Abstract
A DNA duplex can be torn open at a specific position by introducing a branch or bulge to create an asymmetric three-way junction (TWJ). The opened duplex manifests a bent conformation (bending angle ∼60°, relative to the unopened form), which leads to a dramatic decrease in gel electrophoretic mobility. In the presence of a basepair mismatch at the opening position, the DNA backbone becomes less bent and assumes a distorted T-shaped structure, resulting in an increase in polyacrylamide gel electrophoretic mobility. Both conformational changes are confirmed using fluorescence resonance energy transfer experiments and found to be similar to the signature conformational changes of DNA duplex upon MutS protein binding. Our results imply that some structural rearrangements essential for mismatch recognition are achievable without protein interference. The gel electrophoretic mobility data for DNA TWJs with and without base mismatches correlates well with rotational diffusivity, computed by taking into account the conformational change of TWJ induced by base mismatch.
INTRODUCTION
Single nucleotide or point mutations are by far the most common form of genetic sequence polymorphisms in the human genome. These mutations occur at a frequency of ∼1 per 1,000 bp (1,2) and contribute significantly to the genetic diversity within species. In most cases single nucleotide mutations have no known effect on gene expression or gene products and are commonly phenotypically silent (3). There are several notable exceptions, however, where mutations in specific genes have been identified with increased risk for diseases in humans. Point mutations in the p53 tumor suppressor/cell cycle gene, for example, have been found at elevated levels in ovarian and breast cancer tumors (4,5). Single nucleotide mutations in the dystrophin gene are likewise strongly correlated with muscular dystrophy (6).
A large volume of research has been conducted to develop fast, reliable, and economical techniques to screen for single nucleotide mutations. The most successful methods so far all utilize external agents, such as mismatch repair proteins, denaturants (7,8), and synthetic chemical agents (9), to open up the mismatch site, allowing the unpaired bases to be detected and identified. Mismatch repair proteins are believed to first bind to the dsDNA, interrogate DNA basepairing nonspecifically, and then systematically search for the mismatched base site (10). Denaturants and synthetic chemicals selectively melt the double helix at the mismatched site and create local defects that amplify the difference between wild and mutated dsDNA. In either case, formation of a branch or bulge is essential for trigging the mismatch recognition process. This statement is generally true whether the recognition process occurs in vivo or in vitro.
The best understood mismatch repair (MMR) system thus far is that of Escherichia coli. This system utilizes three proteins, MutS, MutL, and MutH (11). Mismatch recognition is achieved by MutS first binding to the DNA backbone then searching progressively for the unpaired bases by probing the local flexibility of the double helix (12). A conformational change induced by protein binding is believed to be essential to tell the difference between homo- and heteroduplex forms of the DNA (13). The detailed mechanism through which mutation repair proteins induce conformational changes in dsDNA is, however, unknown. It is possible that the bound protein triggers formation of more complex functional domains that facilitate opening up of heteroduplex structures or that the protein simply produces a local bulge or branch on dsDNA, which leads to a localized change in stiffness, inducing the conformation change.
DNA three-way junctions (TWJ) possessing an arm or bulge adjacent to a mismatch site can be used to study the latter possibility in detail, without need for mismatch recognition proteins. By examining the impact of basepair mismatch on conformation and properties of TWJs, it should therefore be possible to gain insight into the role that mismatch repair proteins can play in initiating the process. DNA TWJs are the simplest structures of opened dsDNA that can be created by self-assembly of oligonucleotides. Perfectly formed TWJs have been investigated using many experimental methods including short-long arm assays (14), fluorescence resonance energy transfer (FRET) experiments (15), and sticky-end ligation assays (16). From all these studies, TWJs are found to assume a symmetric planar structure without base stacking around the junction and exhibit some degree of interarm flexibility. Structural studies of TWJ containing defects, such as loops (15,17) or base mismatches (18), have also been attempted previously, but the results are less conclusive.
MATERIALS AND METHODS
DNA samples
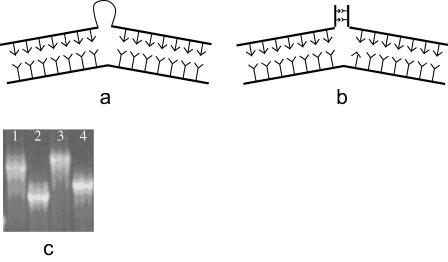
The DNA backbone sequence used for electrophoresis was a 66 bp fragment of p53 tumor suppressor/cell cycle regulator gene spanning exon 7 through intron 7. Point mutations are introduced at codon 260 (TCC→TCT), which is in the middle of the backbone. Complementary oligonucleotide sequences are designed to form a bulge (Fig. 1 a) or branch (Fig. 1 b) with variable length near the mismatched base. All oligos were obtained from Sigma Genosys (Woodlands, TX) and further purified using polyacrylamide gel electrophoresis (PAGE). TWJs were formed by first mixing the backbone oligo, its complements, and annealing buffer (10mM Tris-HCl, 50mM NaCl, 1mM EDTA). The mixture was then denaturated by heating to 95°C for 5 min and then annealed to 40°C in a water bath for 1 h. The samples were slowly cooled to room temperature and then stored at 4°C. The formation of TWJs was verified by running the sample together with low molecular weight ladders (New England Biolabs, Ipswich, MA) on polyacrylamide (PA) gels.
FIGURE 1.
(a) Schematic drawing of TWJ with a single bulge. (b) Schematic drawing of TWJ with a single branch. (c) Representative gel (12.5%), from lanes 1–4 showing mutated TWJ with 4 bp bulge, mutated TWJ with 2 bp branch, wild-type TWJ with 4 bp bulge, and wild-type TWJ with 2 bp branch.
Fluorescein and tetramethyl rhodamine (TAMRA) were chosen as the FRET donor and acceptor, respectively. DNA samples were prepared using ordinary and end-labeled oligos. TWJ of two different sizes were prepared for FRET structural studies: one with the backbone sequence shortened to 29 bp and the branch length kept at 10 bp and the other with branch length of 10 bp each, so that the distance between the ends of each branch is within the Förster radius for the selected dye pair, considering all possible topological conformations. Regular and dye-labeled oligos were obtained from Sigma Genosys. All dye-labeled oligos were purified using HPLC. The labeling efficiency was found to be nearly 100% using mass spectrometry. Oligos were hybridized to form TWJs using the same annealing procedure described above; the stoichiometry used to form each type of TWJ is listed in Table 1. Three types of labeled branched molecules were formed for each distance intended to be measured, molecules containing donors only, acceptors only, and molecules containing A–D pairs. Purification of hybridized TWJs was again performed using PAGE followed by electroelution. The recovered DNA solution concentration was determined using absorption measurement on a Cary 300 Bio-UV-Vis spectrophotometer (Varian, Palo Alto, CA). Fluorescence excitation and emission spectroscopy measurements were performed using a PTI fluorimeter (Photon Technology International, Birmingham, NJ) to confirm the presence of donor and acceptor labels on the purified TWJs.
TABLE 1.
