Abstract
Previous attempts to establish 293cell-based stable and high-titer adeno-associated virus (AAV) packaging cell lines were unsuccessful, primarily due to adenovirus E1-activated Rep gene expression, which exerts cytostatic and cytotoxic effects on the host cells. Control of the two large AAV Rep proteins (Rep78/68) was insufficient to eliminate the adverse effects, because of the leaky expression of the two small Rep proteins (Rep52/40). However, it was unsuccessful to control Rep52/40 gene expression since its promoter is located within the coding sequence of Rep78/68. To tightly regulate all four Rep proteins by using their own promoters, we have developed a novel gene control paradigm termed “dual splicing switch,” which disrupts all four Rep genes by inserting into their shared coding region an intron that harbors transcription termination sequences flanked the LoxP sites. As a result, the structure and activities of the Rep gene promoters, both p5 and p19, are not affected; however, all of the Rep transcripts are prematurely terminated and the genes were inactivated. Removal of the terminator by Cre protein reactivates the transcription of all four Rep proteins derived from their own promoters. This switch system was initially tested in the lacZ gene and a 600-fold induction of β-galactosidase activity was observed. Using the dual splicing switch strategy, we have subsequently established a number of AAV packaging cell lines from 293 cells, which showed a normal growth rate, high stability, and more importantly, high yields of AAV vectors. Such a gene control paradigm is also useful for other viruses, e.g., autonomous parvoviruses. Finally, the high-titer 293-based AAV packaging cell lines should greatly reduce the risk of wild-type adenovirus contamination and provide a scalable AAV vector production method for both preclinical and clinical studies.
The recombinant adeno-associated viral (AAV) vector system is derived from nonpathogenic and defective parvoviruses. AAV vectors have been successfully used to establish efficient and long-term gene transfer in vivo in a variety of tissues without significant cellular immune responses or toxicity. The success of preclinical studies has led to clinical trials with AAV vectors to treat genetic diseases such as cystic fibrosis, hemophilia, and muscular dystrophy (8, 11, 30). Although the use of AAV vectors for human gene therapy is promising, current vector production methods may not meet the demand for AAV vectors in clinical studies involving certain genetic diseases, particularly for ones that require large-quantity and high-quality vectors. Gene therapy for muscular dystrophy (17, 35), for example, requires gene transfer of large groups of muscles and even muscles of the entire body. Such treatment needs an enormous amount of clinical-grade AAV vectors. This prompted us to develop a high-yield and scalable production method to meet the demands for AAV vectors in clinical trials.
Of numerous approaches to improve AAV vector production, two strategies differing in principles are now widely used. One method is based on the adenovirus-free transient transfection of all elements (vector and packaging plasmids, along with helper genes isolated from adenovirus), which are required for AAV production in host cells such as 293 cells (10, 38). The other method relies on wild-type adenovirus infection into the cell lines that stably harbor AAV rep/cap genes, as well as the AAV vector DNA. Although the transient-transfection method generates high-titer AAV vectors that are free of adenovirus, it is labor-intensive and expensive to scale up for clinical studies. On the other hand, the wild-type adenovirus-inducible AAV producer cell lines can be scaled up in cultures and can produce AAV vectors with titers comparable to the transient-transfection method. However, this approach faces a problem of the traditional method, namely, the production of wild-type helper adenovirus. Contamination of wild-type adenovirus is highly undesirable in view of vector safety. As a result, new packaging cell lines are needed for AAV production to meet the demand for high-quality and high-quantity gene vectors in both preclinical and clinical studies.
Previous packaging cell lines featuring both stability and high productivity are almost exclusively derived from cells which do not harbor the adenovirus E1A/E1B genes, including, for instance, HeLa cells (3, 4). Production of AAV vectors from those packaging cell lines requires the infection of wild-type adenovirus. On the other hand, human 293 cells, the most commonly used cells for AAV vector production by the transient-transfection method do contain adenovirus E1A/E1B genes. If stable packaging cell lines were made from 293 cells, the use of E1A/E1B defective adenovirus should suffice in providing helper functions for AAV vector production. In view of the fact that E1A/E1B defective adenovirus has been widely used as a gene therapy vector in humans, its safety profile is much better versus the wild-type adenovirus. Because of this reason, extensive efforts have been made to generate 293 cell-based AAV packaging cell lines. However, no such cell line has been generated to date that has both stability and high productivity (3, 40). The major difficulty of generating 293-based AAV packaging cell lines is the E1A-mediated activation of AAV promoters p5 and p19. These two promoters control the gene expression of four AAV replication proteins (Rep78, Rep68, Rep52, and Rep40), which are well known to be cytostatic (40) and even cytotoxic (28) if expressed at high levels. As a result, both promoters p5 and p19 need to be regulated tightly during packaging cell growth. They also need to be highly induced during AAV vector production. An added layer of difficulty to tightly regulate p19 is due to the location of this promoter, which is situated within the protein coding region of promoter p5 products, Rep78 and Rep68 (2). Manipulation of promoter p19 will inevitably cause mutations in the Rep78 and Rep68 coding sequence and may disrupt the structure and functions of these essential Rep proteins.
In this report we have designed a novel gene expression control strategy named dual splicing switch, where an intron and three polyadenylation [poly(A)] sequences were inserted into the protein coding region of a gene to disrupt its transcription. As a result, the mRNA is prematurely terminated by the insertion of poly(A) sequences, thereby blocking the gene expression. However, when a Cre enzyme is provided in trans, it splices out the inserted DNA fragment containing the poly(A) sequences that are flanked by loxP sites (1). The removal of poly(A) sequences then allows transcription to continue and therefore generating full-length mRNA. After RNA splicing, the remnant of the intron insertion is precisely removed from the full-length mRNA, and the protein coding sequence is restored. Thus, both DNA splicing and RNA splicing (dual splicing) are required to reactivate the controlled target gene. This switch system was initially tested on the β-galactosidase (lacZ) gene. By comparing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) staining and LacZ enzyme activities of a variety of constructs, we have developed the dual-splicing switch system that could tightly block the gene expression in the middle of the β-galactosidase coding sequence. lacZ gene expression can be induced to wild-type levels after the cre gene is introduced into the cells. In an effort to simultaneously block gene expression of all four Rep proteins, this switch system was inserted in the middle of AAV rep gene coding sequences, which are shared by all four Rep proteins. Because of the tight control of all Rep proteins by the dual splicing switch, we have readily obtained 293 cell-based AAV packaging cell lines with both high-stability and high vector yields. Upon induction of AAV rep gene expression by an adenovirus deleted of E1A/E1B/E3 carrying the cre gene, the 293 cell-based AAV packaging cell lines surpassed the yields of transfection method (38), as well as HeLa cell-based cell lines (4, 26).
MATERIALS AND METHODS
Plasmid construction.
To construct plasmid pLacZ-Int-3A (Fig. 1A), a triple simian virus 40 (SV40) poly(A) cassette from plasmid pSVA3 (21) was excised by BamHI and BglII digestion and subcloned into the BamHI site of Bluescript (Stratagene) by sticky-end ligation. Then, a loxP site (CGGGA TCCAT AACTT CGTAT AATGT ATGCT ATACG AAGTT ATCCA GATCTTC) was, respectively, inserted into the SpeI site and the EcoRV site (blunt-end ligation) flanking the triple poly(A) cassette, generating plasmid p2LoxP-SVA3. This plasmid was confirmed by DNA sequencing and revealed direct-repeat orientation of the two loxP sites. Subsequently, the 2LoxP-SVA3 cassette was excised by XhoI and NotI double digestion, filled in by Klenow enzyme, and inserted via blunt-end ligation into the BglII site (Klenow filled in) in the intron of plasmid pLacZ-Int (31), generating plasmid pLacZ-Int-3A (Fig. 1A). Using the same strategy, a hygromycin-resistant (Hygr) gene flanked by the LoxP sites was made and inserted into the BglII site of plasmid pLacZ-Int, generating plasmid pLacZ-Int-Hyg (Fig. 1A). Finally, to combine both the three poly(A) and the Hygr gene into one cassette, the AvaI- and HindIII-digested Hygr fragment (Klenow filled in) from pTK-Hyg (Clontech) was initially cloned into the SmaI site of plasmid p2LoxP-SVA3. Subsequently, the 3A-Hyg cassette flanked by the LoxP sites was digested with XhoI and NotI and inserted into the BglII site of pLacZ-Int by blunt-end ligation, generating plasmid pLacZ-Int-3A-Hyg (Fig. 1A).
