Abstract
Rod outer segment disk membranes are densely packed with rhodopsin. The recent notion of raft or microdomain structures in disk membranes suggests that the local density of rhodopsin in disk membranes could be much higher than the average density corresponding to the lipid/protein ratio. Little is known about the effect of high packing density of rhodopsin on the structure and function of rhodopsin and lipid membranes. Here we examined the role of rhodopsin packing density on membrane dynamic properties, membrane acyl chain packing, and the structural stability and function of rhodopsin using a combination of biophysical and biochemical techniques. We reconstituted rhodopsin into large unilamellar vesicles consisting of polyunsaturated 18:0,22:6n3PC, which approximates the polyunsaturated nature of phospholipids in disk membranes, with rhodopsin/lipid ratios ranging from 1:422 to 1:40. Our results showed that increased rhodopsin packing density led to reduced membrane dynamics revealed by the fluorescent probe 1,6-diphenyl-1,3,5-hexatriene, increased phospholipid acyl chain packing, and reduced rhodopsin activation, yet it had minimal impact on the structural stability of rhodopsin. These observations imply that densely packed rhodopsin may impede the diffusion and conformational changes of rhodopsin, which could reduce the speed of visual transduction.
INTRODUCTION
G-protein coupled receptors (GPCRs) are the largest family of membrane surface receptors and respond to a broad spectrum of extracellular stimuli, e.g., hormones, neurotransmitters, odor and taste molecules, light, etc., and generate intracellular responses by coupling to G-proteins (1,2). A prominent member of the GPCR superfamily is the photoreceptor rhodopsin. A high-resolution structure has been obtained and its function in visual signaling is well characterized (3–5). Upon light activation, rhodopsin rapidly converts into the active conformation, metarhodopsin II (MII). Each MII binds and activates several hundred of the visual G-protein transducin in a process that depends upon lateral diffusion. The activated transducin subsequently activates the effector enzyme, cGMP phosphodiesterase, triggering the hydrolysis of cGMP. The decline of cytosolic cGMP leads to the closure of the cGMP gated Na+/Ca2+ channel and hyperpolarization of the plasma membrane, and eventually triggers the visual response.
Over 90% of the membrane proteins in rod disk membranes are rhodopsin. Rhodopsin is densely packed in disk membranes with an average packing density of ∼25,000 rhodopsin/μm2 (6). Recent high-resolution atomic force microscopy images of disk membranes revealed rhodopsin molecules to be unevenly distributed in disk membranes (7). The packing density of rhodopsin in dense regions is about twice as high as the overall average density. At this point, the physiological relevance of such densely packed rhodopsin molecules observed in atomic force microscopy images is somewhat controversial (8,9), and it is unknown what structural and functional role the high packing density of rhodopsin may play. High packing density of rhodopsin in disk membranes endows the rod cell with high sensitivity, but may also create a slower visual response, which is limited by the diffusion-controlled protein coupling in the visual cascade (10,11). A recent in vivo study established that a reduction of rhodopsin density in disk membranes, which reduces the molecular crowding in membranes, increased the speed of visual transduction (12).
The effect of membrane protein packing density on the dynamic and equilibrium properties of lipid membranes has been studied for many membrane proteins (13–17). Several of these studies were carried out in membranes consisting of phospholipids with disaturated acyl chains, which are not found in abundance in biological membranes. The phospholipid acyl chains in disk membranes consist largely of polyunsaturated fatty acids. About half of the phospholipid acyl chains in rod disk membranes are made of docosahexaenoic acid (22:6n3) (18). These docosahexaenoic acid chains provide a maximal fluidity or maximal free volume of acyl chain packing in disk membranes, which is responsible for optimal visual function (19–23). The effect of membrane protein packing density on membrane physical properties and protein function in membranes consisting of polyunsaturated lipids is largely unknown.
In this study we reconstituted rhodopsin into vesicles consisting of 18:0,22:6n3PC at rhodopsin/lipid ratios varying from 1:422 to 1:40. The membrane physical properties, including acyl chain packing and membrane dynamics, were examined using time-resolved 1,6-diphenyl-1,3,5-hexatriene (DPH) fluorescence, the thermal stability of rhodopsin was measured by differential scanning calorimetry (DSC), and the activity of rhodopsin was quantified by the equilibrium between the metarhodopsin I (MI) and MII conformational states. We found that increased rhodopsin density led to increased acyl chain packing order and reduced rhodopsin activation, whereas it had minimal impact on rhodopsin thermal stability. The importance of these findings for visual transduction and their implications for the effects of membrane crowding in membrane structure and function will be discussed.
MATERIALS AND METHODS
Sample preparations
Concanavalin A sepharose was purchased from Amersham Biosciences (Piscataway, NJ). Phospholipid 18:0,22:6n3PC was obtained from Avanti Polar Lipids (Alabaster, AL). The fluorescent probe DPH was from Molecular Probes (Eugene, OR). Bovine retinas were from James and Wanda Lawson (Lincoln, NE).
Rod outer segments (ROS) were isolated from bovine retinas using the modified Shake-ate method (24). Rhodopsin was purified from the concanavalin A affinity column as described (25). Rhodopsin-containing vesicles consisting of 18:0,22:6n3PC with varied rhodopsin/lipid ratios were prepared using the rapid dilution method (26), which yielded vesicle sizes on the order of 100 nm. The phospholipid 18:0,22:6n3PC contains six double bonds at the sn-2 chain, which is highly susceptible to oxidation. Such oxidation was minimized by addition of antioxidant butylated hydroxytoluene (BHT) and removal of oxygen during preparation. BHT was added into samples at a lipid/BHT ratio of 500:1. All sample preparations were conducted in an argon-filled glove box and all buffers were degassed with argon gas before use. The final samples were suspended in Pipes-buffered solution (PBS) (10 mM Pipes, 30 mM NaCl, 60 mM KCl, 50 μM diethylenetriamine pentaacetic acid, pH 7.0). Rhodopsin concentration was determined by ΔA500 using an extinction coefficient of 40,000 cm−1 M−1; phospholipid was quantified by the Bartlett method (27). The ratio of rhodopsin to phospholipid of each sample was determined by the concentrations of rhodopsin and phospholipids.
