Abstract
Despite their high frequency of RNA recombination, the plus-strand coronaviruses have a characteristic, strictly conserved genome organization with the essential genes occurring in the order 5′-polymerase (pol)-S-E-M-N-3′. We have investigated the significance of this remarkable conservation by rearrangement of the murine coronavirus genome through targeted recombination. Thus, viruses were prepared with the following gene order: 5′-pol-S-M-E-N-3′, 5′-pol-S-N-E-M-3′, 5′-pol-M-S-E-N-3′, and 5′-pol-E-M-S-N-3′. All of these viruses were surprisingly viable, and most viruses replicated in cell culture with growth characteristics similar to those of the parental virus. The recombinant virus with the gene order 5′-pol-E-M-S-N-3′ was also tested for the ability to replicate in the natural host, the mouse. The results indicate that the canonical coronavirus genome organization is not essential for replication in vitro and in vivo. Deliberate rearrangement of the viral genes may be useful in the generation of attenuated coronaviruses, which due to their reduced risk of generating viable viruses by recombination with circulating field viruses, would make safer vaccines.
Presently, the main criteria used in the taxonomy of viruses are the type and character of the viral genome, the strategy of viral replication, and the structure of the virion. In general, therefore, viruses that belong to one order or family have similar genome organizations and gene orders.
Nidovirales is an order of enveloped, positive-strand RNA viruses with nonsegmented genomes that comprises the families Arteriviridae and Coronaviridae. Although their constituent genera differ markedly in virion structure, there are striking resemblances among the viruses in the organization and expression of their genomes (6). They share a genome organization in which the order of the essential genes is fixed: 5′-polymerase (pol) genes-envelope protein genes-nucleocapsid (N) gene-3′. The very large pol1a and pol1b genes constitute about two-thirds of each genome. These are translated directly from the genomic RNA (gRNA), the more downstream pol1b gene being expressed by translational read-through using a ribosomal frameshift mechanism. The genes located downstream of pol1b are expressed from a 3′-coterminal nested set of subgenomic (sg) RNAs, each of which also contains a short 5′ leader sequence derived from the 5′ end of the genome. Transcription regulatory sequences (TRSs) located upstream of each gene serve as signals for the transcription of the sgRNAs. The leader sequence is joined at a TRS to all genomic sequence distal to that TRS by discontinuous transcription, most likely during the synthesis of negative-strand sgRNAs (6, 19, 30).
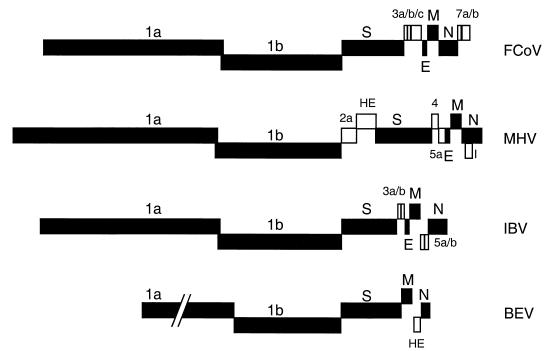
The Coronaviridae comprise two genera, coronaviruses and toroviruses. Viruses belonging to the Coronaviridae have large pleiomorphic virions that bear pronounced surface projections and contain a typical set of structural proteins: the spike (S) glycoprotein, which forms the surface projections; the type III membrane (M) protein; and the N protein. Coronaviruses, but not toroviruses, contain a small envelope (E) protein, and some coronaviruses (members of group 2) and all toroviruses contain a hemagglutinin-esterase (HE) protein. In addition, the Coronaviridae contain, interspersed among the structural protein genes, a varying number of genes that code for nonstructural proteins (Fig. 1). These additional genes appear to be nonessential because of the appearance of natural mutants defective in some of them (21) and the viability of engineered deletion mutants lacking some or all of the genes (3).
FIG. 1.
Genomic organization of the Coronaviridae. A member of each coronavirus subgroup—the group I feline coronavirus (FcoV), group II mouse hepatitis virus (MHV), and group III infectious bronchitis virus (IBV)—and the Berne torovirus (BEV) are represented. The essential genes are represented by solid boxes, while the (presumed) nonessential genes are represented by open boxes.
The genome organization of the essential genes is strictly conserved among the Coronaviridae. Coronaviruses belonging to all three subgroups, which are based on serological and genetic criteria, as well as toroviruses, have their essential genes occurring in the order 5′-pol-S-(E)-M-N-3′ (Fig. 1) (32). This conservation is quite remarkable in view of the frequency of recombination of coronavirus and torovirus genomes (18) (S. Smits and R. J. de Groot, personal communication). Furthermore, since each coronavirus gene is flanked by a stretch of similar TRSs, each viral gene may be regarded as a gene cassette which, at least in theory, can be easily moved to various sites in the genome by recombination among these TRSs. Thus, there seems to be selective pressure to maintain the gene order, which indicates an important functional role.
In this study, we have examined whether the fixed order of the essential genes is a vital property of coronaviruses. To this end, recombinants of mouse hepatitis virus (MHV) with rearranged genomes were constructed using synthetic donor RNAs for targeted recombination with the interspecies chimeric coronavirus fMHV, an MHV derivative in which the spike ectodomain is of feline coronavirus origin (17). The recombinant viruses were viable, and most replicated efficiently in cell culture. In addition, the recombinant virus with the gene order 5′-pol-E-M-S-N-3′ appeared to be viable in the natural host, the mouse. These results indicate that conservation of the coronavirus gene order is not essential.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
The recombinant MHVs (strain A59) were propagated in mouse LR7 cells (17), and plaque assays and plaque purifications were carried out using the same cells. Mouse 17 clone 1 (17Cl1) cells were used for radiolabeling of intracellular viral RNAs. Feline FCWF cells (American Type Culture Collection) were used for infection with the interspecies chimeric coronavirus fMHV (17). The rabbit polyclonal antiserum K134 to MHV A59 (29) has been described previously.
Plasmid constructs.