Stoichiometry of each strand used to form different types of TWJs
| Donor strand | Acceptor strand | Unlabelled strand | |
|---|---|---|---|
| Donor-only junction | 1 | 0 | 3 |
| Acceptor-only junction | 0 | 1 | 3 |
| Acceptor-donor (AD) pair junction | 1 | 3 | 3 |
Gel preparation and running conditions
PA gels in 1 X TAE (40 mM Tris-acetate, 1 mM EDTA) buffer were prepared using the method described previously (19,20). The gel running temperature was maintained constant at 12°C, and a fixed electrical field of 9.6 v/cm is used throughout. Casted gels were prerun for 1 h at 9.6 V/cm to remove ionic impurities before loading samples for electrophoresis measurements. After electrophoresis, the gels were stained with SYBR Green (Molecular Probes, Eugene, OR) dyes under conditions recommended by the manufacturer and visualized on a multiwavelength transilluminator (Ultra-Lum, Claremont, CA). The gel electrophoretic mobility is determined as 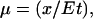 where x is the distance between the bottom of the well and center of the band corresponding to the TWJ. A typical gel picture is shown in Fig. 1 c.
where x is the distance between the bottom of the well and center of the band corresponding to the TWJ. A typical gel picture is shown in Fig. 1 c.
Capillary electrophoresis
Capillary electrophoresis (CE) experiments were performed on a GPA100 unit assembled with UV/Vis detector (Groton Biosystems, Boxborough, MA), using fused silica capillaries, with 50 μm inner diameter, 45 cm effective length, and 70 cm total length (Polymicro Technologies, Phoenix, AZ). To minimize electroosmotic flow, capillaries were covalently coated with linear PA using a procedure similar to that described by Albarghouthi et al. (21). Briefly, in this procedure the capillary is sequentially flushed with 1 M HCl for 45 min, with water for 20 min, and with 1 N NaOH for 45 min, and finally it is rinsed with water for 20 minutes. The capillary surface is subsequently coated with silane using 0.4% v/v methacryloxypropyltrimethoxysilane in 0.4% v/v acetic acid solution for 75 min. A total of 5 ml of a 4 wt % degassed acrylamide solution initiated by 50 μl freshly prepared 10% ammonium persulfate and 5 μl N, N, N′, N′-tetramethylene-diamine solution is pumped into the capillary and the reaction allowed to proceed overnight. The polymer solution is finally flushed out with water and the capillary dried with air. Electroosmotic flow (EOF) mobility was measured in the PA-coated capillary following the protocol of Williams et al. (22). Methoxy benzyl alcohol was used as a neutral marker for these measurements.
Capillary electrophoresis measurements were performed using voltages in the range 100–200 V/cm and a 1× TAE buffer. Electrophoresis measurements in free solution confirmed that the mobility is independent of applied fields in this range. During measurements, capillaries are fan cooled and thermostated at 15°C. An ultraviolet detector set at 254 nm with data acquisition rate of 2 Hz was used throughout. Before each measurement, capillaries were conditioned by rinsing with running buffer at high pressure for 30 s.
Apparent free solution mobility of DNA (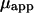 ) was determined from the applied field, E, the known detecting length,
) was determined from the applied field, E, the known detecting length, 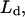 and measured migration time, t, using the relation
and measured migration time, t, using the relation
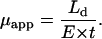 |
(1) |
All results reported are averages from five sequential runs. Errors in the mobility due to residual EOF (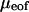 ) can be determined by averaging data obtained from two neutral marker runs before and after measurements using DNA samples. The corrected free solution mobility of the DNA samples is therefore obtained from
) can be determined by averaging data obtained from two neutral marker runs before and after measurements using DNA samples. The corrected free solution mobility of the DNA samples is therefore obtained from
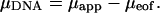 |
(2) |
FRET experiments
FRET has been used by several groups to study the conformation of branched DNA, TWJs, and four-way Holliday junctions (15,23–25). Förster formalism shows that the efficiency of FRET is dependent on the inverse sixth power of the intermolecular separation, 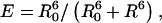 where R0 is the Förster radius, the distance at which energy transfer from donor to acceptor is 50% efficient, and R is the distance between the donor and acceptor dye pair. The Förster radius is a constant for a given acceptor-donor pair and is calculated to be 5.5 nm for the fluorescein-TAMRA dye pair used in this work.
where R0 is the Förster radius, the distance at which energy transfer from donor to acceptor is 50% efficient, and R is the distance between the donor and acceptor dye pair. The Förster radius is a constant for a given acceptor-donor pair and is calculated to be 5.5 nm for the fluorescein-TAMRA dye pair used in this work.
The energy transfer efficiency can be readily determined from the characteristic donor-acceptor and donor decay time constants (lifetimes) using the formula 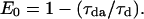 The lifetime of donor in presence (
The lifetime of donor in presence (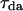 ) and absence (
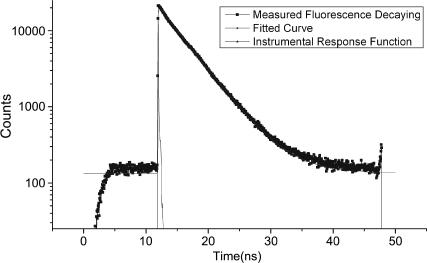
) and absence (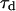 ) of acceptor was determined by measuring the fluorescence decay in the time domain using a homemade lifetime apparatus similar to one described previously (26). Briefly, a 900 nm, 80 MHz repetition rate Ti:Sapphire laser (Spectra Physics, Mountain View, CA) was pulse-picked to 16 MHz using a Pockel cell (Conoptics, Canbury, CT). A 700 nm short-pass dichroic (Chroma, Rockingham, VT) in conjunction with a narrow (515–545 nm) emission filter is used to separate the donor dye fluorescence emission from the two-photon excitation beam as well as to prevent collection of fluorescence emission from the acceptor dye. Fluorescence was detected by a microchannel plate photomultiplier (Hamamatsu, Hamamatsu City, Japan), and the decay curve was calculated using a time-correlated single photon counting card (Becker & Hickl GmbH, Berlin, Germany). The fluorescence decay curve was analyzed using SPC image software (Becker & Hickl GmbH), and lifetime was determined by fitting the decay data to an exponential function (27). A typical plot is shown in Fig. 2 to indicate the quality of the raw data and accuracy of fits obtained. The accuracy of lifetime measurement is determined by measuring the lifetime of Alexa488 in aqueous solution before each measurement. The obtained lifetime shows a maximum 50 ps run-to-run variation.
) of acceptor was determined by measuring the fluorescence decay in the time domain using a homemade lifetime apparatus similar to one described previously (26). Briefly, a 900 nm, 80 MHz repetition rate Ti:Sapphire laser (Spectra Physics, Mountain View, CA) was pulse-picked to 16 MHz using a Pockel cell (Conoptics, Canbury, CT). A 700 nm short-pass dichroic (Chroma, Rockingham, VT) in conjunction with a narrow (515–545 nm) emission filter is used to separate the donor dye fluorescence emission from the two-photon excitation beam as well as to prevent collection of fluorescence emission from the acceptor dye. Fluorescence was detected by a microchannel plate photomultiplier (Hamamatsu, Hamamatsu City, Japan), and the decay curve was calculated using a time-correlated single photon counting card (Becker & Hickl GmbH, Berlin, Germany). The fluorescence decay curve was analyzed using SPC image software (Becker & Hickl GmbH), and lifetime was determined by fitting the decay data to an exponential function (27). A typical plot is shown in Fig. 2 to indicate the quality of the raw data and accuracy of fits obtained. The accuracy of lifetime measurement is determined by measuring the lifetime of Alexa488 in aqueous solution before each measurement. The obtained lifetime shows a maximum 50 ps run-to-run variation.
