 pk-C (100d158), or had an insertion designed to interfere with it (Lpk), were lethal. Moreover, weakening 2 bp (construct dpk) severely compromised RNA synthesis and virus growth (Fig. 2 and 3). The repaired construct (Rdpk) had an increased level of RNA synthesis relative to dpk even though it still had one mismatch. This was probably because of the location of the mismatch in the center of the pseudoknot, allowing a stable structure still to form. A similar effect was observed for central positions of a pseudoknot-like structure in the 3′ UTR of coxsackievirus A9 (19). Despite this increased RNA synthesis, sequence analysis of the RNA from passaged virus showed that a further change had occurred (Fig. 3). This was compensatory and fully repaired the pk-C structure, suggesting that this structure exists and is important for efficient HPeV1 growth The pseudorevertant contained a total of five nucleotide substitutions compared to the wild-type sequence but had wild-type growth properties, indicating that the tertiary structure is more important than the primary sequences which contribute to it. It is possible that the increased level of CAT expression seen in Rdpk compared to dpk is due to the selection of a similar mutant during replication of the mutant replicon, but this was not tested.
Thus, the results described here indicate that HPeV1 requires a number of structures, SL-A, SL-B, and pk-C, for efficient RNA synthesis and replication. In enteroviruses and rhinoviruses the 5′-proximal cloverleaf plays a pivotal role in replication, by binding to virus protein 3CD and cellular protein PCBP2. PCBP2 also binds to the 5′-UTR structure domain IV, a key component of the IRES. Changes in binding of PCBP2 to these elements, brought about by 3CD, are believed to prevent ribosome binding after the first few rounds of translation, thus freeing the template for RNA replication (9). The cloverleaf is also brought close to the 3′ end of the RNA via a protein-protein bridge involving poly(A) binding protein, associated with the 3′ poly(A) tract, and PCBP2 bound to the 5′ cloverleaf (6, 13). This effective circularization of the RNA is needed for both negative- and positive-sense RNA synthesis (6, 9). At present, it is not clear whether the translation-transcription switch and 5′-3′ bridging are mechanistically similar in non-enteroviruses-rhinoviruses, or which RNA sequences are involved in the assembly of protein-RNA complexes. However, the results presented here indicate that 5′-most structures are likely to be involved in the mechanisms operative for RNA replication in HPeV1.
Acknowledgments
A.S.N. wishes to thank the Ministry of Higher Education of Iran for the generous provision of a studentship.
This work was supported by the Wellcome Trust, grant no. 046462.
REFERENCES
- 1.Andino, R., G. E. Rieckhof, D. Trono, and D. Baltimore. 1990. Substitutions in the protease (3Cpro) gene of poliovirus can suppress a mutation in the 5′ noncoding region. J. Virol. 64:607-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andino, R., G. E. Rieckhof, P. L. Achacoso, and D. Baltimore. 1993. Poliovirus RNA synthesis utilizes an rnp complex formed around the 5′ end of viral RNA. EMBO J. 12:3587-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andino, R., N. Boddeker, D. Silvera, and A. V. Gamarnik. 1999. Intracellular determinants of picornavirus replication. Trends Microbiol. 7:76-82. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee, R., A. Echeverri, and A. Dasgupta. 1997. Poliovirus-encoded 2C polypeptide specifically binds to the 3′-terminal sequences of viral negative-strand RNA. J. Virol. 71:9570-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee, R., and A. Dasgupta. 2001. Interaction of picornavirus 2C polypeptide with the viral negative-strand RNA. J. Gen. Virol. 82:2621-2627. [DOI] [PubMed] [Google Scholar]
- 6.Barton, D. J., B. J. O'Donnell, and J. B. Flanegan. 2001. 5′ cloverleaf in poliovirus RNA is a cis-acting replication element required for negative-strand synthesis. EMBO J. 6:1439-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duke, G. M., J. E. Osorio, and A. C. Palmenberg. 1990. Attenuation of Mengo-virus through genetic engineering of the 5′ noncoding poly(C) tract. Nature 343:474-476. [DOI] [PubMed] [Google Scholar]
- 8.Gamarnik, A. V., and R. Andino. 1997. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA 3:882-892. [PMC free article] [PubMed] [Google Scholar]
- 9.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 12:2293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 74:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gamarnik, A. V., N. Boddeker, and R. Andino. 1987. 2000. Translation and replication of human rhinovirus type 14 and mengovirus in Xenopus oocytes. J. Virol. 74:11983-11987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghazi, F., P. J. Hughes, T. Hyypiä, and G. Stanway. 1998. Molecular analysis of human parechovirus type 2 (formerly echovirus 23). J. Gen. Virol. 79:2641-2650. [DOI] [PubMed] [Google Scholar]