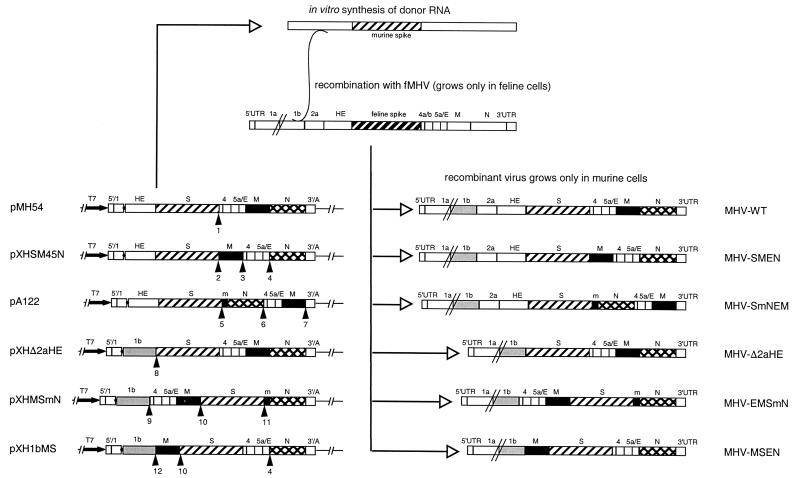
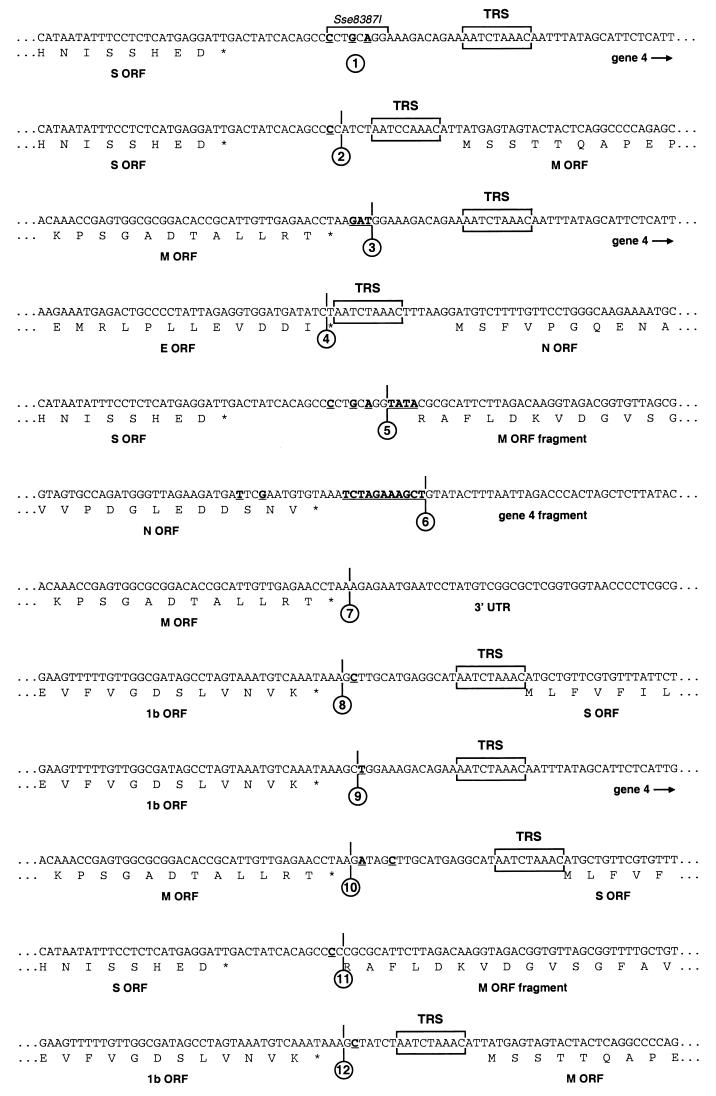
Transcription vectors for the production of donor RNA for targeted recombination were derived from plasmids pMH54 (17) and pXHΔ2aHE (3), which were used for the construction of a wild-type recombinant (MHV-WT) and a recombinant MHV from which the open reading frames (ORFs) located between the pol1b and S genes were deleted (MHV-Δ2aHE), respectively. pMH54 encodes a transcript consisting of the 5′ end of the genome (467 nucleotides [nt]) fused to the 3′ end of the genome starting with codon 28 of the HE gene, while pXHΔ2aHE encodes a transcript consisting of the same 5′ 467 nt ligated to the 3′ end of the genome starting within the pol1b gene some 1,200 nt upstream of the stop codon but lacking the 2a and HE genes (Fig. 2). In transcription vector pXHSM45N, which was used to generate MHV-SMEN, the M gene, including its TRS, was interchanged with a cDNA fragment comprising ORFs 4 and 5. To construct this vector, we first generated a PCR product consisting of the 3′ end of the M gene and the 5′ end of the N gene but with an EcoRV restriction site engineered between the gene fragments. The PCR fragment was prepared by splicing overlap extension PCR using outside primer 1C (5′-GTGTATAGATATGAAAGGTACCGTG-3′), complementary to the region of the M gene where the unique KpnI site is located, and outside primer 1097 (5′-CGAACCAGATCGGCTAGCAG-3′), complementary to the region of the N gene that contains the unique NheI site. Primers 1095 (5′-AGATTAGATATCTTAGGTTCTCAACAATGCGG-3′) and 1096 (5′-GAACCTAAGATATCTAATCTAAACTTTAAGGATG-3′) were used as inside primers. These corresponded to the sequence between the M and the N gene and introduced the EcoRV restriction site. The resulting PCR product was cloned into pGEM-T Easy (Promega), yielding vector pXH0302. As a next step in the construction of pXHSM45N, pMH54 was treated with the restriction enzymes Sse8387I and EcoRV, and the resulting fragment was cloned into the EcoRV site of pXH0302 after having been blunted by T4 DNA polymerase treatment, yielding vector pXH0902. The vector remaining after excision of the Sse8387I-EcoRV fragment from pMH54 was also blunted by T4 DNA polymerase treatment and religated, resulting in plasmid pXH1401. Finally, plasmid pXH0902 was treated with NheI and BssHII, and the resulting fragment was cloned into pXH1401 treated with the same enzymes, yielding pXHSM45N. The construction of vector pA122 will be described elsewhere. Briefly, the N gene, including a duplicated 126-bp fragment of the 3′ end of the M gene, was placed downstream of the S gene, rendering the M gene the most downstream gene. As a result, TRS 4 and most of gene 4 were deleted. In pXHMSmN, the S gene was placed downstream of the M gene in a vector from which the 2a and HE genes had been deleted. For the construction of pXHMSmN, first ORFs 4, 5, and M were removed from pMH54 by restriction with Sse8387I and BssHII, followed by treatment with T4 DNA polymerase and religation of the remaining vector, which yielded pXHΔ45 M5′. Next, a fragment obtained by treatment of pXH0302 with NheI and BssHII was cloned into pMH54, which had been treated with the same enzymes, resulting in pXHMeN. Subsequently, the fragment obtained after restriction of pXHMeN with EcoRV was cloned into pXH1802 (3), which had been digested with HindIII and treated with the Klenow fragment of DNA polymerase I; this yielded pXH0305B. pXH1802 contains the 3′ end of pol1b and the 5′ end of the S gene separated by a HindIII site. The fragment obtained by restriction of pMH54 with the enzymes MluI and EcoRV was cloned into pB59 (8) treated with the same enzymes and resulted in vector pXH2801. This plasmid was digested with the restriction enzymes KpnI and PstI, and the resulting fragment was treated with T4 DNA polymerase and cloned into pXH1802, which had been cut with HindIII and treated with the Klenow fragment of DNA polymerase I; this yielded pXH0806. Subsequently, the fragment obtained by digestion of pXH0305B with SpeI and AflII was cloned into pXH0806 treated with the same enzymes, resulting in pXH1506. Finally, pXHMSmN was obtained by cloning the fragment resulting from restriction of pXH1506 with RsrII and AvrII into pXHΔ45 M5′ treated with the same enzymes. In vector pXH1bMS, the M gene was placed upstream of the S gene, while the genes located between the pol1b and S genes were deleted. For the construction of this vector, vector pXHMeN was restricted with EcoRV, the resulting fragment was removed, and the vector was religated, yielding pXHΔM. Next, the fragment obtained by digestion of pXH0305B with RsrII and AvrII was cloned into pXHΔM treated with the same enzymes, yielding pXH1bMS. All constructs were confirmed by restriction and/or sequence analysis. The sequences of the newly generated junctions indicated in Fig. 2 are shown in Fig. 3.