Fluorescence measurements
Vesicles were suspended in PBS buffer at a phospholipid concentration of 0.1 mM. DPH was dissolved in tetrahydrofuran and added at a phospholipid/DPH ratio of 500:1. Fluorescence lifetime and differential polarization measurements were performed with a K2 multifrequency cross-correlation phase fluorometer (ISS, Urbana, IL). Excitation at 351 nm was provided by an Innova 307 argon ion laser (Coherent, Santa Clara, CA). All measurements were conducted at 37°C in PBS buffer. Fifteen modulation frequencies, logarithmically spaced from 5 to 150 MHz were used for both lifetime and differential polarization measurements. For lifetime measurements, the emission polarizer was set at the magic angle of 54.7° relative to the vertically polarized excitation beam, and p-bis[2-(5-phenyloxazolyl)]benzene in absolute ethanol was used as a lifetime reference. For each differential polarization measurement the instrumental polarization factors were measured, found to be between 1 and 1.05, and the appropriate correction factor applied. Scattered excitation light was removed from the emission beam by a 390-nm highpass filter. At each frequency, data was accumulated until the standard deviations of the phase and modulation ratio were below 0.2° and 0.004, respectively, and these values were used as the standard deviation for the measured phase and modulation ratio in all subsequent analysis. Both total intensity decay and differential polarization measurements were repeated a minimum of three times with each sample.
Total fluorescence intensity decays were analyzed with the sum of two exponential decays. The resulting time constants were averaged with intensity weighting and reported as 〈τ〉. Measured polarization-dependent differential phases and modulation ratios for each sample were combined with the measured total intensity decay to yield the anisotropy decay, r(t). All anisotropy decay data were analyzed using the Brownian rotational diffusion (BRD) model (28), which yields the order parameters 〈P2〉 and 〈P4〉. These two order parameters were used to construct an orientational distribution function, f(θ), of the probe molecule. In general, the orientation of a molecule with cylindrical symmetry in lipid vesicles is completely described by the angle, θ, between its symmetry axis and the local membrane normal. The results of the BRD model-based analysis were interpreted in terms of an angular distribution function, which is symmetric about θ = π/2, f(θ). The extent to which the equilibrium orientational freedom of DPH is restricted by the phospholipid acyl chains was quantified using the parameter, fv (29,30). DPH fluorescence anisotropy decays were also analyzed using an empirical sum-of-three-exponentials model. The results of this analysis are reported as the weighted average of the three rotational correlation times, 〈φ〉 (30). All analyses of differential polarization data were performed with NONLIN, with subroutines specifying the fitting function written by the authors.
Differential scanning calorimetry
Differential scanning calorimetry measurements were performed with a Nano-scan II calorimeter equipped with capillary cells (Calorimetry Sciences, Provo, UT). Rhodopsin-containing vesicles were dialyzed in PBS buffer before loading into the sample cell. The dialysis buffer was loaded into the reference cell as a reference buffer. Samples were loaded under argon in complete darkness using infrared night vision goggles. The cells were sealed in the dark and maintained at a pressure of 2.8 atm to maintain a stable baseline. Samples were scanned at a scan rate of 0.25°C/min from 45°C to 85°C. A second heating scan was repeated after the sample was cooled and equilibrated at 20°C. DSC scans were converted into excess molar heat capacity (Cp) using Cp-Cal 2.1 (Calorimetry Sciences), and the Cp curves were analyzed for Tm and ΔH using Origin 7 (OriginLab, Northampton, MA) as described (31).
MI-MII equilibrium measurements
The equilibrium spectra of MI and MII were collected and deconvolved according to Straume et al (32). Briefly, rhodopsin-containing vesicles were diluted to 0.3 mg/ml in pH 7.0 PBS buffer and equilibrated at 37°C in a thermally regulated sample holder. A set of four absorption spectra were collected sequentially in an HP 8453A diode array spectrophotometer. These included the spectra acquired 1) after the sample was equilibrated at 37°C; 2) 3 s after the sample was 15–20% bleached by a 520-nm flash; 3) 10 min after addition of 30 mM hydroxylamine to remove bleached rhodopsin; and 4) after complete bleach of the sample. Individual MI and MII spectra were deconvolved from their equilibrium mixture using a nonlinear least-squares method, and the equilibrium constant, Keq, was calculated according to Keq = [MII]/[MI].
RESULTS
DPH fluorescence
The fluorescence intensity decay of DPH yielded two exponential time constants (Table 1). The major contribution of DPH fluorescence decay was from the longer lifetime decay. Increasing the rhodopsin/lipid ratio, which increases rhodopsin packing density in membranes, resulted in progressive quenching of the longer lifetime decay of DPH and reduced the overall average lifetime, 〈τ〉. At the lowest rhodopsin/lipid ratio (1:422), the value of 〈τ〉 was 6.92 ± 0.05 ns, which is close to that found in pure 16:0,22:6n3PC membranes measured under similar conditions (30). As the rhodopsin/lipid ratio increased to 1:40, 〈τ〉 was reduced by ∼50%.
TABLE 1.
Summary of DPH florescence lifetime in reconstituted rhodopsin/18:0,22:6n3PC membranes
| Rhodopsin/18:0,22:6n3PC | a1 | τ1 (ns)* | a2 | τ2 (ns)* | 〈τ〉 (ns)* |
|---|---|---|---|---|---|
| 1:422 | 0.87 | 7.05 ± 0.07 | 0.13 | 1.15 ± 0.10 | 6.92 ± 0.05 |
| 1:184 | 0.75 | 5.34 ± 0.16 | 0.25 | 1.11 ± 0.08 | 5.07 ± 0.15 |
| 1:94 | 0.72 | 5.07 ± 0.20 | 0.28 | 1.00 ± 0.11 | 4.78 ± 0.17 |
| 1:40 | 0.65 | 4.02 ± 0.08 | 0.35 | 0.78 ± 0.03 | 3.72 ± 0.07 |
The fluorescence intensity decay of DPH was analyzed with the sum of two exponential decays, which yielded τ1 and τ2. a1 and a2 are the fractions of the two exponential decays. 〈τ〉 is the intensity-weighted average lifetime.
