Abstract
The Escherichia coli lactose repressor protein (LacI) provides a classic model for understanding protein-induced DNA looping. LacI has a C-terminal four-helix bundle tetramerization domain that may act as a flexible hinge. In previous work, several DNA constructs, each containing two lac operators bracketing a sequence-induced bend, were designed to stabilize different possible looping geometries. The resulting hyperstable LacI-DNA loops exist as both a compact “closed” form with a V-shaped repressor and also a more “open” form with an extended hinge. The “9C14” construct was of particular interest because footprinting, electrophoretic mobility shift, and ring closure experiments suggested that it forms both geometries. Previous fluorescence resonance energy transfer (FRET) measurements gave an efficiency of energy transfer (ET) of 70%, confirming the existence of a closed form. These measurements could not determine whether open form or intermediate geometries are populated or the timescale of interconversion. We have now applied single-molecule FRET to Cy3, Cy5 double-labeled LacI-DNA loops diffusing freely in solution. By using multiple excitation wavelengths and by carefully examining the behavior of the zero-ET peak during titration with LacI, we show that the LacI-9C14 loop exists exclusively in a single closed form exhibiting essentially 100% ET.
INTRODUCTION
The Escherichia coli lactose repressor protein (LacI) represses expression of the lac operon in the absence of lactose. Binding to the primary promoter-proximal operator O1 located at +11 relative to the start of transcription prevents transcription, but LacI also binds to two secondary operators, O2 and O3, centered at positions +401 and −82, respectively. At the low cellular LacI concentration, the presence of O2 or O3 or both operators is critical for maximal repression, which occurs via DNA loop formation through binding of two sites by a single LacI (1). LacI is a tetramer composed of a dimer of dimers joined by a C-terminal four-helix bundle tetramerization domain connected to the dimers through flexible linkers that may allow the domain to act as a hinge (2). The resulting structural flexibility enhances the ability of LacI to loop DNA under a variety of conditions. Flexible intersubunit arrangements are also observed in the AraC looping protein (3), and more complex heteromeric looping interactions are often mediated through multivalent or otherwise variable interaction surfaces (4).
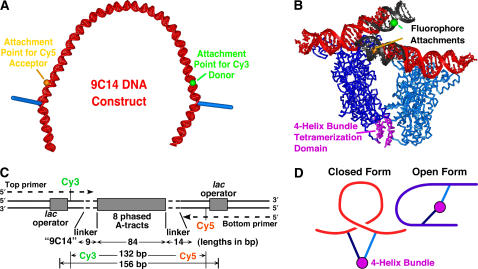
To investigate the geometry of LacI loops, we have prepared and characterized DNA constructs designed to stabilize DNA loops with different possible geometries (5,6). The constructs have two lac operators that bracket a sequence-directed phased A-tract bend (7). The different designs vary in the orientation of the two lac operator dyad axes (and hence LacI dimers) relative to the bend direction. The constructs were characterized by the anticipated geometry of their LacI induced loop as either “wrapping away” (WA) or “wrapping toward/simple loop” (WT/SL). The WA sequence dubbed “9C14” is shown in Fig. 1 and is the focus of these studies.
FIGURE 1.
Design of the fluorophore-labeled 9C14 DNA construct. (A) Model for the looping construct illustrating the orientation of dyad axes of the DNA operators (blue cylinders) relative to the sequence-induced bend. (B) The cocrystal structure of a LacI tetramer-DNA “sandwich” complex, Protein Data Bank entry 1LBG (8). The red duplex DNA is from the cocrystal structure, and the black DNA represents modeled ideal B-DNA extensions. The major groove attachment sites are shown for the donor (green ball) and acceptor (orange ball) fluorophores. The four-helix bundle tetramerization domain (magenta) is connected to the rest of the protein by linkers that are proposed to be flexible. (C) Schematic of the 9C14 sequence showing dimensions in basepairs. The 56-nt PCR primers used to synthesize the full construct are indicated with their internal fluorophores. The total length is 212 bp. (D) Proposed DNA geometries. The closed form loop adopted by 9C14 (red) results from decreasing the radius of curvature of the DNA between the lac operators. The open form conformation is preferred by other constructs with different linkers and was previously proposed to be accessible to 9C14 via DNA twist changes.
The experimental results with designed DNA looping constructs have demonstrated the usefulness of this approach for predicting and constructing hyperstable looping geometries. Biochemical experiments on looping of 9C14 and other constructs showed that different conformations exist, with the WA design adopting a compact “closed form” and the WT/SL design adopting an “open form”. The closed form probably contains a V-shaped repressor in accord with the cocrystal structure of LacI·DNA (8), whereas the open form is proposed to contain an extended LacI tetramer. The 9C14 construct was of special interest in that it was proposed to exist as an equilibrium mixture of the two forms (5). The formation of the two different loop geometries would require distinctly different DNA mechanical strains. The LacI-9C14 open form loop would require twisting the DNA between the bent region and the lac operators to direct the operator dyads inwards, whereas the closed form would result from increased bending of the phased A-tract region to bring outwardly oriented dyads closer in space.
Subsequent bulk fluorescence resonance energy transfer (FRET) measurements confirmed the existence of the closed form for 9C14 but did not resolve conclusively whether or not it exists as a mixture (6). An observed 70% efficiency of energy transfer (ET) could have been due to 70% ET from 100% of the molecules, 100% ET from 70% of the molecules, or any intermediate combination. Preliminary time-resolved measurements on donor emission were consistent with two populations, but the results were inconclusive because a reduced-lifetime population ascribed to the donor undergoing transfer was also observed in the absence of LacI. Elucidation of the equilibrium distribution of these two populations would provide a quantitative comparison between the relative stabilities of the two loop geometries and therefore report on the relative energies involved in twisting versus bending of DNA and also possible protein deformation.
