Abstract
The nonsymbiotic tomato hemoglobin SOLly GLB1 (Solanum lycopersicon) is shown to form a homodimer of ∼36 kDa with a high affinity for oxygen. Furthermore, our combined ultraviolet/visible, resonance Raman, and continuous wave electron paramagnetic resonance (EPR) measurements reveal that a mixture of penta- and hexacoordination of the heme iron is found in the deoxy ferrous form, whereas the ferric form shows predominantly a bis-histidine ligation (F8His-Fe2+/3+-E7His). This differs from the known forms of vertebrate hemoglobins and myoglobins. We have successfully applied our recently designed pulsed-EPR strategy to study the low-spin ferric form of tomato hemoglobin. These experiments reveal that, in ferric SOLly GLB1, one of the histidine planes is rotated 20°(±10°) away from a Nheme-Fe-Nheme axis. Additionally, the observed g-values indicate a quasicoplanarity of the histidine ligands. From the HYSCORE (hyperfine sublevel correlation) measurements, the hyperfine and nuclear quadrupole couplings of the heme and histidine nitrogens are identified and compared with known EPR/ENDOR data of vertebrate Hbs and cytochromes. Finally, the ligand binding kinetics, which also indicate that the ferrous tomato Hb is only partially hexacoordinated, will be discussed in relation with the heme-pocket structure. The similarities and differences with other known nonsymbiotic plant hemoglobins will be highlighted.
INTRODUCTION
Hemoglobins (Hbs) are ubiquitous proteins found in animal, fungi, bacteria, protozoa, and plants. Although the existence of hemoglobin in the animal kingdom has long been common knowledge, the findings of its presence in nonanimal sources, including bacteria, unicellular eukaryotes, and plants, are relatively recent (1). In plants, there are at least three distinct types of hemoglobins: symbiotic, nonsymbiotic, and truncated hemoglobins. All plant hemoglobins, described so far, are either monomers or homodimers without heme-heme interaction (2).
Nonsymbiotic hemoglobins (nsHbs) have recently been identified in many mono- and dicotyledonus plants, including barley, soybean, rice, Arabidopsis, tomato, cotton, chicory, and corn (3–5). They possess the highest reported oxygen affinities among plant hemoglobins. Expression is constitutive in some plants and induced by hypoxia in others. Based on the observations made by Sowa et al. (6), three possible functions have been proposed for nsHbs: i), they might be O2 transport proteins that scavenge oxygen under hypoxic conditions; ii), they might act as terminal oxidases that, by virtue of their high O2 affinity, facilitate glycolytic generation of ATP by removing NADH under microaerobic conditions; iii), they could also be O2 sensing proteins that activate other proteins with regulatory functions (6,7). Recently, it was found that tomato nonsymbiotic hemoglobin gene expression can be influenced also by a broad range of changes in mineral macronutrient and micronutrient status, suggesting a previously unrecognized generic role for nonsymbiotic hemoglobins in processes associated with mineral nutrient nutrition (8).
The gene structure appears the same for symbiotic, nonsymbiotic, and truncated hemoglobins with four exons and three introns at position B12.2, E15.0, and G7.0 (1). There is a conservation of important amino acids such as the CD1 phenylalanine, C2 proline, F8 proximal histidine, and E7 distal histidine, between symbiotic and nonsymbiotic hemoglobins. EST clones from tomato (Solanum lycopersicon) with high homology to Arabidopsis GLB1 and GLB2 were identified (5).
In this study, the cDNA for the tomato SOLly GLB1 gene, which has been suggested to code a nonsymbiotic hemoglobin, was cloned in the expression vector and the corresponding protein was expressed. Next to the cDNA sequence that was already determined by Hunt et al. (5), the gene structure was determined for the first time.