FIG. 2.
Plasmid constructs, targeted recombination, and recombinant viruses. The transcription vectors from which the defective RNAs were produced in vitro by using T7 RNA replicase are indicated on the left. Vector pMH54 has been described before (17). The other vectors were derived from pMH54 as described in Materials and Methods. The arrow at the left end of each vector indicates the T7 promoter; the solid circle represents the polylinker between the 5′-end segment of the MHV genome (labeled 5′/1) and either the HE gene or the 3′ end of the replicase gene (1b), followed by the structural and group-specific genes and the 3′ untranslated region (UTR) and the polyadenylated segment (together labeled 3′/A). “m” indicates a 126-nt segment from the 3′end of the M gene. The positions of the sequences shown in Fig. 3 are indicated by arrowheads (the numbers correspond to those in the circles in Fig. 3). On top, a scheme for targeted recombination using the interspecies chimeric virus fMHV, which grows only in feline cells, is shown. Recombinant viruses generated by the indicated crossover event can be selected on the basis of their ability to grow in murine cells. The genomes of these viruses are represented on the right.
FIG. 3.
Sequences of the numbered gene junctions shown in Fig. 2. The numbers in the circles correspond to those under the arrowheads in Fig. 2. Nucleotides that differ from those in the original wild-type MHV A59 sequence are underlined and in boldface.
Construction of MHV mutants.
Incorporation of mutations into the MHV genome by targeted RNA recombination was carried out as described previously (3, 11). Donor RNAs transcribed from PacI-linearized, pMH54-derived plasmids were transfected by electroporation into feline FCWF cells that had been infected 4 h earlier with fMHV (17). These cells were then plated onto a monolayer of mouse LR7 cells. After 24 h of incubation at 37°C, progeny viruses released into the cell culture medium were harvested and plaque purified twice on LR7 cells before a passage 1 stock was grown. After confirmation of the recombinant genotypes by reverse transcription (RT)-PCR on purified gRNA, a passage 2 stock was grown that was subsequently used in the experiments.
Radiolabeling of viral RNA.
The metabolic labeling of virus-specific infected cell RNAs was carried out essentially as reported previously (12, 23). Briefly, 20-cm2 17Cl1 cell monolayers were infected with each MHV strain at a multiplicity of infection (MOI) of 5 PFU per cell. At 2 or 3 h postinfection, the cells were starved in Eagle's minimal essential medium containing 5% dialyzed fetal bovine serum (FBS) and 1/10 of the normal phosphate concentration. At 7 h postinfection, the cells were labeled for 2 h with either [33P]orthophosphate or [32P]orthophosphate (83 μCi/ml) in phosphate-free Eagle's minimal essential medium containing 5% dialyzed FBS and 20 μg of actinomycin D (Sigma) per ml. Total cytoplasmic RNA was purified using an NP-40 lysis procedure (23) followed by phenol and chloroform extraction and ethanol precipitation. Samples of RNA were denatured with formaldehyde and formamide, separated by electrophoresis through 1% agarose containing formaldehyde, and visualized by fluorography. The radioactivity in RNA bands was quantitated by scanning them with an Image Quant PhosphorImager. The subtracted background intensity for each band was determined from a rectangle of equal size drawn immediately above or below the rectangle for each RNA band.
Metabolic labeling and immunoprecipitation.
LR7 cells were grown and infected with each virus at an MOI of 8 50% tissue culture infective doses (TCID50) per cell. Before being labeled, the cells were starved for 1 h in cysteine- and methionine-free minimal essential medium containing 10 mM HEPES (pH 7.2) and 5% dialyzed FBS. The medium was then replaced by 600 μl of the same medium containing 100 μCi of in vitro cell-labeling mix (Amersham). The cells were labeled from 6 to 8 h postinfection. At the end of the labeling period, total lysates were prepared by the addition of 3 volumes of five-times-concentrated lysis buffer to the culture medium (2). Proteins were immunoprecipitated from the lysates as described before (2) using 2.5 μl of anti-MHV serum per immunoprecipitation. Immune complexes were adsorbed to Pansorbin cells (Calbiochem) for 30 min at 4°C and were subsequently collected by centrifugation. The pellets were washed four times by resuspension and centrifugation using wash buffers as described before (2). The final pellets were suspended in electrophoresis sample buffer. The immunoprecipitates were heated for 2 min at 95°C and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 15% polyacrylamide gels.
One-step growth curve.
LR7 cell monolayers were grown in 35-mm-diameter dishes in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS. Confluent monolayers were infected in parallel with each virus (8 TCID50/cell) and incubated for 1 h at 37°C. After adsorption, the cells were washed with phosphate-buffered saline three times and then fed with 2 ml of DMEM-10% FBS. Viral infectivity in culture media at different times postinfection was determined by a quantal assay on LR7 cells, and the TCID50 were calculated.
Replication in mice.
Eight-week-old MHV-negative female BALB/c mice were used in the experiment. Viruses were diluted in phosphate-buffered saline, and 106 TCID50 in a total volume of 100 μl was used for injection into the peritoneal cavity. Four animals per virus were inoculated. The mice were sacrificed, and their livers were removed on day 4 postinfection. The livers were placed in 1.5 ml of DMEM, weighed, and then frozen at −80°C until their virus titers were determined. Virus titers were determined by plaque assay on LR7 cell monolayers following homogenization of the organs.
RESULTS AND DISCUSSION
Construction of recombinant viruses.
The significance of the conserved arrangement of the coronavirus gene order was examined by preparing viruses in which the essential genes, including their respective upstream TRSs, were rearranged. Thus, the following viruses were prepared (Fig. 2): MHV-SMEN, in which the M gene was exchanged with a genome fragment comprising ORFs 4 and 5; MHV-SmNEM, in which the N gene, including a small part (m) of the 3′ end of the M gene, was positioned immediately downstream of the S gene, thereby deleting most of gene 4; MHV-EMSmN, which lacks ORFs 2a and HE and in which the S gene was placed downstream of the M gene; and MHV-MSEN, which also lacks ORFs 2a and HE and in which the M gene was moved to a position upstream of the S gene. As controls, we used a reconstructed recombinant wild-type MHV A59 (MHV-WT) and a recombinant virus lacking ORFs 2a and HE (MHV-Δ2aHE) which had been constructed previously using transcription vectors pMH54 (17) and pXHΔ2aHE (3), respectively. These transcription vectors encode defective RNAs composed of the genomic 5′ 467 nt fused to the 3′ end of the genome (approximately 8.6 kb) (Fig. 2). The gene rearrangements were engineered in transcription vectors as described in Materials and Methods. All new junctions generated, including the previously described introduction of an Sse8387I site downstream of the S gene in pMH54 (17), are indicated in Fig. 2, while their sequences are shown in Fig. 3. After confirmation of all constructs by restriction and sequence analyses, corresponding recombinant viruses were generated by targeted RNA recombination with fMHV, an MHV derivative in which the S ectodomain is of feline coronavirus origin (17). Thus, feline cells were infected with fMHV and transfected with transcription vector-derived runoff transcripts, each of which carried the MHV S sequence, and recombinant viruses were selected by growth in murine cells, as was detailed earlier (3). After two rounds of plaque purification on murine cells, recombinant viruses were analyzed by RT-PCR on gRNA and were found to have genomes with the expected organization. Obviously, since all rearrangements were tolerated by the virus, none was lethal.