Here we present results from single-molecule FRET (SM-FRET) experiments aimed at providing a quantitative description of DNA-loop conformational distributions for the 9C14 sequence. Detailed consideration of photochemical bleaching effects, which can affect both single-molecule and ensemble FRET measurements, demonstrates clearly that LacI-9C14 exists solely as a closed form loop. This finding reconciles some previous discrepancies among bulk FRET, DNA cyclization kinetics, and electrophoretic mobility shift assay (EMSA) results. Comparison of the biochemical and spectroscopic results suggests that flanking DNA may affect the conformations of LacI-DNA loops.
MATERIALS AND METHODS
Materials
Oligonucleotides labeled with either Cy3 or Cy5 were purchased from Gene Link (Hawthorne, NY) and purified as described below. An expression vector for Lac repressor was generously provided by Dr. Michael Brenowitz (Albert Einstein College of Medicine), and the protein was expressed and purified by L. Edelman essentially as described by Brenowitz et al. (9). Unless otherwise specified, all reagents were purchased from Fisher Scientific (Pittsburgh, PA). All restriction enzymes and T4 DNA ligase were purchased from New England Biolabs (Beverly, MA). The FailSafe PCR system was purchased from Epicentre Technologies (Madison, WI).
Synthesis of fluorescently labeled 9C14
Edelman et al. (6) had used molecular visualization with Insight II (Accelrys) to determine that the optimal labeling position for fluorophores is two basepairs outside the operator sequence. Strategic positioning of the fluorophores ensures that they are oriented away from the face of the DNA bound by the protein and that upon loop formation they are separated by only ∼35 Å in a closed form loop. Assuming random orientations of the transition dipoles, ET should therefore be extremely efficient in the closed form loop. Here we utilized the same optimal positions for the fluorophores in 9C14, but we switched from fluorescein and TAMRA to Cy3 and Cy5 fluorophores to reduce photobleaching.
Two 56 nt fluorescently labeled oligonucleotides were used as PCR primers for synthesis of the 9C14 DNA with a Cy3 (donor) and Cy5 (acceptor) fluorophore as shown in Fig. 1. Oligonucleotides were synthesized on a 1 μM scale with an amino dT C-6 internal modification to which the fluorophore was conjugated. Oligonucleotides were synthesized and purified by Gene Link as follows: unlabeled oligonucleotides were purified on a 12% polyacrylamide 8.3 M urea gel. The full length primers were excised, eluted, and ethanol precipitated. Subsequent conjugation of the Cy3 or Cy5 dye was followed by another round of gel purification. The PCR primer sequences are as follows, where the Cy3 dT modification is indicated as D, the Cy5 dT modification is Q, and the lac operator sequence is underlined: 9C14-Cy3: 5′-CTGCAGGTCAGTCTAGGTAATTGTGAGCGCTCACAATTADATCTCAATTCGTACGG-3′ 9C14-Cy5: 5′-CAAGCTTTACCATCAACGAATTGTGAGCGCTCACAATTAQCTAGCTTCGATTCTAG-3′.
Dual-labeled DNA looping constructs were synthesized using the Failsafe PCR system (Epicentre Technologies). The previously described template for the 9C14 construct (5) was restricted with BstN1 and diluted to 108 molecules/μL. PCR reactions (50 μL) contained 1 μL of template solution, 1 μL each of 50 μM Cy3 and 50 μM Cy5 labeled primers, 1 μL of Failsafe PCR Enzyme Mix, 25 μL of Premix J, and 21 μL of ddH2O. PCR cycling conditions were as follows: 90°C for 30 s, 55°C for 30 s, 72°C for 1 min. The cycle was repeated 25 times, followed by a final extension step of 72°C for 5 min. PCR products were purified in a 6% polyacrylamide native gel (75:1 (w/w) acrylamide: bis-acrylamide) and eluted by shaking overnight in 400 μL of 50 mM NaOAc (pH 7.0, 1 mM EDTA) followed by phenol-chloroform extraction and ethanol precipitation.
Determination of fluorophore labeling efficiencies
Labeling efficiencies were determined by measuring the [dye]/[DNA] of the Cy3 and Cy5 56-nt-labeled oligonucleotides. The concentrations of dyes were calculated using the extinction coefficients reported by Genelink: at their excitation maxima, ɛCy3(552 nm) = 150,000 M−1 cm−1 and ɛCy5(643 nm) = 250,000 M−1 cm−1. The contributions of the dyes to absorption at 260 nm were calculated based on their concentrations and their extinction coefficients at 260 nm, ɛCy3(260) = 4,930 M−1 cm−1 and ɛCy5 (260) = 10,000 M−1 cm−1. The absorbance of the dye at 260 nm was subtracted out, and the concentration of DNA in each sample was then determined using extinction coefficients calculated from the dinucleotide composition (∼33 μg/OD260 unit). The labeling efficiencies for the 9C14Cy3 and 9C14Cy5 labeled primers were initially determined to be 38% and 54%, respectively.
SM-FRET measurements
All SM-FRET measurements were conducted at 1.25 nM DNA concentration with varying LacI concentrations, in 1X LacI buffer: 25 mM bis-Tris pH 7.9, 5 mM MgCl2, 100 mM KCl, 2 mM DTT, 0.02% Nonidet P40 detergent (NP40), 50 μg/ml bovine serum albumin (BSA), and 1X oxygen scavenger mix. The 100X O2 scavenger mix stock is 450 mg/mL glucose, 50% v/v β-mercaptoethanol, 43.2 mg/mL glucose oxidase, and 7.2 mg/ml catalase in glucose oxidase/catalase buffer: 24 mM Pipes pH 6.8, 4 mM MgCl2, 2 mM EGTA. Samples were allowed to equilibrate at room temperature (20°C) for at least 10 min.
