Abstract
Actin is the principal component of microfilaments. Its assembly/disassembly is essential for cell motility, cytokinesis, and a range of other functions. Recent evidence suggests that actin is present in the nucleus where it may be involved in the regulation of gene expression and that cofilin binds actin and can translocate into the nucleus during times of stress. In this report, we combine fluorescence resonance energy transfer and confocal microscopy to analyze the interactions of cofilin and G-actin within the nucleus and cytoplasm. By measuring the rate of photobleaching of fluorescein-labeled actin in the presence and absence of Cy5-labeled cofilin, we determined that almost all G-actin in the nucleus is bound to cofilin, whereas ∼½ is bound in the cytoplasm. Using fluorescence resonance energy transfer imaging techniques we observed that a significant proportion of fluorescein-labeled cofilin in both the nucleus and cytoplasm binds added tetramethylrhodamine-labeled G-actin. Our data suggest there is significantly more cofilin-G-actin complex and less free cofilin in the nucleus than in the cytoplasm.
INTRODUCTION
Actin is the principal component of eukaryotic microfilaments. It is involved in important processes including cell division, motility, and the maintenance of cell shape (reviewed in dos Remedios et al. (1)). Although actin is usually recognized as a cytoplasmic protein, it may also be located in the nucleus. Evidence dating back almost three decades (2) remains controversial. Some authors (3) have dismissed it as a probable contamination with cytoplasmic actin. Furthermore, the presence of actin inside the nucleus was questioned by researchers who failed to detect nuclear actin with fluorescently labeled phalloidin (4). This peptide binds and stabilizes filamentous F-actin with very high specificity (5), lowers the critical concentration of monomers, but does not itself bind to G-actin.
The actin sequence lacks a nuclear translocation signal and, at 42 kDa, is unlikely to enter the nucleus by diffusion. It therefore relies on a transporter protein, such as cofilin, to mediate this entry that might be promoted by a variety of adverse cellular conditions including heat shock (6,7) and ATP depletion (8). Residues 30–34 of cofilin encode an SV-40-type nuclear translocation signal (KKRKK), perhaps enabling a cofilin-actin complex to pass into the nucleus (7).
The ability of adverse cellular conditions to invoke nuclear translocation of actin suggests that the presence of nuclear actin is a consequence of cell stress-induced disorganization. However, actin is also present in the nucleus under normal physiological conditions. It has been identified in the nucleus of differentiated myogenic cells and oocytes by staining with a monoclonal antibody that specifically recognized G-, but not F-actin (9). This suggests there is a pool of actin monomers or short oligomers present in the nucleus. This notion is supported by the absence of nuclear phalloidin staining and by the presence of G-actin sequestering proteins within the nucleus, including profilin (10) and cofilin (11).
Actin appears to be involved in the regulation of gene expression (12,13), chromatin remodeling (14–16), and the nuclear export of protein (17) and mRNA (13). The presence of two functional leucine-rich nuclear export sequences in the middle of the actin sequence (18) suggests that it may act as a shuttle between the nucleus and cytoplasm.
In this report, we combine confocal microscopy with fluorescence resonance energy transfer (FRET, reviewed elsewhere (19,20)) to evaluate the content of cofilin-actin complex and free cofilin in fixed cells. FRET provides a pathway for the transfer of excitation energy from an excited donor probe to a nearby acceptor. This transfer reduces the fluorescence intensity and lifetime of the donor, thereby decreasing its propensity to photobleach. Quantification of this transfer enables us to distinguish proteins in molecular contact from those merely in the same confocal volume.
Binding of exogenous actin to intrinsic cofilin, and endogenous cofilin to endogenous actin was monitored using FRET between donor and acceptor probes on actin and cofilin. FRET efficiency was calculated from either the reduced fluorescence intensity (quenching) or the reduced rate of photobleaching of donor probes (fluorescein isothiocyanate (FITC) or iodoacetamide fluorescein (IAF)) in the presence of excess functional acceptor probes (tetramethylrhodamine (TMR) or Cy5). FRET efficiencies indicate the proportion of labeled proteins in molecular contact.
MATERIALS AND METHODS
Preparation of G-actin
Actin was prepared from an acetone-dried powder of rabbit skeletal muscle according to the method of Spudich and Watt (21), with slight modifications as described in Barden and dos Remedios (22). Monomeric actin concentration was determined from its optical density at 290 nm (OD290), where E0.1% = 0.63 cm−1 (23).
Fluorescent labeling of G-actin
Actin was labeled on cysteine 374 by overnight incubation at 4°C with a threefold excess of TMR conjugated to maleimide. The reaction was stopped by addition of dithiothreitol (DTT) to a final concentration of 5 mM. The actin solution was clarified by centrifugation at 10,000 × g for 5 min and excess label was removed by repeated dialysis and polymerization/depolymerization. Actin was polymerized by dialyzing overnight against F-buffer (2 mM Tris, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, 0.2 mM DTT, 4 mM MgCl2, 0.1 M KCl) at 4°C, pelleted by ultracentrifugation at 40,000 × g for 1 h, followed by dialysis against G-buffer (2 mM Tris, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, 0.2 mM DTT) for 48 h at 4°C, with a change in dialysate after 24 h. The resultant TMR-G-actin solution was clarified at 4°C by ultracentrifugation at 40,000 × g for 1 h.