Cell culture growth characteristics.
First, we evaluated the in vitro growth phenotypes of the recombinant viruses. We found that the passage 2 stocks of all recombinant viruses reached titers close to 107 PFU/ml, except for MHV-SMEN, which yielded only 103 to 104 PFU/ml. Plaque sizes on LR7 cells were not appreciably different among the different viruses and compared to the wild-type MHV A59 strain, with the exception again of MHV-SMEN, which displayed a much smaller plaque size. The recombinant viruses were also analyzed in a one-step growth experiment. Because of its low titer, MHV-SMEN was not examined. All recombinant viruses displayed similar one-step growth kinetics, as shown in Fig. 4, and reached similar titers. The recombinant viruses induced complete syncytium formation in the monolayer during the one-step growth experiment at 9 to 12 h postinfection. The induction of extensive syncytia by MHV-SmNEM and MHV-MSEN was somewhat slower, corresponding to the slightly lower titers observed at 7.5 h postinfection. These results show that rearrangement of the coronavirus gene order is not necessarily detrimental to the virus, as most of the viruses replicated efficiently in cell culture.
FIG. 4.
Single-step growth kinetics of MHV recombinants. LR7 cells were infected with each recombinant MHV at an MOI of 8 PFU per cell. Viral infectivities in culture media at different times postinfection were determined by a quantal assay on LR7 cells, and the TCID50 were calculated.
As most of these genomically rearranged viruses replicated with characteristics quite similar to those of their parental virus, we decided to select one pair for a direct fitness comparison. Thus, a competition experiment was carried out by coinfecting LR7 cells with MHV-EMSmN and MHV-Δ2aHE mixed in 1:100, 1:1, and 100:1 ratios, followed by serial passaging at a low MOI (0.01 to 0.05 PFU/ml). The culture media were harvested 16 to 20 h postinfection. A competitive RT-PCR analysis was used to evaluate the relative yields of the viruses after each passage, as outlined in Fig. 5A. The RT step was performed with primer 1092, which is complementary to the 5′ end of the M gene, while the PCR was performed with a mixture of primers 1261, 990, and 1173, which are complementary to the 3′ ends of the E, S, and 1b genes, respectively (3). To verify the PCR, different ratios of MHV-EMSmN and MHV-Δ2aHE, based on their titers, were mixed and used in the analysis (Fig. 5B). When the viruses were mixed in 1:100 and 100:1 ratios, RT-PCR products were observed that were of sizes similar to those of the PCR products obtained with the plasmid controls pXHΔ2aHE and pXHMSmN, corresponding to the predicted 1,328 and 1,078 bp, respectively. Both RT-PCR products were observed in the same reaction when the viruses were mixed in ratios varying from 1:1 to 25:1. Clearly, the smaller product was amplified more efficiently in the competitive PCR, which was confirmed when the control plasmids mixed in equal amounts were used as templates (data not shown). In some reactions, a slightly larger PCR product was also observed, which was also found in a reaction with the mixed control plasmids. This product, the nature of which is unknown, may be a PCR artifact, but it did not appear to interfere with the competitive RT-PCR analysis. The results of the competition experiment are shown in Fig. 5C. Single RT-PCR products were observed when the viruses mixed in 1:100 and 100:1 ratios were passaged, indicating that the virus originally applied in excess had remained the predominant species throughout the experiment. Upon passaging the 1:1 mixture of the viruses, the initially less abundant larger PCR fragment became predominant. When the band intensities are compared with those of the reference series (Fig. 5B), it appears that the 1:1 ratio gradually shifted to a 25:1 ratio in favor of the parental virus MHV-Δ2aHE. Obviously, MHV-Δ2aHE outcompetes MHV-EMSmN, indicating that under these assay conditions the fitness of the virus with the rearranged genome was reduced relative to its immediate parent.
FIG. 5.
Relative fitness of MHV-Δ2aHE and MHV-EMSmN in cell culture. (A) competitive RT-PCR was used to evaluate the relative yields of MHV-Δ2aHE and MHV-EMSmN after each passage. Primers used in the experiment, their approximate loci in the MHV genome, and the predicted sizes of the PCR products are indicated. Primer 1092 was used in the RT step, while primers 1261, 990, and 1173 were used for the PCR (3). PCR products were analyzed by electrophoresis in 1% agarose gels stained with ethidium bromide. (B) As a reference, MHV-Δ2aHE and MHV-EMSmN were mixed in different ratios, and RT-PCR was performed on purified genomic RNA. As controls, PCR products obtained with plasmids used for the construction of the recombinant viruses and with water are shown. (C) MHV-Δ2aHE and MHV-EMSmN were mixed in 1:100, 1:1, and 100:1 infectivity ratios and inoculated onto LR7 cells. The culture supernatant harvested after 16 h (P1) was passaged four additional times at a low MOI on the same cells (P2 to P5). RT-PCR was performed on genomic RNA purified from aliquots of the successive culture media, and the products were analyzed again in 1% agarose gels. The sizes of relevant DNA fragments of the marker are indicated to the right of each gel.
RNA synthesis.
Coronaviruses express their genomes via a discontinuous transcription mechanism that generates a 3′-coterminal nested set of sgRNAs, each containing a short 5′ leader segment fused at a TRS to all genomic sequence distal to that TRS. Recombinant MHVs with rearranged genome organizations would therefore be predicted to synthesize patterns of viral sgRNAs distinctly different from that of the parent wild-type virus. To examine this, 17Cl1 cells infected with a high MOI of each virus were metabolically labeled with [33P]orthophosphate or [32P]orthophosphate in the presence of actinomycin D for 2 h starting at 7 h postinfection. Purified total cellular RNA was analyzed by electrophoretic separation, as reported previously (12, 23). Representative data are shown in Fig. 6. For the reconstructed wild-type virus (MHV-WT) and for MHV-Δ2aHE, the mobilities and amounts of the gRNA and sgRNA species were essentially identical to those observed previously (3). It should be noted that MHV-Δ2aHE, as well as its derivatives MHV-EMSmN and MHV-MSEN, does not synthesize an sgRNA species containing the 2a and HE genes. For the MHV mutants with rearranged genomes, all variant sgRNAs had mobilities that corresponded to their predicted sizes (Fig. 6), and no obvious additional species were observed. MHV-SMEN grew very poorly, and its RNA was thus labeled only weakly (Fig. 6, right panel).
FIG.6.