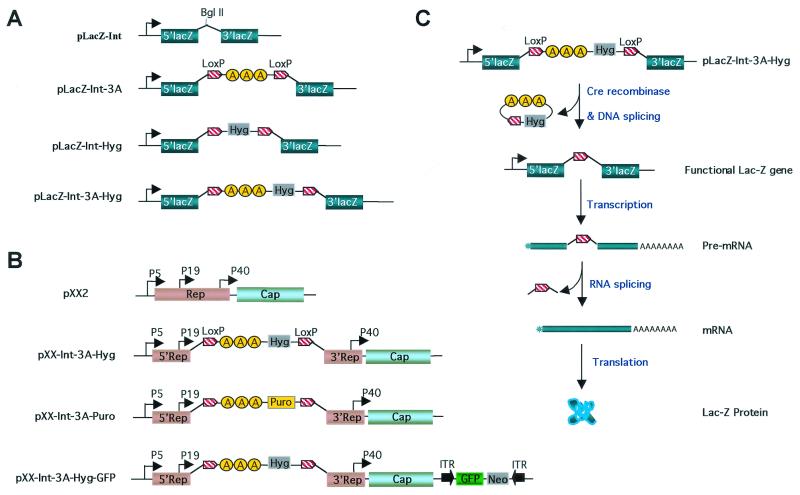
FIG. 1.
Construction of dual-spicing switch plasmids for gene expression control of lacZ and AAV rep genes. (A) Construction of dual-spicing lacZ plasmids. For pLacZ-Int, the lacZ gene contains an hCG intron insertion in the coding sequence (31). The BglII site is for cloning purpose. For pLacZ-Int-3A, a triple SV40 poly(A) cassette (3A) flanked by two loxP sequences was inserted into the BglII site in the hCG intron of pLacZ-Int to block lacZ gene transcription. For pLacZ-Int-Hyg, the Hygr gene flanked by two loxP sequences was inserted into the BglII site of pLacZ-Int. For pLacZ-Int-3A-Hyg, the triple SV40 poly(A), together with the Hygr gene flanked by two loxP sequences, was inserted into the BglII site of the pLacZ-Int. (B) Construction of dual-splicing switch AAV packaging plasmids. pXX2 is an AAV type 2 packaging plasmid (38). For pXX2-Int-3A-Hyg, the 3.2-kb termination cassette (Int-3A-Hyg) from plasmid pLacZ-Int-3A-Hyg was inserted into the shared Rep coding sequence downstream of promoter p19 to block rep gene transcriptions. For pXX2-Int-3A-Puro, the Hygr gene was replaced by Puror gene for additional selection. For pXX2-Int-3A-Hyg-GFP, an AAV GFP vector which contains a CMV promoter-driven EGFP gene and a neomycin-resistant gene was inserted into the SseI site of pXX2-Int-3A-Hyg plasmid. (C) Activation of lacZ gene controlled by the dual-splicing switch. A Cre recombinase is provided in trans by an adenovirus infection. The enzyme recognizes the two loxP sites and splices out the inserted DNA fragment that contains the poly(A) sequences between the loxP sites. The removal of poly(A) sequences then allows transcription to proceed and full-length mRNA is generated. After RNA splicing, the inserted intron is precisely removed from the full-length mRNA, and the coding sequence is restored.
To make the inducible AAV packaging constructs that have the intron, the triple poly(A) and Hygr cassette inserted in the Rep coding sequences, the 3.2-kb Int-3A-Hyg fragment was amplified by PCR from plasmid pLacZ-Int-3A-Hyg with two primers (HCG-Int-forward [5′-GTAAG AAGATCCGAGGTC-3′] and HCG-Int-reverse [5′-CCTTGTCCGGTTACCCTGCAG-3′]). The PCR-amplified intron-poly(A)-Hyg cassette was inserted into the Rep coding sequences at two different sites (AAV-2 nucleotide number 1022 or 1340, respectively), generating plasmids pXX2-Int-A3-Hyg-2 (Fig. 1B) and pXX2-Int-A3-Hyg-8. To construct a similar plasmid that contains a puromycin-resistant (Puror) gene rather than the Hygr gene, a Puror cassette was excised by PvuII and BamHI from plasmid pPUR (Clontech). Plasmid pXX2-Int-A3-Hyg-2 was then digested with MluI and RsrII to delete the Hygr gene cassette. The Puror cassette was then cloned into MluI and RsrII double-digested pXX2-Int-A3-Hyg-2 to replace the Hygr gene, generating plasmid pXX2-Int-A3-Puro (Fig. 1B).
To further combine the AAV vector sequence into the above AAV packaging plamsid pXX2-Int-A3-Hyg-2, an AAV GFP vector which contains a cytomegalovirus (CMV) promoter-driven enhanced green fluorescent protein (EGFP) gene and a neomycin-resistant gene was inserted into the SseI site of pXX2-Int-A3-Hyg-2, generating plasmid pXX2-Int-3A-Hyg-GFP (Fig. 1B).
Cells and viruses.
Ad-GFP, an adenovirus vector carrying an EGFP gene driven by CMV promoter, and Ad-Cre, an adenovirus vector carrying the Cre recombinase gene of P1 phage (1) driven by CMV promoter, were made by a conventional method (15). These adenoviruses were E1A/E1B and E3 defective first-generation adenovirus vectors.
293 cell lines were propagated in Dulbecco modified Eagle medium (Invitrogen) supplemented with heat-inactivated 10% fetal bovine serum (FBS; Invitrogen). Stable transfection was done in 293 cells by the calcium phosphate transfection method as previously described (38). Briefly, right before transfection each well of a six-well plate of 293 cells was fed with 1.5 ml of fresh Iscove modified Dulbecco medium (Invitrogen) containing 10% FBS without antibiotics. A total of 3 μg of linearized plasmid DNA was dissolved in 125 μl of 0.25 M CaCl2 and then quickly mixed with 125 μl of HEPES-buffered saline and added to the cells. At 12 h after transfection, the medium was replaced with fresh Dulbecco modified Eagle medium containing 10% FBS and antibiotics. Two days later, the cells were trypsinized, diluted, and plated onto 15-cm-diameter dishes to allow for outgrowth of single-cell clones. Concentrations of antibiotics for selection and maintenance of drug-resistant cells were, respectively, as follows: hygromycin (Clontech), 200 and 100 μg/ml; G418 (Invitrogen), 800 and 200 μg/ml; puromycin (Sigma), 2 and 1 μg/ml.
To generate 293 cell lines that contain the tightly controlled rep-cap genes, plasmid pXX2-Int-A3-Hyg-2 was transfected into 293 cells and selected by hygromycin. To screen for AAV rep and cap gene expression from each of the 293 cell clones, 105 cells of each individual clone in a well of a 12-well plate were infected with Ad-Cre virus at a multiplicity of infection (MOI) of 5. After infection for 48 to 72 h, the cells were collected by centrifugation. The cell pellets were subjected to Western blot analysis. The cell clones showing a high level of expression of four Rep proteins were selected and further expanded.
To generate 293 cell-based AAV-GFP vector packaging cell lines, plasmid pXX2-int-3A-Hyg-GFP was transfected into a 293 cell line (293-XX2-DS-19) that already contained high copies of pXX2-Int-A3-Hyg-2 from a previous step (see the above section). Single-cell clones were produced by G418 antibiotic selection. To screen for high-titer AAV-GFP vector packaging cell lines, 105 cells of each individual clone in a well of a 12-well plate were infected with Ad-Cre at an MOI of 5. After infection for 48 to 72 h, both the cells and the media (1 ml) were collected, and subjected to four cycles freeze-thaw. The cell debris was removed by centrifugation, and the titers of the cell lysates were determined on 293 cells for AAV-GFP vector yields from each cell line. Clones with yields of AAV-GFP that were >107 transducing units (t.u.) per 105 cells (equivalent to 109 t.u./10-cm plate) were selected for further characterization.
To generate 293 cell-based AAV-minidystrophin vector packaging cell lines, plasmid pXX2-int-3A-puro was cotransfected with an AAV vector plasmid containing the human dystrophin minigene 3990 under the control of an MCK promoter (35) into a 293 cell line (293-XX2-DS-19) that already contained high copies of pXX2-Int-A3-Hyg-2 from a previous step. Single-cell clones were produced by puromycin selection. To screen for high-titer AAV-minidystrophin vector packaging cell lines, 105 cells of each individual clone were infected in a well of a 12-well plate with Ad-Cre at an MOI of 5. After infection for 48 to 72 h, both the cells and the medium (1 ml) were collected, and these were subjected to four cycles of freeze-thaw. The cell debris was removed by centrifugation. Supernatant of the cell lysates were subjected to DNA dot blot analysis to determine the viral genome particle (v.g.) titers of AAV-minidystrophin vector yields from each cell line. Clones with yields of AAV-minidystrophin greater than 1010 v.g. per 105 cells (equivalent to 1012 v.g./10-cm plate) were selected for further characterization.
AAV vector production and purification.
To produce AAV vectors by using the 293-based cell lines, the cells were simply infected with Ad-Cre (MOI = 5). At 2 days after infection, 20 15-cm plates of the cells were pelleted by centrifugation and resuspended in suspension buffer (phosphate-buffered saline [PBS] saline with 25 mM HEPES and 150 mM NaCl). After three freeze-thaw cycles, 0.5% deoxycholic acid and 50 U of Benzonase (Sigma)/ml were added into the cells. The mixture described above was incubated for 30 min at 37°C and centrifuged at 10,000 rpm to remove the debris. The supernatant was further clarified by 0.8-μm (pore-size) filter and directly loaded onto a Hitrap-Heparin column by using the AKTA purifier (Amersham Pharmacia Biotech). The AAV viral particles were eluted with 400 mM NaCl2 in PBS (42). The viral genome particle titer of the AAV vector was determined by DNA dot blot (38).