FIGURE 2.
Typical fluorescence decay curve illustrating instrument response characteristics and exponential fit to the data.
Typical lifetimes for fluorescein attached to DNA, in the absence of any acceptor molecules, were found to be ∼4.1 ns., which is comparable to the 4.2 ns value for free fluorescein dye as provided by the supplier. The 0.1 ns lifetime difference can arise from several factors, including from the quenching effect of phosphate side groups of DNA or restriction of rotational motion of dye molecules due to their attachment to DNA.
Anisotropy of the attached fluorescein and TAMRA fluorescence was measured on single labeled samples. The anisotropy of fluorescein was determined to be 0.20 and the anisotropy of TAMRA 0.17. Segmental mobility of fluorescein attached to the TWJ is captured by measuring the time-domain anisotropy decays. The attached fluorescein features a segmental rotation time of 0.4∼0.5 ns, which is much smaller than its lifetime (4.1 ns), indicating that attached fluorescein rotates rapidly during its fluorescence lifetime. Anisotropy measurements therefore support an orientation factor, 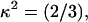 in calculating the Förster distance.
in calculating the Förster distance.
RESULTS AND DISCUSSIONS
Effect of single basepair mismatch on electrophoretic mobility and its dependence on branch and bulge size
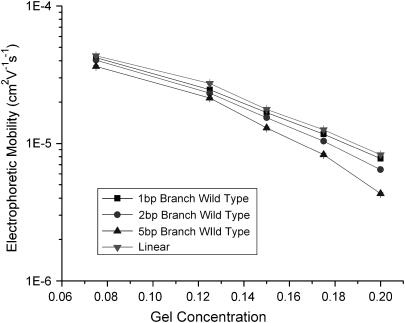
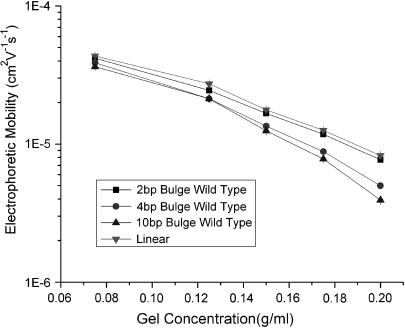
We have reported previously that TWJ (10 bp branch) containing a single mismatched base near the junction point has higher mobilities in PA gels than TWJ with fully complementary sequences (19). By fixing the backbone length at 66 bp, branches varying 1 bp ∼ 10 bp or bulges varying 2 bp ∼ 10 bp were introduced to the mismatched site to quantify the effect of branch/bulge size on the enhanced mobilities. Measurable increases in gel electrophoretic mobility was observed for all TWJs, as depicted in Figs. 3 and 4, and 5 a and 5 b. This result clearly shows that single basepair mismatches near the center of a dsDNA TWJ branch junction universally increase electrophoretic mobility, compared with a corresponding TWJ without basepair mismatch, in PA gels irrespective of the attached branch type and size. Fig. 3, for example, plots the gel electrophoretic mobility of fully complementary TWJs containing branches ranging from 1 bp to 5 bp. It is apparent from the experimental data that even a 2 bp branch can yield a noticeable reduction in mobility (20). Fig. 4 depicts similar data for double-stranded DNA containing bulges of variable size. Again it is evident that even a 2 bp bulge yields a measurable reduction in mobility, implying that even a small branch or bulge can dramatically change the conformation of dsDNA. This observation also provides strong support to the argument that by opening dsDNA at unpaired bases, mutation detection proteins such as MutS induce local conformation changes in DNA that facilitate further processing of the mutated bases (13).
FIGURE 3.
Ferguson plot of TWJs of varying branch length compared with plot for linear molecule.
FIGURE 4.
Ferguson plot of TWJs of varying bulge size compared with plot for linear molecule.
FIGURE 5.
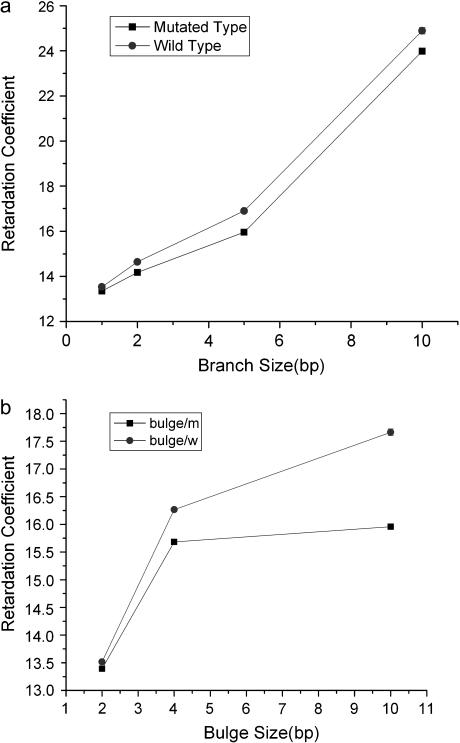
Retardation coefficient, 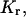 determined by fitting the gel electrophoretic mobility data for wild- and mutated-type TWJ with varying branch or bulge size using the expression
determined by fitting the gel electrophoretic mobility data for wild- and mutated-type TWJ with varying branch or bulge size using the expression 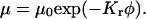
Figs. 3 and 4 also show that gels with higher concentrations of PA are better able to resolve conformational differences between TWJs and linear dsDNA molecules. Specifically, the difference between linear and TWJ is seen to become larger with increasing gel concentration. One explanation of this finding is that whereas the orientation of rigid rod dsDNA can be specified by a single vector during electrophoresis in a gel, a minimum of two vectors are needed to specify the orientation of a branched species during electrophoresis. This means that the branched species is capable of exploring the nanoporous structure of the gel at a higher dimensionality compared with linear molecules. For the branched molecule, its motion is then not only decided by the length scale of geometric hindrances but also by the tortuosity of the path through which it must travel. The tortuosity of the gel is determined by the connectivity of gel pores along the path of the electrophoresing analyte.