Analysis of intracellular RNA synthesis by recombinant MHVs with rearranged genomes. Mouse 17Cl1 cells were mock infected (mock) or infected with recombinant virus (MHV-WT, MHV-SmNEM, MHV-Δ2aHE, MHV-EMSmN, or MHV-MSEN) at an MOI of 5 PFU per cell and were labeled from 7 to 9 h postinfection with [33P]orthophosphate as described in Materials and Methods. In the case of MHV-SMEN, where the maximal MOI that could be achieved was approximately 0.02 PFU per cell, labeling was carried out from 24 to 26 h postinfection. Purified total cytoplasmic RNA was separated by electrophoresis through 1% agarose containing formaldehyde, and the labeled RNA was visualized by fluorography. The genomic composition of each virus is indicated schematically at the top, and each RNA species, with its viral sources in parentheses, is indicated between the panels. Note that in the right-hand gel, the RNA sample in the MHV-SMEN lane was from five times as many cells as that in the MHV-WT lane.
To analyze the relative amounts of sgRNAs synthesized from the rearranged MHV genomes, viral RNA bands were quantitated by PhosphorImager scanning. The poor growth of MHV-SMEN precluded our obtaining quantitative data for that virus. Previous studies have compared the levels of synthesis of MHV sgRNAs by normalizing moles of a given transcript with respect to moles of gRNA (3, 12, 20). The ratio of gRNA to sgRNAs does not vary throughout the infection (30). However, we have observed considerable variation in the values obtained for any given virus by this method owing to variability in the recovery of gRNA from experiment to experiment. Accordingly, we have found that the most consistent basis of comparison of sgRNA levels is the molar fraction of total sgRNA (Tables 1 and 2). The values obtained for this parameter were reproducible in multiple independent determinations for viruses used in this study, and they also compared well with those computed from separate experiments with MHV-WT and MHV-Δ2aHE reported previously (3).
TABLE 1.
RNA synthesis by MHV recombinants with genomic rearrangementsa
| Virus | RNA speciesb | RNA length (nt)c | Molar fraction of sgRNAd | TRS genomic positione | TRS upstream contextf |
|---|---|---|---|---|---|
| MHV-WT | 1a/1b-2a/HE-S-4-5a/E-M-N | 31,528 | |||
| 2a/HE-S-4-5a/E-M-N | 9,852 | 0.005 ± 0.002 | |||
| S-4-5a/E-M-N | 7,677 | 0.015 ± 0.003 | |||
| 4-5a/E-M-N | 3,664 | 0.138 ± 0.018 | |||
| 5a/E-M-N | 3,281 | 0.059 ± 0.009 | |||
| M-N | 2,641 | 0.160 ± 0.016 | |||
| N | 1,944 | 0.623 ± 0.046 | |||
| MHV-SmNEM | 1a/1b-2a/HE-S/m-N/[4]-5a/E-M | 31,383 | |||
| 2a/HE-S/m-N/[4]-5a/E-M | 9,707 | 0.007 ± 0.003 | Same | Same | |
| S/m-N/[4]-5a/E-M | 7,532 | 0.017 ± 0.002 | Same | Same | |
| N/[4]-5a/E-M | 3,401 | 0.559 ± 0.005 | Changed | Same | |
| 5a/E-M | 1,902 | 0.125 ± 0.007 | Changed | Same | |
| M | 1,262 | 0.293 ± 0.003 | Changed | Same | |
| MHV-Δ2aHE | 1a/1b-S-4-5a/E-M-N | 29,370 | |||
| S-4-5a/E-M-N | 7,677 | 0.009 ± 0.003 | Same | Changed | |
| 4-5a/E-M-N | 3,664 | 0.155 ± 0.023 | Same | Same | |
| 5a/E-M-N | 3,281 | 0.060 ± 0.012 | Same | Same | |
| M-N | 2,641 | 0.174 ± 0.029 | Same | Same | |
| N | 1,944 | 0.602 ± 0.063 | Same | Same | |
| MHV-EMSmN | 1a/1b-4-5a/E-M-S/m-N | 29,500 | |||
| 4-5a/E-M-S/m-N | 7,806 | 0.137 ± 0.047 | Changed | Changed | |
| 5a/E-M-S/m-N | 7,423 | 0.046 ± 0.015 | Changed | Same | |
| M-S/m-N | 6,783 | 0.114 ± 0.037 | Changed | Same | |
| S/m-N | 6,065 | 0.026 ± 0.006 | Changed | Changed | |
| N | 1,944 | 0.678 ± 0.103 | Same | Same | |
| MHV-MSEN | 1a/1b-M-S-4-5a/E-N | 29,384 | |||
| M-S-4-5a/E-N | 7,698 | 0.063 ± 0.011 | Changed | Changed | |
| S-4-5a/E-N | 6,980 | 0.014 ± 0.003 | Changed | Changed | |
| 4-5a/E-N | 2,967 | 0.239 ± 0.016 | Changed | Same | |
| 5a/E-N | 2,584 | 0.103 ± 0.014 | Changed | Same | |
| N | 1,944 | 0.581 ± 0.022 | Same | Changed |
Radioactivity in RNA bands was quantitated from dried gels by Phosphorimager scanning.
m, 126-nt segment from the 3′ end of the M gene; [4], 106-nt segment from the 3′ end of gene 4. Each figure or capital letter represents both the gene and its natural TRS.
RNA length includes a polyadenylate tail of 200 nt.
Mean ± standard deviation from three experiments.
Position, relative to the 3′ end of the genome, of the TRS that is located immediately upstream of the gene at the 5′ end of each sgRNA.
Sequence in the viral genome located upstream of the TRS.
TABLE 2.
Comparison of TRS efficiencies in MHV recombinants with genomic rearrangements
| Virus | Genomea | Molar fraction of sgRNAb
|
|||||
|---|---|---|---|---|---|---|---|
| 2a/HE | S | 4 | 5a/E | M | N | ||
| MHV-WT | 1a/1b-2a/HE-S-4-5a/E-M-N | 0.005 | 0.015 | 0.138 | 0.059 | 0.160 | 0.623 |
| MHV-SmNEM | 1a/1b-2a/HE-S/m-N/[4]-5a/E-M | 0.007 | 0.017 | 0.125 (P-3) | 0.293 (P-3) | 0.559 (P-5) | |
| MHV-Δ2aHE | 1a/1b-S-4-5a/E-M-N | 0.009 (C) | 0.155 | 0.060 | 0.174 | 0.602 | |
| MHV-EMSmN | 1a/1b-4-5a/E-M-S/m-N | 0.026 (C, P-3) | 0.137 (C, P-5) | 0.046 (P-5) | 0.114 (P-5) | 0.678 | |
| MHV-MSEN | 1a/1b-M-S-4-5a/E-N | 0.014 (C, P-3) | 0.239 (P-3) | 0.103 (P-3) | 0.063 (C, P-5) | 0.581 (C) | |
m, 126-nt segment from the 3′ end of the M gene; [4], 106-nt segment from the 3′ end of gene 4. Each figure or capital letter represents both the gene and its natural TRS.
Data are the same as in Table 1. Each value is the mean from three experiments. Standard deviations have been omitted to facilitate comparison. Each sgRNA is indicated by its most upstream gene and its respective TRS. C, upstream context of the TRS is changed with respect to the wild type; P-5, genomic position of the TRS is changed in the upstream direction with respect to the wild type; P-3, genomic position of the TRS is changed in the downstream direction with respect to the wild type.
