Abstract
Hepatitis C virus (HCV) replication appears to be restricted to the human hepatoma cell line Huh-7, indicating that a favorable cellular environment exists within these cells. Although adaptive mutations in the HCV nonstructural proteins typically enhance the replicative capacity of subgenomic replicons in Huh-7 cells, replication can only be detected in a subpopulation of these cells. Here we show that self-replicating subgenomic RNA could be eliminated from Huh-7 clones by prolonged treatment with alpha interferon (IFN-α) and that a higher frequency of cured cells could support both subgenomic and full-length HCV replication. The increased permissiveness of one of the cured cell lines allowed us to readily detect HCV RNA and antigens early after RNA transfection, eliminating the need for selection of replication-positive cells. We also demonstrate that a single amino acid substitution in NS5A is sufficient for establishing HCV replication in a majority of cured cells and that the major phosphate acceptor site of subtype 1b NS5A is not essential for HCV replication.
An estimated 3% of the world's population is seropositive for hepatitis C virus (HCV) (32). The acute phase of infection is often subclinical; however, approximately 70% of seropositive individuals develop a chronic infection, predisposing the infected patient to the development of progressive liver pathology, including fibrosis, cirrhosis, and hepatocellular carcinoma (1, 28). The current treatments for HCV infection are alpha interferon (IFN-α) in combination with ribavirin or, more recently, a polyethylene glycol-modified form of IFN-α; however, sustained responses are only observed in ∼50% of treated patients, and effectiveness varies depending on the infecting HCV genotype (19).
HCV has been classified within its own genus, Hepacivirus, within the family Flaviviridae, which comprises three genera of small enveloped positive-strand RNA viruses (27). The 9.6-kb genome consists of a single open reading frame (ORF) flanked by 5′ and 3′ nontranslated regions (NTRs) (reviewed in references 3, 4, and 16). The 5′ NTR contains an internal ribosome entry site (IRES), mediating cap-independent translation of the ORF of ∼3,011 amino acids. The resulting polyprotein is processed co- and posttranslationally into at least 10 individual proteins. Host signal peptidase cleavages within the N-terminal portion of the polyprotein generate the structural proteins core (C), E1, and E2. Two HCV-encoded proteases mediate downstream cleavages, liberating the nonstructural (NS) proteins involved in viral replication. The NS2-3 protease spanning the C-terminal half of NS2 and the N-terminal one-third of NS3 catalyzes autocatalytic cleavage between NS2 and NS3. The N-terminal one-third of NS3 also encodes a serine protease that functions in concert with NS4A to cleave downstream sites, while the C-terminal two-thirds harbors RNA helicase and RNA-stimulated nucleoside triphosphatase activities. The NS5B protein exhibits an RNA-dependent RNA polymerase activity. Although the NS4B and NS5A proteins are membrane associated and form complexes with other HCV NS proteins (10), their biochemical functions remain speculative (29). NS5A is a serine phosphoprotein associated with one or more cellular kinases (23), and at least two distinct phosphorylated forms, p56 (basal phosphorylated form) and p58 (hyperphosphorylated form), have been described (11, 30). For the consensus HCV-H genotype 1a NS5A, Ser-2321 was identified as the major phosphate acceptor site (24), whereas the preferred site for p56 phosphorylation in the genotype 1b HCV-BK isolate was identified as Ser-2194 (12). For the hyperphosphorylated form of NS5A, deletion mapping and site-directed mutagenesis identified Ser-2197, Ser-2201, and Ser-2204 as putative phosphorylation sites (30). We recently demonstrated that Ser-2204 and apparent hyperphosphorylation were not essential for HCV replication in vitro (5); however, the importance and role of NS5A phosphorylation in replication are unknown.
Recently, Lohmann and colleagues reported that bicistronic subgenomic HCV replicons, containing the neomycin phosphotransferase gene (neo) in lieu of the HCV structural genes, were capable of autonomous replication in ∼1 in 106 Huh-7 cells (18). This development facilitated the identification of adaptive mutations in the HCV NS proteins that increased RNA replication as well as the frequency of Huh-7 cells supporting detectable levels of replication (5, 9, 15, 17). Previously, we identified a series of amino acid substitutions and a deletion of 47 amino acids in NS5A that enhanced the initiation of productive replication and G418-resistant colony formation (5). Replacement of the Ser residue with Ile at position 2204 in NS5A permitted HCV RNA replication in ∼10% of transfected Huh-7 cells (a 20,000-fold improvement) and increased replication to a level sufficient for the detection of HCV RNA early after transfection (5). Although the development of adapted subgenomic replicons provided a system to study HCV replication, we were only able to detect HCV replication in ∼10% of transfected Huh-7 cells, suggesting that the cellular environment was a major determinant of HCV replication efficiency.
In order to obtain cell lines more permissive for HCV replication, clonal and population Huh-7 cell lines supporting adapted and nonadapted subgenomic RNA replication were cured of HCV RNA by treatment with IFN-α. In general, a higher percentage of cured cells were able to support HCV replication and facilitated the transient detection of both subgenomic and full-length replication by multiple assays. We also examined the adaptive value of different amino acid substitutions at the NS5A 2204 locus and the effect of combining different adaptive mutations. Furthermore, we demonstrate that phosphorylation of the major phosphate acceptor site in subtype 1b NS5A is not an absolute requirement for replication in vitro. Our findings highlight the importance of the host environment for the establishment of HCV replication and provide a valuable cell line for studying early events in HCV replication.
MATERIALS AND METHODS
Cell culture and IFN treatment.
Huh-7 cell monolayers were propagated in Dulbecco's modified minimal essential medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 0.1 mM nonessential amino acids (DMEM-10% FBS). For cells supporting subgenomic replicons, G418 (Geneticin; Gibco-BRL) at 750 μg/ml was added to the culture medium. Replicon-containing Huh-7 cells were cured of HCV RNA by initially passaging cells twice in the absence of G418. On the third passage cells were cultured with human leukocyte-derived IFN-α (100 IU/ml; Sigma-Aldrich). After 3 to 4 days, confluent monolayers were trypsinized, plated and cultured for 24 h before the addition of IFN-α. Cells were passaged a total of four times in the presence of IFN-α, and prior to the fourth passage cells were grown for 3 days without IFN-α. Cured cell lines were expanded and cryopreserved at early passage levels. Experiments were conducted using cells that been passaged fewer than 20 to 30 times from these cryopreserved seed lots.
Plasmid constructions.
Standard recombinant DNA technology was used to construct and purify all plasmids. Primed DNA synthesis was performed with KlenTaqLA DNA polymerase (kindly provided by Wayne Barnes, Washington University, St. Louis, Mo.), and regions amplified by PCR were confirmed by automated nucleotide sequencing. Plasmid DNAs for in vitro transcription were prepared from large-scale bacterial cultures and purified by centrifugation in CsCl gradients.
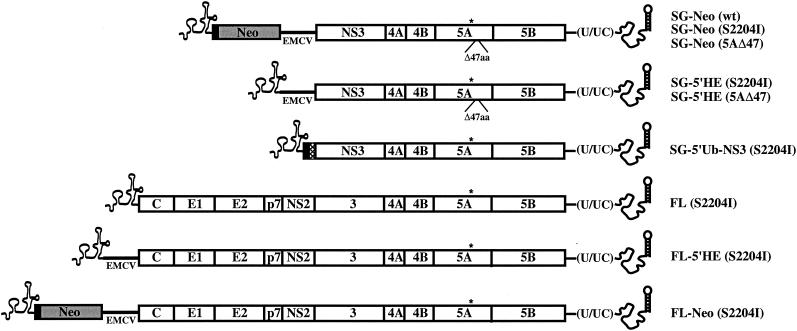
All nucleotide and amino acid numbers refer to the location within the genotype 1b Con1 full-length HCV genome (GenBank accession no. AJ238799) commencing with the core-coding region. This sequence was assembled from chemically synthesized DNA oligonucleotides in a stepwise PCR assay essentially as described previously (5). Briefly, 10 to 12 gel-purified oligonucleotides (60 to 80 nucleotides [nt] long) with unique complementary overlaps of 16 nt were used to synthesize cDNAs spanning 600 to 750 bases. The final PCR products were purified, digested with appropriate restriction enzymes, and ligated into the similarly cleaved pGEM3Zf(+) plasmid vector (Promega). Multiple recombinant clones were sequenced, correct clones were identified, and overlapping cDNA fragments were assembled into the following contiguous genomic sequence: 5′ NTR-C-E1-E2-p7-NS2-3-4A-4B-5A-5B-3′ NTR (pHCVBMFL). The selectable replicon pHCVrep1bBartMan/AvaII (SG-Neo [wild type, [wt]) (Fig. 1) and the derivatives pHCVrep1b/BBVII [SG-Neo (S2204I)] and pHCVrep1b/BBI [SG-Neo (5AΔ47)] containing the NS5A adaptive mutations, S2204I, and an in-frame deletion of 47 amino acids (Δ47aa) between nt 6960 and 7102, respectively have been described previously (5) (Fig. 1). The plasmid pHCVBMFL/S2204I [FL (S2204I)] (Fig. 1) contains the full-length genome with the NS5A adaptive change S2204I. For the genomic and subgenomic constructs, NS5B polymerase-defective derivatives carrying a triple amino acid substitution were generated, changing the Gly-Asp-Asp (GDD) motif in the active site to Ala-Ala-Gly (AAG) (5), and throughout this report these constructs are referred to as pol−.
FIG. 1.
Schematic representation of HCV RNAs used in this study. The 5′ and 3′ NTR structures are shown, and ORFs are depicted as open boxes with the polyprotein cleavage products indicated. The first 12 amino acids of the core-coding region (solid box), the neo gene (Neo; shaded box), the EMCV IRES (EMCV; solid line), and ubiquitin (cross-hatched box) are illustrated. Locations of the NS5A adaptive mutations S2204I (*) and Δ47aa are indicated.