- 13.Herold, J., and R. Andino. 2001. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell 7:581-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hughes, P. J., C. Horsnell, T. Hyypiä, and G. Stanway. 1995. The coxsackievirus A9 RGD motif is not essential for virus infectivity. J. Virol. 69:8035-8040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyypiä, T., C. Horsnell, M. Maaronen, M. Khan, N. Kalkkinen, P. Auvinen, L. Kinnunen, and G. Stanway. 1992. A distinct picornavirus group identified by sequence analysis. Proc. Natl. Acad. Sci. USA 89:8847-8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King, A. M. Q., F. Brown, P. Christian, T. Hovi, T. Hyypiä, N. J. Knowles, S. M. Lemon, P. D. Minor, A. C. Palmenberg, T. Skern, and G. Stanway. 2000. Picornaviridae, p. 657-673. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 17.Martin, L. R., and A. C. Palmenberg. 1996. Tandem mengovirus 5′ pseudoknots are linked to viral RNA synthesis, not poly(C)-mediated virulence. J. Virol. 70:8182-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin, L. R., Z. C. Neal, M. S. McBride, and A. C. Palmenberg. 2000. Mengovirus and encephalomyocarditis virus poly(C) tract lengths can affect virus growth in murine cell culture. J. Virol. 74:3074-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirmomeni, M., P. J. Hughes, and G. Stanway. 1997. An RNA tertiary structure in the 3′ untranslated region of enteroviruses is necessary for efficient replication. J. Virol. 71:2363-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nateri, A. S., P. J. Hughes, and G. Stanway. 2000. In vivo and in vitro identification of structural and sequence elements of the human parechovirus 5′ untranslated region required for internal initiation. J. Virol. 74:6269-6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oberste, M. S., K. Maher, and M. A. Pallansch. 1998. Complete sequence of echovirus 23 and its relationship to echovirus 22 and other human enteroviruses. Virus Res. 56:217-223. [DOI] [PubMed] [Google Scholar]
- 22.Palmenberg, A. C., and Y. J. Sgro. 1997. Topological organization of picornaviral genomes: statistical prediction of RNA structural signals. Semin. Virol. 8:231-241. [Google Scholar]
- 23.Parsley, T. B., J. S. Towner, L. B. Blyn, E. Ehrenfeld, and B. L. Semler. 1997. Poly(rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA 3:1124-1134. [PMC free article] [PubMed] [Google Scholar]
- 24.Paul, A. V., E. Rieder, D. W. Kim, J. H. van Boom, and E. Wimmer. 2000. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 74:10359-10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rieder, E., T. Bunch, F. Brown, and P. W. Mason. 1993. Genetically engineered foot-and-mouth disease viruses with poly(C) tracts of two nucleotides are virulent in mice. J. Virol. 67:5139-5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohll, J. B., N. Percy, R. Ley, D. J. Evans, J. W. Almond, and W. S. Barclay. 1994. The 5′ untranslated regions of picornavirus RNAs contain independent functional domains essential for RNA replication and translation. J. Virol. 68:4384-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sasaki, J., Y. Kusuhara, Y. Maeno, N. Kobayashi, T. Yamashita, K. Sakae, N. Takeda, and K. Taniguchi. 2001. Construction of an infectious cDNA clone of Aichi virus (a new member of the family Picornaviridae) and mutational analysis of a stem-loop structure at the 5′ end of the genome. J. Virol. 75:8021-8030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shaffer, D. R., and S. M. Lemon. 1995. Temperature-sensitive hepatitis A virus mutants with deletions downstream of the first pyrimidine-rich tract of the 5′ nontranslated RNA are impaired in RNA synthesis. J. Virol. 69:6498-6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaffer, D. R., S. U. Emerson, P. C. Murphy, S. Govindarajan, and S. M. Lemon. 1994. A hepatitis A virus deletion mutant which lacks the first pyrimidine-rich tract of the 5′ nontranslated RNA remains virulent in primates after direct intrahepatic nucleic acid transfection. J. Virol. 69:6600-6604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silvera, D., A. V. Gamarnik, and R. Andino. 1999. The N-terminal K homology domain of the poly(rC)-binding protein is a major determinant for binding to the poliovirus 5′-untranslated region and acts as an inhibitor of viral translation. J. Biol. Chem. 274:38163-38170. [DOI] [PubMed] [Google Scholar]
- 31.Stanway, G. 1990. Structure, function and evolution of picornaviruses. J. Gen. Virol. 71:2483-2501. [DOI] [PubMed] [Google Scholar]
- 32.Stanway, G., N. Kalkkinen, M. Roivainen, F. Ghazi, M. Khan, M. Smyth, O. Meurman, and T. Hyypiä. 1994. Molecular and biological characteristics of echovirus 22—a representative of a new picornavirus group. J. Virol. 68:8232-8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stanway, G., and T. Hyypiä. 1999. Parechoviruses. J. Virol. 73:5249-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanway, G., P. Joki-Korpla, and T. Hyypiä. 2000. Human parechoviruses—biological and clinical significance. Rev. Med. Virol. 10:57-69. [DOI] [PubMed] [Google Scholar]
- 35.Stewart, S. R., and B. L. Semler. 1997. RNA determinants of picornavirus cap-independent translation initiation. Semin. Virol. 8:242-255. [Google Scholar]