It has been observed previously that, in general, the efficiency of synthesis of coronavirus sgRNAs increases with TRS proximity to the 3′ end of the genome (10, 15, 31). There are, however, a number of exceptions to this principle, and the influence of the TRS context on transcription efficiency has also been well documented (13, 22, 27, 34). For MHV and bovine coronavirus, the TRS context includes homology with the leader sequence within the 5 nt immediately flanking each side of the 9-nt consensus core 5′-AAUCUAAAC-3′, as well as incompletely defined sequences or structures in the regions further upstream and downstream of the TRS. All prior efforts at manipulation of the contexts and genomic positions of MHV and bovine coronavirus TRSs have been carried out in defective interfering RNAs (13, 14, 16, 22, 27, 34, 35). The MHV rearrangement mutants, although a limited set, provided an opportunity to examine the strengths of position and context effects in the background of the entire genome. In our mutants, the core TRS consensus sequence and the downstream TRS context always remained unaltered. The genomic positions of multiple TRSs, however, changed in each mutant, resulting from both the repositioning of a given gene and the consequent movements of its new and former neighboring genes (Fig. 2 and 3 and Table 1). In addition, there were resultant changes in the upstream contexts of some, but not other, TRSs in the rearrangement mutants (Fig. 3 and Table 1).
With respect to TRS position, a clear trend could be seen in the quantitated transcriptional data. In almost all cases, an effect was noted on transcription efficiency when a TRS was moved relative to the 3′ end of the genome. Movement in the 3′ direction resulted in an increase in the molar fraction of transcripts; movement in the 5′ direction led to a reduction in the molar fraction of transcripts. This was consistent with previous observations that downstream TRSs can have an attenuating effect on upstream TRSs (12, 14, 16, 35). Thus, the upstream repositioning of a given TRS would increase the number of other TRSs between it and the 3′ end of the genome, while conversely, the downstream repositioning of a given TRS would remove intervening TRSs. For some, but not all, of these cases (e.g., the up-regulated 5a/E-M and M sgRNAs of MHV-SmNEM and the down-regulated M-S/m-N sgRNA of MHV-EMSmN), the change in transcription efficiency could be attributed solely to the change in TRS position, because the TRS context remained exactly the same as that of the wild type (Tables 1 and 2). However, all observed positional effects were relatively small, always twofold or less (Tables 1 and 2).
By contrast, for some TRSs, context appeared to play a strong dominant role. This was particularly evident in four cases. First, as has been reported previously (3, 26), the 4-5a/E-M-N sgRNA of MHV-WT is much more abundant than its counterpart in standard (nonrecombinant) wild-type MHV A59. This dramatic up-regulation, which is at least sevenfold (data not shown), is due to one or more of the three nucleotide changes introduced into the transcription vector pMH54 to create an Sse8387I site upstream of the TRS (Fig. 2 and 3, junction 1). It should be noted that these base changes do not increase the homology between the leader and the upstream flanking region of this TRS. A second strong context effect was observed with the 4-5a/E-M-S/m-N sgRNA of MHV-EMSmN. Levels of this transcript were as high as those for the 4-5a/E-M-N sgRNA of MHV-WT (Tables 1 and 2), despite the relocation of the TRS some 4.2 kb upstream of its position in the wild-type genome. In this case, the upstream context of the TRS had been altered by juxtaposition with the terminus of the pol1b gene (Fig. 2 and 3, junction 9). Similarly, in a third example, the N/[4]-5a/E-M sgRNA ([4] indicates a 106-nt segment from the 3′ end of gene 4) in MHV-SmNEM retained a molar fraction comparable to that of the N sgRNA in MHV-WT despite its TRS having been moved 1.5 kb upstream (Tables 1 and 2). If genomic position were the sole determinant of TRS efficiency, then we would have expected the abundance of this sgRNA to have dropped to the levels of those of the 4-5a/E-M-N or 5a/E-M-N sgRNA of MHV-WT. Finally, the most salient effect of context on TRS efficiency was seen with MHV-SMEN, in which synthesis of the M-4-5a/E-N sgRNA was drastically reduced. Only at much longer exposures of the gel, shown in the right-hand gel of Fig. 6, was it possible to detect a band with the predicted mobility of this sgRNA (data not shown). Although the precise reason for the low abundance of this sgRNA is unknown, the consequent rate-limiting synthesis of M protein is likely to be the cause of the severe growth impairment of this virus.
Protein synthesis.
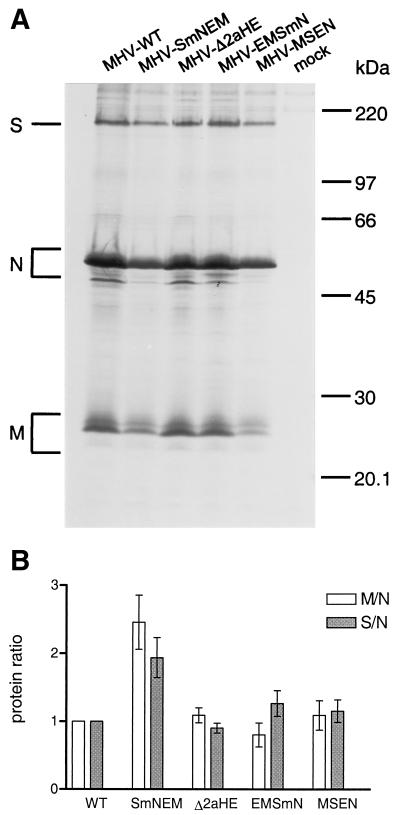
Since eukaryotic mRNAs are functionally monocistronic, each coronavirus sgRNA is the mRNA for the ORF that appears closest to its 5′ end. To analyze the effects that gene rearrangement might have on protein expression, LR7 cells were infected at a high MOI with the different recombinant viruses and labeled for 3 h with 35S-labeled amino acids starting at 5 h postinfection. Subsequently, total lysates of cells and culture media were prepared by the addition of concentrated lysis buffer to the culture supernatant (2). These lysates were processed for immunoprecipitation with an anti-MHV serum followed by SDS-PAGE in a 15% gel. Previously, it was shown that the MHV structural proteins were synthesized in very similar ratios throughout the infection (28). As shown in Fig. 7A, all the major structural proteins, i.e., the S, N, and M proteins, were precipitated in all cases. For the M protein, the O-glycosylated species were predominant. The E protein is not precipitated to detectable levels with the anti-MHV serum and therefore was not included in the analyses. The radioactivity in each protein was quantitated by PhosphorImager scanning of the dried gels from three independent experiments. In order to correct for variations in viral protein synthesis, the relative amounts of the proteins present in the cultures were determined and normalized to those of the wild-type virus (Fig. 7B). This showed that the relative M/N and S/N ratios in cells infected with MHV-Δ2aHE and MHV-MSEN were not significantly altered compared to those in MHV-WT-infected cells. For MHV-EMSmN, the modest decrease in M/N and increase in S/N protein ratios paralleled the changes in the levels of the corresponding mRNAs (Tables 1 and 2). For MHV-SmNEM, the increase observed in the M/N protein ratio was likely to be due to the combined effects of the 1.8-fold increase in M mRNA and a small reduction in N mRNA. However, for the same recombinant, the twofold increase in the S/N protein ratio is hard to attribute to changes in mRNA levels. Similarly, for MHV-MSEN, we did not see the 2.5-fold drop in the M/N protein ratio that would have been expected on the basis of the reduction of the M mRNA level with respect to MHV-WT. These apparent discrepancies may suggest that the translational efficiency of the N gene differs from those of the M and S genes.