DPH lifetime is expressed as ± SD obtained from three independent measures.
Rotational dynamics of DPH probe in membranes was obtained from the analysis of DPH differential polarization data using two different methods: the empirical sum-of-exponential model and the BRD model as described in the Materials and Methods section. The average rotational correlation time, 〈φ〉, was ∼1.0 ns in 18:0,22:6n3PC membranes with the lowest rhodopsin density (Fig. 1 A), in agreement with the value obtained in pure 16:0,22:6n3PC membranes (30), indicating rapid rotational motion of DPH in membranes. Increasing the packing density of rhodopsin in membranes hindered the rotational motion of DPH, as demonstrated by the progressive increase of 〈φ〉. The rotational diffusion coefficient of DPH, D⊥, obtained from the BRD model also showed that an increase of rhodopsin density in membranes led to slower rotational motion of DPH (Fig. 1 B). There was an ∼2-fold reduction of D⊥ between the samples with lowest and highest rhodopsin/lipid ratios (1:422 and 1:40), further demonstrating that the DPH diffusion is impeded by increased rhodopsin density in membranes.
FIGURE 1.
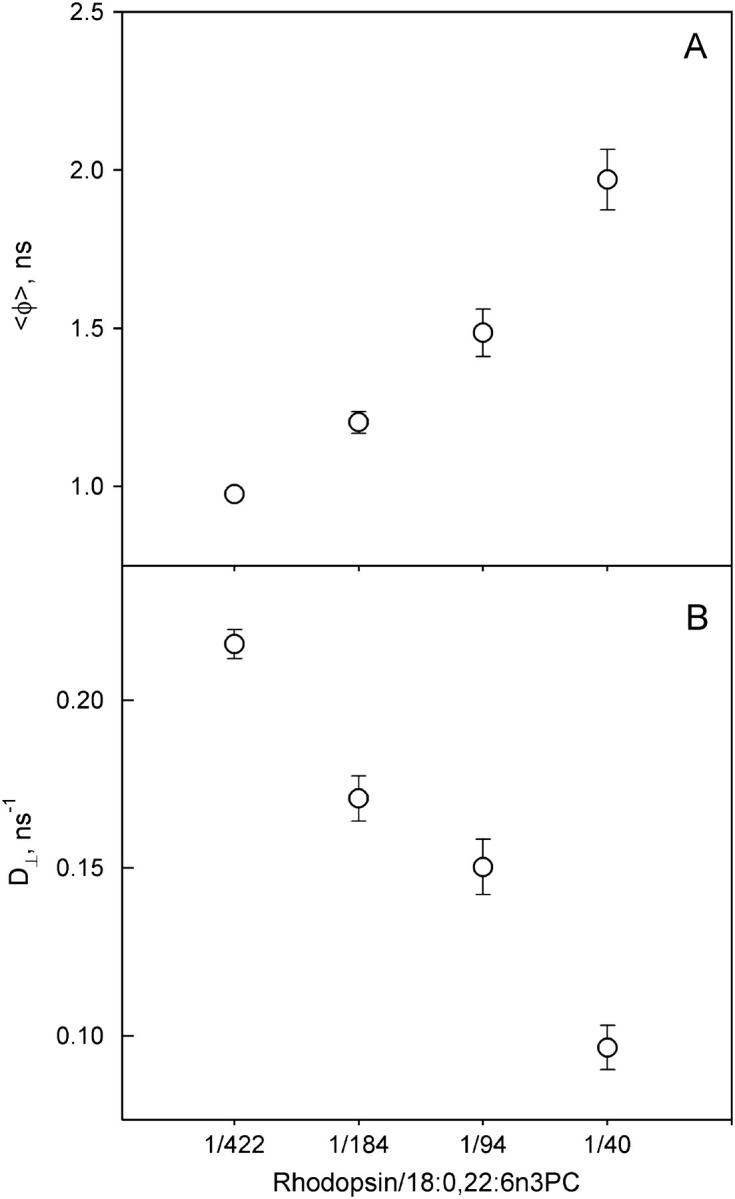
Effect of rhodopsin/18:0,22:6n3PC ratios on DPH rotational dynamics. Time-resolved DPH fluorescence anisotropy was measured at 37°C in pH 7 PBS buffer. (A) The anisotropy decays were analyzed using an empirical sum-of-exponential model. The resulting rotational correlation times were averaged to obtain 〈φ〉. (B) The anisotropy decays were analyzed using the BRD model as described in the Materials and Methods section. D⊥ is the rotational diffusional coefficient of DPH obtained from the BRD analysis. Rhodopsin/lipid ratios are expressed as molar ratios.
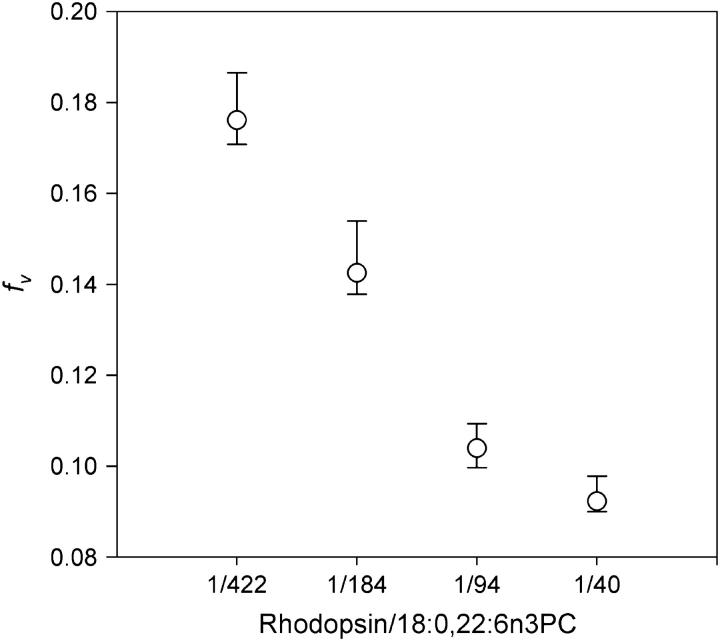
Analysis of DPH fluorescence anisotropy decays in terms of the BRD model yields the orientational distribution function for DPH in membranes, f(θ). Comparison of the orientational probability functions, f(θ)sinθ, for membranes with varying levels of rhodopsin showed the effect of membrane protein density on equilibrium acyl chain order (Fig. 2 A). The BRD model depicts DPH distribution in membranes as two orthogonal populations that are perpendicular and parallel to the membrane normal (30). Increased rhodopsin density in membranes reduced the DPH population perpendicular to the membrane normal and increased the DPH population parallel to the membrane normal. At a rhodopsin/lipid ratio of 1:422, ∼50% of the DPH population is in the population that is distributed perpendicular to the membrane normal (Fig. 2 B). This fraction was reduced to ∼30% at a rhodopsin/lipid ratio of 1:40, indicating the ordering of membranes by rhodopsin. The overall acyl chain packing free volume in membranes, which is summarized by the parameter fv, was sensitive to the rhodopsin density in membranes. As the rhodopsin density increased, a progressive reduction of fv was observed (Fig. 3), indicating the ordering of membrane packing by rhodopsin.