Western analysis of AAV Rep and Cap proteins.
Western blots were carried out by previously published methods with modifications (18). Briefly, the cell pellet from one well of a six-well plate was lysed in 200 μl of radioimmunoprecipitation assay buffer (10 mM Tris-Cl, pH 8.2; 1% Triton X-100; 1% sodium dodecyl sulfate; 150 mM NaCl). The samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% polyacrylamide) and transferred to a nitrocellulose membrane. After being subjected to blocking in 10% nonfat dry milk in Tris-buffered saline buffer (TBS; 50 mM Tris-Cl, pH 7.5; 200 mM NaCl) for 1 h, the membranes were incubated at room temperature for 1 h with primary antibodies in TBS containing 0.5% Tween 20. The primary antibody for Rep was a monoclonal antibody that recognizes all four Rep proteins (13). It was used at a 1:75 dilution. The primary antibody for Cap proteins was a guinea pig polyclonal antibody against AAV-2 (Braton Biotech, Inc.). It was used at a dilution of 1: 400. After primary antibody incubation and rinses, the membranes were incubated with the secondary antibodies at room temperature for 1 h. The secondary antibody for Rep was a goat anti-mouse polyclonal antibody conjugated to horseradish peroxidase (sigma) at a 1:4,000 dilution. The secondary antibody for Cap was a rabbit anti-guinea pig polyclonal antibody conjugated with horseradish peroxidase. It was used at a 1:4,000 dilution. All of the antibodies were diluted with 2% dry milk in TBS buffer. After three washes with TBS, the specific protein bands were visualized by chemiluminescence reagent and exposed to X-ray film.
X-Gal staining and β-galactosidase enzyme activity assay.
The X-Gal staining and β-galactosidase enzyme activity assay were performed as previously described (31, 37).
Infection-based viral amplification assay for rcAAV detection.
Human 293 cells were seeded at 5 × 106 cells per 10-cm plate. The next day, the cultures were infected with adenovirus (MOI = 5) and infected with various concentrations of AAV vector or wild-type AAV. After 72 h, the cells were scraped into the medium and collected by centrifugation. The cell pellets were resuspended in 1 ml of PBS and subjected to three freeze-thaw cycles in a dry ice-ethanol bath and centrifuged to pellet debris. The clarified crude lysate was incubated at 56°C for 1 h to inactivate the adenovirus, and half amount of the crude lysate virus (500 μl) was added to a fresh well of 293 cells with fresh adenovirus (MOI =5) for a second round of amplification. After an additional 72 h, the cells were scraped into the medium and pelleted, and episomal DNA was isolated by the method of Hirt extraction (12). The DNA was subjected to Southern analysis and hybridized with a biotinylated PstI fragment of the pXX2 plasmid containing the cap gene sequence. A DNA detection kit North2South chemiluminescent nucleic acid hybridization and detection (Pierce) was used to detect the AAV DNA after exposure to X-ray films.
RESULTS
Design of a dual-splicing switch system by using lacZ gene as an example.
Previous efforts to make AAV packaging cell lines in 293 cells were focused on regulation of the expression of promoter p5 products Rep78 and Rep68. There has been success by substituting p5 with exogenous inducible promoters that partially resolve the problem of Rep-mediated toxicity. However, Rep52/40 produced by promoter p19 could not be regulated and remained problematic in obtaining stable 293 cell-based packaging cell lines (23, 40). In order to simultaneously control the expression of all four AAV Rep proteins derived from both promoters p5 and p19, we disrupted the coding sequences of all four Rep proteins at their overlapping region, reasoning that this should effectively stop rep gene expression altogether, while keeping both promoters p5 and p19 intact and active. It is important to keep these two endogenous promoters in place because they are proven to be the most effective for AAV vector production (38). However, after gene disruption we faced the problem of how to restore the coding sequences when needed. We designed a novel strategy to achieve the above goal by taking advantage of an intron, three polyadenylation sequences and DNA recombinase Cre and its recognition site loxP.
First, we used the β-galactosidase(lacZ) reporter gene as a model to test our dual-splicing switch strategy. As illustrated in Fig. 1A, a small intron was inserted in the middle of the lacZ coding sequence (31). Subsequently, three SV40 poly(A) sequences in tandem were inserted in the middle of the intron. The insertion of three poly(A) signals would effectively terminate the transcription, yield a truncated mRNA, and render the gene nonfunctional. To restore the transcription of full-length mRNA, the poly(A) tandem must be subsequently removed. For this purpose, we utilized the Cre/loxP system where the site-specific recombinase Cre of bacteriophage P1 recognizes its target sequence loxP sites and loops out the DNA sequences in between (1). A loxP site was placed both upstream and downstream to flank the poly(A) tandem, generating plasmid pLacZ-Int-3A (Fig. 1A). As expected, this plasmid had much lower (∼80-fold) lacZ gene expression compared to its parental plasmid pLacZ-Int (Fig. 2). However, low levels of leaky gene expression were still seen. This could be due to the very strong CMV promoter activity in 293 cells and the high copy numbers of plasmid transfected into the cells. It is also possible that the poly(A) tandem was not sufficient to completely terminate all transcription activity, perhaps because it was not long enough to allow for full termination (27, 33). As a result, a 2.1-kb Hygr gene cassette [including a thymidine kinase promoter and a poly(A) site] was further inserted downstream of the poly(A) tandem, generating plasmid pLacZ-Int-3A-Hyg (Fig. 1A). As a comparison, a similar plasmid pLacZ-Int-Hyg was constructed without the poly(A) tandem but had the Hygr gene cassette (Fig. 1A). Transfection of the above constructs in 293 cells demonstrated synergistic effects between the poly(A) tandem and Hygr cassette in terminating lacZ gene transcription. The addition of the Hygr cassette in pLacZ-Int-3A plasmid further lowered the expression of the lacZ gene by another ∼8-fold, a total of ∼640-fold decrease compared to the original parental plasmid pLacZ-Int (Fig. 2). The Hygr cassette alone also decreased the lacZ gene expression by ∼80-fold in the pLacZ-Int-Hyg plasmid (Fig. 2). In summary, insertion of our best transcription termination unit [three poly(A) sequences plus a Hygr cassette] diminished lacZ gene expression by as many as 640-fold (Fig. 2B).
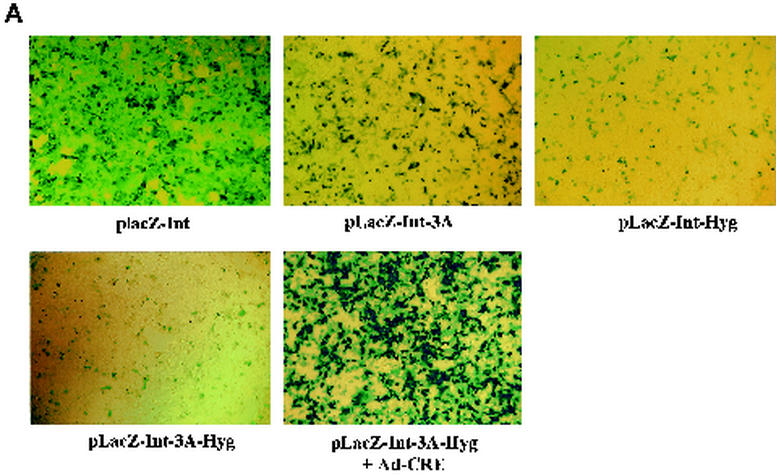
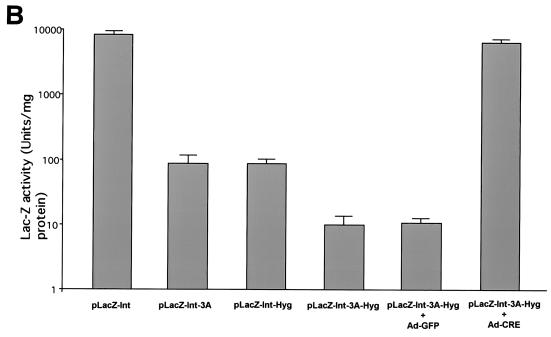
FIG. 2.
(A) lacZ gene expression from dual-splicing controlled lacZ constructs. Different plasmids were transfected into the 293 cells. X-Gal staining was performed 36 h after transfection. For LacZ-Int-3A-Hyg plasmid, the Ad-Cre was added at an MOI of 5 during transfection to restore the lacZ gene expression. (B) β-Galactosidase enzyme activity in 293 cells from dual-splicing controlled lacZ constructs. The data represented three parallel experiments. The standard deviation bar is shown in the figure. For plasmid pLacZ-Int-3A-Hyg, the induction experiment was achieved by infection with Ad-Cre (MOI = 5), and the control group was infected with Ad-GFP (MOI = 5).