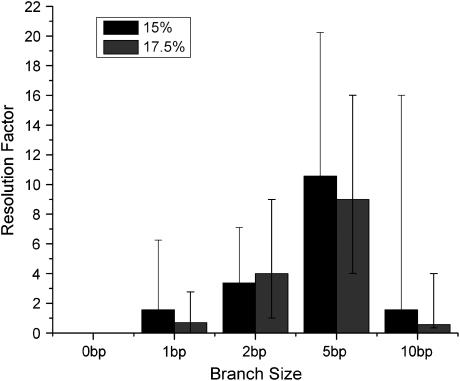
For all TWJs studied in this work, we found that introducing even a single basepair branch increases mobility in PA gels. The retardation coefficient for mutated and wild-type TWJ with varying branch lengths is shown in Fig. 5 a, and a similar trend can be observed for TWJs containing bulges of various size (Fig. 5 b). As the attached branch size is increased, the difference in mobility of the fully complementary and mutated TWJ increases slowly and reaches a maximum value for branches around 5 bp. To quantify this effect we introduce a resolution factor,  and plot it as a function of branch size in Fig. 6. Here,
and plot it as a function of branch size in Fig. 6. Here, 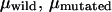 are the electrophoreitc mobility of wild- and mutated-type DNA and
are the electrophoreitc mobility of wild- and mutated-type DNA and 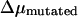 is the uncertainty of the electrophoreitc mobility data for the mutated-type DNA, as reflected in the gel band width. For all the gel data analyzed in this work,
is the uncertainty of the electrophoreitc mobility data for the mutated-type DNA, as reflected in the gel band width. For all the gel data analyzed in this work, 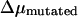 remains roughly constant. Combining these observations with previous findings (19) that electrophoretic mobility of branched structures containing TWJs is independent of the actual position of the branch, it is safe to conclude that introducing a single basepair mismatch to a TWJ near the site of a branch or bulge will increase its gel electrophoreitc mobility.
remains roughly constant. Combining these observations with previous findings (19) that electrophoretic mobility of branched structures containing TWJs is independent of the actual position of the branch, it is safe to conclude that introducing a single basepair mismatch to a TWJ near the site of a branch or bulge will increase its gel electrophoreitc mobility.
FIGURE 6.
Resolution factor and its dependence on the attached branch size observed in 15% and 17.5% PA gels.
This mobility increase is believed to reflect the ability of the attached branch or bulge to magnify the difference between a homo- and heteroduplex. It also implies that there must be some molecular level difference between wild-type and mutated TWJs. The fundamental molecular origin of this effect, i.e., whether this difference in gel electrophoretic mobility directly implicates the conformation difference between a homo- and heteroduplex with a small branch or bulge, is presently unknown.
The size of all TWJs used in our work is <100 bp, which places them firmly in the conventional Ogston sieving regime. In this regime, the gel electrophoreitc mobility can be expressed as
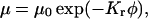 |
(3) |
where 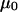 is the free solution mobility, and
is the free solution mobility, and 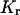 is the retardation coefficient (28). Within the context of Ogston sieving, several mechanisms can be proposed to explain the observed increase in mobility of branched DNA structures containing unpaired bases:
is the retardation coefficient (28). Within the context of Ogston sieving, several mechanisms can be proposed to explain the observed increase in mobility of branched DNA structures containing unpaired bases:
The mismatched basepair causes a small net increase of surface charge on the DNA molecules near the junction. This effect will be reflected in
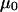 of Eq. 3.
of Eq. 3.A single basepair mismatch at the junction will cause a conformational change from the conventional Y-shaped planar structure expected for a fully complementary TWJ and will thereby have a similar effect as reported for unpaired loops on TWJs (15). Such a conformational change will influence all hydrodynamic properties of the TWJ, namely, translational and rotational diffusivity, and free solution mobility, which can all influence the electrophoretic mobility in gels.
The mismatch at the junction will decrease the bending stiffness of DNA TWJ, allowing the interarm angle to achieve lower values in PA gels under the action of electric fields.
We consider each of these mechanisms in detail in the following sections.
Mismatch-induced change in surface charge
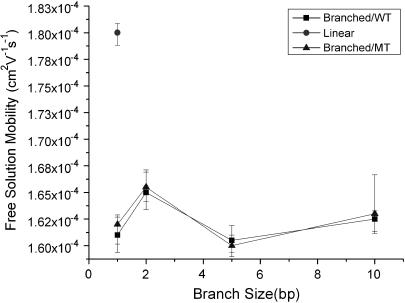
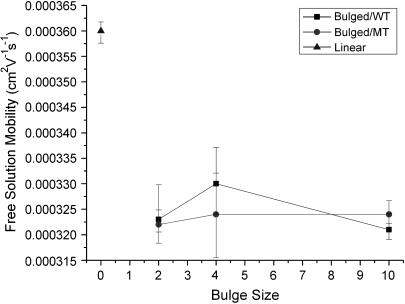
A significant change in surface charge is detectable from the migration time difference between TWJs with and without base mismatches under sufficiently high electric fields. The free solution mobility of TWJs with and without a single basepair mismatch was determined using capillary electrophoresis. Fig. 7 shows the free solution mobility of TWJs without point mutations as a function of branch length. It is apparent from this figure that the free solution mobility of all TWJs is ∼10% lower than that of linear dsDNA of comparable size. However, the difference in free solution mobility between wild- and mutated-type TWJs is negligible and well within the experimental error for CE measurements. In addition, the free solution mobility is seen to have no systematic dependence on the branch size. Similar trends are also observed for TWJs with varying bulge sizes (Fig. 8). Taken together, these results appear to conclusively show that in the Ogston sieving region, the effective separation of wild- and mutated-type TWJs in PA gel is not due to their net surface charge difference. Dissimilarities between the hydrodynamics of TWJs without base mismatches, reflected in the retardation coefficient, 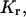 are then the most plausible source of their mobility differences.
are then the most plausible source of their mobility differences.
FIGURE 7.
Free solution mobility of wild- and mutated-type TWJs with varying branch length.
FIGURE 8.
Free solution mobility of wild- and mutated-type TWJs with varying bulge size.
Mismatch-induced conformational change
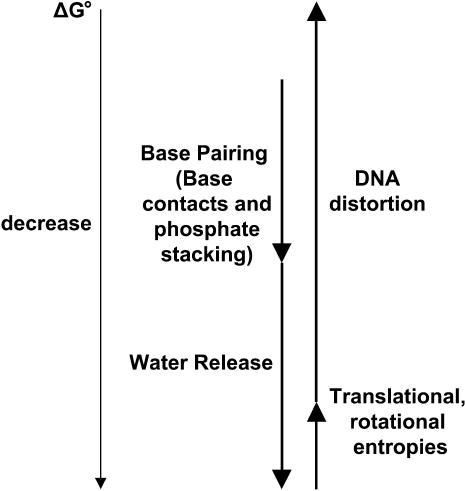
Several factors contribute to the free energy change in forming the TWJs. Some of these factors are illustrated in Fig. 9. The free energy of formation of the TWJ junction is expected to change when unpaired bases are present. If only a single basepair mismatch is present, the free-energy change due to basepairing alone is relatively modest (ΔΔG° = 2.4 kcal/mol for a mismatched duplex (18)). However at the branch point, DNA is highly bent on length scales well below its persistence length (∼50 nm, 150 bp). This means that even a single basepair mismatch may produce a rather large change in the contribution made by the bending free energy of the junction to the overall free energy of formation of the TWJ. Furthermore, the bending energy is sequence dependent, which means that this contribution will vary depending upon details such as what sequences flank the mismatch and possibly even what bases are unpaired. The bending constant for (AT)n is, for example, just 80% of the average value for DNA (28), and a single-stranded region in DNA can readily facilitate downstream bending (29). It is then apparent that a mismatch near the branch point can lead to a significant difference in final yield of TWJ and/or TWJs with very different branch conformations. Because the reduced electrophoretic mobility is not a function of DNA sample concentration, the yield of the final TWJ product will clearly not affect the dynamic properties of TWJs of interest in this study. We therefore focus on mismatch-induced conformational changes.