The plasmid pC-Ubi-NS3/HCVrepBBVII [SG-5′Ub-NS3 (S2204I)] (Fig. 1) containing ubiquitin instead of the neo gene and encephalomyocarditis virus (EMCV) IRES was constructed as follows. An AscI-SacI-digested PCR fragment amplified from pHCVrep12/Neo (K. J. Blight et al., unpublished results) with primers 1289 and 1290 (Table 1) and the SacI-BsrGI portion of a second PCR product generated using the primer pair 1291-1292 (Table 1) with pHCVrep1b/BBVII were ligated between the XbaI and BsrGI sites of HCVrep1b/BBVII together with the XbaI-AscI fragment from HCVrep1b/BBVII. To delete the neo gene from pHCVrep1b/BBVII, synthetic overlapping oligonucleotides 1287 and 1288 (Table 1) were hybridized and extended to create the junction between the 5′ NTR and the EMCV IRES. This product was digested with ApaLI and AclI and inserted, together with XbaI-ApaLI and AclI-EcoRI fragments from pHCVrep1b/BBVII, into XbaI-EcoRI digested pHCVrep1b/BBVII. This construct was named p5′NTR-EMCV/HCVrepBBVII [SG-5′HE (S2204I)] (Fig. 1). To replace S2204I with NS5AΔ47, the EcoRI-XhoI fragment from pHCVrep1b/BBI was ligated into similarly cleaved p5′NTR-EMCV/HCVrepBBVII, generating p5′NTR-EMCV/HCVrepBBI [SG-5′HE (5AΔ47)] (Fig. 1).
TABLE 1.
Oligodeoxynucleotides used in this studya
| Name | Sequence |
|---|---|
| 885 | (−)CCCTCTAGAACGCCCCGAAACCTAGGGTGGCG |
| 1030 | (−)CCCTCTAGACTCGAGGGAATTTCCTGGAC |
| 1184 | (+)GACGGCTAAGCGTAGGCTGGCCAGGGGATCTCCCCCCTCCTTGGCCAGCTCATCAGCTGACCAGCTGTCTGCGCCTTCC |
| 1287 | (+)AGACCGTGCACCAGACCACAACGGTTTCCCTCTAGCGGGATCAATTCCG |
| 1288 | (−)CCAGTAACGTTAGGGGGGGGGGAGGGAGAGGGGCGGAATTGATCCCGCT |
| 1289 | (+)CCAAAGGGCGCGCCATGCAGATCTTCGTGAAGACC |
| 1290 | (−)AATAGGAGCTCCACCGCGGAGACGC |
| 1291 | (+)CGGTGGAGCTCCTATTACGGCCTACTCCCAAC |
| 1292 | (−)ATTGGTGTACATTTGGGTGATTGG |
| 1293 | (+)TCTGGAAGCTTCTTGAAGACA |
| 1294 | (−)GGCTTGACGTCCTGTGGGCGGCGGTTGGTGTTACGTTTGGTTTTTCTTTGAGGTTTAGGATTCGTGCTCATTATTATCGTGTTTTTCAAAGG |
| 1319 | (+)AGACGGCTAAGCGTAGGCTGGCCAGGGGATCTCCCCCCTCCTTGGCCAGCTCATCAGCTGTACAGCTGTCTGCGCCTTCC |
| 1320 | (+)AGACGGCTAAGCGTAGGCTGGCCAGGGGATCTCCCCCCTCCTTGGCCAGCTCATCAGCTGCCCAGCTGTCTGCGCCTTCC |
| 1322 | (+)AGACGGCTAAGCGTAGGCTGGCCAGGGGATCTCCCCCCTCCTTGGCCAGCTCATCAGCTTACCAGCTGTCTGCGCCTTCC |
| 1324 | (+)AGACGGCTAAGCGTAGGCTGGCCAGGGGATCTCCCCCCTCCTTGGCCAGCTCATCAGCTGAACAGCTGTCTGCGCCTTCC |
| 1325 | (+)AGACGGCTAAGCGTAGGCTGGCCAGGGGATCTCCCCCCTCCTTGGCCAGCTCATCAGCTACACAGCTGTCTGCGCCTTCC |
| 1326 | (+)AGACGGCTAAGCGTAGGCTGGCCAGGGGATCTCCCCCCTCCTTGACCAGCTCATCAGCTATCCAGCTGTCTGCGCCTTCG |
| 1327 | (+)AGACGGCTAAGCGTAGGCTGGCCAGGGGATCTCCCCCCCCCTTGACCAGCTCATCAGCTATCCAGCTGTCTGCGCCTTCC |
| 1356 | (−)CCGCTCTAGATACGTGATGGGGGCACCCGTGGTGATGGTCCTTACCCCGATTCTGATGTTAGGGTCGATAC |
| 1358 | (+)CCGATGTACACCAATGTGGACCAGGACCTCGTCGGCTGGCGAGCGCCCCCCGGGGCGCGTTCC |
| 1359 | (+)CCGCGTGCACCCGAGGGGTTGCGAAGGCGGTGGACTTTGTACCCGTCGAGTCTATGGGAACCACTATGCGGTCCCCGGTC |
| 5′Ala | (+)CCACGCTAAGCGTAGGCTGGCCAGGGGAGCACCCCCCTCCTTGGCCAGCTC |
| 5′Asp | (+)CCACGCTAAGCGTAGGCTGGCCAGGGGAGATCCCCCCTCCTTGGCCAGCTC |
Nucleotide changes are highlighted in boldface type, and the resultant codon is underlined. Restriction sites used for cDNA cloning are underlined. The polarities of oligonucleotides are indicated as either the HCV genome RNA sense (+) or its complement (−).
The plasmid p5′NTR-EMCV/HCVFLBM(S2204I) [FL-5′HE (S2204I)] (Fig. 1) was created by ligating the XbaI-HindIII fragment from p5′NTR-EMCV/HCVrepBBVII, the HindIII-AatII fragment of a PCR product amplified from p5′NTR-EMCV/HCVrepBBVII using primers 1293 and 1294, and the AatII-NotI fragment from pHCVBMFL/S2204I into pHCVBMFL/S2204I previously digested with XbaI and NotI. The selectable bicistronic full-length HCV clone pHCVBMFL(S2204I)/Neo [FL-Neo (S2204I)] (Fig. 1) was assembled by ligating the XbaI-HindIII fragment from pHCVrep1b/BBVII and the HindIII-EcoRI fragment from p5′NTR-EMCV/HCVBMFL(S2204I) between the XbaI and EcoRI sites of pHCVrep1b/BBVII.
To obtain plasmids with mutations at position 2204, and to introduce single A2199T or double S2197P + A2199T mutations into p5′NTR-EMCV/HCVrepBBVII, PCRs were first performed using p5′NTR-EMCV/HCVrepBBVII as a template with the reverse primer 1030 and one of the following mutant forward primers: 1319 (S2204V), 1320 (S2204A), 1322 (S2204Y), 1324 (S2204E), 1325 (S2204T), 1184 (S2204D), 1326 (A2199T + S2204I), or 1327 (S2197P + A2199T S2204I) (Table 1). PCR-amplified products were digested with BlpI and XhoI and cloned into these sites in p5′NTR-EMCV/HCVrepBBVII. S2204 was engineered by insertion of the EcoRI-XhoI fragment from pHCVrep1bBartMan/AvaII into similarly cleaved p5′NTR-EMCV/HCVrepBBVII.
To engineer the mutation Q1112R into p5′NTR-EMCV/HCVrepBBVII in order to create p5′NTR-EMCV/HCVrepCloneA (Q1112R + S2204I), nt 3640 to 3991 of NS3 were PCR amplified from p5′NTR-EMCV/HCVrepBBVII using mutant primer 1358 and oligonucleotide 885 (Table 1). The resulting product was digested with BsrGI and EagI and combined in a ligation reaction mixture with the EagI-EcoRI and BsrGI-EcoRI fragments from p5′NTR-EMCV/HCVrepBBVII. The double mutation (E1202G + T1280I) in NS3 was created via a multistep cloning procedure. First, a PCR fragment amplified from p5′NTR-EMCV/HCVrepBBVII with forward primer 1359 and reverse primer 1356 (Table 1) was digested with ApaLI and XbaI and cloned into EcoRI-XbaI-digested pGEM3Zf(+) together with the EcoRI-ApaLI fragment from pGEM3Zf(+)/HCV1bnt1796-2524, which contains nt 3420 to 4124 in NS3 (K. J. Blight and C. M. Rice, unpublished results), generating the intermediate plasmid pGEM3Zf(+)/HCV1bnt1796-2524NS3*. Second, in a four-part cloning strategy, the BsrGI-BsaAI fragment, excised from pGEM3Zf(+)/HCV1bnt1796-2524NS3*, was inserted, together with fragments BsaAI-BssHII and BssHII-EcoRI from p5′NTR-EMCV/HCVrepBBVII, into p5′NTR-EMCV/HCVrepBBVII cleaved with BsrGI and EcoRI. The resultant plasmid was named p5′NTR-EMCV/HCVrepBBVII+NS3* (E1202G + T1280I + S2204I).
The mutations S2194A and S2194D were introduced by using primer pairs 5′Ala-1030 and 5′Asp-1030 (Table 1), respectively, to PCR amplify nt 6897 to 7186 in NS5A from pHCVrep1b/BBVII. These mutations were incorporated into pHCVrep1b/BBVII by replacing the BlpI-XhoI portion with the corresponding BlpI-XhoI-digested PCR product.
RNA transcription.