Expression and purification of recombinant DNase I
The clone for recombinant DNase I was a kind gift from Dr. B. A. Connolly (University of Newcastle, Newcastle, UK). JM105 Escherichia coli were transformed with plasmid (pAW4) in a Micropulser electroporator (Bio-Rad, Hercules, CA) and transformants were grown at 37°C in LB medium (Gibco, Carlsbad, CA) containing 50 μg/mL ampicillin. Protein expression was induced when the cells were in log phase of growth (OD600 ≈ 0.6/cm) by addition of isopropyl-β-D-thiogalactopyranoside to a final concentration of 1 mM. After 3 h growth, the cells were harvested by centrifugation, resuspended in 100 mL of DNase-lysis buffer (10 mM Tris, pH 8.0, 50 mM NaCl, 2 mM CaCl2), and disrupted using a French press. The lysate was clarified by centrifugation at 10,000 × g for 1 h and applied to a Q-Sepharose column (Pharmacia, Uppsala, Sweden) and eluted with a 5–350 mM NaCl gradient in lysis buffer. Fractions containing DNase I were concentrated using 10K Omega centrifugal concentrators (Pall, Ann Arbor, MI), followed by passage through a Sephacryl S-200 (Pharmacia) column in lysis buffer. Concentrated DNase I was dialyzed for 24 h against G-buffer and its concentration was determined from the OD280, where E0.1% = 1.1 cm−1 (24).
Fluorescent labeling of DNase I
Fluorescently labeled DNase I is used to identify actin monomers in fixed cells. Labeling of a Cys residue was achieved by overnight incubation at 4°C with a threefold excess of IAF. The reaction was stopped by addition of DTT to a final concentration of 5 mM. Excess label was removed by overnight dialysis against 10 mM PIPES (pH 6.8) followed by passage through a disposable PD-10 (Sephadex G-25) desalting column (Pharmacia).
Cell culture and immunohistochemistry
Vero African green monkey fibroblasts (ACTC, Manassas, VA) were suspended in 10% fetal calf serum (FCS) in RPMI-1640 medium (Sigma-Aldrich; St. Louis, MO) and incubated overnight in a 35-mL culture flask under standard culture conditions (37°C in a humidified atmosphere of 5% CO2 and air). After incubation, culture medium was removed and cells were washed three times in phosphate-buffered saline (PBS; 0.15 M NaCl, 2 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.2) and resuspended in T/E solution (PBS containing 0.5% w/v trypsin and 1 mM EDTA). Cells were centrifuged at 4000 × g for 5 min and the pellet was washed in PBS and resuspended in 10% FCS in RPMI-1640 to a final density of 106 cells/mL. Cells were subsequently seeded on 18-mm glass coverslips and incubated overnight under standard culture conditions. Cells were then washed three times in PBS, fixed in freshly prepared 3% paraformaldehyde in PBS for 20 min at room temperature and permeabilized in 100% acetone at −20°C for 5 min. Paraformaldehyde was favored over formaldehyde as a fixative because it does not require methanol to improve solubility. Methanol causes protein coagulation resulting in decreased antigenicity and loss of cell architecture (25). Cells were subsequently washed and incubated in 10% FCS in RPMI-1640 for 45 min to reduce nonspecific interactions.
For FRET imaging studies, cofilin was labeled by incubating in a 1:200 dilution of rabbit anticofilin primary antibody (Cytoskeleton, Denver, CO) for 45 min followed by a 1:200 dilution of a sheep antirabbit antibody conjugated to FITC (Silenus, Hawthorn, Australia) for 1 h. Cells were subsequently incubated with 0.25 μM TMR-actin monomers that labeled all available actin-binding proteins. Cells were washed three times in PBS between each step.
For fluorescence decay experiments, cofilin was labeled by incubating in a 1:200 dilution of rabbit anticofilin antibody for 45 min followed by a 1:200 dilution of a donkey antirabbit antibody conjugated to Cy5 (Silenus) for 1 h. Endogenous G-actin was labeled with 0.25 μM DNase I conjugated to donor IAF (IAF-DNase I). Cells were washed three times in PBS between each step.
Confocal microscopy and FRET imaging
Cells were viewed with a BioRad Radiance 2100 confocal microscope (Hemel Hempstead, Hertfordshire, UK). Distribution of intracellular FITC-labeled cofilin was observed by excitation with an Ar-488 laser and viewing through an HQ 515/30 bandpass filter. Distribution of free actin-binding protein labeled with TMR-actin was observed by excitation with a He-Ne-543 laser and viewing through a 570LP bandpass filter; ∼1 μm optical sections were taken in the z axis.