FIGURE 9.
Free energy change on forming a TWJ.
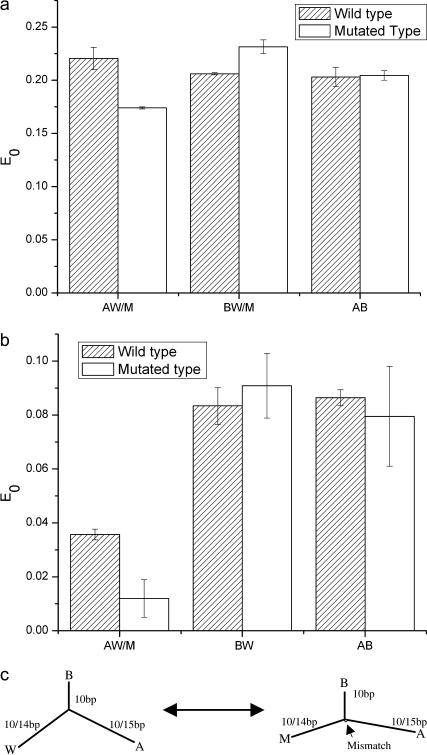
FRET experiments can be used to quantify conformational changes of TWJs with and without single base mismatches. The energy transfer efficiencies for TWJs of two different sizes are shown in Fig. 10, a and b. The energy transfer efficiency can be readily converted to the actual interarm distance (and angles), making use of the fact that the R0 value calculated from measured fluorescein emission and TAMRA excitation spectra is 5.5 nm. For the symmetric TWJ with a 10 bp (3.4 nm) branch (Fig. 10 a), the energy transfer efficiency is almost identical for all arms, varying 0.20 ∼ 0.22. The interarm angle is calculated to be 112° ∼ 115°, by assuming that the fluorescein extending from the DNA helix contributes 0.25 nm to the branch length (15). This result indicates that the perfectly formed TWJ assumes a nearly planar symmetric conformation similar to that reported previously by Lilley et al. (15).
FIGURE 10.
(a) Energy transfer efficiency of symmetric wild- and mutant-type TWJs with 10 bp branch. (b) Energy transfer efficiency of asymmetric TWJs. (c) Schematic drawing of how a single base mismatch can induce the conformational transition.
For the symmetric TWJ with a point mutation, the energy transfer efficiency varies considerably among arms. The lowest energy transfer efficiency, 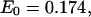 corresponding to a distance of 7.13 nm, indicates two essentially collinear aligned branches (7.05 nm). It is therefore apparent that introducing a point mutation to the TWJ breaks its structural symmetry and converts it from a nearly planar Y-shaped structure to a distorted T-shaped structure, by reducing the bending angle from 60° to nearly 0° between two branches, one branch with a fully complementary sequence and the other containing a point mutation. It is of course possible that two conformers of mutated TWJ exist, since there are two options in selecting the perfectly formed branch. However, only one of these conformers is observed in our measurement. This high selectivity is believed to originate from the different energy required to bend specific DNA sequences.
corresponding to a distance of 7.13 nm, indicates two essentially collinear aligned branches (7.05 nm). It is therefore apparent that introducing a point mutation to the TWJ breaks its structural symmetry and converts it from a nearly planar Y-shaped structure to a distorted T-shaped structure, by reducing the bending angle from 60° to nearly 0° between two branches, one branch with a fully complementary sequence and the other containing a point mutation. It is of course possible that two conformers of mutated TWJ exist, since there are two options in selecting the perfectly formed branch. However, only one of these conformers is observed in our measurement. This high selectivity is believed to originate from the different energy required to bend specific DNA sequences.
All the molecules used in the electrophoresis study are asymmetric, with one branch length (10 bp) significantly smaller than the other two (33 bp). Although there is no obvious reason why this asymmetry should change the interarm angles, as compared to the symmetric TWJ, how the mutated TWJ selects its dominant conformer remains unknown. Two branches of the TWJ are extended from 10 bp to 14 bp and 15 bp to explore whether the actual size of the arms plays any role in determining the dominant conformer. The interarm distance is again maintained below 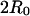 to ensure measurable FRET efficiencies. Let AW/M represent distance between the ends of the longest backbone (29 bp) and AB, BW represent the end-to-end distance between the other two smaller strands (24, 25 bp, respectively). Despite the lower energy transfer efficiency due to the increase in branch length, the interarm angle for the fully complementary TWJ can still be readily calculated and found to be ∼120° (AW: 115 ± 1°, BW: 118 ± 4°, AB: 110 ± 3°). In the case of TWJ containing a single basepair mismatch, the interarm angle computed in the same manner is 180°, 101 ± 3°, 100 ± 5° (AM, BM, AB), which are essentially identical to the interarm angles obtained from FRET analysis of the smaller symmetric TWJs (AM: 180; BM: 96 ± 1°; and AB: 99 ± 2°). In both cases the summation of the three angles yields an unphysical value larger than 360°, reflecting uncertainty in the length contribution from the dye pair and the unknown junction size, i.e., the region where three branches interchange its complementary strand. Again, only one dominant conformer is found for the mutated TWJ. Responding to the introduction of a point mutation, the longest backbone (AW) changes from a 60° bent structure to a nearly linear unbent structure. This dramatic change in conformation is very similar to what is observed in the TWJ containing a 3 bp loop (15).
to ensure measurable FRET efficiencies. Let AW/M represent distance between the ends of the longest backbone (29 bp) and AB, BW represent the end-to-end distance between the other two smaller strands (24, 25 bp, respectively). Despite the lower energy transfer efficiency due to the increase in branch length, the interarm angle for the fully complementary TWJ can still be readily calculated and found to be ∼120° (AW: 115 ± 1°, BW: 118 ± 4°, AB: 110 ± 3°). In the case of TWJ containing a single basepair mismatch, the interarm angle computed in the same manner is 180°, 101 ± 3°, 100 ± 5° (AM, BM, AB), which are essentially identical to the interarm angles obtained from FRET analysis of the smaller symmetric TWJs (AM: 180; BM: 96 ± 1°; and AB: 99 ± 2°). In both cases the summation of the three angles yields an unphysical value larger than 360°, reflecting uncertainty in the length contribution from the dye pair and the unknown junction size, i.e., the region where three branches interchange its complementary strand. Again, only one dominant conformer is found for the mutated TWJ. Responding to the introduction of a point mutation, the longest backbone (AW) changes from a 60° bent structure to a nearly linear unbent structure. This dramatic change in conformation is very similar to what is observed in the TWJ containing a 3 bp loop (15).