Next, we wanted to see whether the gene expression could be induced back to its original levels upon delivery of the cre gene by an adenovirus vector (Ad-Cre). Plasmid pLacZ-Int-3A-Hyg was transfected into 293 cells, followed by Ad-Cre infection. X-Gal staining of the cells after plasmid transfection and Ad-Cre infection showed dramatic induction of lacZ expression (Fig. 2A) and revealed no significant difference from its parental plasmid pLacZ-Int. Quantitative LacZ enzyme activity assay also showed consistent results (Fig. 2B). The lacZ activity from plasmid pLacZ-Int-3A-Hyg was induced by ca. 600-fold by Ad-Cre infection but not by Ad-GFP control virus infection (Fig. 2B). In summary, we have successfully designed and tested our dual-splicing switch system, which effectively stopped the gene expression from the middle of the coding sequence. In addition, the diminished gene expression could also be effectively restored upon induction by Ad-Cre infection.
Tight control of all four Rep proteins by the dual-splicing switch.
Equipped with the same strategy utilized in regulating the lacZ gene expression, we have inserted the transcription termination unit (Int-3A-Hyg cassette) into the Rep coding region downstream from the p19 promoter (Fig. 1B), disrupting all four Rep coding sequences. To compare the effect of different insertion loci, we selected two sites in the shared Rep coding region for the insertion of the transcription termination unit, respectively. Two constructs, pXX2-Int-3A-Hyg-2 and pXX2-Int-3A-Hyg-8, were generated (see Materials and Methods for details). Western analysis of Rep protein expression showed similar results from both plasmids (Fig. 3, lanes 2 and 4). All four Rep proteins, particularly the two smaller Rep proteins, were all induced from both plasmids after Ad-Cre infection. These two plasmids were further examined for their packaging function in producing an AAV-GFP vector (38). Transient-transfection experiments showed no difference in packaging functions between the two inducible plasmids and a commonly use AAV packaging plasmid pXX2 (38) (data not shown). These results convinced us that gene expression of all four Rep proteins could be tightly controlled simultaneously and also be readily inducible by using the dual-splicing switch system.
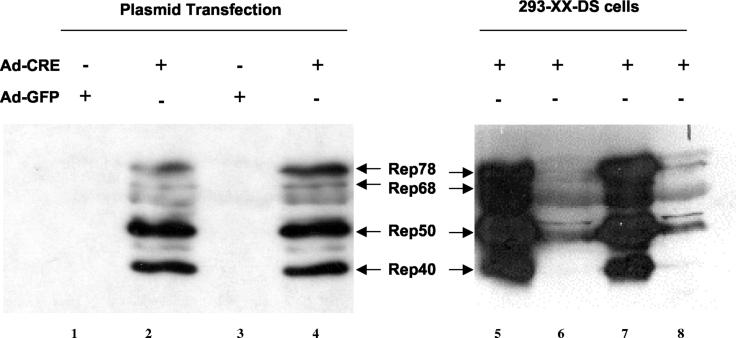
FIG. 3.
Western analysis of Rep gene induction by Ad-Cre infection. rep gene induction by Ad-Cre infection either in plasmid transfection experiments (lanes 1 to 4) or in cell lines (lanes 5 to 8). Plasmids pXX2-Int-A3-Hyg-2 (lanes 1 and 2) and plasmid pXX2-Int-A3-Hyg-8 (lanes 3 and 4) had no rep gene expression in transfection (lanes 1 and 3) but showed high-level Rep gene expression at 48 h after Ad-Cre infection (lanes 2 and 4). Lanes 5 to 8 are examples of Cre-inducible 293 cell-based AAV packaging cell lines. After Ad-Cre infection for 48 h, two clones showed very high rep gene expression (lanes 5 and 7), and two clones showed modest rep gene expression (lanes 6 and 8).
To further investigate if the dual-splicing switch could tightly control the AAV genes to generate stable 293 cell lines, plasmid pXX2-Int-3A-Hyg-2 was transfected into 293 cells and selected for Hygr colonies. Strikingly, a large number of drug-resistant colonies formed after selection. Among numerous clones, two of them (293-XX2-DS-19 and 293-XX2-DS-34) were identified as highly inducible for Rep expression (Fig. 3, lanes 5 and 7). We chose clone 293-XX2-DS-19 as a packaging cell line precursor for the next step.
Generation of stable and high-titer 293 cell-based AAV packaging cell lines.
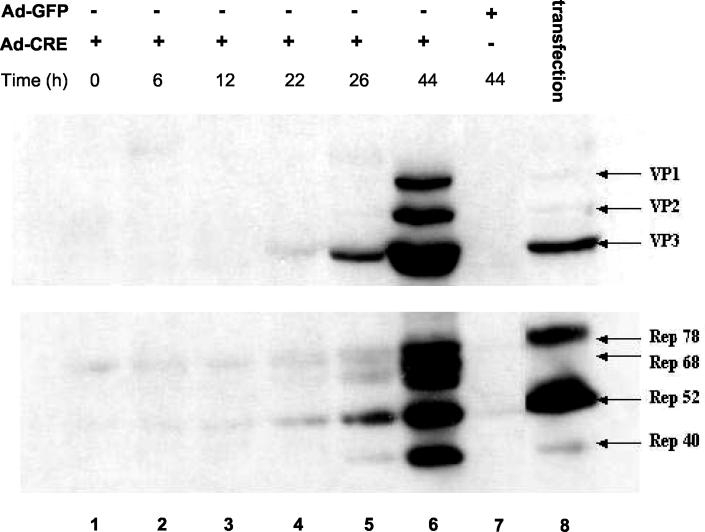
To establish a final AAV packaging cell line, we stably delivered an AAV-GFP vector into the above 293-XX2-DS-19 cells. In order to acquire a higher copy number of rep and cap genes in the final cell line, we elected to deliver the AAV-GFP vector sequence along with an additional copy of a packaging plasmid pXX2-Int-3A-Hyg into the 293-XX2-DS-19 cells. Thus, the AAV-GFP vector was first inserted into pXX2-Int-3A-Hyg plasmid to generate pXX2-Int-3A-Hyg-GFP (Fig. 1B). This plasmid also contained a neomycin-resistant gene as a marker for G418 selection. After transfection into 293-XX2-DS-19 cells and G418 antibiotic selection, a large number of drug-resistant colonies again formed, suggesting that the rep genes were tightly controlled and that Rep-mediated toxicity was successfully averted. A time course of rep and cap gene expression after Ad-Cre helper virus infection was monitored at different intervals. The results showed that Rep and Cap expression started from 22 h (Fig. 4, lane 4) and reached high levels at 44 h after Ad-Cre infection (Fig. 4, lane 6). As expected, AAV gene expression could not be induced by a control adenovirus vector Ad-GFP even after 44 h of infection (Fig. 4, lane 7). Compared to the adenovirus-free transfection method (Fig. 5, lane 8), the cell line had significantly higher expression of capsid genes and Rep40 protein (Fig. 4, lane 6), which are particularly important for AAV vector packaging (9, 16, 18).
FIG. 4.
Time course of rep and cap gene expression in 293-based AAV-GFP packaging cell line after Ad-Cre infection. Stable AAV packaging cell line 293-GFP-145 was infected with Ad-Cre at an MOI of 5. Cell pellets were harvested at different time intervals and analyzed by Western blot with anti-Cap antibody (top panel) or anti-Rep antibody (bottom panel). A control virus Ad-GFP was also used to infect the cells (lane 7). A transfection experiments was done as another control with packaging plasmid pXX2 and mini-Ad plasmid pXX6 (lane 8) (38).
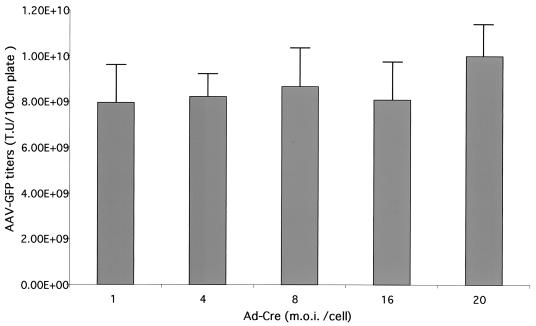
FIG. 5.
MOI by Ad-Cre showed minimal effects on the yields of AAV-GFP vector from cell line 293-GFP-145. The data represented three independent experiments, and the standard deviation bar was shown in the figure.
Among the 293 cell-based AAV-GFP cell lines, more than 10 of them demonstrated vector yields of >109 transducing units (t.u.) per 10-cm plate. One cell line, 293-GFP-145, reached as high as 8.6 × 109 t.u./10-cm plate. Optimization of helper Ad-Cre mutiplicity of infection (MOI) ranging from 1 to 20 showed little improvement in AAV vector yields from 293-GFP-145 cells (Fig. 5), suggesting a broad range of tolerance for Ad-Cre helper function. This may be due to the replication of Ad-Cre in 293-based cells. Furthermore, the AAV packaging cell lines created by the dual-splicing strategy have been very stable. For example, the 293-GFP-145 cell line has been continuously cultured for more than 6 months. The growth rate was very similar to the parental 293 cells. Importantly, the capacity of producing AAV-GFP vector remained the same after consecutive passages (data not shown).