The presence of endogenous cofilin with an exposed actin-binding domain was determined by assessing the ability of FITC-immunolabeled cofilin to bind, and therefore undergo FRET, with exogenously added TMR-G-actin. FRET was determined by observing enhancement of FITC donor fluorescence after selective photobleaching of the TMR acceptor within the region of interest. TMR acceptor was selectively photobleached by prolonged, wavelength-specific (He-Ne-543) high-energy laser excitation for 30 s. FITC fluorescence intensity before and after photobleaching of TMR was quantified by Image Pro-Plus 4.5 (Scitech, Victoria, Australia) and FRET efficiency was calculated by using the following equation:
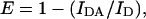 |
(1) |
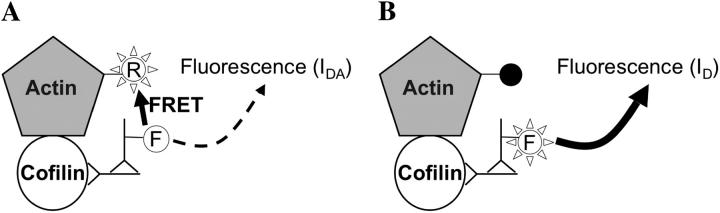
where E is FRET efficiency, IDA is the intensity of donor fluorescence in the presence of functional acceptor, and ID is the intensity of donor fluorescence where the acceptor was photodestroyed by laser irradiation (Fig. 1). The intensity was calculated by integration over a region of interest to determine an average intensity rather than a point value.
FIGURE 1.
Fluorescence intensity of FITC donor immunolabeled cofilin before (A) and after (B) selective photobleaching of TMR-G-actin acceptor. Abolition of FRET increases FITC fluorescence intensity from IDA to ID.
Photobleaching kinetics
The content of cofilin-G-actin complex was determined by affinity labeling endogenous G-actin with IAF-DNase I acceptor probe and immunolabeling endogenous cofilin with FITC donor probe. Endogenous cofilin-actin complex would thus be part of multiprotein complexes containing IAF and Cy5 probes that would undergo FRET. FRET between IAF and Cy5 probes would decrease the lifetime of the IAF donor probe and reduce its propensity to photobleach. FRET between the IAF-DNase I affinity label bound to G-actin and Cy5-immunolabeled cofilin was quantified from the rate of fluorescence decay of the IAF donor in the presence and absence of the Cy5 acceptor. The IAF donor was excited with an Ar-488 laser and emission was monitored through an HQ 515/30 bandpass filter. FRET efficiency was calculated from decay rates using the following equation:
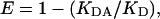 |
(2) |
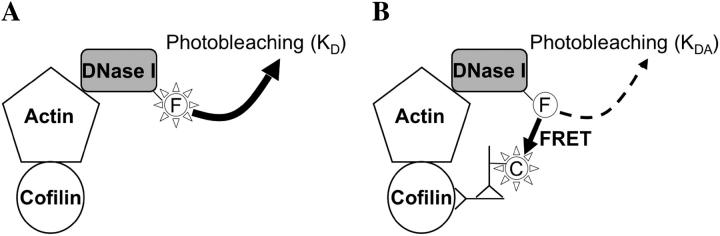
where E is FRET efficiency, KDA is the decay rate of donor fluorescence in the presence of acceptor, and KD is the decay rate of donor fluorescence in the absence of acceptor (Fig. 2).
FIGURE 2.
IAF-DNase I affinity-labeled actin photobleaches with an exponential rate constant of KD (A). In the presence of Cy5 immunolabeled cofilin (B), FRET between IAF and Cy5 decreases fluorescence lifetime of the IAF donor probe and reduces its rate of photobleaching to KDA.
RESULTS
Determination of free cofilin
In the nucleus and cytoplasm of paraformaldehyde-fixed cells, the amount of cofilin with an available actin-binding domain was determined by FRET. We immunolabeled endogenous cofilin with a donor probe (anticofilin primary antibody, FITC-conjugated secondary antibody) and assessed its ability to bind and undergo FRET to exogenous TMR-G-actin acceptor. Cofilin already bound to endogenous actin would not bind the TMR-G-actin. FRET efficiency was determined by monitoring the increase in donor fluorescence intensity after selective photodestruction of the acceptor, as described above.
Fig. 3 demonstrates the photobleaching of exogenous TMR-G-actin in the nuclear and cytoplasmic regions of interest (circles) of representative cells and its effect on endogenous FITC-immunolabeled cofilin.
FIGURE 3.
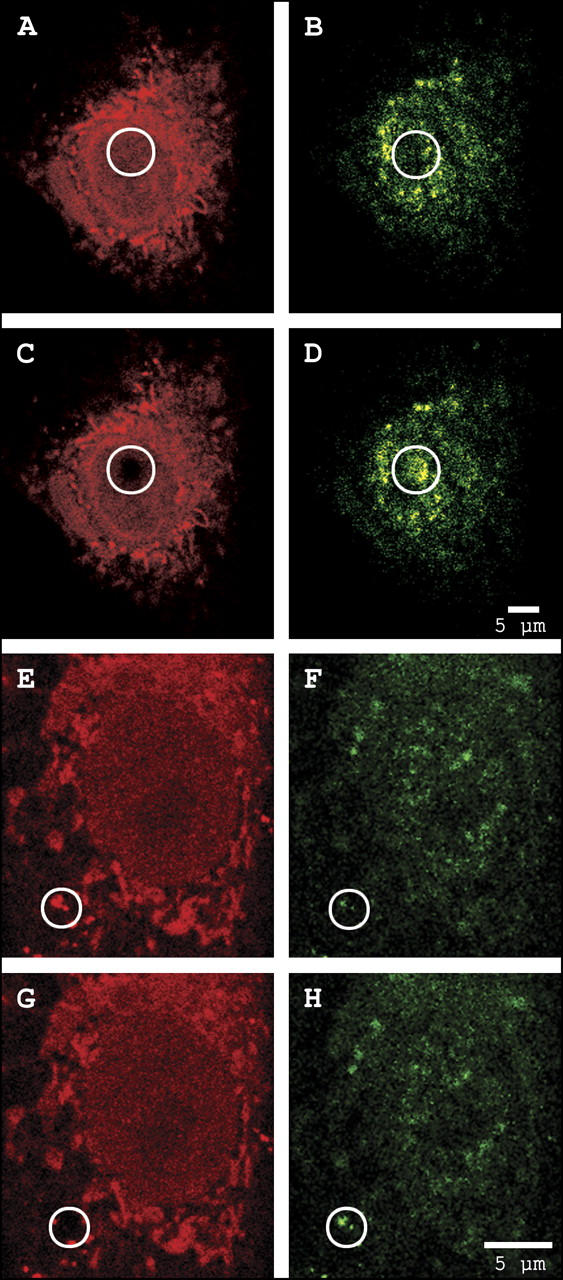
Observation of FRET between exogenous TMR-labeled actin (red) and endogenous FITC-immunolabeled cofilin (green) in the nucleus (A–D) and cytoplasm (E–H). Photodestruction of TMR in the region of interest (circles) results in a corresponding increase in FITC fluorescence intensity. See text for details.