Plasmid DNAs containing full-length and subgenomic HCV sequences were linearized with ScaI and a poliovirus subgenomic replicon digested with BamHI. The linearized DNAs were phenol-chloroform (1:1) extracted and precipitated with ethanol. Pelleted DNAs were washed in 80% ethanol and resuspended in 10 mM Tris-HCl (pH 8.0)-1 mM EDTA (pH 8.0). RNA transcripts were synthesized at 37°C for 90 min in a 100-μl reaction mixture containing 40 mM Tris-HCl (pH 7.9), 10 mM NaCl, 12 mM MgCl2, 2 mM spermidine, 10 mM dithiothreitol (DTT), a 3 mM concentration of each nucleoside triphosphate, 0.025 U of inorganic pyrophosphatase (Roche Applied Science), 100 U of RNasin (Promega), 100 U of T7 RNA polymerase (Epicentre Technologies), and 2 μg of linearized DNA. RNA was extracted with phenol-chloroform (1:1) and ethanol precipitated, and the pellet was washed in 80% ethanol before resuspension in double-distilled H2O. DNA template was removed by three serial DNase digestions for 20 min at 37°C in a solution of 33 mM Tris-HCl (pH 7.8), 66 mM KCl, 10 mM MgCl2, and 5 mM dithiothreitol containing 10 U of DNase I (Roche Applied Science). DNase-digested RNAs were extracted with phenol-chloroform (1:1) and ethanol precipitated, and the RNA pellet was resuspended in double-distilled H2O after washing in 80% ethanol. The RNA concentration was determined by measurement of the optical density at 260 nm, and the integrity and concentration were confirmed by 1% agarose gel electrophoresis and ethidium bromide staining.
Transfection of cultured cells.
In vitro-transcribed RNA was transfected into Huh-7 and IFN-α-cured cells by electroporation. Briefly, subconfluent Huh-7 cells were detached by trypsin treatment, collected by centrifugation (500 × g, 5 min), washed three times in ice-cold RNase-free phosphate-buffered saline (PBS), and resuspended at 1.25 × 107 cells/ml in PBS. RNA transcripts (1 μg) were mixed with 0.4 ml of washed Huh-7 cells in a 2-mm gap cuvette (BTX) and immediately pulsed (0.92 kV; pulse-length, 99 μs; five pulses) using a BTX ElectroSquarePorator. Pulsed cells were left to recover for 10 min at room temperature and then diluted into 10 ml DMEM-10% FBS. Cells were plated in (i) 35-mm-diameter wells for quantifying HCV RNA and for metabolic labeling experiments, (ii) eight-well chamber slides (Becton Dickinson) for immunofluorescence studies, or (iii) 100-mm-diameter dishes for fluorescence-activated cell sorting (FACS) analysis and G418 selection. To determine the efficiency of G418-resistant colony formation, transfected cells were plated at multiple densities (between 1 × 103 and 2 × 105 cells), together with cells transfected with pol− RNA transcripts such that the total cell number was maintained at 2 × 105 cells per 100-mm-diameter dish. Forty-eight hours after plating, medium was replaced with DMEM-10% FBS supplemented with G418 at 1 mg/ml. Three weeks later, G418-resistant foci were fixed with 7% formaldehyde and stained with 1% crystal violet in 50% ethanol to facilitate colony counting. The G418 transduction efficiency was calculated based on the number of G418-selected colonies relative to the number of Huh-7 cells plated after electroporation.
Transfection efficiency was monitored for each series of RNAs by electroporating in parallel a poliovirus subgenomic replicon expressing green fluorescent protein (GFP) (A. A. Kolykhalov and C. M. Rice, unpublished results). Transfected cells were observed for poliovirus replicon-induced cytopathic effect, and GFP expression was visualized using a fluorescent inverted microscope at 12 to 16 h posttransfection. After 24 h, the surviving attached cells (presumably not transfected with the poliovirus replicon) were trypsinized and mixed with trypan blue and viable cells were counted to determine the percentage of cells electroporated.
Viral RNA analysis.
Total cellular RNA was isolated using TRIZOL reagent (Gibco-BRL) according to the manufacturer's protocol. One-tenth of each RNA sample was used to quantify HCV-specific RNA levels using an ABI PRISM 7700 sequence detector (Applied Biosystems). Real-time reverse transcription (RT)-PCR amplifications were performed using the TaqMan EZ RT-PCR core reagents (Applied Biosystems) and primers specific for the HCV 5′ NTR: 5′-CCTCTAGAGCCATAGTGGTCT-3′ (sense, 50 μM), 5′CCAAATCTCCAGGCATTGAGC-3′ (antisense, 50 μM), and FAM-CACCGGAATTGCCAGGACGACCGG (probe, 10 μM; Applied Biosystems). RT reactions were incubated for 30 min at 60°C, followed by inactivation of the reverse transcriptase coupled with activation of Taq polymerase for 7 min at 95°C. Forty cycles of PCR were performed with cycling conditions of 15 s at 95°C and 1 min at 60°C. Synthetic HCV RNA standards of known concentration were included with each set of reactions and used to calculate a standard curve. The real time PCR signals were analyzed using SDS software (version 1.6.3; Applied Biosystems).
FACS analysis.
Transfected cell monolayers were removed from 100-mm-diameter culture dishes by Versene-EDTA treatment and a single-cell suspension prepared by passing cells through a 16-gauge needle and a 74-μm-pore-size membrane. Cells were resuspended at 2 × 106 per ml, and an equal volume of 4% paraformaldehyde added to the cell suspensions and incubated for 20 min at room temperature. Fixed cells were washed twice with PBS, and the resultant cell pellet was resuspended at 2 × 106 cells per ml in 0.1% saponin-PBS. After incubation for 20 min at room temperature, cells were stained (1 h at room temperature) with HCV-specific monoclonal antibodies (MAbs) (core [C750], NS3 [1B6], and NS5B [12B7] [all generously provided by Darius Moradpour, University of Freiburg, Freiburg, Germany]) diluted to 10 μg/ml in 3% FBS-0.1% saponin-PBS. Cells were washed three times with 0.1% saponin-PBS, and bound MAb was detected by incubation for 1 h at room temperature with anti-mouse immunoglobulin G (IgG) conjugated to Alexa 488 (Molecular Probes) diluted 1:1,000 in 3% FBS-0.1% saponin-PBS. Stained cells were washed three times with 0.1% saponin-PBS, resuspended in FACSflow buffer (BD Biosciences), and analyzed immediately using a FACS Calibur apparatus (BD Biosciences).
Indirect immunofluorescence.
Electroporated Huh-7.5 cells seeded in eight-well chamber slides were washed with PBS and fixed in 4% paraformaldehyde for 20 min at room temperature. Cells were washed twice with PBS, permeabilized by incubation with 0.1% saponin-PBS for 20 min at room temperature, and blocked with 3% goat serum for 20 min at room temperature. The NS5B MAb (12B7) was diluted to 10 μg/ml in 0.1% saponin-3% goat serum-PBS and incubated for 1 h at room temperature, followed by three washes with 0.1% saponin-PBS. Bound MAbs were detected by incubating for 1 h at room temperature with anti-mouse IgG conjugated to Alexa 488 diluted 1:1,000 in 0.1% saponin-3% goat serum-PBS. Nuclei were stained for 20 min at room temperature with Hoechst 33342 (10 μg/ml; Sigma-Aldrich) in PBS. Unbound fluorescent conjugate was removed by three washes with 0.1% saponin-PBS, and cells were mounted in Vectashield (Vector Laboratories) and viewed with a fluorescence microscope (Eclipse TE300; Nikon).
Metabolic labeling of proteins and immunoprecipitation.
Cell monolayers in 35-mm-diameter wells were incubated for 0.5 to 10 h in methionine- and cysteine-deficient minimal essential medium containing 1/40 the normal concentration of methionine, 5% dialyzed FBS, and Express 35S-protein labeling mix (140 μCi/ml; NEN). Labeled cells were washed once with cold PBS and harvested in 200 μl of sodium dodecyl sulfate (SDS) lysis buffer (0.1 M sodium phosphate buffer [pH 7.0], 1% SDS, 1× complete protease inhibitor cocktail [Roche Applied Science], 80 μg of phenylmethylsulfonyl fluoride [PMSF] per ml), and cellular DNA was sheared by repeated passage through a 27-gauge needle. Equal amounts of protein lysates (50 μl) were heated at 75°C for 10 min and clarified by centrifugation prior to mixing with 200 μl of TNA (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.67% bovine serum albumin, 1 mM EDTA, 0.33% Triton X-100, 80 μg of PMSF per ml). One-μl of HCV-positive serum (8) was added, and immune complexes allowed to form by incubation overnight at 4°C with rocking. Immune complexes were collected by adding 50 μl of prewashed Pansorbin cells (Calbiochem) and incubating for 1 to 2 h at 4°C with rocking. Immunoprecipitates were collected by centrifugation and washed three times in TNAS (TNA containing 0.125% SDS) and once with TNE (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 80 μg of PMSF per ml), solubilized by heating at 80°C for 20 min in protein sample buffer, and separated on an SDS-10% polyacrylamide gel. Metabolically labeled proteins were visualized by fluorography.
RESULTS
Cell lines highly permissive for HCV replication.
To date, productive HCV subgenomic RNA replication is restricted to the Huh-7 human hepatoma cell line, indicating that a favorable cellular environment exists within these cells. Although adaptive mutations in the HCV NS proteins are required to establish HCV replication in a higher frequency of transfected Huh-7 cells (5, 9, 15, 17), we identified several Huh-7 clones harboring subgenomic replicons with no consensus amino acid changes in the HCV polyprotein, as determined by sequencing the population of RT-PCR amplified HCV RNAs (5). We reasoned that these cells might provide a more permissive environment for HCV replication and a system to study HCV replication in the absence of adaptive changes.