FIGURE 2.

Effect of rhodopsin/18:0,22:6n3PC ratios on the orientational distribution of DPH. (A) The orientational probability distribution for DPH in reconstituted rhodopsin/18:0,22:6n3PC membranes at protein/lipid ratio of 1:40 (solid line);1:94 (long-dashed line); 1:184 (dash-dotted line); and 1:422 (short-dashed line). (B) Plot of fraction of DPH in the distribution centered about the perpendicular-to-bilayer normal (f⊥) as a function of rhodopsin/lipid ratios. Increasing rhodopsin/lipid ratios resulted in reduced f⊥ values.
FIGURE 3.
Effect of rhodopsin/18:0,22:6n3PC ratios on membrane acyl chain packing free volume (fv). fv is calculated from the DPH orientational distribution function as described in the Materials and Methods section. Increasing rhodopsin/lipid ratios led to lower fv values.
Rhodopsin thermal stability
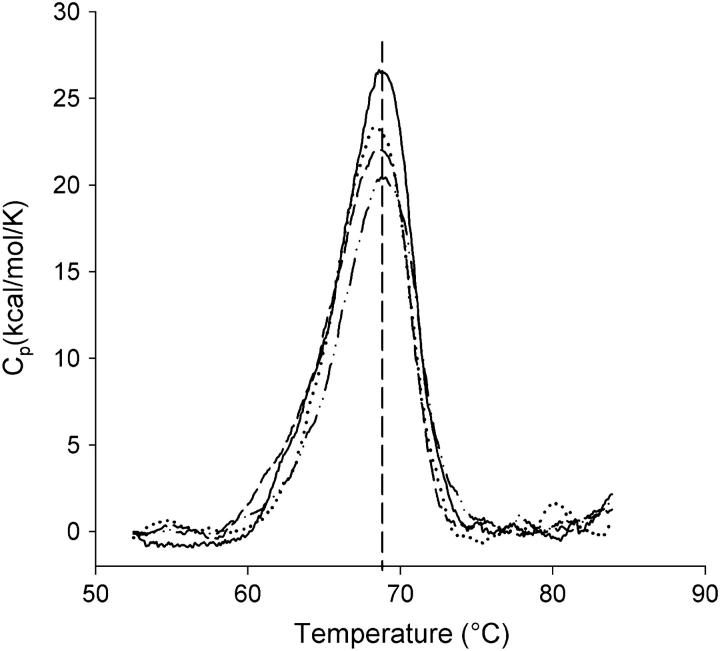
Previous studies show that the thermal stability of rhodopsin is regulated by the composition of lipids in membranes (31,33,34). Rhodopsin is stabilized in membranes consisting of more saturated phospholipids or in the presence of cholesterol. The stabilization of rhodopsin is manifested by the increase of the denaturation temperature of rhodopsin. For all reconstituted rhodopsin samples studied, the thermal denaturation of rhodopsin exhibited a single thermal transition peak (Fig. 4). The transition temperature of rhodopsin denaturation was centered around 68.5°C for all rhodopsin/lipid ratios, indicating that the number of lipid molecules surrounding rhodopsin had no influence on the structural stability of rhodopsin. The denaturation enthalpy was slightly increased by the increase of rhodopsin density in membranes (Table 2). There is an ∼14% difference of ΔH between the samples with the lowest and highest rhodopsin/lipid ratios. The ΔH at the highest rhodopsin/lipid ratio is close to that in native disk membranes (35,36).
FIGURE 4.
Effect of rhodopsin/18:0,22:6n3PC ratios on thermal denaturation of rhodopsin. Shown here are DSC thermograms of reconstituted rhodopsin/18:0,22:6n3PC at protein/lipid ratios of 1:40 (solid line); 1:94 (dotted line); 1:184 (dashed line); and 1:422 (dash-dotted line). Varying rhodopsin/18:0,22:6n3PC ratios did not shift the transition temperature of rhodopsin denaturation.
TABLE 2.
Summary of the thermal denaturation measurements of rhodopsin
| Rhodopsin/18:0,22:6n3PC | Tm (°C) | ΔH (kcal/mol) |
|---|---|---|
| 1:422 | 68.5 ± 0.1 | 137.9 ± 0.5 |
| 1:184 | 68.0 ± 0.1 | 144.1 ± 0.8 |
| 1:94 | 68.1 ± 0.1 | 148.4 ± 0.6 |
| 1:40 | 68.4 ± 0.1 | 157.9 ± 0.8 |
Values are expressed as ± SD propagated from the analysis of DSC thermograms.