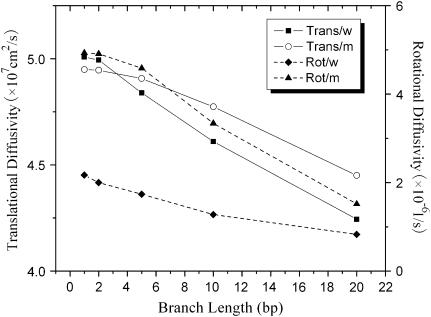
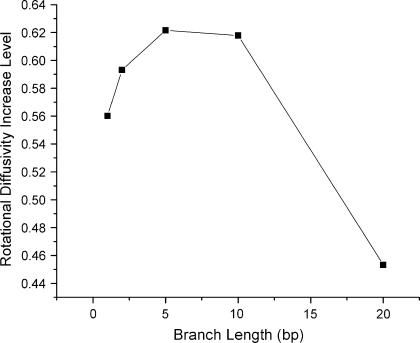
It is therefore apparent that introducing even a single basepair mismatch at a TWJ will change its conformation from a planar symmetric Y-shaped structure to a slightly distorted T-shaped structure by unbending the longest backbone. Since molecular conformation is directly associated with hydrodynamic properties, such as translational and rotational diffusivity, these conformational changes can yield large changes in the dynamics of TWJs in gels. To quantify these effects, the program HydroPix (30) was used to model the DNA molecules and to calculate their hydrodynamics properties. The normal TWJ was modeled as three cylinders connected to form a planar structure with an interarm angle of 120°, whereas the TWJ with a single basepair mismatch was constructed as three cylinders connected with interarm angles of 90°, 90°, and 180°. The calculated rotational and translational diffusivity of these structures are shown in Fig. 11 for varying branch lengths. Although the calculated translational diffusivity shows a negligible difference between the Y-shaped and T-shaped representations of the TWJ, translational diffusivity decreases by 8% when the attached branch length varies from 1 bp to 10 bp, and the rotational diffusivity shows a strong dependence on the molecular conformation and branch length. When the planar symmetry of molecular conformation is broken, the rotational diffusivity increases in some cases by >50% (Fig. 12). The increased rotational diffusivity implies a higher pore sampling rate when DNA electrophoreses in PA gels. The well-known heterogeneity of these gels means that the DNA must continually readjust its orientation to meander from large to small gel pores, normally ∼10–20 nm (31). The increase of rotational diffusivity due to conformational change also manifests a local maxima 2∼20 bp arm length (Fig. 12), which nicely coincides with the trend observed for reduced electrophoretic mobility in PA gels (Fig. 6) at two different gel concentrations. This observation suggests a strong correlation between conformation-induced changes in rotational diffusivity and gel electrophoretic mobility of TWJs.
FIGURE 11.
Rotational and translational diffusivity of wild- and mutated-type TWJ with variable branch length calculated using HydroPix (30).
FIGURE 12.
Increase of rotational diffusivity when TWJ conformation changes from Y-shaped to T-shaped 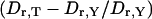 and its dependence on attached branch length.
and its dependence on attached branch length.
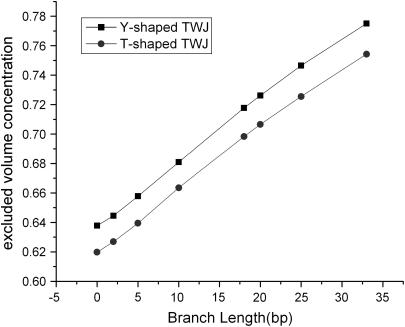
Both the Ogston model and the revised Slater-Guo model suggest that the reduced mobility of TWJs in gels could be determined exclusively by free volume effects (31). If correct, this means that the reduced mobility of branched DNA in gels should not correlate with any dynamic property of the analyte. To evaluate this point, a three-dimensional gel lattice model is constructed in the same way as described in Allison et al. (32) and the excluded volume concentration calculated for T-shaped and Y-shaped TWJs with varying branch length. At various gel fiber spacing (equivalent to gel concentration), the T conformation is found to have a slightly lower excluded volume concentration (see Fig. 13), which can be directly interpreted as higher reduced mobility. Thus, the best separation of T-shaped and Y-shaped structures is anticipated when the gel fiber spacing is close to the largest dimension of TWJs, which is ∼20 nm for the systems studied here. However, the difference in excluded volume concentration between T-shaped and Y-shaped TWJs remains almost constant with increasing branch length. If the excluded volume effect is the decisive factor, we would therefore expect similar resolution independence of arm length, which obviously contradicts our experimental observations.
FIGURE 13.
Excluded volume concentration versus branch length of T-shaped and Y-shaped TWJs for a gel fiber spacing of 20 nm.
Our result therefore raises concerns that the gel pore size may not be the most important variable in determining how TWJs are resolved, but rather the tortuosity of the analyte path in the gel. In the case of a spherical analyte particle, the tortuosity can be neglected since the particles are isotropic. However, for the TWJs discussed here, when pore size becomes comparable to the TWJ size, especially when the pore size is small enough to strongly favor certain orientations of the molecule, how the gel fiber and TWJ are orientated relative to each other will play a progressively more significant role in setting the mobility. In that case, how fast the favorable orientation(s) can be reached, i.e., the rotational diffusivity of the analyte, will be explicitly expressed in the gel electrophoretic mobility.
Mismatch-induced bending stiffness change
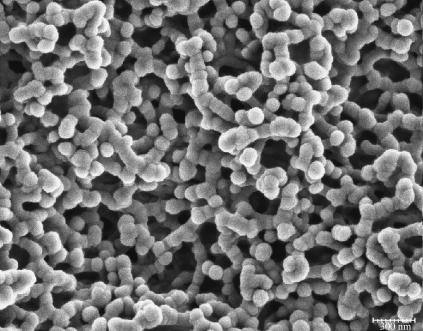
The third possible explanation for the increased mobility of TWJs with single base mismatches is that the mismatch basepair softens the junction and makes the TWJ more squeezable when it encounters geometric hindrance during gel electrophoresis. This should allow structures containing mutations to transition more easily from large to small gel pores, even if the arms are not properly aligned. The force applied by the external electrical field (Eq. 4) on DNA is about an order of magnitude smaller than the thermal force that attempts to extract a TWJ trapped in a small gel pore (see Eq. 5). E is the applied electrical field; q is the surface charge of the TWJ; l is the typical size of a small gel pore, taken to be ∼15 nm. The smallest pore size shown in Fig. 14 for 7.5% PA gel is ∼20 nm, and the smallest gel concentration that shows distinctive difference between wild and mutant TWJ is larger than 12.5%. Small pore size in PA gels is reported to vary with gel concentration (c) as 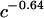 (33), supporting the value of 15 nm used in this calculation.
(33), supporting the value of 15 nm used in this calculation.
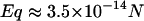 |
(4) |
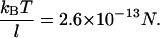 |
(5) |
FIGURE 14.
Scanning electron micrograph of 7.5% (w/v) PA gel. The scale bar corresponds to a distance of 300 nm. Gel specimens were prepared using freeze drying followed by carbon coating. Images were obtained at 3 kV on a Hitachi S4500.