Since HCV replication can be readily blocked by IFN-α (5, 6, 9), several Huh-7 lines harboring subgenomic HCV replicons were cured of HCV RNA by prolonged treatment with IFN-α (see Materials and Methods). From 22 G418-resistant clones (5), we cured clones Huh-7.5 and Huh-7.8, harboring SG-Neo subgenomic replicons with no amino acid changes within the HCV NS region, as well as clone Huh-7.4, containing a replicon with the Ser-to-Ile change at position 2204 in NS5A. Uncloned population lines Huh-7/S2204I and Huh-7/5AΔ47 (5), selected with G418 after transfection of subgenomic replicons containing either S2204I in NS5A [SG-Neo (S2204I)] (Fig. 1) or the 47-amino-acid NS5A deletion [SG-Neo (5AΔ47)] (Fig. 1), were also treated with IFN-α. To exclude the possibility that IFN-α treatment alone may alter the ability of Huh-7 cells to support HCV replication, the parental Huh-7 cells were treated with IFN-α in parallel. Following IFN-α treatment, cells were shown to lack HCV RNA by a nested RT-PCR specific for the 3′ NTR in which the detection limit was ∼10 molecules of HCV RNA (14) and by sensitivity to G418.
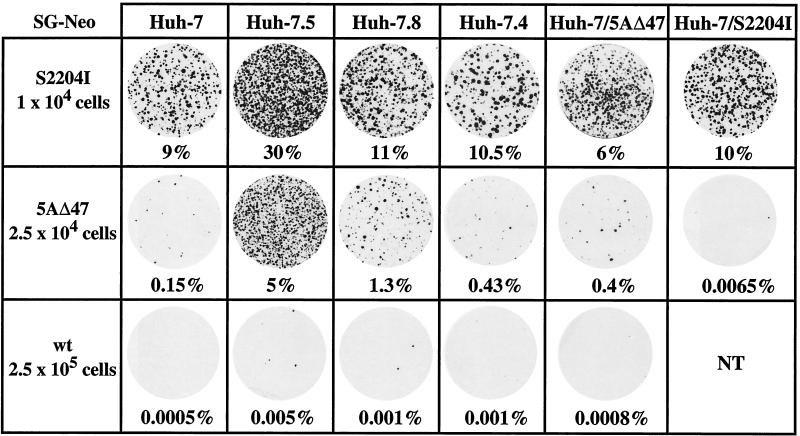
To examine the ability of IFN-α-cured cell lines to support HCV replication, three G418-selectable replicons, SG-Neo (S2204I), SG-Neo (5AΔ47), and SG-Neo (wt) (Fig. 1), with G418 transduction efficiencies in parental Huh-7 cells of 10, 0.2, and 0.0005%, respectively, were used (5). In vitro-synthesized RNA was electroporated into IFN-α-cured cells; after 48 h, G418 selection was imposed, and the resulting colonies were counted after fixing and staining. The transduction efficiencies were calculated on the basis of the number of G418-selected colonies relative to the number of Huh-7 cells plated after electroporation. The frequency of Huh-7.5 cells able to support SG-Neo (S2204I) replication was approximately threefold higher than that of the parental Huh-7 cells (Fig. 2). For cell lines Huh-7.5 and Huh-7.8, the number of G418-resistant colonies obtained after transfection of SG-Neo (5AΔ47) was significantly higher than that for the parental Huh-7 cells (∼33- and 9-fold increases, respectively; Fig. 2). The same was true for Huh-7.4, although the increased frequency of colony formation was not as great (∼3-fold; Fig. 2). The SG-Neo (wt) replicon also showed an enhanced replicative capacity in Huh-7.5, Huh-7.8, and Huh-7.4 cells (10-, 2-, and 2-fold increases, respectively; Fig. 2).
FIG. 2.
Identification of Huh-7 lines highly permissive for HCV replication. Huh-7 cells that had been cured of self-replicating subgenomic RNAs by extended IFN-α treatment were electroporated with 1 μg of the subgenomic replicons SG-Neo (S2204I), SG-Neo (5AΔ47), and SG-Neo (wt). Forty-eight hours later, cells were subjected to G418 selection, and the resulting colonies were fixed and stained with crystal violet. Representative plates are illustrated, with the number of transfected cells seeded per 100-mm-diameter dish shown on the left. Percentages below each dish refer to the calculated G418 transduction efficiency of the replicon. To determine the G418 transduction efficiency, transfected cells were serially titrated from 5 × 105 to 103 cells per 100-mm-diameter dish, together with feeder cells electroporated with the pol− replicon. The resulting G418-resistant foci were counted for at least three cell densities, and the relative G418 transduction efficiency was expressed as a percentage, after dividing the number of colonies by the number of electroporated cells initially plated. Similar transduction efficiencies were obtained in two independent transfections. A poliovirus subgenomic replicon expressing GFP (see Materials and Methods) was electroporated in parallel. Based on both the fraction of GFP-positive cells and replicon-induced cytopathogenicity, ∼90% of cells were routinely transfected. NT, not tested
The two cured cell populations, Huh-7/5AΔ47 and Huh-7/S2204I, showed either comparable or modest increases in G418 transduction efficiencies after transfection of the adapted replicon RNA originally present within the population line (Fig. 2). The frequency of G418-resistant colonies increased ∼2.5-fold when Huh-7/5AΔ47 cells were electroporated with SG-Neo (5AΔ47), whereas transfection with SG-Neo (S2204I) resulted in a slight decrease in the G418 transduction efficiency (Fig. 2). However, a 23-fold reduction in colony formation was observed after transfection of Huh-7/S2204I cells with SG-Neo (5AΔ47) (Fig. 2). No significant differences in G418-resistant colony formation were noted between the parental Huh-7 cells and IFN-α-treated Huh-7 cells (data not shown), indicating that the IFN-α-mediated curing protocol did not stably influence the ability of these cells to support HCV replication. G418-resistant colonies were not observed when the polymerase-defective replicon RNA, pol−, was transfected in parallel (data not shown). Hence, a higher frequency of cells in the cured clonal lines, in particular those originally able to support replication of RNAs without adaptive mutations (Huh-7.5 and Huh-7.8), are permissive for HCV replication.
HCV replication in unselected Huh-7.5 and Huh-7 cells.
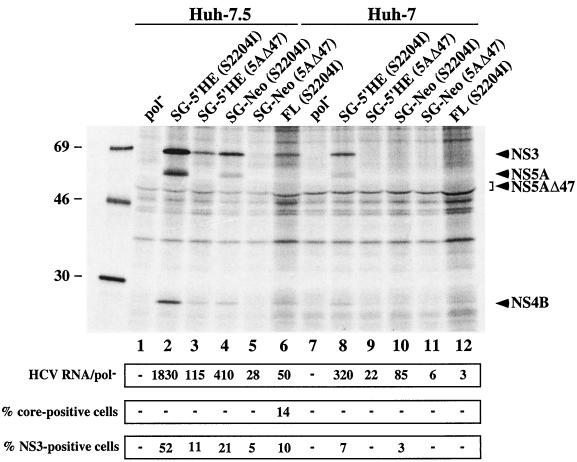
Since the cured Huh-7.5 line was the most permissive of those tested, we examined HCV replication in this subline compared with the parental Huh-7 cells using a number of different methods. We focused on transient assays that would allow an assessment of HCV replication early after transfection without the need for G418 selection. Ninety-six hours after transfection with SG-Neo (S2204I) and SG-Neo (5AΔ47) RNA (Fig. 1), total RNA was extracted from Huh-7.5 and IFN-α-treated Huh-7 cells, and the HCV RNA levels were quantified by RT-PCR. The replication-defective replicon, pol−, was transfected in parallel to allow discrimination between input RNA and RNA generated by productive replication. As shown in Fig. 3, the levels of HCV RNA relative to the pol− control were consistently higher in the transfected Huh-7.5 cells. Transfection with SG-Neo (S2204I) and SG-Neo (5AΔ47) RNAs resulted in 410- and 28-fold increases, respectively, in Huh-7.5 cells (Fig. 3, lanes 4 and 5), compared to only 85- and 6-fold increases in Huh-7 cells (Fig. 3, lanes 10 and 11). Since these increases are measured relative to the replication-defective pol− control, they reflect accumulation of newly synthesized RNA versus degradation of input RNA. In Huh-7.5 and Huh-7 cells, the level of residual pol− RNA declined by about 10-fold at each time point. In Huh-7.5 cells, RNAs with good replicative abilities [like SG-Neo (S2204I)] tended to accumulate over time such that a 10-fold increase was observed by 96 h. Those with lower replicative ability [like SG-Neo (5AΔ47)] remained constant or declined slightly, but never to the level of the pol− control. For Huh-7 cells, the picture was somewhat different. For example, SG-Neo (S2204I) RNA remained relatively constant, whereas SG-Neo (5AΔ47) RNA decreased over time, but again, not to the extent of the pol− control.
FIG. 3.
Detection of HCV proteins and RNA in Huh-7.5 and Huh-7 cells transiently transfected with HCV RNA. Top panel, Huh-7.5 and Huh-7 cells were transfected with the subgenomic replicons pol− (lanes 1 and 7), SG-5′HE (S2204I) (lanes 2 and 8), SG-5′HE (5AΔ47) (lanes 3 and 9), SG-Neo (S2204I) (lanes 4 and 10), SG-Neo (5AΔ47) (lanes 5 and 11), and FL (S2204I) HCV RNA (lanes 6 and 12). At 96 h posttransfection, monolayers were incubated for 10 h in the presence of [35S]methionine and [35S]cysteine. Labeled cells were lysed and immunoprecipitated with HCV-positive human serum (anti-NS3, NS4B, and NS5A), and labeled proteins were separated by SDS-10% PAGE. Note that twice the amount of immunoprecipitated sample was loaded in lanes 6 and 12. The mobilities of molecular weight standards (in thousands) are indicated on the left, and the migration of NS3, NS4B, NS5A, and 5AΔ47 is shown on the right. For data in the panel directly below the gel, total cellular RNA was extracted at 96 h posttransfection and quantified for HCV RNA levels as described in the Materials and Methods. The ratio of HCV RNA relative to the pol− defective replicon is shown (HCV RNA/pol−). HCV RNA levels relative to the pol− control were comparable in three independent experiments. For data in the bottom two panels, 96 h after transfection cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% saponin, stained for either HCV core or NS3 antigens, and analyzed by FACS. The percentage of cells expressing core and NS3 relative to an isotype matched irrelevant IgG is displayed. Values <1.5% were considered negative (−).