Rhodopsin activation
The effect of rhodopsin packing density on rhodopsin activation was evaluated by the level of activated rhodopsin, which is quantified by the equilibrium constant, Keq, for the MI-MII equilibrium. Rhodopsin was functional at all rhodopsin/lipid ratios, as demonstrated by the ability to form the active MII conformation (Fig. 5 A). However, the level of MII formation was influenced by the packing density of rhodopsin in membranes; rhodopsin is most active in membranes with the lowest rhodopsin packing density. Increased rhodopsin density in membranes led to reduced Keq values. The reduction of Keq was more dramatic at higher rhodopsin density. An ∼2-fold increase of rhodopsin/lipid ratio from 1:422 to 1:184 resulted in a 9% reduction of Keq, whereas a similar increase of rhodopsin/lipid ratio from 1:94 to 1:40 resulted in a 19% reduction of Keq.
FIGURE 5.

Effect of rhodopsin/18:0,22:6n3PC ratios on rhodopsin activation. Keq was obtained at 37°C in pH 7.0 PBS buffer. (A) Plot of Keq versus rhodopsin/lipid ratios. Increasing rhodopsin/lipid ratios led to a lower level of rhodopsin activation. (B) Plot of Keq versus fv. Values of fv obtained from Fig. 3. Increasing fv led to a nonlinear increase of Keq.
The decline of Keq induced by the increase of rhodopsin density in membranes correlated with the decline of fv (Fig. 5 B). Such correlation between Keq and fv was observed in many previous studies (34,37,38). Since the conversion of MI to MII involves large conformational change (39), membranes with lower fv would impose higher energy barrier for such conformational change, thus a lower level of MII formation is expected. However, the correlation between Keq and fv was nonlinear, which differs from previous observations. Keq values approached a plateau at higher values of fv. This suggests that changes in membrane acyl chain ordering produced by increasing protein density and by changes in acyl chain composition are not identical.
DISCUSSION
Recent studies suggest that disk membranes are laterally heterogeneous and contain microdomains or lipid rafts (7,40,41). In rhodopsin-dense regions, the average rhodopsin density is ∼2-fold higher than the overall average rhodopsin density in the ROS disk; 48,300 rhodopsin/μm2 compared to 25,000 rhodopsin/μm2 (7). At present, it is not clear how such a heterogeneous structure is formed in disk membranes or whether such regions of densely packed, paracrystalline rhodopsin are physiologically relevant (8). An earlier DSC study observed a broad phase transition in ROS disks and lipid extract from ROS disks (42). This leads to the possibility that laterally segregated microdomains may be induced by a nonphysiological temperature below the broad phase transition observed by Miljanich et al. (42). In this study, we determined the structural and functional consequences of densely packed rhodopsin in polyunsaturated membranes. Our results showed that high packing density of rhodopsin slows the dynamics of the fluorescence probe DPH and increases the order of phospholipid acyl chain packing, producing reduced rhodopsin activation. This suggests that densely packed rhodopsin in the ROS disk membranes would lower visual transduction by retarding lateral diffusion and reducing the amount of rhodopsin in the active MII conformation.
Effect on membrane properties
A 10-fold increase of the rhodopsin/18:0, 22:6n3PC ratio resulted in an ∼50% reduction of the DPH rotational diffusion coefficient and an ∼50% reduction in fv. This is in general agreement with the reduced membrane dynamics and increased membrane acyl chain packing order observed in previous studies, which used DPH fluorescence anisotropy to assess ensemble acyl chain packing (13,14). There are 22–24 phospholipid molecules located in the annular lipid layer surrounding rhodopsin (43,44). Those phospholipids have hindered motion due to direct interaction with rhodopsin (44–47). At rhodopsin/lipid ratios ranging from 1:422 to 1:40, ∼6% to 60% of the phospholipids are located in the annular layer. This increase in the percentage of acyl chains in contact with rhodopsin with increasing protein content would be expected to produce more ordered ensemble acyl chain packing and slower membrane dynamics detected by DPH. However, there is evidence that the effects of transmembrane protein on acyl chain order may depend upon the degree of acyl chain unsaturation. A recent 2H-NMR study of rhodopsin in 16:0,18:1PC recombinant membranes (rhodopsin/lipid ratio of 1:100) reported that rhodopsin slightly decreased the order of segments C8–C15 of the sn-1 chain in 16:0,18:1PC (48). The observed difference between 16:0,18:1PC and 18:0,22:6n3PC suggests that a membrane containing 22:6n3 acyl chains responds differently to the presence of a transmembrane protein than a membrane composed of monounsaturated acyl chains. The difference could also be due to the different nature of the DPH fluorescent technique and the 2H-NMR measurement. The DPH fluorescence measurements used in this study reflect the dynamic property of DPH in membranes on a nanosecond timescale, whereas the 2H-NMR order parameter measurement reports the averaged correlation time of the C-D bond on a microsecond timescale.
The change of DPH rotational diffusion induced by increased rhodopsin density in 18:0,22:6n3PC membranes is small compared to the changes in membrane diffusional properties observed in other reconstituted membranes consisting of disaturated phospholipids (13,14,49). This suggests that polyunsaturated lipids may dampen the reduction of membrane diffusional properties induced by increased rhodopsin density.
Effect on receptor stability
Rhodopsin is a transmembrane protein that has direct contact with lipid membranes, thus it is not difficult to envision that lipid membranes would influence its stability. Previous studies show that membrane composition modulates rhodopsin thermal stability (33,34). Membranes consisting of more saturated phospholipids or cholesterol, which have more ordered acyl chain packing, stabilize rhodopsin with respect to thermal denaturation. Thus, the denaturation temperature of rhodopsin is higher in these membranes. In this study we observed that increased rhodopsin density in membranes caused reduction of ensemble acyl chain packing free volume, fv. However, the denaturation temperature of rhodopsin remained unchanged. Raising the number of lipids per rhodopsin increases the number of nonannular or bulk lipids that are not in contact with rhodopsin, but rhodopsin is surrounded by the same lipid, 18:0,22:6n3PC. The nature of the rhodopsin-lipid interaction remains the same regardless of the variation of rhodopsin/lipid ratio. The thermal denaturation measurements demonstrate that the thermal stability of rhodopsin is independent of the acyl chain packing properties of the membrane or how many lipids are in the bulk. Rather, rhodopsin thermal stability is dependent upon the composition of the phospholipids that interact directly with rhodopsin.