To demonstrate that the success in AAV-GFP cell lines can be reproduced in other cell lines carrying AAV vectors of different transgenes, we used a similar strategy to establish another packaging cell line containing a human dystrophin minigene (35). Because there is no drug-resistant marker gene in the AAV-minidystrophin vector, we cotransfected into the 293-XX2-DS-19 cell the vector plasmid with the packaging plasmid pXX-Int-3A-Puro (Fig. 1B), which is identical to plasmid pXX-Int-3A-Hyg except that the hygromycin gene was replaced by the Puror gene for selection (6). Reproducibly, a large number of colonies formed after puromycin selection of the AAV-minidystrophin vector packaging cell lines. Numerous clones were isolated and challenged with Ad-Cre helper virus and then monitored for vector production by the DNA dot blot method. Again, more than 10 clones yielded AAV vectors of >1012 viral particles (v.g.) per 10-cm plate (data not shown). Some cell clones were further subcloned and expanded for larger-scale AAV vector production. Satisfactory vector yields were obtained from those packaging cell lines. For example, large-scale vector preparation from the AAV-GFP producer cell line 293-GFP-145 (20-by-15-cm plates) generated 6.8 × 1013 vector genome particles after heparin affinity column chromatography purification (42). The AAV-minidystrophin vector cell line 293-AAV-3999-70 (20-by-15-cm plates) generated 1.24 × 1014 vector genome particles after column purification.
Characterization of AAV vector produced in the 293-GFP-145 cells.
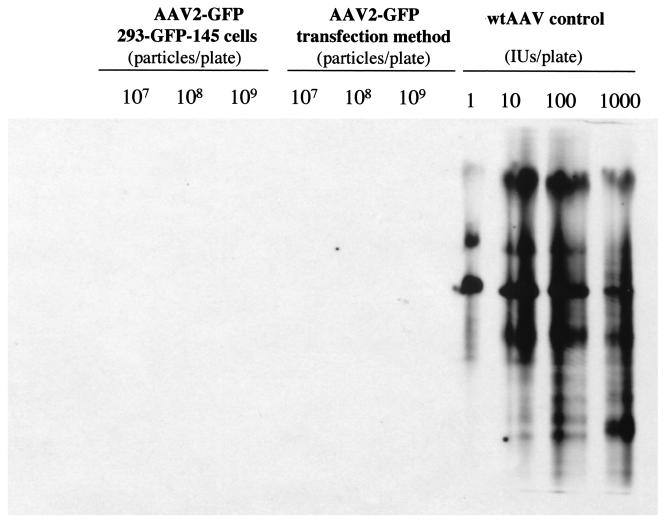
A clinically useful cell line should produce vectors free of replication-competent AAV (rcAAV, also termed wild-type-like AAV) (36). Therefore, we examined whether the 293-GFP-145 cell line could generate rcAAV, which would most probably be derived from nonhomologous recombination between the vector and packaging sequences during AAV production. We chose to use an infection-based viral amplification assay to detect the rcAAV, because this method is highly sensitive. Up to 109 viral genome particles of highly purified AAV-GFP, generated either from the 293-GFP-145 cells or from the triple plasmid transfection method (38), were used to infect 293 cells, which were coinfected with wild-type adenovirus to provide helper functions for rcAAV propagation. As a positive control, wild-type AAV was also used similarly to infect the 293 cells at doses ranging from 1 to 1,000 infectious units per 10-cm plate of 293 cells along with wild-type adenovirus coinfection. The viruses were harvested after full cytopathic effect and amplified again by infecting fresh 293 cells. Southern analysis of the viral DNA isolated after the second amplification revealed no detectable AAV coding sequences from the above two individual AAV-GFP vector stocks (Fig. 6). However, strong signals of AAV monomer and dimer replication intermediates were detected in the wild-type AAV positive control samples at all doses from 1 to 1,000 infectious units per plate of 293 cells (Fig. 6). These results indicated that there is no detectable rcAAV in up to 109 viral genome particles in our AAV-GFP vectors produced either by the 293-GFP-145 cell line or by the triple plasmid transfection method (38).
FIG. 6.
Southern analysis of replication-competent AAV (rcAAV) in different AAV-GFP vectors. Up to 109 viral genome particles of highly purified AAV-GFP, generated either from the 293-GFP-145 cells or from the triple-plasmid transfection method, were used to infect one 10-cm plate, which were coinfected with wild-type adenovirus at an MOI of 5 to provide helper functions for rcAAV amplification. As a positive control, wild-type AAV was also used similarly to infect the 293 cells at a total dose ranging from 1 to 1,000 infectious units per 10-cm plate along with wild-type adenovirus coinfection (see Materials and metHods). Low-molecular-weight DNA samples from the second round of amplification were isolated and subjected to Southern analysis with an AAV cap gene fragment as the probe.
We next compared the ratios of viral genome particles versus the transducing units (v.g./t.u.), which is an indicator of AAV vector infectivity. In our experiments, the AAV-GFP viruses, derived from an identical vector backbone but produced by three different production methods, were subjected to the comparison. When the vector titers were measured by t.u. (each t.u. generates one GFP-positive green cell after infection), packaging cell line 293-GFP-145 produced the highest titer (>8 × 109 t.u./10-cm plate), followed by triple plasmid transfection (38) (1.5 × 109 t.u./10-cm plate), and then a HeLa cell-based packaging cell line XX-GFP-53 (109 t.u./10-cm plate) (26). Moreover, the AAV-GFP vector v.g./t.u. ratios of the above three methods were, respectively, 80, 500, and 2,000 for the 293-GFP-145 cell line, triple transfection, and XX-GFP-53 cell, suggesting that AAV vector generated by the 293-GFP-145 cell line had the highest infectivity.
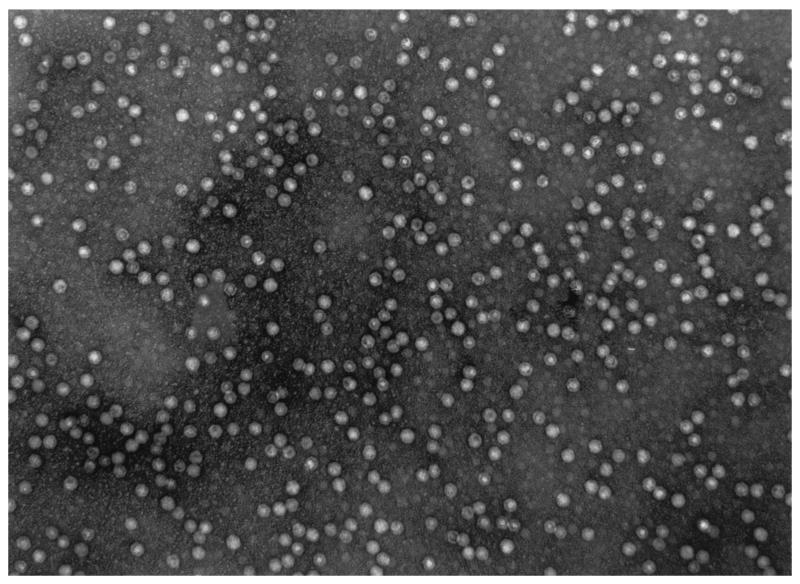
Transmission electron microscopy was used to study the morphology of the AAV virions produced from the 293-GFP-145 cells (Fig. 7). The viral particles were purified by heparin affinity column chromatography method without separation of empty particles from full particles by any density gradient centrifugation. Quantitation of a large number of virions revealed that approximately one-half of the virions were dense full particles, whereas the other half were empty or defective particles with an electron-lucent inner sphere.
FIG. 7.
Electron microscopy of AAV viral particles produced in the 293-GFP-145 cell line and purified by heparin sulfate column purification. Original magnification, ×50,000. The particles were diluted to 2 × 1012 vector genomes per ml for detection.
Molecular characterization of 293-based AAV packaging cell lines.