This finding was mirrored by the frequency of NS3-positive cells measured by FACS analysis. The percentage of NS3-positive cells was consistently higher in Huh-7.5 cells [21% for SG-Neo (S2204I) and 5% for SG-Neo (5AΔ47)] (Fig. 3, lanes 4 and 5) compared to Huh-7 cells [3% for SG-Neo (S2204I) and undetectable for SG-Neo (5AΔ47) and pol− RNAs] (Fig. 3, lanes 7, 10, and 11). These results confirm our earlier conclusion that a larger fraction of Huh-7.5 cells support detectable levels of HCV replication. The lower frequency of HCV antigen-positive cells quantified by FACS compared to the G418 transduction efficiency is attributable to the sensitivity of FACS analysis, which varies with different HCV-specific antibodies (unpublished observations).
We also examined HCV protein accumulation by metabolically labeling cells 96 h after transfection. Cell monolayers were labeled with 35S-labeled methionine and cysteine for 10 h, followed by SDS-mediated lysis and immunoprecipitation of HCV proteins with an HCV-positive patient serum recognizing NS3, NS4B, and NS5A (8). After separation of labeled proteins by SDS-polyacrylamide gel electrophoresis (PAGE), NS3, NS4B, and NS5A were only visible in Huh-7.5 cells transfected with SG-Neo (S2204I) (Fig. 3, lane 4). HCV proteins were never detected in Huh-7.5 and Huh-7 cells transfected with SG-Neo (5AΔ47), pol−, or SG-Neo (S2204I) RNA-electroporated Huh-7 cells (Fig. 3, lanes 1, 5, 7, 10, and 11). Similar results were obtained after metabolic labeling of HCV RNA in the presence of actinomycin D (data not shown). Taken together, these analyses demonstrate the advantages of using Huh-7.5 cells for rapid analysis of HCV replication by RNA accumulation, FACS analysis, and metabolic labeling of viral proteins.
Replicative efficiencies of subgenomic and genomic HCV RNAs.
The ability to monitor HCV replication without selection eliminated the need for bicistronic replicons and allowed constructs with minimal heterologous elements to be tested. We engineered a subgenomic replicon in which the HCV 5′ NTR and 12 amino acids of core were fused to ubiquitin followed by the NS3-5B coding region (including the S2204I adaptive mutation in NS5A) and the 3′ NTR [SG-5′Ub-NS3 (S2204I)] (Fig. 1). In this polyprotein, cellular ubiquitin carboxyl-terminal hydrolase cleaves at the ubiquitin/NS3 junction to produce NS3 with an authentic N-terminal Ala residue (2, 21). In vitro-synthesized RNA was electroporated into Huh-7.5 and Huh-7 cells, and the level of HCV RNA was quantified 96 h later by RT-PCR. To our surprise, HCV RNA levels did not differ from the pol− control (data not shown), indicating that SG-5′Ub-NS3 (S2204I) RNA failed to replicate. It is possible that ubiquitin may interfere with the production of a functional NS3 protein. However, a bicistronic derivative, where expression of ubiquitin/NS3-5B was under the control of the EMCV IRES, replicated as efficiently as SG-Neo (S2204I) RNA (data not shown), suggesting that HCV IRES-driven translation may be sensitive to RNA elements present within the core-ubiquitin coding sequence.
We also tested a replicon lacking neo but retaining the EMCV IRES (SG-5′HE derivatives; Fig. 1). SG-5′HE (S2204I) and SG-5′HE (5AΔ47) RNA levels relative to those in the pol− control were measured 96 h after transfection and found to be higher than the selectable versions in both Huh-7.5 and Huh-7 cells (Fig. 3). Moreover, the levels of SG-5′HE (S2204I) and SG-5′HE (5AΔ47) RNA were significantly higher in Huh-7.5 compared to Huh-7 cells (Fig. 3, lanes 2, 3, 8, and 9). Approximately 52% of Huh-7.5 cells stained positive for the NS3 antigen after transfection of SG-5′HE (S2204I) RNA, compared to 21% for SG-Neo (S2204I) RNA (Fig. 3, lanes 2 and 4). Similarly, a higher frequency of Huh-7.5 cells expressed NS3 after electroporation with SG-5′HE (5AΔ47) compared to SG-Neo (5AΔ47) (11 versus 5% [Fig. 3, lanes 3 and 5]). As expected, lower frequencies of NS3-positive cells were observed for transfected Huh-7 cells (Fig. 3). The relative amounts of immunoprecipitated 35S-labeled HCV proteins from Huh-7.5 and Huh-7 cells paralleled both the frequency of NS3-positive cells and relative HCV RNA levels (Fig. 3). Taken together, these data demonstrate that replicons lacking the neo gene initiate RNA replication more efficiently. These constructs, together with the highly permissive Huh-7.5 subline, are valuable tools for genetic studies on HCV RNA replication, some of which are described later in this report.
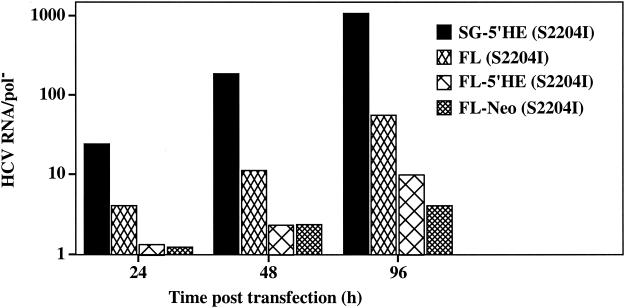
We next assessed the ability of Huh-7.5 cells to support replication of full-length HCV RNA containing S2204I in NS5A [FL (S2204I)] (Fig. 1). Ninety-six hours after transfection of Huh-7.5 and Huh-7 cells, the relative levels of HCV RNA and protein were measured as described above. A 50-fold increase in HCV RNA relative to that in pol− cells was observed after transfection of Huh-7.5 cells, compared to only a 3-fold increase in Huh-7 cells (Fig. 3, lanes 6 and 12). Similarly, FACS analysis and immunoprecipitation of metabolically labeled proteins failed to detect HCV antigen expression in FL (S2204I) RNA-transfected Huh-7 cells, whereas 14 and 10% of Huh-7.5 cells expressed core and NS3 antigens, respectively, and 35S-labeled NS3 was detectable (Fig. 3, lanes 6 and 12). The frequency of core antigen-positive cells was consistently higher than that seen for NS3, possibly reflecting differences in antibody affinity. The ability of full-length HCV RNA to establish replication in Huh-7.5 cells demonstrates that replication is not dependent upon EMCV IRES-driven translation of HCV-encoded replicase components. In fact, inclusion of the EMCV IRES downstream of the HCV 5′ NTR [FL-5′HE (S2204I)] (Fig. 1) or creation of a bicistronic construct with the neo gene added [FL-Neo (S2204I)] (Fig. 1) impaired replication relative to FL (S2204I) RNA (Fig. 4). It is interesting that all of the constructs containing the complete HCV coding sequence (S2204I-containing FL, FL-5′HE and FL-Neo) were less efficient at establishing replication than subgenomic replicons lacking the structural-NS2 coding region [e.g., SG-5′HE (S2204I)] (Fig. 4). This suggests that cis RNA elements or proteins encoded in this region of the genome may downregulate the efficiency of HCV replication in this system. Nonetheless, the ability of Huh-7.5 cells to support replication of both FL and FL-Neo RNAs provides systems that may be useful for studying steps in particle assembly and examining the impact of the entire HCV protein complement on host cell biology.
FIG. 4.
HCV RNA accumulation after transfection of Huh-7.5 cells with full-length HCV RNA. One microgram of in vitro-transcribed RNA was electroporated into Huh-7.5, and 2 × 105 cells were plated into 35-mm-diameter wells. Total cellular RNA was isolated at 24, 48, and 96 h posttransfection, and HCV RNA levels were quantified as described in the Materials and Methods. The ratio of HCV RNA relative to the pol− defective subgenomic RNA (HCV RNA/pol−) was plotted against the time posttransfection, and similar results were obtained when this experiment was repeated
Effect(s) of mutations in NS3 and NS5A on HCV RNA replication.
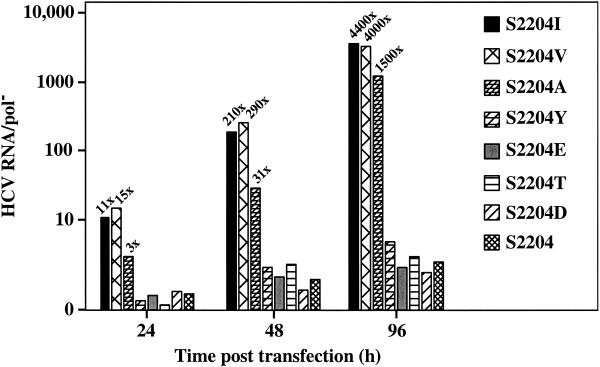
Besides the cellular environment, adaptive mutations selected in Huh-7 cells exhibit a wide spectrum of different efficiencies (5, 9, 15, 17). Thus far, the best single mutation we have identified was the S2204I substitution in NS5A (5). To examine the importance of Ile at this position and to see if replication efficiency could be improved further, we tested a number of other amino acids at this position and compared the replication efficiency of these replicons to SG-5′HE (S2204I) or the unmodified parent, SG-5′HE (S2204) (Fig. 5). Replicative ability was assessed in RNA-transfected Huh-7.5 cells by comparing the HCV RNA levels to that in a SG-pol− control. Comparable levels of HCV RNA were observed at 96 h for replicons containing Ile or Val at position 2204, whereas an Ala substitution resulted in a threefold reduction in HCV RNA compared to SG-5′HE (S2204I) (Fig. 5). In contrast, the remaining amino acid substitutions dramatically reduced HCV RNA to levels similar to the unmodified parental replicon, SG-5′HE (S2204) (∼1,400-fold decrease; Fig. 5). As expected, the relative HCV RNA levels were lower at 24 and 48 h after transfection; however, the levels were sufficient to assess replicative ability at 48 h (Fig. 5). Although substitutions that enhance subgenomic replication above that observed with SG-5′HE (S2204I) were not found, Val and Ala at position 2204 in NS5A allowed efficient RNA replication.