Effect on receptor function
The initial step in rhodopsin activation is the transient formation of the MI and MII equilibrium. The conformational change from MI to MII involves a volume expansion (39), which is restricted by the surrounding lipid matrix. Previous studies demonstrate that the conformation change from MI to MII is modulated by phospholipid acyl chain composition (21–23,38,50–53), membrane cholesterol (32,34,37), and phospholipid headgroup composition (53–55). The effects of phospholipid acyl chain composition and cholesterol in a PC membrane on MII formation are related to ensemble acyl chain packing, as indicated by the linear correlation between MII formation and fv (32,37,38). This empirical relationship between ensemble acyl chain packing and MII formation in bilayers with a low level of curvature stress may represent a special case of the more generalized flexible surface model proposed by Brown and co-workers (48,53,55). By considering bilayer deformation in terms of the curvature of the individual monolayer, the flexible surface model provides an elegant description of the energetics involved in the stabilization of MII by lipids with a tendency to form nonlamellar phases, such as phosphatidylethanolamines.
CONCLUSIONS
In this study, we focused on the effect of rhodopsin density on rhodopsin activation in PC-containing membranes. Our results showed that the energy barrier is higher in membranes with lower fv values; thus, the formation of MII is reduced. In this study, increased rhodopsin density resulted in reduction of fv in membranes and a reduced level of MII formation, in general agreement with the previously observed correlation between MII formation and fv (21,23,34,38,56). However, these earlier studies always found a strong linear relationship between Keq and fv for changes induced by temperature or membrane composition. In this study, changes in rhodopsin density produced a nonlinear correlation between Keq and fv, Fig. 5 B. This difference in the Keq-fv relationship is likely due to the different environments where DPH is located. The linear correlation between Keq and fv was observed in membranes with fixed protein/lipid ratios, whereas in this study the protein/lipid ratio was varied. At higher protein/lipid ratios, the majority of DPH probes are located in the vicinity of rhodopsin, whereas at lower protein/lipid ratios, a large fraction of the DPH fluorescence signal originates from probe molecules distant from rhodopsin. Thus, in membranes with a high number of lipids per rhodopsin, DPH may detect a greater increase in fv than is sensed by the MI-MII transition, and the plot of Keq versus fv approaches a plateau, as seen in Fig. 5 B.
Our results show that there is a clear difference in how rhodopsin packing density affects rhodopsin stability and rhodopsin function. The structural stability of rhodopsin is determined by the nature of the lipid-protein interaction at the lipid-protein interface, dependent on lipid composition and independent of protein/lipid ratio. In contrast, the conformational change from MI to MII is strongly correlated with membrane free volume, dependent on both lipid composition and the protein/lipid ratio. This study also demonstrates that increased membrane protein density reduces membrane dynamic properties, increases membrane acyl chain packing order, and lowers rhodopsin activation. These changes were generally less than changes observed in membranes with disaturated phospholipids, suggesting that the presence of 22:6n3 attenuates these changes. Calvert et al. (12) demonstrated that reduced rhodopsin density in vivo increased the speed of visual transduction, and interpreted the increase in speed as an enhancement in lateral diffusion. Our results support their interpretation and suggest that the enhanced production of MII could be an additional factor in their finding of increased speed of visual transduction.
References
- 1.Kroeze, W. K., D. J. Sheffler, and B. L. Roth. 2003. G-protein-coupled receptors at a glance. J. Cell Sci. 116:4867–4869. [DOI] [PubMed] [Google Scholar]
- 2.Gainetdinov, R. R., R. T. Premont, L. M. Bohn, R. J. Lefkowitz, and M. G. Caron. 2004. Desensitization of G protein-coupled receptors and neuronal functions. Annu. Rev. Neurosci. 27:107–144. [DOI] [PubMed] [Google Scholar]
- 3.Lagnado, L., and D. Baylor. 1992. Signal flow in visual transduction. Neuron. 8:995–1002. [DOI] [PubMed] [Google Scholar]
- 4.Palczewski, K., T. Kumasaka, T. Hori, C. A. Behnke, H. Motoshima, B. A. Fox, I. Le Trong, D. C. Teller, T. Okada, R. E. Stenkamp, M. Yamamoto, and M. Miyano. 2000. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 289:739–745. [DOI] [PubMed] [Google Scholar]
- 5.Filipek, S., R. E. Stenkamp, D. C. Teller, and K. Palczewski. 2003. G protein-coupled receptor rhodopsin: a prospectus. Annu. Rev. Physiol. 65:851–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pugh, E. N., Jr., and T. D. Lamb. 1993. Amplification and kinetics of the activation steps in phototransduction. Biochim. Biophys. Acta. 1141:111–149. [DOI] [PubMed] [Google Scholar]