FIG. 7.
Analysis of synthesis of recombinant MHV proteins. (A) LR7 cells were mock infected (mock) or infected with recombinant viruses as indicated on the top of the gel. The cells were labeled for 2 h with 35S-labeled amino acids starting 6 h postinfection. Total lysates were prepared and used for immunoprecipitation with the anti-MHV serum K134, and the precipitates were analyzed by SDS-15% PAGE. The positions of the different proteins are indicated on the left, while the molecular mass marker is indicated on the right of the gel. (B) For quantitative analysis, PhosphorImager scanning of the amounts of radioactivity in the M, S, and N proteins in the dried gels from three independent experiments (as shown in panel A) was performed. The ratios of the amounts of M and N (M/N) and S and N (S/N) proteins were calculated relative to those of MHV-WT-infected cells, and standard deviations are indicated.
Replication in mice.
Having analyzed their in vitro growth characteristics, the replication of MHV-WT, MHV-Δ2aHE, and MHV-EMSmN was studied in their natural host, the mouse. MHV-EMSmN was chosen, since it appeared to replicate slightly more efficiently in cell culture than the other viruses with rearranged genomes. While the 50% lethal dose of MHV-WT in mice was previously determined to be 2.7 × 104 PFU, MHV-Δ2aHE was not virulent enough for a 50% lethal dose determination (3). Therefore, we decided to analyze the in vivo replication of the recombinant viruses. Mice inoculated intraperitoneally with 106 TCID50 were euthanized on day 4 postinfection, and the extent of viral replication in the liver was determined. The results are shown in Fig. 8. Surprisingly, MHV-Δ2aHE and MHV-EMSmN replicated in the liver to similar extents, albeit to much lower levels than MHV-WT. Deletion of ORFs 2a and HE generated a recombinant virus (MHV-Δ2aHE) that was attenuated in the natural host, as shown previously (3), while additional rearrangement of the coronavirus gene order (MHV-EMSmN) did not appear to result in a more attenuated phenotype in this assay. The results indicate that the typical coronavirus genome organization is not essential for replication in vivo.
FIG. 8.
Replication of recombinant viruses in BALB/c mice. Eight-week-old mice were inoculated intraperitoneally with a 100-μl volume containing 106 TCID50 of recombinant virus (n = 4 per virus). The mice were sacrificed, and the livers were removed on day 4 postinfection. Virus titers were determined by plaque assay on LR7 cell monolayers following homogenization of the livers. The error bars indicate standard deviations.
Flexibility of the coronavirus genome.
The coronavirus genome is highly flexible. It not only tolerates very large deletions (3) and the stable insertion of foreign genes (7, 8) (C. A. M. de Haan, unpublished data) but also the rearrangement of the genome organization (this study). The flexibility of the coronavirus genome is also demonstrated by the heterogeneity of the nonessential, coronavirus group-specific genes (Fig. 1) (19, 21, 32). Together with the high frequency of recombination, this genomic flexibility may provide coronaviruses with a mechanism to counteract potential deleterious effects of the high mutation rate of their very large RNA genome and to evolve and adapt their genome more easily to changing conditions. This is in contrast to the related arteriviruses, which appear to be much less flexible. All of the genes of the arterivirus equine arteritis virus were demonstrated to be essential (25), and hence, the virus could not tolerate deletion, while insertion of a foreign gene resulted in genetic instability of the recombinant virus (5). For porcine reproductive and respiratory syndrome virus, which also belongs to the Arteriviridae, only a few mutations were found to be tolerated (24). The reasons for these differences are unclear. Arteriviruses contain icosahedral nucleocapsid structures rather than the extended helical nucleocapsids that coronaviruses exhibit. It is conceivable that the icosahedral nucleocapsid limits the arterivirus genome size (13 to 15 kb), resulting in and allowing much less genomic flexibility. Another factor preventing arterivirus genome rearrangement may be the overlap of all adjacent ORFs and the consequent embedding of each TRS within the preceding gene (4, 33).
In view of the high frequency of RNA recombination, the observed tolerance of gross genome rearrangements raises the question of why the coronavirus gene order is so strictly conserved. If such viable rearranged genomes are generated in nature, there are apparently strong selection mechanisms acting against these viruses: they are outcompeted by “normal” viruses. This was indeed observed when a virus with rearranged gene order was tested directly in an in vitro competition against its immediate nonrearranged parent (Fig. 5). We can only speculate about the selective advantages of the canonical coronavirus genome organization. It may be required for providing coronaviruses with an efficient mechanism to synthesize their proteins in optimal relative quantities during the course of infection. Alternatively, the coronavirus gene order may have become arbitrarily fixed in an early ancestor of the present members of this family. Despite the high rate of recombination, the generation of a rearranged gene order would require that a single genome undergo multiple nonhomologous crossover events, the combined probability of which may be vanishingly small.
Rearrangement of the genome organization of viruses provides a new approach to the generation of live attenuated vaccines. This was recently shown for the nonsegmented negative-strand vesicular stomatitis virus. Gene expression of this virus is controlled by the highly conserved order of genes relative to the single transcriptional promoter. Similar to our findings with MHV, for vesicular stomatitis virus it was found that gene rearrangement is not lethal (1). However, rearrangement resulted in viruses with altered gene expression levels that appeared to attenuate virulence and to protect infected animals against wild-type virus challenge (9, 36). Similarly, it will be of interest to study the phenotypic consequences of the MHV genome rearrangements in mice in more detail. In the single case we examined in vivo, rearrangement per se did not appear to be attenuating. However, in conjunction with other mutations, deliberate rearrangement of the viral genes may be useful in the generation of attenuated coronaviruses, which, due to their reduced risk of generating viable viruses by recombination with circulating field viruses, would create safer vaccines.
Acknowledgments
We thank Lili Kuo and Xiaolan Shen for advice and assistance in the RNA-labeling experiments. We gratefully acknowledge Marèl de Wit for assistance with the animal experiments.
This work was carried out in part with financial support from the Commission of the European Communities specific RTD program Quality of Life and Management of Living Resources, QLK2-CT-1999-00002, “Generic coronavirus vaccine vectors for protection of farm animals against mucosal infections,” to C.A.M.D.H. and P.J.M.R. This work was also supported in part by Public Health Service grant AI 45695 from the National Institutes of Health to P.S.M.
This study does not necessarily reflect the views of the Commission of the European Communities and in no way anticipates the Commission's future policy in this area.