Fig. 3, A and E, demonstrates the distribution of G-actin binding proteins (ABPs) with exposed actin-binding domains labeled by exogenous TMR-G-actin. ABPs are present in excess throughout the cell but a smaller excess is present within the nucleus. The perinuclear region contains discrete clusters of TMR-G-actin bound to free ABPs. These clusters are ideal for FRET analysis because of the presence of excess TMR acceptor.
Fig. 3, B and F, demonstrates the distribution of intracellular cofilin immunolabeled with FITC. Cofilin is dispersed throughout the cell, with a higher concentration in the perinuclear region.
Fig. 3, C and G, demonstrates selective photobleaching of the TMR acceptor in the region of interest, with a corresponding increase in fluorescence intensity of the FITC donor probe demonstrated in Fig. 3, D and G, respectively. This increased intensity of FITC-immunolabeled cofilin is due to the loss of FRET resulting from selective photodestruction of TMR acceptor probe conjugated to exogenous actin.
These data confirm the presence of free cofilin within the nucleus and the cytoplasm. FRET efficiency, and therefore the fraction of cofilin capable of binding G-actin, was determined from FITC fluorescence intensities before and after selective photobleaching of TMR and was calculated to be 37% in the nucleus and 69% in the cytoplasm.
Omission of primary anticofilin antibody produced no increase in background FITC donor fluorescence upon TMR acceptor photobleaching (results not shown), demonstrating the validity of the FRET signal.
Analysis of cofilin-G-actin complex
The prevalence of a cofilin-G-actin complex was analyzed by affinity-labeling endogenous actin monomers with IAF-DNase I and immunolabeling cofilin with Cy5 acceptor (unlabeled anticofilin primary antibody, Cy5-conjugated secondary antibody). Fluorescence of IAF was measured as a function of time to determine the level of this molecular interaction.
The distinction between molecular interaction (binding) and molecular proximity was assessed by quantifying the protective effect of FRET on the rate of photobleaching of IAF. The photobleaching process and FRET represent alternative pathways for the loss of excitation energy of a donor probe. Consequently, endogenous G-actin affinity-labeled with IAF undergoing FRET to Cy5-immunolabeled cofilin displays a reduced rate of photobleaching.
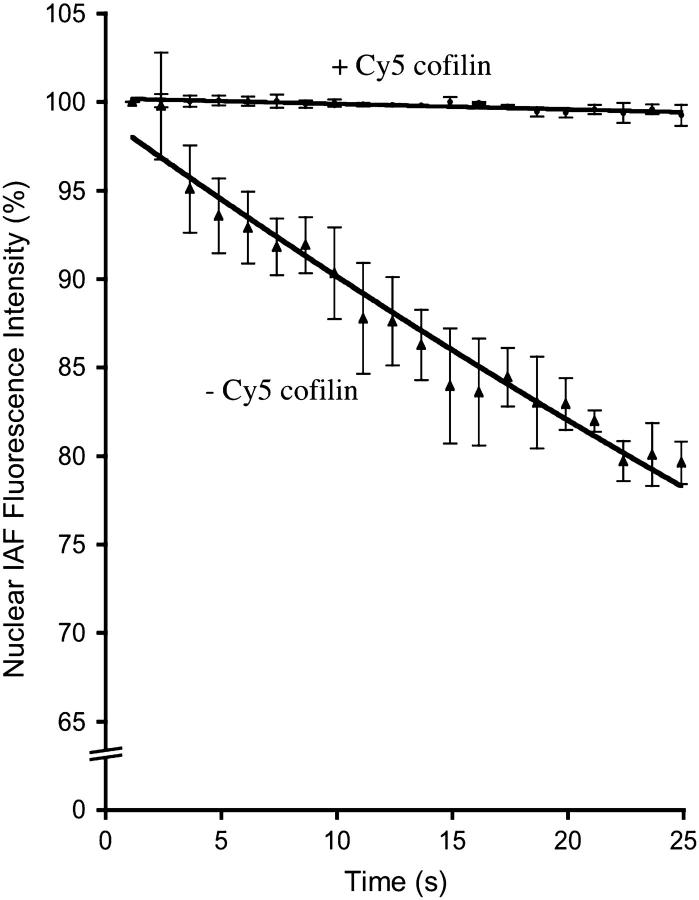
The average rate constant (KD) for exponential decay of the IAF-DNase I affinity probe on actin in the absence of a functional acceptor probe was calculated to be 0.0095 (Fig. 4, upper curve, where r2 = 0.96 and n = 3). In the presence of Cy5-immunolabeled cofilin, the average rate of photobleaching of IAF decreased (Fig. 4, lower curve) by more than an order of magnitude (KDA = 0.0003; r2 = 0.76, n = 3). FRET efficiency was calculated to be 97%, suggesting that almost all G-actin in the nucleus is sequestered by cofilin.