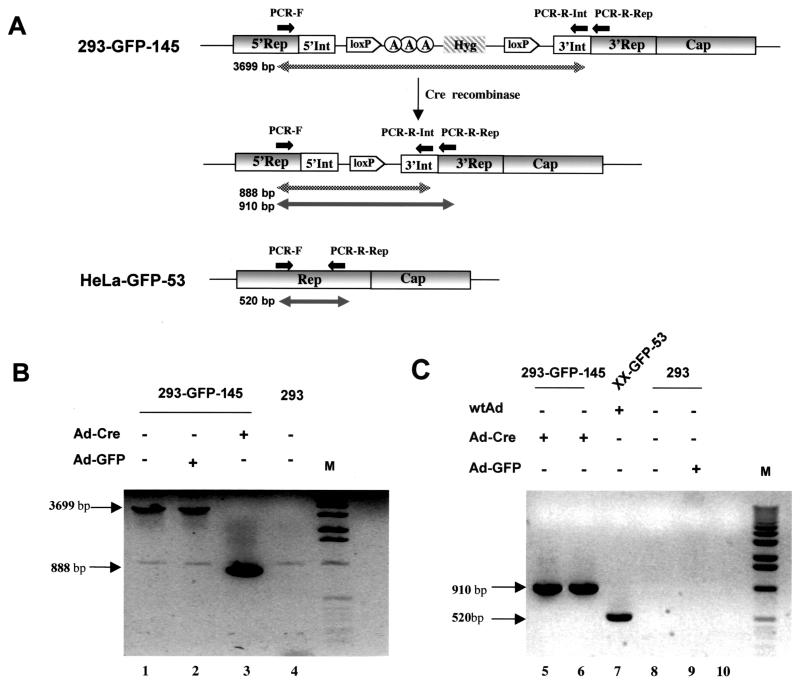
Since induction of AAV rep genes is accomplished by the removal of the transcription termination cassette [three poly(A) signals plus a drug-resistant gene] by the Cre enzyme expressed from the Ad-Cre vector, we sought to investigate how efficient Ad-Cre infection could lead to the excision of the termination cassette in the Rep coding sequence. PCR primers flanking the termination cassette were designed to detect the DNA before and after the deletion of the Int-3A-Hyg cassette (Fig. 8A). The forward primer (PCR-F) was in the rep gene, whereas the reverse primer (PCR-R-Int) was in the 3′ region of the inserted intron (Fig. 8A). DNA isolated from 293-GFP-145 cells before Ad-Cre infection revealed a 3,699-bp PCR product (Fig. 8A and B, lane 1). Control Ad-GFP virus infection did not result in any deletion and therefore yielded the same 3,699-bp PCR product (Fig. 8B, lane 2). As expected, Ad-Cre infection of this cell line yielded an 888-bp PCR product as a result of the deletion of the 2.8-kb termination cassette by the Cre enzyme (Fig. 8B, lane 3). In addition, the Cre-mediated excision was so efficient that no parental DNA (3,699 bp) could be detected (Fig. 8B, lane 3). To further confirm the absence of wild-type Rep sequence in our cell line, another reverse primer (PCR-R-Rep) located in the 3′rep gene was paired with the forward primer (PCR-F) located in the 5′ Rep sequence, flanking the artificial intron insertion (Fig. 8A). Wild-type Rep sequence from HeLa cell-based cell line XX-GFP-53 generated a 520-bp PCR product (Fig. 8C, lane 7), whereas DNA isolated from a Ad-Cre-infected 293-GFP-145 cell generated a 910-bp PCR product (Fig. 8C, lanes 5 and 6), which was the sum of the 520-bp wild-type Rep sequence plus the artificial intron (338 bp) and the remaining loxP site (52 bp) after excision of the termination cassette. Finally, PCR products from low-molecular-weight DNA (Hirt extraction; Fig. 8C, lane 5), as well as total cellular DNA (Fig. 8C, lane 6), reveal the same intensity, an indication of episomal amplification of AAV coding sequence (see below for Southern analysis results).
FIG. 8.
PCR analysis of AAV Rep gene structure before and after Ad-Cre and loxP-mediated DNA splicing. (A) Schematic illustration of the rep gene structure and expected sizes of PCR products. Primers PCR-F and PCR-R-Rep were located in the rep gene before and after, respectively, the inserted intron. Primer PCR-R-Int was located in the 3′ end of the intron. (B) Gel electrophoresis of PCR products with primers PCR-F and PCR-R-Int. Total cellular DNA was isolated from cell line 293-GFP-145 (lanes 1, 2, and 3) or control 293 cells (lane 4) with or without the indicated adenovirus infection. The DNA was subjected to PCR amplification and gel separation. (C) Gel electrophoresis of PCR products with primers PCR-F and PCR-R-Rep. Total cellular DNA (lanes 5, 7, 8, and 9) or episomal DNA (lane 6) was isolated from different cells with or without indicated adenovirus infection. Cell line XX-GFP-53 (lane 7) contain wild-type Rep sequence and yielded a 520-bp PCR product, whereas cell line 293-GFP-145 contains an inserted intron sequence and a loxP site (after Cre-mediated splicing) and yielded a 910-bp PCR product from both total cellular DNA (lane 5) and episomal DNA (lane 6).
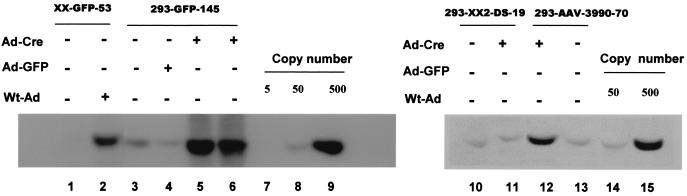
Since selective rep-cap gene amplification is a mechanism for high-titer AAV production from HeLa cell-based stable cell lines (3, 22, 34), we sought to examine whether this phenomenon is also responsible for the high-titer vector production in our 293-based producer cell lines. Southern analysis was performed on both total DNA and episomal DNA (Hirt extraction) to see whether the rep-cap genes were amplified during vector production. As shown in Fig. 9, the HeLa cell-based XX-GFP-53 cell line had its rep-cap genes amplified from four copies per cell (Fig. 9, lane 1, and J. Li et al., unpublished results) to approximately 200 copies (Fig. 9, lane 2). Similarly, the 293-GFP-145 cell line had its rep-cap genes amplified from ca. 50 copies (Fig. 9, lane 3) to more than 500 copies per cell after Ad-Cre infection in both fractions of total DNA (Fig. 9, lane 5) and episomal DNA (Fig. 9, lane 6). This amplification was Rep dependent because Ad-GFP infection in the 293-GFP-145 cell could not result in any amplification of the AAV genes (Fig. 9, Lane 4). Amplification of rep-cap genes was also observed in the mini-dystrophy producer cell line 293-AAV-3A990-70. The initial Rep-Cap copy number in this cell line was also ca. 50 (Fig. 9, lane 13), and was amplified to ca. 250 copies after Ad-Cre infection (Fig. 9, lane 12).
FIG. 9.
Southern analysis of AAV rep-cap gene amplification in AAV packaging cell lines. Total cellular DNA (15 μg) was isolated from different cell lines 48 h after the indicated adenovirus infection. Episomal DNA (lane 6) was isolated from 293-GFP-145 cells after Ad-Cre infection. All DNA samples were digested with PstI to drop a 2.3-kb internal fragment of AAV cap gene. The same DNA fragment purified from AAV plasmid was used as a probe for Southern hybridized. The DNA copy number standards were set at 5, 50, and 500 rep-cap genomes per cell by spiking 0.13, 1.3, and 13 ng of plasmid pXX2-Int-3A-Hyg into 15 μg of plain 293 cell DNA. The detected band corresponds to the expected 2.3-kb cap gene fragment.
DISCUSSION
AAV vectors have been increasingly used in gene therapy studies for a variety of genetic as well as acquired diseases. The vector system is also being used as a gene transfer tool for functional genomics research (39). Due to a high demand for AAV vectors in both basic science and preclinical and clinical applications, continued efforts are being made to improve the vector production methodology. Because of the lack of a practical in vitro viral packaging system (41), all AAV vectors are currently produced from tissue culture cells. The human embryonic kidney cell line 293 is predominantly the cell of choice for transient transfection-based production (5, 7, 38), which generates adenovirus-free AAV vectors. Likewise, the human cervical epithelial cell line HeLa is the most commonly used cell for AAV production through stable packaging cell lines with wild-type adenovirus as the helper (4, 14, 26). Recently, we made an effort to generate HeLa cell-based AAV packaging cell lines that contained the inducible E1A and E1B genes of adenoviruses (26). However, the AAV vector yields were not robust, and the cells needed tetracyclin and trichostatin A induction for E1 gene expression. A major advantage of using 293 cells to generate an AAV packaging cell line is its capability of supporting AAV production without the requirement of using wild-type Ad as a helper. Since 293 cells already stably express E1A and E1B genes, which are two of the five essential adenovirus helper genes (E1A, E1B, E2a, E4, and VA RNA), an adenovirus deleted of E1A/E1B, rather than a wild-type adenovirus, would provide sufficient helper functions for an efficient AAV production. In addition, 293 cells are highly receptive for infection and transfection, are very well characterized, and have been broadly used to produce adenovirus vectors for clinical trials. Despite these attractive features, numerous previous efforts have failed to generate stable yet high-titer AAV packaging cell lines from 293 cells (4, 23, 40).