The SM-FRET experiments were conducted on freely diffusing molecules in solution following the methods of Weiss and others (10–12). Briefly, a microscope objective (Fluar, 100X, N.A. = 1.3, Carl Zeiss, Oberkochen, Germany) is used to focus a laser beam into a DNA solution, with the focal spot located 10 μm from the glass-liquid interface formed by a microscope coverslip and the DNA solution. An argon ion laser was used for excitation at 514 nm or a HeNe laser for 543 nm. Typical radiant flux densities were on the order of 2–5 kW cm−2 in the laser focal volume to provide excitation rates sufficient for producing detectable bursts during the brief transit time of a single molecule in the detection volume (<1 ms). Fluorescence bursts are detected when molecules traverse the beam. The emission is collected by the same microscope objective and directed through a holographic notch filter (Kaiser Optical, Ann Arbor, MI) to remove scattered excitation light. The collected emission is then directed to a dichroic beam splitter (625DCLP, Chroma, Rockingham, VT), which transmits long wavelength emission from the acceptor fluorophore onto one avalanche photodiode detector (APD) and reflects shorter wavelength emission (<625 nm) from the donor fluorophore onto a second APD. Photon counts from the two photodiodes are recorded in 1 ms time bins on separate channels of a counter/timer board (PCI 6602, National Instruments, Austin, TX) controlled by a personal computer running Labview 6.0 (National Instruments).
Fig. 2 A shows an example of burst data acquired from freely diffusing doubly labeled DNA. Every burst that had a combined donor and acceptor intensity above a certain threshold was analyzed, and the results were binned to construct a histogram describing the probability distribution of FRET values (Fig. 2 B). The arrows in Fig. 2 A denote bursts that have total intensities above 30 counts per ms. This threshold value was chosen to fall within a regime where the FRET histogram shape is unchanged by threshold choice and was used consistently throughout the work (13). The ET efficiency, Eff, of each burst falling above the threshold is calculated from the following equation:
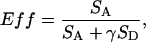 |
(1) |
where SA and SD are the acceptor and donor signals and γ is a correction factor to account for differences in the dye quantum yields and photodiode detection efficiencies. As in previous studies using Cy3 and Cy5, we consider γ to be approximately unity (14). SA and SD are corrected for background counts, and SA is also corrected for the presence of donor-emitted photon counts appearing in the acceptor channel as follows:
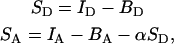 |
(2) |
where IA and ID are the intensities for the acceptor and donor channels, respectively, and BA and BD are the background counts in the two channels. The α-factor is used to correct for donor photons that “leak” into the acceptor channel detector as a result of spectral overlap. α is experimentally determined using bursts collected from a sample of donor-only labeled DNA. Correcting the acceptor channel for donor leak-through improves the FRET efficiency accuracy but degrades the precision, since the noise in the donor channel is added to the acceptor channel. As a result the zero-ET peak has a width that is about twice the expected value at our signal levels.
FIGURE 2.
SM-FRET data from diffusing 9C14 molecules. (A) A typical burst sequence acquired with the 9C14 DNA construct (1 nM) excited at 514 nm. The acceptor counts are shown in red, and donor counts are in green. The LacI concentration is 2.5 nM, and a mix of bursts exhibiting both high and low ET values are observed. The bin size for the transient is 1 ms. Arrows denote bursts that exceed the intensity threshold for analysis. The bursts located near 0.5 s appear principally in the acceptor channel and originate from LacI/9C14 complexes forming a closed loop with an ET efficiency of nearly 1.0. Bursts that appear principally in the donor channel such as the one near 0.15 s belong to free or mislabeled 9C14 molecules. (B) Typical FRET histograms acquired with 514 nm laser excitation for 9C14 both as free DNA (blue) and fully occupied with LacI (gray). Negative efficiency values in the zero-ET peak are due to bursts in which the acceptor channel intensity was less than the average background. The width of the zero-ET peak is further broadened by the subtraction of donor counts from the acceptor channel, which is necessary to correct for nonideal optical filtering, as described in Materials and Methods. The histogram for fully bound 9C14 has a significant zero-ET peak, which is due to a combination of donor-only labeled 9C14 and 9C14 with photobleached acceptors.
RESULTS
Previous experiments, including DNA cyclization kinetics, EMSAs, and ensemble FRET measurements (5,6), have shown that the 9C14 construct forms a hyperstable LacI-DNA loop, with t1/2 > 24 h. However, the precise geometry and possible heterogeneity of the loop remain unresolved. DNA cyclization products of 9C14-LacI loops with extended DNA tails included minicircles with both relaxed and positively supercoiled loop topologies, and on this basis it was suggested that 9C14 can assume a closed form loop configuration but that the most abundant conformation may be an open form bridged by an extended LacI tetramer (5). However, these results were at odds with EMSAs of 9C14-LacI complexes. Only one conformer was resolved in the gel, and based on its mobility it was believed to be the closed form loop. The observations could be reconciled if the electrophoretic gel acts as a stabilizing medium for the closed form loop or if the conformers are in rapid exchange in the gel and the closed form migrates very rapidly. In an effort to better determine the conformational makeup of the 9C14-LacI complex and resolve the apparent discrepancy, a fluorescently labeled version of 9C14 was synthesized for use in steady-state and time-resolved FRET measurements (6). The outcome of ensemble FRET measurements was the definitive verification of the closed form LacI-9C14 loop complex by observation of significant ET, in accordance with the predicted model. Due to complications including apparent non-Förster quenching of the donor molecule as well as donor-dependent quenching of the acceptor, the ensemble FRET data were not suitable for definitive determination of the number and relative abundance of 9C14 conformations. For this reason we have undertaken single-molecule experiments to resolve the equilibrium composition of the LacI/9C14 complex directly.
Single-molecule 9C14 DNA FRET results upon titration with LacI
The 9C14 system is ideally suited for study with SM-FRET since it is expected to exhibit a much larger FRET efficiency for the closed form loop conformation than for free 9C14 or the open form loop. The predicted interdye distance for the closed form loop is ∼35 Å, which should yield a FRET efficiency of 0.9 based on the measured Förster distance of 60 Å for Cy3-Cy5 coupled to DNA (15). Hence the closed-loop geometry should be easily distinguished from unbound 9C14 or from an open form looped complex.