FIGURE 4.
Fluorescence intensity as a function of time of endogenous actin affinity-labeled with IAF-DNase I in the presence and absence of Cy5-immunolabeled endogenous cofilin. IAF affinity-labeled G-actin donor undergoes exponential decay with KD = 0.0095. In the presence of Cy5-immunolabeled cofilin, the rate of photobleaching of the IAF affinity label on actin is reduced with KDA = 0.0003.
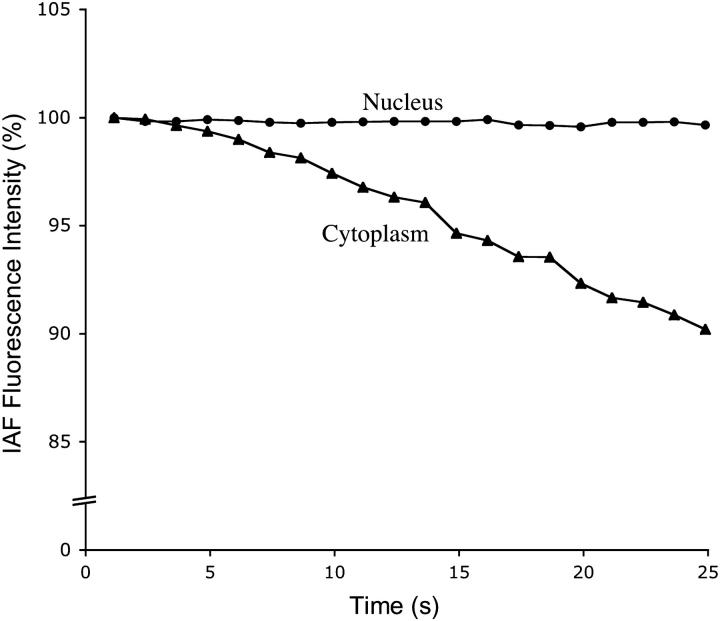
Simultaneous measurements of IAF-DNase I-labeled G-actin fluorescence in the presence of Cy5-immunolabeled cofilin were taken in the nucleus and cytoplasm of a representative cell to compare the content of cofilin-G-actin complexes in the two compartments (Fig. 5). The KDA for exponential decay of IAF donor probe was 0.0001 (r2 = 0.35) in the nucleus and 0.0046 (r2 = 0.98) in the cytoplasm, corresponding to FRET efficiencies of 52% and 99%, respectively. Thus, ∼½ the G-actin in the cortex appears to be bound to cofilin.
FIGURE 5.
Comparison of fluorescence intensity as a function of time of nuclear (upper curve) and cytoplasmic (lower curve) actin affinity-labeled with IAF-DNase I in the presence of Cy5-immunolabeled endogenous cofilin. The IAF donor probe undergoes exponential decay with KD = 0.0001 and 0.0046 in the nucleus and cytoplasm, respectively.
DISCUSSION
The role of cofilin in depolymerizing actin, as well as its regulation of treadmilling and nucleotide exchange of actin, is well understood (26). The binding of cofilin to actin is primarily controlled by phosphorylation of Ser-3 by LIM kinase (27), although pH (28) and the binding of specific phosphoinositides, particularly PIP2 (29), are also involved.
In this report, we identified the cofilin-G-actin complex in the nucleus and cytoplasm by comparing the rate of fluorescence loss of the IAF-DNase I affinity label bound to actin monomers in the presence and absence of Cy5-immunolabeled cofilin. We also quantified cofilin that was not complexed to actin by comparing fluorescence intensity of FITC-immunolabeled cofilin in the presence of added actin conjugated to a functional or nonfunctional TMR probe.
For FRET to occur, the interprobe distance must be within 1.5 times the Förster distance (R0) (i.e., <100 Å) and the probes must be able to precess freely (30). Probe motion (IAF, TMR) around the covalent bonds linking them to Cys residues in actin or DNase I allows some degree of molecular flexibility. In addition, a much larger motion arises from the flexible links between the probes (FITC and Cy5) that are indirectly attached to cofilin by a pair of antibodies. This combined motion of probes allows them to transiently approach and undergo FRET.
FRET efficiency is related to the fourth power of the donor-acceptor distance. When probe pairs approach to within 1.5 × R0, FRET quenches donor fluorescence and reduces fluorescence lifetime, thus imparting a resistance to photobleaching (31). Measurement of this quenching and photobleaching resistance allows calculation of FRET efficiency and determination of whether proteins indirectly labeled by FRET probes are in molecular contact. The calculated FRET efficiency is best described as semiquantitative. It does, however, allow comparison between two cells or cellular compartments. As the cells observed in these experiments are fixed, cytoskeletal remodeling is inhibited. Thus, we are in effect monitoring a static population of actin and cofilin.