Clark et al. have attempted to generate AAV packaging cell lines in parallel from both HeLa cells and 293 cells (4). To their credit, they have succeeded with HeLa cells in generating stable and high-titer AAV packaging cell lines. However, they were unable to generate such cell lines with 293 cells. A similar effort were also made by Chadeuf et al. in an attempt to generate AAV packaging cell line from both HeLa cells and 293 cells (3). Again, they succeeded with the HeLa cells and also partially succeeded in obtaining a 293 cell-based AAV packaging cell line named 293RC21. However, this cell line gave rise to very low titers of AAV vectors (4 × 1010 v.g. particles from 20-by-15-cm cell plates). Furthermore, the capability for AAV vector production decreased by >10-fold after the cell line was passaged in culture for 1 month. Subcultivation of 293RC21 turned out unstable with low vector yield, in sharp contrast to their HeLa cell-based packaging cell line HeRC32, which remained stable and high yield (3). Previous failure to generate 293 cell-based AAV packaging cell lines was primarily due to the constitutive expression of the adenovirus E1A gene in 293 cells that activates the AAV rep gene promoters p5 and p19 (29), which in turn produced four Rep proteins. The latter have been proven to be cytostatic (40) and cytotoxic (28); therefore, preventing the formation of stable cell lines. On the other hand, both AAV p5 and p19 promoters in HeLa cell-based packaging cell lines are silenced, similar to the latent AAV infection, where no AAV gene expression could be detected. Although silencing of the AAV promoters made it possible for HeLa cell-based packaging cell line to grow, such promoter silencing could not be achieved in 293 cells because of the transcription activator E1A acting on AAV promoters.
Several attempts to overcome Rep-mediated cytostatic and cytotoxic effects in 293 cells have been focused on the control of gene expression of the two larger Rep proteins, Rep78 and Rep68, because manipulation of p5 promoter was relatively convenient. Yang et al. made a 293 cell line with inducible rep78 and rep68 gene expression by replacing the p5 promoter with an inducible promoter (40). However, in that cell line the promoter p19 products Rep52 and Rep40, which are essential for virus packaging, were barely detectable even under induced conditions or after adenovirus infection. Furthermore, there was no AAV capsid gene in the cell line. Another effort to control Rep78 and Rep68 expression in 293 cells made by Ozawa and coworkers (23) had also replaced the p5 promoter with an exogenous promoter, a strong promoter followed by a “stuffer” sequence before the Rep coding sequence. The removal of the stuffer sequence by Ad-Cre infection turned on rep78 and rep68 gene expression. Nonetheless, rep52 and rep40, as well as cap genes, could not be induced in the cell line even after Ad-Cre infection or after a different adenovirus infection. The authors of that study attributed the failed gene expression of the essential AAV proteins to their deleterious effects, which prevented the growth of 293 cells that could have functional Rep and Cap proteins (23). An antisense DNA strategy designed by the same group was also unable to generate a stable and high-titer 293-based AAV packaging cell line, and it was concluded that long-term leaky expression of the rep gene products may confer a growth disadvantage on cells (24).
Aiming at simultaneous and tight control of all four Rep protein gene expression, we have devised a novel dual-splicing strategy that blocked the gene expression in the middle of the protein coding sequence. Because the four Rep proteins share the majority of their coding sequences and the promoter p19 is located in the middle of the Rep78 and Rep68 coding sequences, we reasoned that disruption of the rep genes downstream of promoter p19 should stop expression of all four Rep protein genes. An intron was inserted into the shared coding sequence of the four Rep proteins to disrupt the gene (Fig. 1B). The advantage of inserting an intron is that it can in theory be inserted anywhere in the transcribed region, coding or noncoding, and also can be readily and accurately removed through RNA splicing. As a result, the risk of introducing mutations into the coding sequence is minimal. We have shown previously that the intron itself can tolerate insertions (31). The insertion of a polyadenylation tandem into the intron effectively terminated transcription and decreased gene expression by 80-fold. Moreover, an addition of a drug-resistant gene cassette further decreased the gene expression by another 8-fold for a total of >640-fold diminution (Fig. 2). Two loxP sites flanking the transcription termination cassette [poly(A) sites plus the Hygr gene] enabled the alleviation of the transcription blockade after Cre recombinase-mediated DNA splicing (Fig. 1C), resulting in effective restoration of full-length transcription and gene expression (Fig. 2). The key advantage of our dual splicing switch system in controlling AAV rep gene expression is that we could simultaneously control expression of all four Rep protein genes. This was not the case in previous efforts by others in making 293 cell-based AAV packaging cell lines. Merely controlling promoter p5 products could not eliminate the cytostatic and cytotoxic effects from leaky expression of p19 products Rep52 and Rep40 (3, 23, 24, 40). Second, a very tight control of rep gene expression is also important. In our study, besides the triple polyadenylation sites, a drug-resistant gene as part of the termination cassette was also used. It not only rendered synergistic effects in terminating the rep gene transcription with the three poly(A) sites but also served as a selection marker for stable cell line establishment. The stringency of rep gene expression control in our system was partially reflected by the ease of obtaining stable cell clones after transfection and antibiotics selection. We have consistently obtained hundreds of drug-resistant colonies during the process of cell line development. Most of the clones exhibited normal growth rate compared to the parental 293 cells. A large majority of those cell clones retained their inducibility of rep gene expression after Ad-Cre infection, suggesting that the rep gene under the control of the dual splicing switch was very tight and nontoxic. For example, cell line 293-XX2-DS-19 harbors ca. 50 copies of the dual-splicing-controlled AAV rep-cap genes. Nonetheless, no toxicity was observed and the cell line has been very stable throughout the long-term passage and subcloning process. Its progeny cell lines also exhibited similar stability after obtaining additional copies of the controlled rep-cap genes along with the AAV vector sequences, e.g., AAV-GFP and AAV-minidystrophin vectors. It is interesting that AAV rep and cap gene amplification occurred in our GFP and minidystrophin cell lines by 5- to 10-fold during vector production. The amplification phenomenon has been observed in a number of HeLa cell-based AAV packaging cell lines, which showed high-titer AAV vector production (3, 19, 22, 34) (Fig. 9, lanes 1 and 2). In our 293 cell-based cell lines, the cells already had ca. 50 copies of the rep and cap gene. Further amplification of these genes may contribute to further boost AAV vector titers.
An additional advantage of the dual splicing control is the preservation of the AAV endogenous promoters (p5 and p19), which have been proven to be the best regulatory elements by ensuring optimal viral gene expression both temporally and quantitatively during AAV vector production (25, 38). A very important distinction between our strategy and other strategies is that in our system there is no selection pressure to inactivate both p5 and p19 promoters in order to obtain a packaging cell line (3, 4, 14, 20, 26, 32). In contrast, promoter shutoff is a prerequisite for p5 and p19 in previous studies, where these two promoters are expected to turn back on in the packaging cell lines after adenovirus infection. However, it is well known that long-term promoter shutoff may lead to DNA methylation which makes reactivation difficult. This phenomenon seems true in previous 293 cell-based cell lines, especially for promoter p19 (23, 24, 40). This phenomenon may also explain in part why some HeLa cell-based AAV packaging cell lines required very high doses of wild-type adenovirus infection to reactivate AAV gene expression (19). In our 293 cell-based cell lines, however, promoters p5 and p19 are expected to remain constitutively active without causing a deleterious effect to the cells, because the mRNAs were terminated prematurely and rendered nonfunctional until Ad-Cre infection and dual splicing, which restored the coding regions. Thus, the promoters maintained their activities and their responsiveness to adenovirus helper infection throughout. This may be a contributing factor for the modest dose requirement of Ad-Cre infection and the resulting high-titer AAV vector production in our cell lines. Having succeeded in controlling both lacZ and AAV rep gene expression, this technology should also be useful for other applications, for example, for genes with multiple overlapping open reading frames found in other viruses and for genes that require regulation by its endogenous promoters in animal and plant models.
REFERENCES
- 1.Anton, M., and F. L. Graham. 1995. Site-specific recombination mediated by an adenovirus vector expressing the Cre recombinase protein: a molecular switch for control of gene expression. J. Virol. 69:4600-4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berns, K. I. 1990. Parvoviridae and their replication, 2nd ed., vol. 2. Raven Press, New York, N.Y.