The data in Fig. 3 are the results from LacI titration experiments conducted with 1.2 nM 9C14 DNA. The binding of LacI results in a decrease in the number of molecules showing zero FRET and a concomitant increase in single molecules displaying high ET, corresponding to the formation of a closed form 9C14-LacI loop complex. The average FRET efficiency (〈Eff〉) of the high-ET population is 0.9, in good agreement with predicted values. The titration results are summarized in Fig. 3 B, which plots the fraction of molecules possessing FRET efficiency >50% as a function of LacI concentration. The trend saturates at a LacI concentration of 2.5 nM or less, no more than twice the DNA concentration. At saturation, the proportion of closed-loop molecules was typically 50–60%. Small variations between samples or between experiments conducted on different days are attributed to variability in the PCR process or to sample degradation. In all our experiments, the population with high ET efficiency was absent in the free DNA and increased with LacI concentration until saturation was reached, after which a small decrease in the population was observed. This saturation and subsequent quenching or conversion to other forms agrees with results from gel mobility shift assays and bulk FRET (5). An obvious advantage of the single-molecule experiments is the ability to track both populations throughout the titration process rather than observing an average value. In bulk steady-state titration experiments, a weighted average of the two populations is observed throughout the titration process (6).
FIGURE 3.
SM-FRET results from titration experiments acquired with 543 nm excitation. (A) Histograms acquired with three different LacI concentrations illustrating the decrease in the zero-ET peak and the simultaneous increase in the proportion of molecules appearing near 90% ET efficiency. Without LacI (dashed line), only the zero-ET peak is observed. Upon addition of LacI (solid line) a population near 90% ET efficiency appears. This population is assigned to the closed form 9C14-LacI loop conformation. In the fully bound histogram (dotted line) 60% of the recorded single-molecule bursts show high ET efficiency. The remaining bursts are assigned to the zero-ET peak. (B) Graph of the fraction of bursts with ET values above 50% versus LacI concentration. The trend illustrates the direct relationship between the number of bursts with efficient ET and the LacI concentration. Note that at 5.0 nM LacI, a decrease in the fraction with high ET efficiency is observed, which may be due to quenching of the acceptor by nonspecific binding of LacI to 9C14.
Comparison of SM-FRET results with ensemble results
Steady-state bulk FRET experiments had previously established an apparent average ET efficiency of 74% for the bound LacI-9C14 complex, based on donor quenching (6). This placed a lower limit on the FRET efficiency for the closed form loop, since the bulk experiments report a weighted average of all values present in the sample. The single-molecule histograms shown in Figs. 2 B and 3 A show that this average value is in fact very rarely observed for any single molecule. Our results definitively establish 90% as the closed form loop ET efficiency calculated from Eq. 1. The single-molecule results also establish a lower limit of 60% as the fraction of molecules appearing with 90% ET efficiency in the fully bound sample. We emphasize that at this point in the analysis 60% was only a lower limit due to the lack of information on the identity of the molecules which compose the zero-energy transfer peak (henceforth the “zero-ET peak”). Molecules displaying zero ET could arise from several sources, such as 9C14-LacI complexes with open form loop geometries, 9C14 molecules missing the Cy5 acceptor fluorophore, or Cy5 fluorophores which have undergone the photophysical changes described below. To investigate the composition of the zero-ET peak, we conducted oxygen scavenging and power dependence experiments to determine the role of laser-induced photobleaching or transient dark acceptor states, and we used experiments conducted at two different excitation wavelengths to reveal compositional heterogeneity.
Role of excited state processes on the size of the zero-ET peak
The intense laser power necessary to produce detectable emission from single molecules can lead to undesired excited-state processes. The power dependences of excited state photophysical and photochemical processes affecting single-molecule measurements have been investigated by a number of groups (16–18). Irreversible photobleaching is of particular relevance for single-molecule measurements since this process limits the observation time for immobilized single molecules (17) and introduces artifacts in experiments with single diffusing molecules (11,12). In our experiments, irreversible photobleaching of the donor fluorophore has no effect on the FRET histogram since direct excitation of the acceptor is weak. Photoinduced processes affecting the acceptor fluorophore present a more serious difficulty, since they introduce artifacts in the SM-FRET results (11,12). When the acceptor fluorophore undergoes irreversible photobleaching, the donor emission is still present. Thus, if the acceptor fluorophore bleaches before reaching the observation volume the apparent ET efficiency will always be zero, regardless of the actual conformation of the molecule. If acceptor bleaching occurs during the acquisition of a single-molecule burst, the apparent ET efficiency will be a weighted average of the true ET efficiency value (prephotobleaching) and zero (postphotobleaching). In addition to irreversible photobleaching, an acceptor fluorophore may enter a transient, reversible dark state through intersystem crossing or by assuming a nonemissive conformation. These transient states may also introduce artifacts in the SM-FRET histogram depending on their rate of production and lifetimes. Importantly, the excited state photophysics of the acceptor fluorophore are only relevant when there is efficient ET, since otherwise the acceptor predominantly occupies its ground state.
To evaluate the effects of excited state processes here, we have conducted excitation-power dependence studies to investigate the role of light-induced bleaching of Cy5 on the observed SM-FRET histogram. To evaluate the effect of oxygen, experiments were conducted on samples of fully bound 9C14 under ambient conditions or in the presence of an oxygen scavenging system. Under these conditions, Cy5 photobleaching should appear as a decrease in the fraction of molecules showing efficient ET with a concomitant increase in the size of the zero-ET peak. The results from these studies are summarized in Fig. 4, in which the percentage of total molecules with ET efficiency above 50% is plotted against excitation laser power. There is clearly a strong power dependence under ambient conditions (open circles). The addition of an oxygen scavenging system (solid circles) decreases the power dependence markedly, but a decrease of ∼20% to a constant value of ∼40% is still observed.
FIGURE 4.
Effect of excitation power on the appearance of molecules with high ET efficiency. This plot demonstrates the strong laser excitation power dependence on the observed SM-FRET histogram. The fraction of molecules in the fully bound sample that exhibit ET efficiency above 50% is plotted as a function of laser excitation power under ambient conditions (○) and with the addition of an oxygen scavenging system (•). Under ambient conditions, as the power is increased there is a steep decrease in the fraction of molecules exhibiting efficient ET. With the addition of the glucose oxidase- and catalase-based oxygen scavenger, the power dependence is significantly diminished, leveling off above 50 μW.