Our results demonstrate that cofilin and monomeric actin are present in the nucleus under normal physiological conditions and that almost all of this actin is sequestered by cofilin. In contrast, ∼½ the G-actin is associated with cofilin in the cytoplasm. The observed excess of cofilin in both the nucleus and cytoplasm is consistent with a previous report suggesting that there is an excess of cofilin over actin in vivo (32).
Despite the excess of cofilin in the cytoplasm, a smaller proportion of cytoplasmic actin is associated with cofilin. This may be attributed to the inactivation of cofilin by phosphorylation (33). Phosphorylated cofilin is known to be dispersed throughout the cytoplasm (34) with dephosphorylation, mediated by the Slingshot family of proteases (35), restoring its affinity for actin (36) and correlating with its translocation into the nucleus in many (27,37), but not all (38), cell types. The presence of a large number of competing ABPs within the cytosol may also explain the reduced interaction of cofilin and actin.
Cofilin displays ∼2 orders of magnitude greater affinity for actin containing bound ADP rather than ATP (39) and therefore preferentially binds to ADP-actin (40). This allows cofilin to selectively recycle mature actin filaments that primarily contain ADP.
Our results together with previous reports are consistent with the suggestion that actin may continually shuttle between the nucleus and cytoplasm under normal physiological conditions (4). We propose a model in which cofilin transports ADP-actin subunits into the nucleus. Once in the nucleus, the threefold higher ATP concentration in the nucleoplasm over the cytoplasm (41) promotes the exchange of ADP for ATP. Further disruption of the cofilin-actin complex in the nucleus could be promoted by phosphorylation of cofilin by nuclear LIM-kinase (8). We speculate that the net effect of this would be to increase the concentration of ATP-actin monomers in the nucleus capable of assembling to form short oligomers that could influence chromatin remodeling or gene expression (15). Free actin may also interact with nuclear profilin (42) to enhance exportin-6-mediated nuclear export (10).
It is probable that a population of actin monomers exists within the nucleus bound to profilin. However profilin and DNase I bind with negative cooperativity to actin (43) and thus profilin-actin complexes could not be identified with our IAF-DNase I affinity label.
Actin is a well-known inhibitor of DNase I activity in vitro (44) and the complex can be stabilized by cofilin (45). In addition to inhibiting the hydrolytic activity of DNase I, it has been suggested that actin sequesters DNase I in the cytoplasm and prevents its entry into the nucleus (44).
Although DNase I is primarily considered a secretory glycoprotein, a growing body of evidence suggests that it may mediate DNA degradation during apoptosis (reviewed in Mannherz et al. and Oliveri et al. (46,47)). In addition to delivering actin monomers into the nucleus during times of cell stress (8), cofilin may also transport DNase I into the nucleus by forming a tight cofilin-actin-DNase I ternary complex, as demonstrated in vitro (45). However, the labeling of endogenous actin requires an exposed DNase-I binding locus. Thus intracellular actin already complexed to endogenous DNase I alone, or in a ternary complex with cofilin, remained unlabeled upon probing with IAF-DNase I.
In conclusion, this is the first report, to our knowledge, to demonstrate that actin primarily exists in a complex with cofilin in the nucleus. We also demonstrate that a significantly higher proportion of cytoplasmic actin is not bound to cofilin despite a significant excess of cofilin over actin.
Acknowledgments
The authors thank Dr. Neil J Nosworthy for his assistance with protein preparation, and Dr. Maria Luz Paje (Bio-Rad Laboratories, Sydney, Australia) for her assistance with confocal microscopy.