This means that the field cannot contribute significantly to squeezing modes in gel electrophoresis runs and that, to the extent such modes are important in setting the mobility, they will be observable in free solution with greater probabilities for TWJ containing point mutations. Time-resolved FRET experiments of TWJ in buffer show just one dominant subpopulation of conformational modes for TWJ with and without single basepair mismatches. If a broad array of conformations were present, then we would expect complex decay curves with multiple exponential components, rather than the single-exponent decays measured (25). The dominant conformation observed through any macroscopic experiment is recognized to be an ensemble average over a large number of subpopulations with different conformations. So the subpopulation of drastically squeezed TWJ conformation must be marginally small based on our FRET data, and TWJ is more likely to reorient itself instead of changing its conformation to fit through small gel pores.
Role of mismatch protein in mismatch recognition
Based on the discussion above, the most plausible explanation for the increased mobility of TWJ with base mismatches in PA gels is that unpaired bases near the junction lead to a conformational transition from a Y-shaped to a T-shaped structure. The rotational diffusivity, DR, is sensitive to such a conformational transition in heterogeneous gels because the predicted changes in DR due to Y- to T-shaped conformational change provides a direct and convincing linkage between these two properties.
Thus, we can conclude from the data that once a branch or bulge is introduced to the DNA backbone, as in the most common mechanism for mismatch repair proteins (10,11), it triggers a first order conformational transition of dsDNA from a straight linear conformation to a bent structure. The 60° bend in the backbone of the TWJ is in exact accordance with experimental observations for protein-induced DNA bending (12). This conformation can only be stabilized if the protein forms favorable interactions with the DNA surface to compensate for the energy penalty for forming the bend. To dock the protein and trigger the downstream repair mechanism, a second order conformational transition is needed to differentiate the mismatched type from wild-type DNA. For example, in the E. coli mismatch repair system, once the MutS-heterduplex complex is stabilized, it is believed to recruit another protein, MutL, to form a MutS-MutL-DNA complex to activate the cleavage activity of MutH, which can nick the mismatch-included strand in both directions. This recruiting process requires a structural rearrangement of the MutS-heterduplex complex (11). Our work indicates that this second order structural transition can actually be achieved by the DNA molecule itself if the duplex is torn open on the mismatch site. Although there is no direct evidence to rule out the possibility that the second order transition may be protein assisted, the conformational transition state suggested by Wang et al. (13) is readily achievable by DNA without any protein interference. The characteristic bend angle suggested for wild- and mutated-type backbones are ∼120° and ∼180°, respectively; in agreement with the trend observed from our FRET measurements on DNA TWJs.
CONCLUSIONS
Introducing a single basepair mismatch to the junction site of three-way branched DNA will increase its gel electrophoretic mobility. This phenomenon is universal for TWJ of various branch length (1 bp ∼ 10 bp) and bulge sizes (2 bp ∼ 6 bp). The mobility difference between wild-type and mutated-type TWJ manifests a local maximum with increasing branch length; the resolution in PA gel electrophoresis will be lost completely when the attached branch size becomes comparable to the other two arms of the TWJ (16). Free solution mobility is found to be insensitive to the presence of single basepair mismatch and independent of the attached branch or bulge size.
A conformational change induced by single basepair mismatch is observed and quantified using FRET experiments. A single basepair mismatch is capable of inducing similar interarm length change in a TWJ, with 10, 14, and 15 bp arms, similar to the 3 bp loop-induced conformational change in symmetric DNA TWJ (14). The associated change of hydrodynamic properties was calculated using computer simulations. Rotational diffusivity, DR, is found to be sensitive to the conformational change. Specifically, DR increases when the TWJ changes from a Y-shaped to a T-shaped structure. This increase varies with branch length in an analogous manner to the variation of gel electrophoretic mobility with branch length. The rotational diffusivity difference decreased to just 8% for a symmetric TWJ with equal arm length and can be used to explain the loss of resolution between wild- and mutated-type TWJ as branch length increases. The excluded volume concentration was also calculated based on a cubic lattice model. Although the conformational transition form Y-shaped structure to T-shaped structure does cause a slight decrease in calculated excluded volume concentration, the effect is independent of attached arm length.
The extra bending flexibility induced by base mismatch might also contribute to the increased mobility of mutated TWJs in gels. However, only one dominant conformation can be extracted from time-resolved FRET experiments, negating the existence of a subpopulation of TWJ with largely squeezed structure in PA gels. The conformational change in DNA TWJs observed in this study coincides with the conformational change of dsDNA when mismatch repair proteins, such as MutS, bind to it (13). This suggests that after the mismatch repair protein binds to dsDNA and opens the DNA duplex nonspecifically (6), the conformational difference between the hetero- and homoduplex can be triggered by internal conformational adjustment of the DNA itself. When the DNA duplex is disrupted by introducing a small bulge or branch, the backbone manifests a ∼60° bending angle, similar to the bending angle of DNA duplex responding to MutS binding. The bent structure is stable when the sequence on the “disrupted” region is fully complementary. However, the presence of a single basepair mismatch in this region will cause the duplex backbone to release this bending and favors a nearly linear conformation, a conformational transition essential for MutS mismatch recognition (13). Observation of this conformational change in TWJ with a point mutation suggests that mismatch repair proteins can achieve their mismatch recognition function by simply disrupting the base pairing locally and opening up the duplex.
Acknowledgments
We are grateful to the National Science Foundation (DMR0237052) and Department of Energy (DE-FG02-02ER4600) for supporting this study.