The strong power dependence observed under ambient conditions is most probably due to oxygen-induced photobleaching. This occurs through the interaction of the fluorophore's triplet state with ground state molecular oxygen, leading to the formation of singlet oxygen (19,20) followed by oxidation of the fluorophore by singlet oxygen or one of its byproducts. This process, whether occurring in or out of the confocal detection volume, will cause a depletion of the fraction of molecules displaying efficient ET, since photochemical oxidation of the acceptor is irreversible. In contrast, for transient dark states to significantly impact the FRET histogram, they must be rapidly formed in or near the observation volume and persist long enough to have a measurable effect on the observed FRET value.
From Fig. 4 it is clear that under reduced-oxygen conditions the power dependence is greatly diminished. It is unlikely that the observed power dependence is due solely to residual oxygen, since the saturating power at ∼50 μW would correspond to the region where photobleaching becomes diffusion limited and would be the same for both sets of experiments. Therefore, the observed effect is more probably due to the FRET-induced formation of a nonemissive acceptor fluorophore triplet state. Under ambient conditions triplet state lifetimes are on the order of 10 μs and do not appreciably affect the measured FRET efficiency value, since the triplet lifetime is a small fraction of the average observation time of 9C14 (several hundred microseconds). However, at reduced oxygen concentrations it is well known that cyanine fluorophores such as Cy5 form long-lived triplet states (21,22). Previous single-molecule studies with the carbocyanine dye DiIC18 have shown that triplet lifetimes can exceed 100 ms under vacuum (21,23–25). Sauer and co-workers have shown that Cy5 can display millisecond triplet lifetimes under a nitrogen-purged environment (21). Hence, under the conditions used here, it is reasonable to assume that the triplet lifetime will be on the millisecond timescale.
The power dependence data can all be reconciled by considering the long-lived triplet acceptor state 3A. We also must assume that it acts as an ET acceptor, as shown previously (21), but with R0 for the D-3A pair shorter than that for D-A. At very low excitation powers, the time spent in the singlet manifold (i.e., before triplet formation) will be longer than the observation time due to the low quantum yield of triplet formation, and hence the triplet state will have no effect on the FRET histogram. As the laser intensity increases, the excitation rate increases linearly and so does the rate of triplet formation. As the probability of observing the nonemissive triplet state increases, apparent ET decreases, because the decreased R0 of the D-3A pair, as opposed to D-A, is much more likely to lead to donor emission. The leveling off of ET as a function of power suggests one additional property of the triplet: it must undergo light-activated conversion to the singlet state. Our reasoning is that if the triplet state persisted during the entire observation period for a single molecule, then we would expect a continued decrease in observed efficiency. However, if the triplet lifetime is also power dependent, then a photostationary state is established in which the singlet and triplet manifolds of states are in dynamic equilibrium. The establishment of a steady state requires that Cy5 undergo a reverse intersystem crossing process that is induced by ET from the Cy3 singlet state. The recent work of Zhuang and co-workers has shown that direct excitation of Cy3 can dramatically increase the rate of back-conversion of Cy5 from a dark state to an emissive state. This effect is strongest when the two chromophores are separated by 30 Å or less and requires β-mercaptoethanol and a glucose oxidase oxygen scavenging system (22). We believe that our ET power dependence under reduced oxygen conditions reflects the same phenomenon.
In summary, we propose that the depletion of efficient ET observed at low power in the absence of oxygen is due to formation of a single-molecule photostationary state at laser powers above 40 μW. The observed Eff is the weighted average of the efficiencies of the singlet and triplet states of the acceptor. At lower power where the triplet is not created or not reexcited during the time the molecule is in the beam, we observe the Eff characteristic of the singlet manifold of the acceptor. These results illustrate the necessity for using oxygen scavenging to reduce irreversible photobleaching but also show that the resulting increased triplet lifetimes can impact the FRET histogram.
Excitation wavelength dependence studies
The discussion above illustrates how photobleaching and other excited state processes can lead to a decrease in observed FRET. The fits for the power-dependent data in Fig. 4 suggest that at minimal laser power up to 70% of the observed molecules are undergoing efficient ET in the fully bound sample. To determine the actual distribution of loop geometries for fully bound 9C14, evaluation of the 30–40% of molecules comprising the zero-ET peak is necessary. To achieve this, we have employed a two-color excitation scheme.
Single-molecule experiments conducted with multiple excitation wavelengths have proven useful for elucidating a variety of photophysical processes (24,26,27). More recently, rapidly alternating, multiwavelength excitation has been demonstrated as a powerful technique for sorting freely diffusing single molecules and molecular complexes in solution (28,29). We employ a simpler technique in which data acquired at two different excitation wavelengths are analyzed to determine the composition of molecules that contribute to the zero-ET peak at each LacI concentration. In these experiments the two excitation wavelengths were 514 nm and 543 nm. Both wavelengths efficiently excite the donor fluorophore, Cy3, but only excitation with 543 nm can significantly excite the acceptor fluorophore, Cy5. At 543 nm Cy5 is excited ∼12 times more efficiently than at 514 nm, as shown in Fig. 5.
FIGURE 5.
Corrected excitation spectra of the donor (dotted line) and acceptor (solid lines). The excitation spectra are plotted in units of M−1 cm−1 rather than emission intensity to highlight the disparity in the absorptivity of the two dyes at the excitation wavelengths used. The inset provides an expanded view of the acceptor excitation spectrum in the region near the two laser wavelengths to illustrate the large increase in absorbance (an order of magnitude) with 543 nm excitation relative to 514 nm excitation.