FIG. 5.
Effect(s) of alternative substitutions at position 2204 in NS5A on HCV RNA replication. Huh-7.5 cells were transfected with 1 μg of the SG-5′HE replicons carrying the indicated amino acid substitutions and 2 × 105 cells plated in 35-mm-diameter wells. After 24, 48, and 96 h in culture, total cellular RNA was extracted and HCV RNA levels were measured as described in Materials and Methods. The ratio of HCV RNA relative to the pol− defective subgenomic RNA (HCV RNA/pol−) was plotted against the time posttransfection. The increase in HCV RNA above pol− is indicated above each bar. In this figure the levels of HCV RNA relative to the pol− are the highest we have achieved so far. When these RNAs were transfected into Huh-7.5 cells a second time, a similar trend in HCV RNA accumulation was observed.
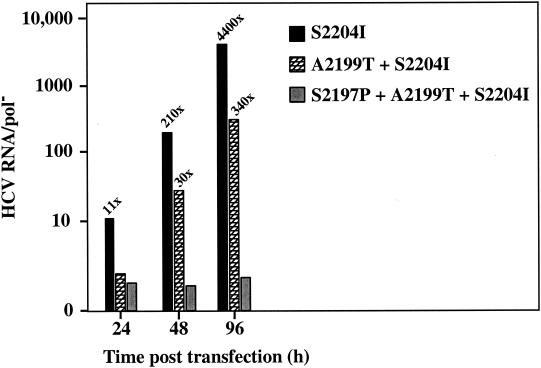
We investigated the replication efficiency of subgenomic replicons carrying multiple adaptive mutations in NS5A. NS5A mutations S2197P, A2199T, and S2204I independently enhance G418-resistant colony formation approximately 2,500-, 15,000-, and 20,000-fold, respectively (5). SG-5′HE replicons (Fig. 1) carrying S2204I together either with A2199T or with A2199T and S2197P were constructed, and HCV RNA levels in Huh-7.5 cells were measured by RT-PCR. Combining these NS5A mutations led to a reduction in HCV RNA levels compared to SG-5′HE (S2204I), with a 13-fold decrease for the combination of A2199T and S2204I and negligible replication when all three were combined (Fig. 6). Despite the observation that each of these NS5A adaptive mutations alone enhanced replication, when combined, the replicative ability of subgenomic RNAs declined, suggesting that these combinations are incompatible.
FIG. 6.
Effect(s) of combining NS5A adaptive mutations on HCV RNA replication. Subgenomic replicons (SG-5′HE) carrying the indicated mutations were transfected into Huh-7.5 cells and HCV RNA levels were quantitated as described in Fig. 5. The ratio of HCV RNA relative to the pol− defective subgenomic RNA (HCV RNA/pol−) was plotted against the time posttransfection, and the increase in HCV RNA above pol− is indicated above each bar. An additional transfection experiment yielded HCV RNA/pol− ratios similar to those illustrated here.
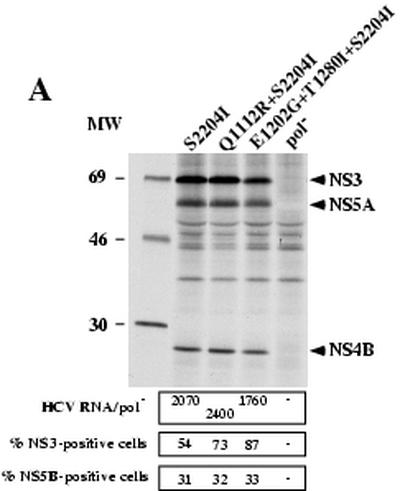
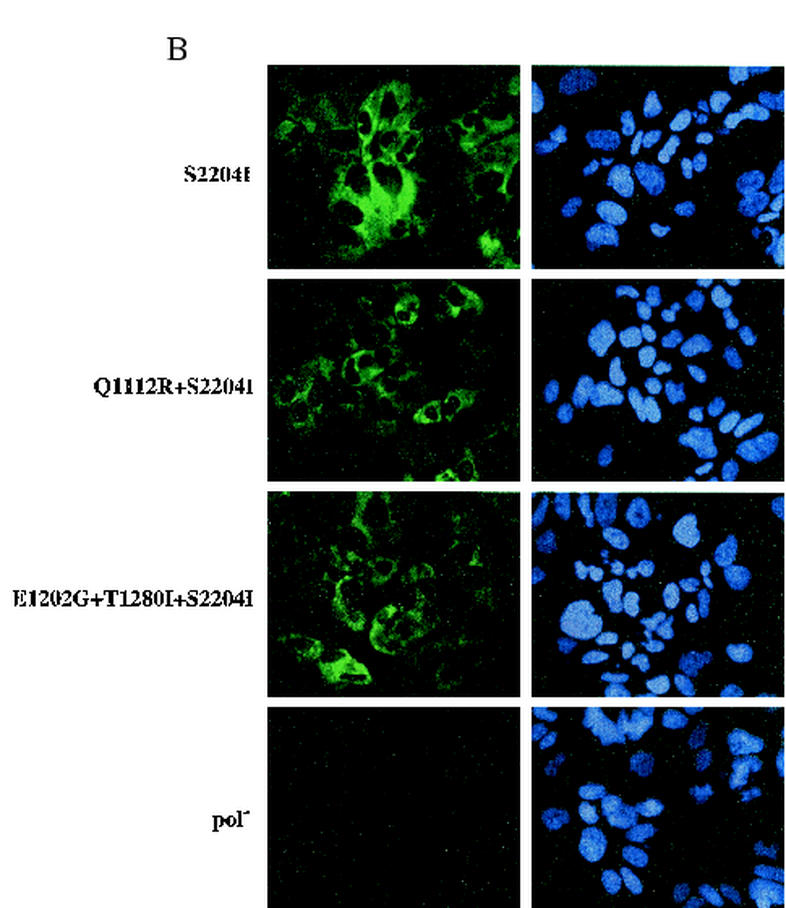
We previously isolated a G418-resistant cell clone harboring replicon RNA with the S2204I mutation in combination with a substitution in the protease domain of NS3 (Q1112R) (5). In light of recent observations by Krieger and colleagues (15) in which mutations in NS3 and NS5A were reported to enhance RNA replication in a synergistic manner, we examined the effect(s) of combining NS3 mutation(s) with S2204I. NS3 changes at positions 1112 (Q to R) (5), 1202 (E to G) (15), and 1280 (T to I) (15) were engineered into SG-5′HE (S2204I) (Fig. 1), and their replication in Huh-7.5 cells was determined by measuring HCV RNA levels, by measuring the frequency of antigen-positive cells, and by detection of 35S-labeled proteins at 96 h following transfection. Equivalent levels of HCV RNA relative to the pol− RNA control were observed for each replicon (Fig. 7A). The percentages of NS5B-positive cells detected by FACS (∼30%) (Fig. 7A) and immunofluorescence (Fig. 7B) were also similar. However, the frequency of NS3-positive cells was higher for replicons carrying the NS3 mutations (∼73 to 87%; Fig. 7A), which may simply reflect altered affinity of the NS3-specific antibody for these NS3 mutants. Finally, the levels of immunoprecipitated NS3, NS4B, and NS5A were comparable (Fig. 7A). Although we did not verify that the Q1112R change alone was adaptive, Krieger and coworkers previously reported that the E1202G and T1280I changes alone or together increased the replication efficiency by ∼13-, 6-, and 25-fold, respectively (15). Hence, these NS3 adaptive mutations do not further enhance replication when combined with the S2204I mutation in NS5A.
FIG. 7.
Effect(s) of combining NS3 and NS5A mutations on HCV RNA replication. Subgenomic replicons lacking neo (SG-5′HE) were generated carrying S2204I with further mutations in NS3. (A) For the gel shown at top, 96 h after RNA transfection of Huh-7.5 cells, monolayers were labeled with 35S-protein labeling mixture; cells were lysed; and NS3, NS4A, and NS5A were analyzed by immunoprecipitation, SDS-10% PAGE, and autoradiography. Positions of the molecular weight standards (in thousands) are given on the left, and HCV-specific proteins are indicated to the right. For data in the panel directly below the gel, total cellular RNA was extracted at 96 h posttransfection and HCV RNA levels were quantified as described in Materials and Methods. The ratio of HCV RNA relative to the pol− negative control is shown (HCV RNA/pol−). Comparable ratios were obtained in two independent experiments. For data in the bottom two panels, 96 h after transfection, cells were fixed with 4% paraformaldehyde, permeabilized with 0.1% saponin, stained for HCV NS3 and NS5B antigens, and analyzed by FACS. The percentage of cells expressing NS3 and NS5B relative to an isotype-matched irrelevant IgG is displayed. Values <1.5% were considered negative (−). (B) Transfected cells seeded in eight-well chamber slides were fixed, permeabilized, and stained for NS5B by immunofluorescence after 96 h in culture. Nuclei were counterstained with Hoechst 33342, and stained cells visualized by fluorescence microscopy (magnification, × 40).