- 7.Fotiadis, D., Y. Liang, S. Filipek, D. A. Saperstein, A. Engel, and K. Palczewski. 2003. Atomic-force microscopy: rhodopsin dimers in native disc membranes. Nature. 421:127–128. [DOI] [PubMed] [Google Scholar]
- 8.Chabre, M., R. Cone, and H. Saibil. 2003. Biophysics: is rhodopsin dimeric in native retinal rods? Nature. 426:30–31. [DOI] [PubMed] [Google Scholar]
- 9.Fotiadis, D., Y. Liang, S. Filipek, D. A. Saperstein, A. Engel, and K. Palczewski. 2004. The G protein-coupled receptor rhodopsin in the native membrane. FEBS Lett. 564:281–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kahlert, M., and K. P. Hofmann. 1991. Reaction rate and collisional efficiency of the rhodopsin-transducin system in intact retinal rods. Biophys. J. 59:375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruckert, F., M. Chabre, and T. M. Vuong. 1992. Kinetic analysis of the activation of transducin by photoexcited rhodopsin. Influence of the lateral diffusion of transducin and competition of guanosine diphosphate and guanosine triphosphate for the nucleotide site. Biophys. J. 63:616–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calvert, P. D., V. I. Govardovskii, N. Krasnoperova, R. E. Anderson, J. Lem, and C. L. Makino. 2001. Membrane protein diffusion sets the speed of rod phototransduction. Nature. 411:90–94. [DOI] [PubMed] [Google Scholar]
- 13.Heyn, M. P., R. J. Cherry, and N. A. Dencher. 1981. Lipid–protein interactions in bacteriorhodopsin–dimyristoylphosphatidylcholine vesicles. Biochemistry. 20:840–849. [DOI] [PubMed] [Google Scholar]
- 14.Peters, R., and R. J. Cherry. 1982. Lateral and rotational diffusion of bacteriorhodopsin in lipid bilayers: experimental test of the Saffman-Delbruck equations. Proc. Natl. Acad. Sci. USA. 79:4317–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rehorek, M., N. A. Dencher, and M. P. Heyn. 1983. Fluorescence energy transfer from diphenylhexatriene to bacteriorhodopsin in lipid vesicles. Biophys. J. 43:39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Straume, M., and B. J. Litman. 1988. Equilibrium and dynamic bilayer structural properties of unsaturated acyl chain phosphatidylcholine-cholesterol-rhodopsin recombinant vesicles and rod outer segment disk membranes as determined from higher order analysis of fluorescence anisotropy decay. Biochemistry. 27:7723–7733. [DOI] [PubMed] [Google Scholar]
- 17.Williams, B. W., A. W. Scotto, and C. D. Stubbs. 1990. Effect of proteins on fluorophore lifetime heterogeneity in lipid bilayers. Biochemistry. 29:3248–3255. [DOI] [PubMed] [Google Scholar]
- 18.Stinson, A. M., R. D. Wiegand, and R. E. Anderson. 1991. Fatty acid and molecular species compositions of phospholipids and diacylglycerols from rat retinal membranes. Exp. Eye Res. 52:213–218. [DOI] [PubMed] [Google Scholar]
- 19.Neuringer, M., W. E. Connor, C. Van Petten, and L. Barstad. 1984. Dietary omega-3 fatty acid deficiency and visual loss in infant rhesus monkeys. J. Clin. Invest. 73:272–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birch, E. E., S. Garfield, D. R. Hoffman, R. Uauy, and D. G. Birch. 2000. A randomized controlled trial of early dietary supply of long-chain polyunsaturated fatty acids and mental development in term infants. Dev. Med. Child Neurol. 42:174–181. [DOI] [PubMed] [Google Scholar]
- 21.Niu, S. L., D. C. Mitchell, and B. J. Litman. 2001. Optimization of receptor-G protein coupling by bilayer lipid composition II: formation of metarhodopsin II-transducin complex. J. Biol. Chem. 276:42807–42811. [DOI] [PubMed] [Google Scholar]
- 22.Mitchell, D. C., S. L. Niu, and B. J. Litman. 2001. Optimization of receptor-G protein coupling by bilayer lipid composition I: kinetics of rhodopsin-transducin binding. J. Biol. Chem. 276:42801–42806. [DOI] [PubMed] [Google Scholar]
- 23.Niu, S. L., D. C. Mitchell, S. Y. Lim, Z. M. Wen, H. Y. Kim, N. Salem, Jr., and B. J. Litman. 2004. Reduced G protein-coupled signaling efficiency in retinal rod outer segments in response to n-3 fatty acid deficiency. J. Biol. Chem. 279:31098–31104. [DOI] [PubMed] [Google Scholar]
- 24.McDowell, J. H., and H. Kuhn. 1977. Light-induced phosphorylation of rhodopsin in cattle photoreceptor membranes: substrate activation and inactivation. Biochemistry. 16:4054–4060. [DOI] [PubMed] [Google Scholar]
- 25.Litman, B. J. 1982. Purification of rhodopsin by concanavalin A affinity chromatography. Methods Enzymol. 81:150–153. [DOI] [PubMed] [Google Scholar]
- 26.Jackson, M. L., and B. J. Litman. 1985. Rhodopsin-egg phosphatidylcholine reconstitution by an octyl glucoside dilution procedure. Biochim. Biophys. Acta. 812:369–376. [DOI] [PubMed] [Google Scholar]
- 27.Barlett, G. R. 1959. Phosphorus assay in column chromatography. J. Biol. Chem. 234:466–468. [PubMed] [Google Scholar]
- 28.Levine, Y. K., and G. van Ginkel. 1994. Molecular dynamics in liquid-crystalline systems studied by fluorescence depolarization techniques. In The Molecular Dynamics of Liquid Crystals. G. R. Luckhurst and C. A. Veracini, editors. Kluwer Academic, Amsterdam. 537–71.