Our two-color scheme is easily implemented using conventional continuous wave excitation. Comparison of data acquired at the two different excitation wavelengths provides information about the composition of the zero-ET peak. To illustrate, we have constructed zero ET-difference histograms for data acquired at each wavelength by subtracting the histograms obtained for free DNA from those for 9C14 completely bound by LacI. The difference histogram for the zero-ET peak acquired with 514 nm excitation is shown in Fig. 6 A (negative-going bars). This represents the portion of the signal which has been shifted to the peak at Eff = 0.9 by loop formation. The difference peak is symmetric about zero, indicating homogeneous depletion of the zero-ET peak. In contrast to the symmetric depletion seen with 514 nm excitation, the difference histogram for data acquired with 543 nm excitation in Fig. 6 B shows asymmetric depletion of the zero-ET peak. Preferential removal of signal from the right side of the zero-ET peak with 543 nm excitation shows that an inhomogeneous composition exists within the zero-ET peak. Since this heterogeneity is observed only at a wavelength where the acceptor is excited, it suggests that the subpopulation appearing on the right side of the 543 nm zero-ET peak is composed of molecules containing an unbleached acceptor fluorophore. This assignment is possible because a donor-only labeled DNA sample was used to determine the correction factor, α, used in Eq. 2, ensuring that singly labeled, donor-only molecules are distributed symmetrically about zero in the FRET histogram regardless of the excitation wavelength. This correction procedure also has the consequence that with 543 nm excitation, weak direct excitation of the acceptor fluorophore results in a small apparent ET value (centered at 9%) for DNA molecules possessing both fluorophores, even though no actual ET is occurring.
FIGURE 6.
Analysis of SM-FRET histograms. SM-FRET histograms from free 9C14 (dotted line) and fully bound 9C14 (solid line) were acquired with (A) 514 nm and (B) 543 nm excitation. A difference histogram was constructed for the zero-ET peak at each wavelength by subtracting the histogram of the free 9C14 sample (dotted line) from that of the fully bound sample (solid line). The difference histograms (negative-going bars) for each wavelength were fit to Gaussian distributions (negative-going solid lines). With 514 nm excitation, the difference histogram is symmetrically positioned around zero. In contrast, the difference histogram obtained with 543 nm excitation is shifted toward positive values due to selective depletion of the zero-ET peak on its right side. The parameters obtained from the Gaussian fit in (B) along with parameters obtained from fitting data acquired with donor-only labeled DNA are used to decompose the zero-ET peak at all LacI concentrations (Table 1). (C) SM-FRET histogram of free 9C14 DNA (bars) acquired with 543 nm excitation. The fit to the zero-ET peak (black line) is a sum of Gaussian distributions representing donor-only labeled 9C14 (shaded line) and double-labeled 9C14 (dotted line). (D) Histogram from fully bound 9C14 acquired with 543 nm excitation. The zero-ET peak is fit with a single Gaussian distribution, and the high efficiency peak is fit with a β-distribution function. The Gaussian has the same center and width as those required to obtain a good fit to a FRET histogram acquired with a donor-only labeled sample. The β-distribution function has been used previously to describe SM-FRET histograms (11).
A quantitative decomposition of the zero-ET peak was performed for each LacI concentration. To determine the relative contributions to the zero-ET peak from DNA molecules with functional acceptor fluorophores versus those with damaged fluorophore, we have used the position and widths of both the zero-ET difference peak and the donor-only peak to provide a set of basis parameters for decomposing the zero-ET peak into two Gaussian distributions. In Fig. 6 B, a Gaussian distribution with an average ET of 9% and a variance of 22% is shown superimposed on the observed difference histogram, and these parameters were used to describe the contribution to the zero-ET peak from doubly labeled molecules. A FRET histogram collected from donor-only labeled molecules using 543 nm excitation was fit with a Gaussian distribution to provide the position (0%) and variance (28%) used to describe the population with missing or nonemissive acceptor fluorophores. Fig. 6 C shows the resulting fit (black line) to the zero-ET peak (bars) of free 9C14, obtained by holding the widths and means of the two-component Gaussians (shaded line and dotted line) constant while varying their amplitudes. Omission of either of the Gaussians from the fit significantly increases the χ2 value. Fig. 6 D gives the results from fitting the histogram of the fully bound sample. The β-distribution function (11) has been used to fit the peak at 90% efficiency. Only the Gaussian distribution centered at zero was necessary to fit the zero-ET peak.
The results from the decomposition above are shown in Table 1. The amplitude of the Gaussian component centered at zero was constant throughout the range of LacI concentrations. This invariance with respect to LacI concentration agrees with the assignment of the population at zero as molecules lacking a functional acceptor fluorophore. The contribution of the second Gaussian, describing the population with an intact and emissive acceptor fluorophore, steadily decreases with addition of LacI protein. The decrease of this population is accompanied by the appearance of molecules with high ET, as shown in the titration data in Figs. 2 B and 3 A.
TABLE 1.
Summary of best-fit parameters used to decompose the zero-ET peak at different LacI concentrations
| [LacI] (nM) | A1 | A2 |
|---|---|---|
| 0.0 | 0.014 | 0.0087 |
| 0.05 | 0.014 | 0.0086 |
| 0.5 | 0.012 | 0.0054 |
| 2.5 | 0.014 | 0.0003 |
The zero-ET peak (of the probability P(Eff) of observing an ET efficiency Eff) was fit by the sum of two Gaussian distributions: 
The good fit obtained with a single Gaussian for the fully bound sample is observed because the zero-ET peak is symmetrically centered about zero: all signal from doubly labeled DNA has been shifted to the high FRET efficiency region of the histogram. Therefore we assign the zero-ET peak of the histogram for fully bound 9C14 to molecules with either bleached or missing acceptor fluorophore. These molecules, although still presumably existing in a “closed” complex, cannot show ET. Thus, all of the 9C14 molecules capable of ET appear in the peak at 90% ET. We conclude that the 9C14 molecule behaves as predicted for a WA configuration and that the bound species is a closed loop stabilized by a V-shaped LacI tetramer.
DISCUSSION
Previous DNA cyclization kinetics, EMSAs, and bulk FRET studies on the 9C14-LacI protein-DNA loop demonstrated the existence of a closed form WA loop in which the two operators are held close to each other in space. It was also shown that the related 11C12 molecule, which has different helical phasing between the operators and the central bend, gives an open form loop proposed to result from opening of the LacI C-terminal domain hinge. The previous work could not, however, resolve whether the 9C14-LacI complex also assumes an open form loop. EMSA results suggested a single conformation whose rapid mobility was consistent with a compact closed form loop. DNA cyclization kinetics on constructs with extended tails gave a mixture of positively supercoiled products consistent with the closed form and relaxed products consistent with the open form. Bulk FRET measurements were interpreted as being consistent with a mixed population of open form and closed form 9C14-LacI loops.