- 3.Chadeuf, G., D. Favre, J. Tessier, N. Provost, P. Nony, J. Kleinschmidt, P. Moullier, and A. Salvetti. 2000. Efficient recombinant adeno-associated virus production by a stable rep-cap HeLa cell line correlates with adenovirus-induced amplification of the integrated rep-cap genome. J. Gene Med. 2:260-268. [DOI] [PubMed] [Google Scholar]
- 4.Clark, K. R., F. Voulgaropoulou, D. M. Fraley, and P. R. Johnson. 1995. Cell lines for the production of recombinant adeno-associated virus. Hum. Gene Ther. 6:1329-1341. [DOI] [PubMed] [Google Scholar]
- 5.Collaco, R. F., X. Cao, and J. P. Trempe. 1999. A helper virus-free packaging system for recombinant adeno-associated virus vectors. Gene 238:397-405. [DOI] [PubMed] [Google Scholar]
- 6.de la Luna, S., I. Soria, D. Pulido, J. Ortin, and A. Jimenez. 1988. Efficient transformation of mammalian cells with constructs containing a puromycin-resistance marker. Gene 62:121-126. [DOI] [PubMed] [Google Scholar]
- 7.Drittanti, L., C. Jenny, K. Poulard, A. Samba, P. Manceau, N. Soria, N. Vincent, O. Danos, and M. Vega. 2001. Optimised helper virus-free production of high-quality adeno-associated virus vectors. J. Gene Med. 3:59-71. [DOI] [PubMed] [Google Scholar]
- 8.Flotte, T. R., and B. L. Laube. 2001. Gene therapy in cystic fibrosis. Chest 120:124S-131S. [DOI] [PubMed] [Google Scholar]
- 9.Grimm, D., A. Kern, K. Rittner, and J. A. Kleinschmidt. 1998. Novel tools for production and purification of recombinant adenoassociated virus vectors. Hum. Gene Ther. 9:2745-2760. [DOI] [PubMed] [Google Scholar]
- 10.Grimm, D., and J. A. Kleinschmidt. 1999. Progress in adeno-associated virus type 2 vector production: promises and prospects for clinical use. Hum. Gene Ther. 10:2445-2450. [DOI] [PubMed] [Google Scholar]
- 11.High, K. A. 2001. AAV-mediated gene transfer for hemophilia. Ann. N. Y. Acad. Sci. 953:64-74. [DOI] [PubMed] [Google Scholar]
- 12.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 13.Hunter, L. A., and R. J. Samulski. 1992. Colocalization of adeno-associated virus Rep and capsid proteins in the nuclei of infected cells. J. Virol. 66:317-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue, N., and D. W. Russell. 1998. Packaging cells based on inducible gene amplification for the production of adeno-associated virus vectors. J. Virol. 72:7024-7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, X. C., S. Qin, C. Qiao, K. Kawano, M. Lin, A. Skold, X. Xiao, and A. R. Tall. 2001. Apolipoprotein B secretion and atherosclerosis are decreased in mice with phospholipid-transfer protein deficiency. Nat. Med. 7:847-852. [DOI] [PubMed] [Google Scholar]
- 16.King, J. A., R. Dubielzig, D. Grimm, and J. A. Kleinschmidt. 2001. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 20:3282-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, J., D. Dressman, Y. P. Tsao, A. Sakamoto, E. P. Hoffman, and X. Xiao. 1999. rAAV vector-mediated sarcogylcan gene transfer in a hamster model for limb girdle muscular dystrophy. Gene Ther. 6:74-82. [DOI] [PubMed] [Google Scholar]
- 18.Li, J., R. J. Samulski, and X. Xiao. 1997. Role for highly regulated rep gene expression in adeno-associated virus vector production. J. Virol. 71:5236-5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, X., F. Voulgaropoulou, R. Chen, P. R. Johnson, and K. R. Clark. 2000. Selective Rep-Cap gene amplification as a mechanism for high-titer recombinant AAV production from stable cell lines. Mol. Ther. 2:394-403. [DOI] [PubMed] [Google Scholar]
- 20.Liu, X. L., K. R. Clark, and P. R. Johnson. 1999. Production of recombinant adeno-associated virus vectors using a packaging cell line and a hybrid recombinant adenovirus. Gene Ther. 6:293-299. [DOI] [PubMed] [Google Scholar]
- 21.Maxwell, I. H., G. S. Harrison, W. M. Wood, and F. Maxwell. 1989. A DNA cassette containing a trimerized SV40 polyadenylation signal which efficiently blocks spurious plasmid-initiated transcription. BioTechniques 7:276-280. [PubMed] [Google Scholar]
- 22.Nony, P., J. Tessier, G. Chadeuf, P. Ward, A. Giraud, M. Dugast, R. M. Linden, P. Moullier, and A. Salvetti. 2001. Novel cis-acting replication element in the adeno-associated virus type 2 genome is involved in amplification of integrated rep-cap sequences. J. Virol. 75:9991-9994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogasawara, Y., H. Mizukami, M. Urabe, A. Kume, Y. Kanegae, I. Saito, J. Monahan, and K. Ozawa. 1999. Highly regulated expression of adeno-associated virus large Rep proteins in stable 293 cell lines using the Cre/loxP switching system. J. Gen. Virol. 80(Pt. 9):2477-2480. [DOI] [PubMed] [Google Scholar]
- 24.Okada, T., H. Mizukami, M. Urabe, T. Nomoto, T. Matsushita, Y. Hanazono, A. Kume, K. Tobita, and K. Ozawa. 2001. Development and characterization of an antisense-mediated prepackaging cell line for adeno-associated virus vector production. Biochem. Biophys. Res. Commun. 288:62-68. [DOI] [PubMed] [Google Scholar]
- 25.Pereira, D. J., D. M. McCarty, and N. Muzyczka. 1997. The adeno-associated virus (AAV) Rep protein acts as both a repressor and an activator to regulate AAV transcription during a productive infection. J. Virol. 71:1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiao, C., J. Li, A. Skold, X. Zhang, and X. Xiao. 2002. Feasibility of generating adeno-associated virus packaging cell lines containing inducible adenovirus helper genes. J. Virol. 76:1904-1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rassa, J. C., G. M. Wilson, G. A. Brewer, and G. D. Parks. 2000. Spacing constraints on reinitiation of paramyxovirus transcription: the gene end U tract acts as a spacer to separate gene end from gene start sites. Virology 274:438-449. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt, M., S. Afione, and R. M. Kotin. 2000. Adeno-associated virus type 2 Rep78 induces apoptosis through caspase activation independently of p53. J. Virol. 74:9441-9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi, Y., E. Seto, L. S. Chang, and T. Shenk. 1991. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell 67:377-388. [DOI] [PubMed] [Google Scholar]
- 30.Stedman, H., J. M. Wilson, R. Finke, A. L. Kleckner, and J. Mendell. 2000. Phase I clinical trial utilizing gene therapy for limb girdle muscular dystrophy: alpha-, beta-, gamma-, or delta-sarcoglycan gene delivered with intramuscular instillations of adeno-associated vectors. Hum. Gene Ther. 11:777-790. [DOI] [PubMed] [Google Scholar]
- 31.Sun, L., J. Li, and X. Xiao. 2000. Overcoming adeno-associated virus vector size limitation through viral DNA heterodimerization. Nat. Med. 6:599-602. [DOI] [PubMed] [Google Scholar]
- 32.Tamayose, K., Y. Hirai, and T. Shimada. 1996. A new strategy for large-scale preparation of high-titer recombinant adeno-associated virus vectors by using packaging cell lines and sulfonated cellulose column chromatography. Hum. Gene Ther. 7:507-513. [DOI] [PubMed] [Google Scholar]
- 33.Tantravahi, J., M. Alvira, and E. Falck-Pedersen. 1993. Characterization of the mouse beta maj globin transcription termination region: a spacing sequence is required between the poly(A) signal sequence and multiple downstream termination elements. Mol. Cell. Biol. 13:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tessier, J., G. Chadeuf, P. Nony, H. Avet-Loiseau, P. Moullier, and A. Salvetti. 2001. Characterization of adenovirus-induced inverted terminal repeat-independent amplification of integrated adeno-associated virus rep-cap sequences. J. Virol. 75:375-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, B., J. Li, and X. Xiao. 2000. Adeno-associated virus vector carrying human minidystrophin genes effectively ameliorates muscular dystrophy in mdx mouse model. Proc. Natl. Acad. Sci. USA 97:13714-13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, X. S., B. Khuntirat, K. Qing, S. Ponnazhagan, D. M. Kube, S. Zhou, V. J. Dwarki, and A. Srivastava. 1998. Characterization of wild-type adeno-associated virus type 2-like particles generated during recombinant viral vector production and strategies for their elimination. J. Virol. 72:5472-5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xiao, X., J. Li, and R. J. Samulski. 1996. Efficient long-term gene transfer into muscle tissue of immunocompetent mice by adeno-associated virus vector. J. Virol. 70:8098-8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiao, X., J. Li, and R. J. Samulski. 1998. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 72:2224-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xiao, X., J. Li, Y. P. Tsao, D. Dressman, E. P. Hoffman, and J. F. Watchko. 2000. Full functional rescue of a complete muscle (TA) in dystrophic hamsters by adeno-associated virus vector-directed gene therapy. J. Virol. 74:1436-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, Q., F. Chen, and J. P. Trempe. 1994. Characterization of cell lines that inducibly express the adeno-associated virus Rep proteins. J. Virol. 68:4847-4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou, X., and N. Muzyczka. 1998. In vitro packaging of adeno-associated virus DNA. J. Virol. 72:3241-3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zolotukhin, S., B. J. Byrne, E. Mason, I. Zolotukhin, M. Potter, K. Chesnut, C. Summerford, R. J. Samulski, and N. Muzyczka. 1999. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 6:973-985. [DOI] [PubMed] [Google Scholar]