- 29.Straume, M., and B. J. Litman. 1987. Equilibrium and dynamic structure of large, unilamellar, unsaturated acyl chain phosphatidylcholine vesicles. Higher order analysis of 1,6-diphenyl-1,3,5-hexatriene and 1-[4-(trimethylammonio)phenyl]- 6-phenyl-1,3,5-hexatriene anisotropy decay. Biochemistry. 26:5113–5120. [DOI] [PubMed] [Google Scholar]
- 30.Mitchell, D. C., and B. J. Litman. 1998. Molecular order and dynamics in bilayers consisting of highly polyunsaturated phospholipids. Biophys. J. 74:879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niu, S. L., D. C. Mitchell, and B. J. Litman. 2005. Trans fatty acid derived phospholipids show increased membrane cholesterol and reduced receptor activation compared to their cis-analogs. Biochemistry. 44:4458–4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Straume, M., D. C. Mitchell, J. L. Miller, and B. J. Litman. 1990. Interconversion of metarhodopsins I and II: a branched photointermediate decay model. Biochemistry. 29:9135–9142. [DOI] [PubMed] [Google Scholar]
- 33.Polozova, A., and B. J. Litman. 2000. Cholesterol dependent recruitment of di22:6-PC by a G protein-coupled receptor into lateral domains. Biophys. J. 79:2632–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Niu, S. L., D. C. Mitchell, and B. J. Litman. 2002. Manipulation of cholesterol levels in rod disk membranes by methyl-beta-cyclodextrin: effects on receptor activation. J. Biol. Chem. 277:20139–20145. [DOI] [PubMed] [Google Scholar]
- 35.Khan, S. M., W. Bolen, P. A. Hargrave, M. M. Santoro, and J. H. McDowell. 1991. Differential scanning calorimetry of bovine rhodopsin in rod-outer-segment disk membranes. Eur. J. Biochem. 200:53–59. [DOI] [PubMed] [Google Scholar]
- 36.Landin, J. S., M. Katragadda, and A. D. Albert. 2001. Thermal destabilization of rhodopsin and opsin by proteolytic cleavage in bovine rod outer segment disk membranes. Biochemistry. 40:11176–11183. [DOI] [PubMed] [Google Scholar]
- 37.Mitchell, D. C., M. Straume, and B. J. Litman. 1992. Role of sn-1-saturated,sn-2-polyunsaturated phospholipids in control of membrane receptor conformational equilibrium: effects of cholesterol and acyl chain unsaturation on the metarhodopsin I in equilibrium with metarhodopsin II equilibrium. Biochemistry. 31:662–670. [DOI] [PubMed] [Google Scholar]
- 38.Litman, B. J., and D. C. Mitchell. 1996. A role for phospholipid polyunsaturation in modulating membrane protein function. Lipids. 31(Suppl):S193–S197. [DOI] [PubMed] [Google Scholar]
- 39.Lamola, A. A., T. Yamane, and A. Zipp. 1974. Effects of detergents and high pressures upon the metarhodopsin I–metarhodopsin II equilibrium. Biochemistry. 13:738–745. [DOI] [PubMed] [Google Scholar]
- 40.Seno, K., M. Kishimoto, M. Abe, Y. Higuchi, M. Mieda, Y. Owada, W. Yoshiyama, H. Liu, and F. Hayashi. 2001. Light- and guanosine 5′-3-O-(thio)triphosphate-sensitive localization of a G protein and its effector on detergent-resistant membrane rafts in rod photoreceptor outer segments. J. Biol. Chem. 276:20813–20816. [DOI] [PubMed] [Google Scholar]
- 41.Nair, K. S., N. Balasubramanian, and V. Z. Slepak. 2002. Signal-dependent translocation of transducin, RGS9–1-Gβ5L complex, and arrestin to detergent-resistant membrane rafts in photoreceptors. Curr. Biol. 12:421–425. [DOI] [PubMed] [Google Scholar]
- 42.Miljanich, G. P., M. F. Brown, S. Mabrey-Gaud, E. A. Dratz, and J. M. Sturtevant. 1985. Thermotropic behavior of retinal rod membranes and dispersions of extracted phospholipids. J. Membr. Biol. 85:79–86. [DOI] [PubMed] [Google Scholar]
- 43.Dratz, E. A., J. F. Van Breemen, K. M. Kamps, W. Keegstra, and E. F. Van Bruggen. 1985. Two-dimensional crystallization of bovine rhodopsin. Biochim. Biophys. Acta. 832:337–342. [DOI] [PubMed] [Google Scholar]
- 44.Ryba, N. J., L. I. Horvath, A. Watts, and D. Marsh. 1987. Molecular exchange at the lipid-rhodopsin interface: spin-label electron spin resonance studies of rhodopsin-dimyristoylphosphatidylcholine recombinants. Biochemistry. 26:3234–3240. [DOI] [PubMed] [Google Scholar]
- 45.Watts, A., J. Davoust, D. Marsh, and P. F. Devaux. 1981. Distinct states of lipid mobility in bovine rod outer segment membranes. Resolution of spin label results. Biochim. Biophys. Acta. 643:673–676. [DOI] [PubMed] [Google Scholar]
- 46.Albert, A. D., and P. L. Yeagle. 1983. Phospholipid domains in bovine retinal rod outer segment disk membranes. Proc. Natl. Acad. Sci. USA. 80:7188–7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pates, R. D., and D. Marsh. 1987. Lipid mobility and order in bovine rod outer segment disk membranes. A spin-label study of lipid-protein interactions. Biochemistry. 26:29–39. [DOI] [PubMed] [Google Scholar]
- 48.Huber, T., A. V. Botelho, K. Beyer, and M. F. Brown. 2004. Membrane model for the G-protein-coupled receptor rhodopsin: hydrophobic interface and dynamical structure. Biophys. J. 86:2078–2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jacobson, K., A. Ishihara, and R. Inman. 1987. Lateral diffusion of proteins in membranes. Annu. Rev. Physiol. 49:163–175. [DOI] [PubMed] [Google Scholar]
- 50.O'Brien, D. F., L. F. Costa, and R. A. Ott. 1977. Photochemical functionality of rhodopsin-phospholipid recombinant membranes. Biochemistry. 16:1295–1303. [DOI] [PubMed] [Google Scholar]
- 51.Baldwin, P. A., and W. L. Hubbell. 1985. Effects of lipid environment on the light-induced conformational changes of rhodopsin. 2. Roles of lipid chain length, unsaturation, and phase state. Biochemistry. 24:2633–2639. [DOI] [PubMed] [Google Scholar]
- 52.Wiedmann, T. S., R. D. Pates, J. M. Beach, A. Salmon, and M. F. Brown. 1988. Lipid-protein interactions mediate the photochemical function of rhodopsin. Biochemistry. 27:6469–6474. [DOI] [PubMed] [Google Scholar]
- 53.Gibson, N. J., and M. F. Brown. 1993. Lipid headgroup and acyl chain composition modulate the MI-MII equilibrium of rhodopsin in recombinant membranes. Biochemistry. 32:2438–2454. [DOI] [PubMed] [Google Scholar]
- 54.Brown, M. F. 1994. Modulation of rhodopsin function by properties of the membrane bilayer. Chem. Phys. Lipids. 73:159–180. [DOI] [PubMed] [Google Scholar]
- 55.Botelho, A. V., N. J. Gibson, R. L. Thurmond, Y. Wang, and M. F. Brown. 2002. Conformational energetics of rhodopsin modulated by nonlamellar-forming lipids. Biochemistry. 41:6354–6368. [DOI] [PubMed] [Google Scholar]
- 56.Mitchell, D. C., J. T. Lawrence, and B. J. Litman. 1996. Primary alcohols modulate the activation of the G protein-coupled receptor rhodopsin by a lipid-mediated mechanism. J. Biol. Chem. 271:19033–19036. [DOI] [PubMed] [Google Scholar]