The SM-FRET results described here demonstrate that the closed form 9C14-LacI loop exhibits an ET efficiency of at least 90%. This is strong evidence in favor of the proposed WA geometry. Adhya and co-workers have explored the consequences of the four possible loop connectivities for a V-shaped repressor (30). Their experiments show that the Gal and Lac repressors form antiparallel loops on longer DNA molecules (31). The closed form loop we have focused on here is described as one of two possible parallel loops in their terminology. With the labeling positions used here, molecular modeling suggests that the other three possible geometries would exhibit interfluorophore separations of at least 80 Å given the V-shaped repressor. Therefore, the SM-FRET histograms eliminate structural possibilities that bulk FRET, due to its measurement only of average efficiencies, could not conclusively resolve.
Several complementary single-molecule methods have been applied to protein-DNA loops. Finzi and Gelles (32) demonstrated LacI-DNA loop formation using the decreased amplitude of Brownian motion of a bead tethered by the DNA upon looping. Magnetic tweezers experiments on the DNA-Gal repressor-HU protein loop were used to measure the approximate loop size and its stability (33). Both of these experiments have the advantage that a single molecule can be studied over an extended period of time. Finally, SM-FRET experiments similar to ours on the DNA loop induced by the restriction enzyme NgoMIV show that the technique can distinguish among different populations of loops or other shapes (34). Sample immobilization will allow SM-FRET to be used to probe the stability and dynamics of protein-DNA loops at high resolution.
Careful analysis of the zero-ET peak using two excitation wavelengths in our SM-FRET histograms suggests that the 9C14-LacI loop exclusively adopts the closed form, in agreement with previous EMSA results but in apparent contradiction to bulk FRET and DNA cyclization. We suspect that the previous bulk FRET results may have been influenced by photobleaching during the course of DNA synthesis or experiments carried out over a period of time. Labeling efficiency is measured on a concentrated sample of oligonucleotide primer. If the labeling efficiency of the acceptor in the experimental sample is overestimated (i.e., if photobleaching has occurred after the measurement), bulk FRET will give an underestimate for the actual efficiency of ET. It remains to explain the difference between SM-FRET and DNA cyclization results. We suggest that the long DNA tails needed to allow cyclization may influence the population distribution. For example, wrapping of the DNA outside the operators around the protein (35) could preferentially stabilize an open form, and this effect would depend on the length of flanking DNA. In principle, this could be tested using double labeled molecules with the fluorophores in the middle of extended constructs, but these are technically challenging to prepare.
In conclusion, we have demonstrated that SM-FRET on DNA loops can conclusively identify the geometry of protein-DNA loops where other methods fail to do so, and we suggest that population distributions among different loop conformations will also be accessible using single-molecule methods. Correct interpretation of the single-molecule results requires careful consideration of the dark states of the acceptor fluorophores.
Acknowledgments
We are grateful to Laurence Edelman for help with initial experiments.
The work was supported in part by a National Science Foundation Career award and a National Institutes of Health grant to J.D.K., and University of Maryland, College Park, startup funds to D.S.E.
Michael A. Morgan and Kenji Okamoto contributed equally to this work.
Kenji Okamoto's present address is Dept. of Chemistry, Graduate School of Science, Kyoto University, Kyoto, Japan.
Michael A. Morgan's present address is Naval Research Laboratory, Washington, D.C. 20375.
References
- 1.Oehler, S., M. Amouyal, P. Kolkhof, B. von Wilcken-Bergmann, and B. Müller-Hill. 1994. Quality and position of the three lac operators of E. coli define efficiency of repression. EMBO J. 13:3348–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Friedman, A. M., T. O. Fischmann, and T. A. Steitz. 1995. Crystal structure of lac repressor core tetramer and its implications for DNA looping. Science. 268:1721–1727. [DOI] [PubMed] [Google Scholar]
- 3.Harmer, T., M. Wu, and R. Schleif. 2001. The role of rigidity in DNA looping-unlooping by AraC. Proc. Natl. Acad. Sci. USA. 98:427–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lilja, A. E., J. R. Jenssen, and J. D. Kahn. 2004. Geometric and dynamic requirements for DNA looping, wrapping and unwrapping in the activation of E. coli glnAp2 transcription by NtrC. J. Mol. Biol. 342:467–478. [DOI] [PubMed] [Google Scholar]
- 5.Mehta, R. A., and J. D. Kahn. 1999. Designed hyperstable Lac repressor·DNA loop topologies suggest alternative loop geometries. J. Mol. Biol. 294:67–77. [DOI] [PubMed] [Google Scholar]
- 6.Edelman, L. M., R. Cheong, and J. D. Kahn. 2003. Fluorescence resonance energy transfer over ∼130 basepairs in hyperstable Lac repressor-DNA loops. Biophys. J. 84:1131–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crothers, D. M., T. E. Haran, and J. G. Nadeau. 1990. Intrinsically bent DNA. J. Biol. Chem. 265:7093–7096. [PubMed] [Google Scholar]
- 8.Lewis, M., G. Chang, N. C. Horton, M. A. Kercher, H. C. Pace, M. A. Schumacher, R. G. Brennan, and P. Lu. 1996. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science. 271:1247–1254. [DOI] [PubMed] [Google Scholar]