References
- 1.dos Remedios, C. G., D. Chhabra, M. Kekic, I. V. Dedova, M. Tsubakihara, D. A. Berry, and N. J. Nosworthy. 2003. Actin binding proteins: Regulation of cytoskeletal microfilaments. Physiol. Rev. 83:433–473. [DOI] [PubMed] [Google Scholar]
- 2.Clarke, T. G., and R. W. Merriam. 2003. Diffusible and bound actin in nuclei of Xenopus laevis oocytes. Cell. 12:883–891. [DOI] [PubMed] [Google Scholar]
- 3.Fukui, Y., and H. Katsumaru. 1979. Nuclear actin bundles in Amoeba, Dictyostelium and human HeLa cells induced by dimethyl sulfoxide. Exp. Cell Res. 120:451–455. [DOI] [PubMed] [Google Scholar]
- 4.Rando, O. J., K. Zhao, and G. R. Crabtree. 2000. Searching for a function for nuclear actin. Trends Cell Biol. 10:92–97. [DOI] [PubMed] [Google Scholar]
- 5.McGough, A., B. Pope, W. Chiu, and A. Weeds. 1997. Cofilin changes the twist of F-actin: Implications for actin filament dynamics and cellular function. J. Cell Biol. 138:771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iida, K., S. Matsumoto, and I. Yahara. 1992. The KKRKK sequence is involved in heat shock-induced nuclear translocation of the 18-kDa actin-binding protein, cofilin. Cell Struct. Funct. 17:39–46. [DOI] [PubMed] [Google Scholar]
- 7.Nishida, E., K. Iida, N. Yonezawa, S. Koyasu, I. Yahara, and H. Sakai. 1987. Cofilin is a component of intranuclear and cytoplasmic actin rods induced in cultured cells. Proc. Natl. Acad. Sci. USA. 84:5262–5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pendleton, A., B. Pope, A. Weeds, and A. Koffer. 2003. Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J. Biol. Chem. 278:14394–14400. [DOI] [PubMed] [Google Scholar]
- 9.Gonsior, S. M., S. Platz, S. Buchmeier, U. Scheer, B. N. Jockusch, and H. Hinssen. 1999. Conformational difference between nuclear and cytoplasmic actin as detected by a monoclonal antibody. J. Cell Sci. 112:797–809. [DOI] [PubMed] [Google Scholar]
- 10.Stuven, T., E. Hartmann, and D. Gorlich. 2003. Exportin 6: a novel nuclear export receptor that is specific for profilin-actin complexes. EMBO J. 22:5928–5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chhabra, D., S. Bao, and C. G. dos Remedios. 2002. The distribution of cofilin and DNase I in vivo. Cell Res. 12:207–214. [DOI] [PubMed] [Google Scholar]
- 12.Egly, J. M., N. G. Miyamoto, V. Moncollin, and P. Chambon. 1984. Is actin a transcription initiation factor for RNA polymerase B? EMBO J. 3:2363–2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheer, U., H. Hinssen, W. W. Franke, and B. M. Jockusch. 1984. Microinjection of actin-binding proteins and actin antibodies demonstrates involvement of nuclear actin in transcription of lampbrush chromosomes. Cell. 39:111–122. [DOI] [PubMed] [Google Scholar]
- 14.Jamil, K. 1984. Decondensation of human spermatozoal chromatin by nuclear actin polymerization. Arch. Androl. 13:137–146. [DOI] [PubMed] [Google Scholar]
- 15.Olave, I. A., S. I. Reck-Peterson, and G. R. Crabtree. 2002. Nuclear actin and actin-related proteins in chromatin remodelling. Annu. Rev. Biochem. 71:755–781. [DOI] [PubMed] [Google Scholar]
- 16.Rungger, D., E. Rungger-Brandle, C. Chaponnier, and G. Gabbiani. 1979. Intranuclear injection of anti-actin antibodies into Xenopus oocytes blocks chromosome condensation. Nature. 282:320–321. [DOI] [PubMed] [Google Scholar]
- 17.Hoffman, W., B. Reichart, A. Ewald, E. Muller, I. Schmitt, R. H. Stauber, F. Lottspeich, B. M. Jockusch, U. Scheer, J. Hauber, and M. C. Dabauvalle. 2001. Cofactor requirements for nuclear export of Rev response element (RRE)- and constitutive transport element (CTE)-containing retroviral RNAs. An unexpected role for actin. J. Cell Biol. 152:895–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wada, A., M. Fukuda, M. Mishima, and E. Nishida. 1998. Nuclear export of actin: a novel mechanism regulating the subcellular localization of a major cytoskeletal protein. EMBO J. 17:1635–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenworthy, A. K. 2001. Imaging protein-protein interactions using fluorescence resonance energy transfer. Methods. 24:289–296. [DOI] [PubMed] [Google Scholar]
- 20.Chhabra, D., and C. G. dos Remedios. 2005. Fluorescence resonance energy transfer. Nature Encyc. Life Science. In press. [DOI] [PMC free article] [PubMed]
- 21.Spudich, J. A., and S. Watt. 1971. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies on the interaction of tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246:4866–4871. [PubMed] [Google Scholar]
- 22.Barden, J. A., and C. G. dos Remedios. 1984. The environment of the high-affinity cation binding site on actin and the separation between cation and ATP sites as revealed by proton NMR and fluorescence spectroscopy. J. Biochem. (Tokyo) 96:913–921. [DOI] [PubMed] [Google Scholar]
- 23.Houk, W., and K. Ue. 1974. The measurement of actin concentration in solution: A comparison of methods. Anal. Biochem. 62:66–74. [DOI] [PubMed] [Google Scholar]
- 24.Lindberg, U. 1967. Molecular weight and amino acid composition of deoxyribonuclease I. Biochemistry. 6:335–342. [DOI] [PubMed] [Google Scholar]
- 25.Hoetelmans, R. W., F. A. Prins, I. Cornelese-ten Velde, J. van der Meer, C. J. van de Velde, and J. H. van Dierendonck. 2001. Effects of acetone, methanol, or paraformaldehyde on cellular structure, visualized by reflection contrast microscopy and transmission and scanning electron microscopy. Appl. Immunohistochem. Mol. Morphol. 9:346–351. [DOI] [PubMed] [Google Scholar]
- 26.Bamburg, J. R. 1999. Proteins of the ADF/cofilin family: Essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15:185–230. [DOI] [PubMed] [Google Scholar]
- 27.Nebl, G., S. C. Meuer, and Y. Samstag. 1996. Dephosphorylation of serine 3 regulates nuclear translocation of cofilin. J. Biol. Chem. 271:26276–26280. [DOI] [PubMed] [Google Scholar]
- 28.Yonezawa, N., E. Nishida, and H. Sakai. 1985. pH control of actin polymerization by cofilin. J. Biol. Chem. 260:14410–14412. [PubMed] [Google Scholar]
- 29.Yonezawa, N., E. Nishida, K. Iida, I. Yahara, and H. Sakai. 1990. Inhibition of the interactions of cofilin, destrin and deoxyribonuclease I with actin by phosphoinositides. J. Biol. Chem. 265:8382–8386. [PubMed] [Google Scholar]
- 30.dos Remedios, C. G., and P. D. J. Moens. 1999. Fluorescence resonance energy transfer—applications in protein chemistry. In Resonance Energy Transfer. D. L. Andrews and A. A. Demidov, editors. John Wiley & Sons, Chichester, UK. 1–64.