REFERENCES
- 1.Ball, L. A., C. R. Pringle, B. Flanagan, V. P. Perepelitsa, and G. W. Wertz. 1999. Phenotypic consequences of rearranging the P, M, and G genes of vesicular stomatitis virus. J. Virol. 73:4705-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Haan, C. A. M., L. Kuo, P. S. Masters, H. Vennema, and P. J. M. Rottier. 1998. Coronavirus particle assembly: primary structure requirements of the membrane protein. J. Virol. 72:6838-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Haan, C. A. M., P. S. Masters, X. Shen, S. Weiss, and P. J. M. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Vries, A. A. F., A. L. Glaser, M. J. Raamsman, C. A. M. de Haan, S. Sarnataro, G. J. Godeke, and P. J. M. Rottier. 2000. Genetic manipulation of equine arteritis virus using full-length cDNA clones: separation of overlapping genes and expression of a foreign epitope. Virology 270:84-97. [DOI] [PubMed] [Google Scholar]
- 5.de Vries, A. A. F., A. L. Glaser, M. J. Raamsman, and P. J. M. Rottier. 2001. Recombinant equine arteritis virus as an expression vector. Virology 284:259-276. [DOI] [PubMed] [Google Scholar]
- 6.de Vries, A. A. F., M. C. Horzinek, P. J. M. Rottier, and R. J. de Groot. 1997. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin. Virol. 8:33-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enjuanes, L., I. Sola, F. Almazan, J. Ortego, A. Izeta, J. M. Gonzalez, S. Alonso, J. M. Sanchez, D. Escors, E. Calvo, C. Riquelme, and C. Sanchez. 2001. Coronavirus derived expression systems. J. Biotechnol. 88:183-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer, F., D. Peng, S. T. Hingley, S. R. Weiss, and P. S. Masters. 1997. The internal open reading frame within the nucleocapsid gene of mouse hepatitis virus encodes a structural protein that is not essential for viral replication. J. Virol. 71:996-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flanagan, E. B., J. M. Zamparo, L. A. Ball, L. L. Rodriguez, and G. W. Wertz. 2001. Rearrangement of the genes of vesicular stomatitis virus eliminates clinical disease in the natural host: new strategy for vaccine development. J. Virol. 75:6107-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofmann, M. A., R. Y. Chang, S. Ku, and D. A. Brian. 1993. Leader-mRNA junction sequences are unique for each subgenomic mRNA species in the bovine coronavirus and remain so throughout persistent infection. Virology 196:163-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hsue, B., T. Hartshorne, and P. S. Masters. 2000. Characterization of an essential RNA secondary structure in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 74:6911-6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsue, B., and P. S. Masters. 1999. Insertion of a new transcriptional unit into the genome of mouse hepatitis virus. J. Virol. 73:6128-6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeong, Y. S., J. F. Repass, Y. N. Kim, S. M. Hwang, and S. Makino. 1996. Coronavirus transcription mediated by sequences flanking the transcription consensus sequence. Virology 217:311-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joo, M., and S. Makino. 1995. The effect of two closely inserted transcription consensus sequences on coronavirus transcription. J. Virol. 69:272-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konings, D. A., P. J. Bredenbeek, J. F. Noten, P. Hogeweg, and W. J. Spaan. 1988. Differential premature termination of transcription as a proposed mechanism for the regulation of coronavirus gene expression. Nucleic Acids Res. 16:10849-10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishnan, R., R. Y. Chang, and D. A. Brian. 1996. Tandem placement of a coronavirus promoter results in enhanced mRNA synthesis from the downstream-most initiation site. Virology 218:400-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuo, L., G. J. Godeke, M. J. Raamsman, P. S. Masters, and P. J. M. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lai, M. C. 1996. Recombination in large RNA viruses: coronaviruses. Semin. Virol. 7:381-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai, M. M., and D. Cavanagh. 1997. The molecular biology of coronaviruses. Adv. Virus Res. 48:1-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leibowitz, J. L., K. C. Wilhelmsen, and C. W. Bond. 1981. The virus-specific intracellular RNA species of two murine coronaviruses: MHV-a59 and MHV-JHM. Virology 114:39-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luytjes, W. 1995. Coronavirus gene expression, p. 33-54. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 22.Makino, S., and M. Joo. 1993. Effect of intergenic consensus sequence flanking sequences on coronavirus transcription. J. Virol. 67:3304-3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masters, P. S., C. A. Koetzner, C. A. Kerr, and Y. Heo. 1994. Optimization of targeted RNA recombination and mapping of a novel nucleocapsid gene mutation in the coronavirus mouse hepatitis virus. J. Virol. 68:328-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meulenberg, J. J. 2000. PRRSV, the virus. Vet Res. 31:11-21. [DOI] [PubMed] [Google Scholar]
- 25.Molenkamp, R., H. van Tol, B. C. Rozier, Y. van der Meer, W. J. Spaan, and E. J. Snijder. 2000. The arterivirus replicase is the only viral protein required for genome replication and subgenomic mRNA transcription. J. Gen. Virol. 81:2491-2496. [DOI] [PubMed] [Google Scholar]
- 26.Ontiveros, E., L. Kuo, P. S. Masters, and S. Perlman. 2001. Inactivation of expression of gene 4 of mouse hepatitis virus strain JHM does not affect virulence in the murine CNS. Virology 289:230-238. [DOI] [PubMed] [Google Scholar]
- 27.Ozdarendeli, A., S. Ku, S. Rochat, G. D. Williams, S. D. Senanayake, and D. A. Brian. 2001. Downstream sequences influence the choice between a naturally occurring noncanonical and closely positioned upstream canonical heptameric fusion motif during bovine coronavirus subgenomic mRNA synthesis. J. Virol. 75:7362-7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raamsman, M. J. B., J. Krijnse Locker, A. de Hooghe, A. A. F. de Vries, G. Grifiths, H. Vennema, and P. J. M. Rottier. 2000. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 74:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rottier, P. J. M., M. C. Horzinek, and B. A. van der Zeijst. 1981. Viral protein synthesis in mouse hepatitis virus strain A59-infected cells: effect of tunicamycin. J. Virol. 40:350-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawicki, S. G., and D. L. Sawicki. 1998. A new model for coronavirus transcription. Adv. Exp. Med. 280:215-218. [DOI] [PubMed] [Google Scholar]
- 31.Sethna, P. B., S. L. Hung, and D. A. Brian. 1989. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc. Natl. Acad. Sci. USA 86:5626-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Siddell, S. G. 1995. The Coronaviridae: an introduction, p. 1-10. In S. G. Siddell (ed.), The Coronaviridae. Plenum Press, New York, N.Y.
- 33.Snijder, E. J., and J. J. Meulenberg. 1998. The molecular biology of arteriviruses. J. Gen. Virol. 79:961-979. [DOI] [PubMed] [Google Scholar]
- 34.van der Most, R. G., R. J. de Groot, and W. J. Spaan. 1994. Subgenomic RNA synthesis directed by a synthetic defective interfering RNA of mouse hepatitis virus: a study of coronavirus transcription initiation. J. Virol. 68:3656-3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Marle, G., W. Luytjes, R. G. van der Most, T. van der Straaten, and W. J. Spaan. 1995. Regulation of coronavirus mRNA transcription. J. Virol. 69:7851-7856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wertz, G. W., V. P. Perepelitsa, and L. A. Ball. 1998. Gene rearrangement attenuates expression and lethality of a nonsegmented negative strand RNA virus. Proc. Natl. Acad. Sci. USA 95:3501-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]