- 9.Brenowitz, M., A. Pickar, and E. Jamison. 1991. Stability of a Lac repressor mediated “looped complex”. Biochemistry. 30:5986–5998. [DOI] [PubMed] [Google Scholar]
- 10.Deniz, A. A., T. A. Laurence, M. Dahan, D. S. Chemla, P. G. Schultz, and S. Weiss. 2001. Ratiometric single-molecule studies of freely diffusing biomolecules. Annu. Rev. Phys. Chem. 52:233–253. [DOI] [PubMed] [Google Scholar]
- 11.Dahan, M., A. A. Deniz, T. Ha, D. S. Chemla, P. G. Schultz, and S. Weiss. 1999. Ratiometric measurement and identification of single diffusing molecules. Chem. Phys. 247:85–106. [Google Scholar]
- 12.Schuler, B., E. A. Lipman, and W. A. Eaton. 2002. Probing the free-energy surface for protein folding with single-molecule fluorescence spectroscopy. Nature. 419:743–747. [DOI] [PubMed] [Google Scholar]
- 13.Deniz, A. A., M. Dahan, J. R. Grunwell, T. Ha, A. E. Faulhaber, D. S. Chemla, S. Weiss, and P. G. Schultz. 1999. Single-pair fluorescence resonance energy transfer on freely diffusing molecules: observation of Förster distance dependence and subpopulations. Proc. Natl. Acad. Sci. USA. 96:3670–3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cosa, G., E. J. Harbron, Y. N. Zeng, H. W. Liu, D. B. O'Connor, C. Eta-Hosokawa, K. Musier-Forsyth, and P. F. Barbara. 2004. Secondary structure and secondary structure dynamics of DNA hairpins complexed with HIV-1 NC protein. Biophys. J. 87:2759–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ha, T., I. Rasnik, W. Cheng, H. P. Babcock, G. H. Gauss, T. M. Lohman, and S. Chu. 2002. Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature. 419:638–641. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez, E. J., L. Novotny, G. R. Holtom, and X. S. Xie. 1997. Room-temperature fluorescence imaging and spectroscopy of single molecules by two-photon excitation. J. Phys. Chem. A. 101:7019–7023. [Google Scholar]
- 17.Deschenes, L. A., and D. A. Vanden Bout. 2002. Single molecule photobleaching: increasing photon yield and survival time through suppression of two-step photolysis. Chem. Phys. Lett. 365:387–395. [Google Scholar]
- 18.Fleury, L., J. M. Segura, G. Zumofen, B. Hecht, and U. P. Wild. 2000. Nonclassical photon statistics in single-molecule fluorescence at room temperature. Phys. Rev. Lett. 84:1148–1151. [DOI] [PubMed] [Google Scholar]
- 19.Turro, N. J. 1991. Modern Molecular Photochemistry. University Science Books, Mill Valley, CA.
- 20.Kasche, V., and L. Lindqvist. 1964. Reactions between the triplet state of fluorescein and oxygen. J. Phys. Chem. 68:817–823. [Google Scholar]
- 21.Tinnefeld, P., V. Buschmann, K. Weston, and M. Sauer. 2003. Direct observation of collective blinking and energy transfer in a bichromophoric system. J. Phys. Chem. A. 107:323–327. [Google Scholar]
- 22.Bates, M., T. R. Blosser, and X. W. Zhuang. 2005. Short-range spectroscopic ruler based on a single-molecule optical switch. Phys. Rev. Lett. 94:108101–108104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.English, D. S., A. Furube, and P. F. Barbara. 2000. Single-molecule spectroscopy in oxygen-depleted polymer films. Chem. Phys. Lett. 324:15–19. [Google Scholar]
- 24.English, D. S., E. J. Harbron, and P. F. Barbara. 2000. Probing photoinduced intersystem crossing by two-color, double resonance single molecule spectroscopy. J. Phys. Chem. A. 104:9057–9061. [Google Scholar]
- 25.Weston, K. D., P. J. Carson, J. A. DeAro, and S. K. Buratto. 1999. Single-molecule detection fluorescence of surface-bound species in vacuum. Chem. Phys. Lett. 308:58–64. [Google Scholar]
- 26.Fukaminato, T., T. Sasaki, T. Kawai, N. Tamai, and M. Irie. 2004. Digital photoswitching of fluorescence based on the photochromism of diarylethene derivatives at a single-molecule level. J. Am. Chem. Soc. 126:14843–14849. [DOI] [PubMed] [Google Scholar]
- 27.Kulzer, F., S. Kummer, R. Matzke, C. Brauchle, and T. Basche. 1997. Single-molecule optical switching of terrylene in p-terphenyl. Nature. 387:688–691. [Google Scholar]
- 28.Kapanidis, A. N., N. K. Lee, T. A. Laurence, S. Doose, E. Margeat, and S. Weiss. 2004. Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proc. Natl. Acad. Sci. USA. 101:8936–8941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, H., X. Ren, L. Ying, S. Balasubramanian, and D. Klenerman. 2004. Measuring single-molecule nucleic acid dynamics in solution by two-color filtered ratiometric fluorescence correlation spectroscopy. Proc. Natl. Acad. Sci. USA. 101:14425–14430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Semsey, S., K. Virnik, and S. Adhya. 2005. A gamut of loops: meandering DNA. Trends Biochem. Sci. 30:334–341. [DOI] [PubMed] [Google Scholar]
- 31.Virnik, K., Y. L. Lyubchenko, M. A. Karymov, P. Dahlgren, M. Y. Tolstorukov, S. Semsey, V. B. Zhurkin, and S. Adhya. 2003. “Antiparallel” DNA loop in gal repressosome visualized by atomic force microscopy. J. Mol. Biol. 334:53–63. [DOI] [PubMed] [Google Scholar]
- 32.Finzi, L., and J. Gelles. 1995. Measurement of lactose repressor-mediated loop formation and breakdown in single DNA molecules. Science. 267:378–380. [DOI] [PubMed] [Google Scholar]
- 33.Lia, G., D. Bensimon, V. Croquette, J. F. Allemand, D. Dunlap, D. E. Lewis, S. Adhya, and L. Finzi. 2003. Supercoiling and denaturation in gal repressor/heat unstable nucleoid protein (HU)-mediated DNA looping. Proc. Natl. Acad. Sci. USA. 100:11373–11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katiliene, Z., E. Katilius, and N. W. Woodbury. 2003. Single molecule detection of DNA looping by NgoMIV restriction endonuclease. Biophys. J. 84:4053–4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saecker, R. M., and J. M. T. Record. 2002. Protein surface salt bridges and paths for DNA wrapping. Curr. Opin. Struct. Biol. 12:311. [DOI] [PubMed] [Google Scholar]