Mutagenesis of the S2194 NS5A phosphorylation site.
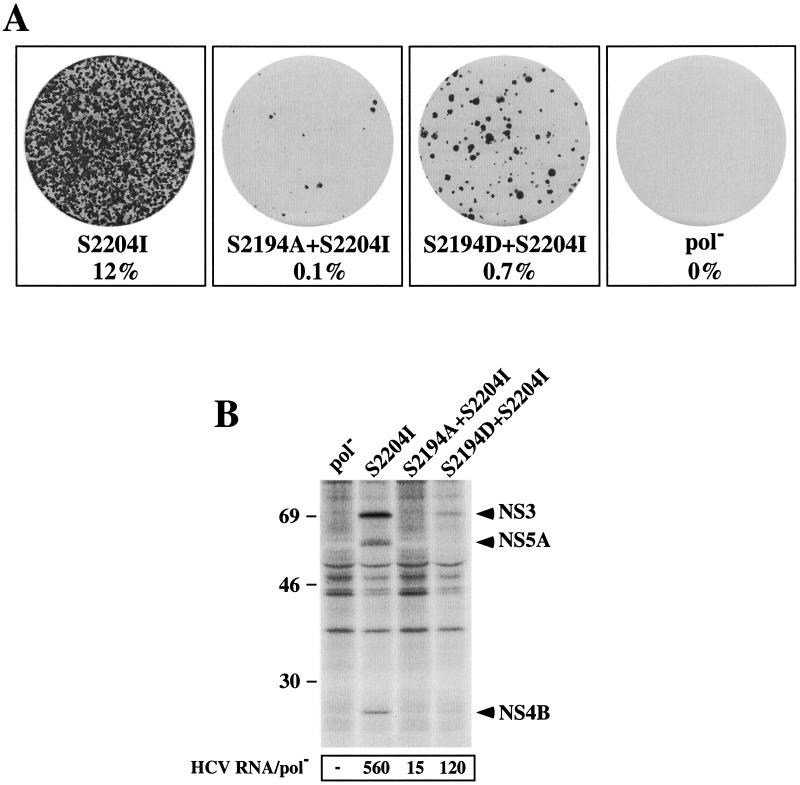
The role of NS5A phosphorylation in HCV replication remains a mystery. Previously we noted differences in the extent of NS5A phosphorylation between replicons with different adaptive mutations in NS5A (5). For example, replicons with S2197C, S2197P, or S2204I expressed minimal or no p58 as assessed by one-dimensional SDS-PAGE separation of immunoprecipitated NS5A, suggesting that NS5A hyperphosphorylation is not essential for HCV replication. Recently, S2194 in NS5A of a subtype 1b isolate was identified as the primary site of p56 phosphorylation (12). To assess the possible requirement for phosphorylation of NS5A S2194, this residue was mutated in SG-Neo (S2204I) (Fig. 1) to either Ala (S2194A + S2204I) or Asp (S2194D + S2204I), to eliminate or mimic phosphorylation, respectively. G418 transduction efficiencies of these replicons in Huh-7 cells were significantly lower than those of SG-Neo (S2204I) (120- and 17-fold lower [Fig. 8A ]). To rule out the possibility that G418-resistant foci were generated by reversion at this locus, the NS5A coding region was amplified from total cellular RNA by RT-PCR and directly sequenced. The original Ala and Asp substitutions at position 2194 were confirmed (data not shown). To minimize the impact of possible second-site compensating changes, we also measured HCV RNA and protein expression 96 h after RNA transfection of Huh-7.5 cells. The HCV RNA levels in replicons containing either S2194A with S2204I or S2194D with S2204I relative to those in the pol− control were ∼37- and 5-fold lower than those in SG-Neo (S2204I) (Fig. 8B), consistent with their reduced ability to render Huh-7 cells G418 resistant. In addition, a lower frequency of NS5B-positive cells was evident in S2194A + S2204I than in S2194D + S2204I mutants (data not shown), and 35S-labeled NS3 and NS4B were only detectable in Huh-7.5 cells transfected with SG-Neo (S2204I) and SG-Neo (S2194D + S2204I) (Fig. 8B). We were unable to directly study the phosphorylation status of NS5A expressed from cells with S2194A + S2204I and S2194D + S2204I changes since the levels of NS5A expressed in transiently transfected cells were below our detection limit (Fig. 8B and data not shown). Although the quantitative differences in G418 transduction and replication efficiencies are difficult to interpret given the possible incompatibility of combining the S2194 substitutions with the S2204I adaptive change, these data show that phosphorylation of S2194 is not an absolute requirement for HCV replication.
FIG. 8.
Effect(s) of S2194A and S2194D mutations on HCV RNA replication. S2194 was replaced with Ala or Asp in the selectable bicistronic replicon SG-Neo (S2204I), and RNA was transcribed in vitro. (A) RNA transcripts were transfected into Huh-7 cells, and G418-selected colonies were fixed and stained with crystal violet. The relative G418 transduction efficiencies are indicated below each dish. (B) Ninety-six hours posttransfection Huh-7.5 cells were labeled with [35S]methionine and [35S]cysteine for 10 h. Cells were lysed, and HCV proteins were isolated by immunoprecipitation using a patient serum specific for NS3, NS4B, and NS5A. HCV proteins and the positions of protein molecular weight standards (in thousands) are shown. The ratio of HCV RNA relative to the pol− negative control at 96 h posttransfection is shown below each track (HCV RNA/pol−). The results illustrated are representative of two independent transfections.
DISCUSSION
Several groups have demonstrated that single amino acid substitutions in the HCV replicase can dramatically increase the efficiency with which subgenomic replicons initiate replication and persist in Huh-7 cells (5, 9, 15, 17). In this study we show that Huh-7 sublines, in particular Huh-7.5 cells, possess a cellular environment that is more permissive for the initiation of HCV RNA replication. For the replicon containing the highly adaptive NS5A S2204I mutation, at least 30% of the Huh-7.5 cells can be transduced to G418 resistance. A comparable fraction was positive for the NS3 antigen by FACS. Similarly, for the SG replicons lacking neo (SG-5′HE in Fig. 1), at least 50% initiation efficiency was achieved. It is likely that these are low estimates, since G418 transduction efficiency is based on the number of cells used for electroporation (and not all cells survive). Indeed, more sensitive FACS analysis suggests that >75% of the cells that survive the transfection procedure harbor replicating HCV RNAs (unpublished results). While it is unclear at present how these cells will compare to the natural host cell for HCV infection, liver resident hepatocytes, they nonetheless provide a useful substrate for future genetic and biochemical studies of HCV.
These highly permissive cells were obtained by curing replicon-containing cell clones with IFN-α. While we cannot rule out the possibility that IFN-α-mediated changes persist in the cellular environment, allowing Huh-7.5 cells to be more permissive for HCV replication, the fact that IFN-α treatment of parental Huh-7 cells did not alter the ability of HCV RNA to replicate suggests otherwise. Interestingly, the most highly permissive sublines (Huh-7.5 and Huh-7.8) were obtained from G418-selected clones that harbored replicons without adaptive changes in the NS3-5B region (at least at the population sequence level). These cells may represent a subpopulation of the original Huh-7 parental line that are permissive for replication of unmodified replicons as well as more permissive for replicons with adaptive mutations. Interestingly, curing of other replicon-containing cell lines did not always yield a cell population that was more permissive for the replicons tested. For instance, curing the Huh-7 population containing the SG-Neo (S2204I) replicon, yielded a cell substrate that was unchanged in its ability to support SG-Neo (S2204I) but less efficient (23-fold) for initiation of SG-Neo (5AΔ47) (Fig. 2). These results suggest a complex interplay between the cellular environment and particular adaptive mutations to achieve productive replication.
The ability to study HCV replication directly after transfection, without the need for G418 selection, allowed us to examine replication of subgenomic replicons lacking neo as well as full-length HCV genome RNAs. Our initial attempt to create a monocistronic replicon by fusing cellular ubiquitin in-frame between the first 12 amino acids of core and NS3 [SG-5′Ub-NS3 (S2204I)] (Fig. 1) was unsuccessful. A bicistronic derivative with ubiquitin fused to NS3-5B was viable, suggesting that the failure of SG-5′Ub-NS3 (S2204I) to replicate was not due to a defect in processing at the ubiquitin/NS3 junction (data not shown). Rather, the fusion of the ubiquitin-coding sequence near the HCV 5′ NTR may have interfered with translation due to the formation of deleterious RNA secondary structures (20, 26) or RNA replication, by disrupting RNA elements that lie in the HCV 5′ NTR or its complement (7, 13). Fusion of the HCV 5′ NTR to the EMCV IRES yielded a subgenomic replicon (SG-5′HE) that replicated better than SG-Neo replicons (Fig. 3). Why deletion of the first cistron (sequences encoding C-Neo fusion) from the bicistronic SG-Neo (S2204I) stimulated replication is unknown, but the stimulation may result from enhanced translation of the replicase due to eliminated binding of the 40S ribosomal subunit to the usual HCV translation initiation site and diminished competition between the EMCV and HCV IRES elements (J. Marcotrigiano and C. M. Rice, unpublished results).
A similar picture was observed for replication of the full-length constructs containing the NS5A S2204I adaptive change (Fig. 4). In Huh-7.5 cells, the bicistronic construct containing the C-Neo cistron [FL-Neo (S2204I)] (Fig. 1) initiated replication less efficiently than the RNA with the HCV 5′ NTR fused to the EMCV IRES [FL-5′HE (S2204I)] (Fig. 1). Interestingly, the FL construct with the unmodified HCV genome (except for the S2204I substitution in NS5A) was better at initiating replication than RNA whose translation was mediated by the EMCV IRES (Fig. 4), demonstrating that EMCV IRES-driven translation is not required for HCV replication in Huh-7.5 cells, thus allowing the study of unmodified HCV genomic RNAs. This point could certainly impact the ability of HCV RNAs to be packaged into infectious particles. However, in our hands (Blight and Rice, unpublished results) and in a recent report (22) selective packaging of these unmodified FL RNAs was not observed in Huh-7 cells. Interestingly, full-length HCV RNAs were less efficient at establishing replication than the corresponding adapted subgenomic replicons, suggesting that addition of the structural-NS2 coding region inhibits HCV replication initiation. Whether this is due to the encoded proteins or RNA elements that lie in this region (or both) (31) is currently unclear.