- 31.Tramier, M., I. Gautier, T. Piolot, S. Ravalet, K. Kemnitz, J. Coppey, C. Durieux, V. Mignotte, and M. Coppey-Moisan. 2002. Picosecond-hetero-FRET microscopy to probe protein-protein interactions in live cells. Biophys. J. 83:3570–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yonezawa, N., E. Nishida, S. Koyasu, S. Maekawa, Y. Ohta, I. Yahara, and H. Sakai. 1987. Distribution among tissues and intracellular localization of cofilin, a 21kDa actin-binding protein. Cell Struct. Funct. 12:443–452. [DOI] [PubMed] [Google Scholar]
- 33.Morgan, T. E., R. O. Lockerbie, L. S. Minamide, M. D. Browning, and J. R. Bamburg. 1993. Isolation and characterization of a regulated form of actin depolymerizing factor. J. Cell Biol. 122:623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arai, H., and Y. Atomi. 2003. Suppression of cofilin phosphorylation in insulin-stimulated ruffling membrane formation in KB cells. Cell Struct. Funct. 28:41–48. [DOI] [PubMed] [Google Scholar]
- 35.Niwa, R., K. Nagata-Ohashi, M. Takeichiz, K. Mizuno, and T. Uemura. 2002. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 108:233–246. [DOI] [PubMed] [Google Scholar]
- 36.Agnew, B. J., L. S. Minamide, and J. R. Bamburg. 1995. Reactivation of phosphorylated actin depolymerizing factor and identification of the regulatory site. J. Biol. Chem. 270:17582–17587. [DOI] [PubMed] [Google Scholar]
- 37.Ohta, Y., E. Nishida, H. Sakai, and E. Miyamoto. 1989. Dephosphorylation of cofilin accompanies heat shock-induced nuclear accumulation of cofilin. J. Biol. Chem. 267:16143–16148. [PubMed] [Google Scholar]
- 38.Abe, H., R. Nagaoka, and T. Obinata. 1993. Cytoplasmic localization and nuclear transport of cofilin in cultured myotubes. Exp. Cell Res. 206:1–10. [DOI] [PubMed] [Google Scholar]
- 39.Carlier, M. F., V. Laurent, J. Santolini, R. Melki, D. Didry, G. X. Xia, Y. Hong, N. H. Chua, and D. Pantaloni. 1997. Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implications for actin-based motility. J. Cell Biol. 136:1307–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chhabra, D., N. J. Nosworthy, and C. G. dos Remedios. 2000. The role of ATP, ADP and divalent cations in the formation of binary and ternary complexes of actin, cofilin and DNase I. Electrophoresis. 21:3863–3869. [DOI] [PubMed] [Google Scholar]
- 41.Miller, D. S., and S. B. Horowitz. 1986. Intracellular compartmentalization of adenosine triphosphate. J. Biol. Chem. 261:13911–13915. [PubMed] [Google Scholar]
- 42.Goldschmidt-Clermont, P. J., L. M. Machesky, J. J. Baldassare, and T. D. Pollard. 1991. The mechanism of interaction of human platelet profilin with actin. J. Cell Biol. 113:1081–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ballweber, E., K. Giehl, E. Hannappel, T. Huff, B. M. Jockusch, and H. G. Mannherz. 1998. Plant profilin induces actin polymerization from actin:β-thymosin complexes and competes directly with β-thymosins and with negative co-operativity with DNase I for binding to actin. FEBS Lett. 425:251–255. [DOI] [PubMed] [Google Scholar]
- 44.Lazarides, E., and U. Lindberg. 1974. Actin is the naturally occurring inhibitor of deoxyribonuclease I. Proc. Natl. Acad. Sci. USA. 71:4742–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nosworthy, N. J., M. Kekic, and C. G. dos Remedios. 2001. The affinity of chick cofilin for actin increases when actin is complexed with DNase I: Formation of a cofilin-actin-DNase I ternary complex. Proteomics. 1:1513–1518. [DOI] [PubMed] [Google Scholar]
- 46.Mannherz, H. G., M. C. Peitsch, S. Zanotti, R. Paddenberg, and B. Polzar. 1995. A new function for an old enzyme: The role of DNase I in apoptosis. Curr. Top. Microbiol. Immunol. 198:161–174. [DOI] [PubMed] [Google Scholar]
- 47.Oliveri, M., A. Daga, C. Cantoni, C. Lunardi, R. Millo, and A. Puccetti. 2001. DNase I mediates internucleosomal DNA degradation in human cells undergoing drug-induced apoptosis. Eur. J. Immunol. 31:743–751. [DOI] [PubMed] [Google Scholar]