References
- 1.Carlson, C. S., M. A. Eberle, M. J. Rieder, J. D. Smith, L. Kruglyak, and D. A. Nickerson. 2003. Additional SNPs and linkage-disequilibrium analyses are necessary for whole-genome association studies in humans. Nat. Genet. 33:518–521. [DOI] [PubMed] [Google Scholar]
- 2.Cooper, D. N., B. A. Smith, H. J. Cooke, S. Niemann, and J. Schmidtke. 1985. An estimate of unique DNA-sequence heterozygosity in the human genome. Hum. Genet. 69:201–205. [DOI] [PubMed] [Google Scholar]
- 3.Hart, J. R., M. D. Johnson, and J. K. Barton. 2004. Single-nucleotide polymorphism discovery by targeted DNA photocleavage. Proc. Natl. Acad. Sci. USA. 101:14040–14044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hainaut, P., T. Soussi, B. Shomer, M. Hollstein, M. Greenblatt, E. Hovig, C. Harris, and R. Montesano. 1997. Database of p53 gene somatic mutations in human tumors and cell lines: updated compilation and future prospects. Nucleic Acids Res. 25:151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malkin, D., F. P. Li, L. C. Strong, J. F. Fraumeni Jr., C. E. Nelson, D. H. Kim, J. Kassel, M. A. Gryka, F. Z. Bischoff, M. A. Tainsky, and S. H. Friend. 1990. Germ line p53 mutations in a familial syndrome of breast-cancer, sarcomas and other neoplasms. Science. 250:1233–1238. [DOI] [PubMed] [Google Scholar]
- 6.Prior, T. W., A. C. Papp, P. J. Snyder, M. S. Sedra, L. M. Western, C. Bartolo, R. T. Moxley, and J. R. Mendell. 1994. Heteroduplex analysis of the gene: application to point mutation and carrier detection. Am. J. Med. Genet. 50:68–73. [DOI] [PubMed] [Google Scholar]
- 7.Nollau, P., and C. Wagener. 1997. Methods for detection of point mutations: performance and quality assessment. Clin. Chem. 43:1114–1128. [PubMed] [Google Scholar]
- 8.Korkko, J., S. Annunen, T. Pihlajamaa, D. J. Prockop, and L. Ala-Kokko. 1998. Conformation sensitive gel electrophoresis for simple and accurate detection of mutations: comparison with denaturing gradient gel electrophoresis and nucleotide sequencing. Proc. Natl. Acad. Sci. USA. 95:1681–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Junicke, H., J. R. Hart, J. Kisko, O. Glebov, I. R. Kirsch, and J. K. Barton. 2003. Bioinorganic chemistry special feature: a rhodium(III) complex for high-affinity DNA base-pair mismatch recognition. Proc. Natl. Acad. Sci. USA. 100:3737–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Banerjee, A., W. Yang, M. Karplus, and G. L. Verdine. 2005. Structure of a repair enzyme interrogating undamaged DNA elucidates recognition of damaged DNA. Nature. 434:612–638. [DOI] [PubMed] [Google Scholar]
- 11.Sixma, T. K. 2001. DNA mismatch repair: MutS structures bound to mismatches. Curr. Opin. Struct. Biol. 11:47–52. [DOI] [PubMed] [Google Scholar]
- 12.Obmolova, G., C. Ban, P. Hsieh, and W. Yang. 2000. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 407:703–710. [DOI] [PubMed] [Google Scholar]
- 13.Wang, H., Y. Yang, M. J. Schofield, C. Du, Y. Fridman, S. D. Lee, E. D. Larson, J. T. Drummond, E. Alani, P. Hsieh, and D. A. Erie. 2003. DNA bending and unbending by MutS govern mismatch recognition and specificity. Proc. Natl. Acad. Sci. USA. 100:14822–14827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo, Q., N. C. Seeman, and N. R. Kallenbach. 1989. Site-specific interaction of intercalating drugs with a branched DNA molecule. Biochemistry. 28:2355–2359. [DOI] [PubMed] [Google Scholar]
- 15.Stuhmeier, F., J. B. Welch, A. I. H. Murchie, D. M. J. Lilley, and R. M. Clegg. 1997. Global structure of three-way DNA junctions with and without additional unpaired bases: a fluorescence resonance energy transfer analysis. Biochemistry. 36:13530–13538. [DOI] [PubMed] [Google Scholar]
- 16.Ma, R. I., N. R. Kallenbach, R. D. Sheardy, M. L. Petrillo, and N. C. Seeman. 1986. Three-arm nucleic acid junctions are flexible. Nucleic Acids Res. 14:9745–9753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, H. D., G. U. Nienhaus, T. Ha, J. W. Orr, J. R. Williamson, and S. Chu. 2002. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc. Natl. Acad. Sci. USA. 99:4284–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong, M., M. S. Rashes, and N. R. Kallenbach. 1993. Effects of unpaired bases on the conformation and stability of three-arm DNA junctions. Biochemistry. 32:6898–6907. [DOI] [PubMed] [Google Scholar]
- 19.Heuer, D. M., C. Yuan, S. Saha, and L. A. Archer. 2005. Effect of topological asymmetry on the electrophoretic mobility of branched DNA structures with single base mismatches. Electrophoresis. 26:64–70. [DOI] [PubMed] [Google Scholar]
- 20.Heuer, D. M., S. Saha, A. T. Kusumo, and L. A. Archer. 2004. Influence of branch length asymmetry on the electrophoretic mobility of rigid rod-like DNA. Electrophoresis. 25:1772–1783. [DOI] [PubMed] [Google Scholar]
- 21.Albarghouthi, M. N., T. M. Stein, and A. E. Barron. 2003. Poly-N-hydroxyethylacrylamide as a novel, adsorbed coating for protein separation by capillary electrophoresis. Electrophoresis. 24:1166–1175. [DOI] [PubMed] [Google Scholar]
- 22.Williams, B. A., and C. Vigh. 1996. Fast, accurate mobility determination method for capillary electrophoresis. Anal. Chem. 68:1174–1180. [DOI] [PubMed] [Google Scholar]
- 23.Clegg, R. M., A. I. Murchie, A. Zechel, C. Carlberg, S. Diekmann, and D. M. Lilley. 1992. Fluorescence resonance energy transfer analysis of the structure of the four-way DNA junction. Biochemistry. 31:4846–4856. [DOI] [PubMed] [Google Scholar]
- 24.Clegg, R. M., A. I. Murchie, and D. M. Lilley. 1994. The solution structure of the four-way DNA junction at low-salt conditions: a fluorescence resonance energy transfer analysis. Biophys. J. 66:99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gohlke, C., A. I. H. Murchie, D. M. J. Lilley, and R. M. Clegg. 1994. Kinking of DNA and RNA helices by bulged nucleotides observed by fluorescence resonance energy-transfer. Proc. Natl. Acad. Sci. USA. 91:11660–11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vishwasrao, H. D., A. A. Heikal, K. A. Kasischke, and W. W. Webb. 2005. Conformational dependence of intracellular NADH on metabolic state revealed by associated fluorescence anisotrophy. J. Biol. Chem. 280:25119–25126. [DOI] [PubMed] [Google Scholar]
- 27.Lakowicz, J. R. 1999. Principles of Fluorescence Spectroscopy, 2nd ed. Kluwer Academic/Plenum Publishers, New York. 368–371.
- 28.Okonogi, T. M., S. C. Alley, A. W. Reese, P. B. Hopkins, and B. H. Robinson. 2000. Sequence-dependent dynamics in duplex DNA. Biophys. J. 78:2560–2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yan, J., and J. F. Marko. 2004. Localized single-stranded bubble mechanism for cyclization of short double helix DNA. Phys. Rev. Lett. 93:108108. [DOI] [PubMed] [Google Scholar]
- 30.Torre, J. G. 2001. Building hydrodynamic bead-shell models for rigid bioparticles of arbitrary shape. Biophys. Chem. 94:265–274. [DOI] [PubMed] [Google Scholar]
- 31.Viovy, J.-L. 2000. Electrophoresis of DNA and other polyelectrolytes: physical mechanisms. Rev. Mod. Phys. 72:813–872. [Google Scholar]
- 32.Allison, S. A., Z. Li, D. Reed, and N. C. Stellwagen. 2002. Modeling the gel electrophoresis of short duplex DNA by Brownian dynamics: cubic gel lattice with direct interaction. Electrophoresis. 23:2678–2689. [DOI] [PubMed] [Google Scholar]
- 33.Park, I. H., C. S. Johnson Jr., and D. A. Gabriel. 1990. Probe diffusion in polyacrylamide gels as observed by means of holographic relaxation methods: search for a universal equation. Macromolecules. 23:1548–1553. [Google Scholar]