In an attempt to further enhance HCV replication in cell culture, we also examined the effect of other amino acid substitutions at position 2204 in NS5A. Efficient subgenomic RNA replication was observed for Ile and Val and, to a much lesser extent, Ala at position 2204 (Fig. 5). Val or Ile are small, β-branched, nonpolar residues, whereas Ala has similar properties but is not β-branched. In contrast, polar residues such as Tyr, Glu, Thr, Ser, or Asp at position 2204 severely impaired HCV replication (Fig. 5), suggesting that replication favors nonpolar residues at this locus. It is interesting that Ser is found naturally at position 2204 for this genotype 1b isolate (18) and is conserved between other HCV genotypes (30), suggesting that this residue may be important for HCV replication and/or pathogenesis in vivo.
Combining NS5A adaptive mutations resulted in replicons that were either impaired (A2199T + S2204I) or unable to replicate (S2197P + A2199T + S2204I) in Huh-7.5 cells (Fig. 6). Incompatibility of adaptive mutations elsewhere in the HCV NS coding region has been previously described (17). For example, combining an adaptive mutation in NS5B (R2884G) with either NS4B (P1936S) or NS5A (E2163G) drastically reduced the efficiency of G418-resistant colony formation. On the other hand, combining certain NS3 and NS5A adaptive mutations can increase replication efficiency (15). However, despite the observation that mutations E1202G and T1280I in NS3 act synergistically with S2197P in NS5A to increase the replication efficiency (15; Blight and Rice, unpublished results), engineering these NS3 changes into SG-5′HE (S2204I) did not enhance replication in our system (Fig. 7). These results again underscore the empirical nature of optimizing adaptive mutations with different Huh-7 cellular environments.
The phosphorylation of NS5A is conserved among divergent HCV genotypes (25), suggesting that it plays an important role in the virus life cycle. We previously showed that NS5A hyperphosphorylation is not essential for HCV replication (5). Following the recent identification of S2194 as a major phosphate acceptor site for subtype 1b (12), we substituted Ala or Asp at this position and examined the effect on HCV replication in the context of the S2204I adaptive mutation. Given the incompatibilities observed when combining NS5A mutations, the absolute replication efficiencies of the different mutants could not be evaluated; however, replicating RNAs were recovered that harbored these substitutions at the 2194 locus. These results show that phosphorylation at S2194 is not an absolute requirement for replication of this subtype 1b isolate. While this observation deserves further study in the context of adaptive mutations outside of NS5A, it should be noted that NS5A phosphorylation is complex and only a few of the potential serine acceptor sites have been identified (12, 24). Hence, additional phosphorylation sites need to be defined and coupled with mutagenesis of these sites as well as a thorough mutational analysis of the NS5A protein.
In conclusion, we have isolated a Huh-7 subline (Huh-7.5) that is highly permissive for replication of subgenomic and full-length HCV RNAs. These cells provide a valuable substrate for future genetic studies on HCV proteins and RNA elements. Finally, Huh-7.5 and other cured Huh-7 cells that differ in their ability to support HCV replication may prove useful for defining cellular parameters that affect the efficiency of HCV RNA replication initiation by gene array and other approaches.
Acknowledgments
We are grateful to many colleagues for helpful discussions during the course of this work and to Peter Balfe and Tim Tellinghuisen for critical reading of the manuscript. We also thank Darius Moradpour for providing the HCV-specific MAbs.
This work was supported in part by grants from the Public Health Service (CA57973 and AI40034) and the Greenberg Medical Research Institute.
REFERENCES
- 1.Alter, H. J., and L. B. Seeff. 2000. Recovery, persistence and sequelae in hepatitis C virus infection: a perspective on the long-term outcome. Semin. Liver Dis. 20:17-25. [DOI] [PubMed] [Google Scholar]
- 2.Bachmair, A., D. Finley, and A. Varshavasky. 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234:179-186. [DOI] [PubMed] [Google Scholar]
- 3.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 4.Blight, K. J., A. Grakoui, H. L. Hanson, and C. M. Rice. 2002. The molecular biology of hepatitis C virus, p. 81-108. In J.-H. J. Ou (ed.), Hepatitis viruses. Kluwer Academic Publishers, Boston, Mass.
- 5.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 6.Frese, M., T. Pietschmann, D. Moradpour, O. Haller, and R. Bartenschlager. 2001. Interferon-α inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol. 82:723-733. [DOI] [PubMed] [Google Scholar]
- 7.Friebe, P., V. Lohmann, N. Krieger, and R. Bartenschlager. 2001. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 75:12047-12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grakoui, A., C. Wychowski, C. Lin, S. M. Feinstone, and C. M. Rice. 1993. Expression and identification of hepatitis C virus polyprotein cleavage products. J. Virol. 67:1385-1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hugle, T., F. Fehrmann, E. Bieck, M. Kohara, H.-G. Krausslich, C. M. Rice, H. E. Blum, and D. Moradpour. 2001. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology 284:70-81. [DOI] [PubMed] [Google Scholar]
- 11.Kaneko, T., Y. Tanji, S. Satoh, M. Hijikata, S. Asabe, K. Kimura, and K. Shimotohno. 1994. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem. Biophys. Res. Commun. 205:320-326. [DOI] [PubMed] [Google Scholar]
- 12.Katze, M. G., B. Kwieciszewski, D. R. Goodlett, C. M. Blakely, P. Neddermann, S. L. Tan, and R. Aebersold. 2000. Ser2194 is a highly conserved major phosphorylation site of the hepatitis C virus nonstructural protein NS5A. Virology 278:501-513. [DOI] [PubMed] [Google Scholar]
- 13.Kim, Y. K., C. S. Kim, S. H. Lee, and S. K. Jang. 2002. Domains I and II in the 5′ nontranslated region of the HCV genome are required for RNA replication. Biochem. Biophys. Res. Commun. 290:105-112. [DOI] [PubMed] [Google Scholar]
- 14.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krieger, N., V. Lohmann, and R. Bartenschlager. 2001. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J. Virol. 75:4614-4624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindenbach, B. D., and C. M. Rice. 2001. Flaviviridae: the viruses and their replication, p. 991-1041. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott-Raven Publishers, Philadelphia, Pa.
- 17.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohmann, V., F. Korner, J. O. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 19.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 20.Myers, T. M., V. G. Kolupaeva, E. Mendez, S. G. Baginski, I. Frolov, C. U. T. Hellen, and C. M. Rice. 2001. Efficient translation initiation is required for replication of bovine viral diarrhea virus subgenomic replicons. J. Virol. 75:4226-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pickart, C. M., and I. A. Rose. 1985. Ubiquitin carboxyl-terminal hydrolase acts on ubiquitin carboxyl-terminal amides. J. Biol. Chem. 260:7903-7910. [PubMed] [Google Scholar]
- 22.Pietschmann, T., V. Lohmann, A. Kaul, N. Krieger, G. Rinck, G. Rutter, D. Strand, and R. Bartenschlager. 2002. Persistent and transient replication of full-length hepatitis C virus genomes in cell culture. J. Virol. 76:4008-4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reed, K. E., A. E. Gorbalenya, and C. M. Rice. 1998. The NS5A/NS5 proteins of viruses from three genera of the family Flaviviridae are phosphorylated by associated serine/threonine kinases. J. Virol. 72:6199-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reed, K. E., and C. M. Rice. 1999. Identification of the major phosphorylation site of the hepatitis C virus H strain NS5A protein as serine 2321. J. Biol. Chem. 274:28011-28018. [DOI] [PubMed] [Google Scholar]
- 25.Reed, K. E., J. Xu, and C. M. Rice. 1997. Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: properties of the NS5A-associated kinase. J. Virol. 71:7187-7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rijnbrand, R., P. J. Bredenbeek, P. C. Haasnoot, J. S. Kieft, W. J. M. Spaan, and S. M. Lemon. 2001. The influence of downstream protein-coding sequence on internal ribosome entry on hepatitis C virus and other flavivirus RNAs. RNA 7:585-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robertson, B., G. Myers, C. Howard, T. Brettin, J. Bukh, B. Gaschen, T. Gojobori, G. Maertens, M. Mizokami, O. Nainan, S. Netesov, K. Nishioka, T. Shin i, P. Simmonds, D. Smith, L. Stuyver, and A. Weiner. 1998. Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. Arch. Virol. 143:2493-2503. [DOI] [PubMed] [Google Scholar]
- 28.Shimotohno, K. 2000. Hepatitis C virus and its pathogenesis. Semin. Cancer Biol. 10:233-240. [DOI] [PubMed] [Google Scholar]
- 29.Tan, S. L., and M. G. Katze. 2001. How hepatitis C virus counteracts the interferon response: the jury is still out on NS5A. Virology 284:1-12. [DOI] [PubMed] [Google Scholar]
- 30.Tanji, Y., T. Kaneko, S. Satoh, and K. Shimotohno. 1995. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J. Virol. 69:3980-3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walewski, J. L., T. R. Keller, D. D. Stump, and A. D. Branch. 2001. Evidence for a new hepatitis C virus antigen encoded in an overlapping reading frame. RNA 7:710-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.World Health Organization. 1997. Hepatitis C: global prevalence. Wkly. Epidemiol. Rec. 72:341-344. [PubMed] [Google Scholar]