The SOLly GLB1 protein shows high sequence homology with other nonsymbiotic plant hemoglobins, (Fig. 1). In this work, we want to determine if and to what extent this sequence homology is reflected in the heme-pocket structure and the ligand-binding properties of the tomato hemoglobin. Resonance Raman (RR) spectroscopy and continuous-wave (CW) and pulsed electron paramagnetic resonance (EPR) spectroscopy is therefore applied to analyze the heme-pocket structure of the nonsymbiotic tomato Hb, SOLly GLB1. In the past, CW-EPR spectroscopy has been used extensively to study the ferric form of heme proteins (9). Furthermore, RR spectroscopy is a widely used tool for the analysis of the heme-pocket structure in both the ferric and ferrous form of heme proteins (10). Recently, high-resolution pulsed EPR techniques have been developed that can reveal detailed information on the environment of the paramagnetic transition metals (11). These techniques have only rarely been applied to the study of heme proteins and the majority of these applications involve the use of one-dimensional (1D) pulsed EPR techniques (12–14), rather than the more advanced two-dimensional (2D) techniques (15). In our recent work, we showed how advanced one- and two-dimensional pulsed EPR techniques can be combined to reveal structural information on low-spin ferric porphyrin complexes (16). Here, we apply this strategy for the first time to the study of the heme-pocket structure of a ferric globin. In addition, the dimerization of SOLly GLB1 is studied using analytical gel chromatography and the O2- and CO-binding kinetics is determined. All the results are then compared to the earlier observations for rice GLB1, the only nonsymbiotic plant Hb for which an x-ray structure is available (17), and for Barley hemoglobin, for which an extended RR analysis is available (18).
FIGURE 1.
Alignment of Solanum lycopersicon nonsymbiotic hemoglobin, SOLly GLB1, with globin sequences of different plant and vertebrate hemoglobins. Phys Mb, Physeter catodon myoglobin (accession No. P02185); HA, human α-chain (accession No. J00153); HB, human β-chain (accession No. M36640); Ho.vu, Hordeum vulgare hemoglobin (accession No. U94968); Or.sa 1, Oryza sativa hemoglobin1 (accession No. U76029); Gl.ma, Glycine max hemoglobin (accession No. U47143); Ca.gl, Casuarina glauca hemoglobin I (accession No. L28826), hemoglobin II (accession No. X53950); Ar.th I, Arabidopsis thaliana hemoglobin 1 (accession No. NM127165); Tr.to, Trema tomentosa hemoglobin (accession No. Y00296); Tr.or, Trema orientalis hemoglobin (accession No. AF027215); So.ly, Solanum lycopersicon hemoglobin (accession No. AY026343). The conserved amino acids are highlighted.
EXPERIMENTAL PROCEDURES
Cloning
The SOLly GLB1 DNA sequence was found in the online database GenBank (EST sequence, accession No. AW035687, J. Alcala, J. Vrebalov, R. White, A. L. Matern, T. Vision, I. E. Holt, F. Liang, J. Upton, M. B. Craven, C. L. Bowman, S. Ahn, C. M. Ronming, et al., Clemson University, Clemson, SC), as “Lycopersicon esculentum hemoglobin 1”. To clone the cDNA, the full-length coding sequence was amplified with specific primers containing the correct adaptors. The forward primer (5′-GCGCATATGAGTAGCTTTAGTGAAGAAC-3′) contains a NdeI restriction site that covers the initiating Met codon of the globin gene. The reverse primer 95′-GCGGGATCCCTACTTCATCTCAGTCTTGATAGC-3′) contains the BamHI restriction site.
Total RNA was isolated using the LiCl method, from young tomato leaves previously frozen and ground in a mortar. Then first-strand cDNA was synthesized by reverse transcriptase polymerase chain reaction (PCR). The full coding sequence of SOLly GLB1 was amplified by PCR (5 min at 95°C (1 min at 94°C, 1 min at 50°C, 1 min at 72°C) × 25, and 10 min at 72°C) in a total volume of 25 μl containing 1 μl of cDNA, 100 ng of each primer, 1× polymerase buffer, 2.5 mM dNTPs, 1.5 mM MgCl2, and 1 μl of Taq polymerase.
The amplified product corresponding to SOLly GLB1 was cleaned and cut with NdeI and BamHI and subsequently ligated into the equivalently cleaved expression vector pET3a. Recombinants obtained in the Escherichia coli strain XL1-Bleu were tested by PCR and restriction digests. The complete sequence and orientation were determined by dideoxy sequencing.
Expression and purification of the protein
The expression plasmid (SOLly GLB1cDNA) was transformed into the E. coli strain BL21(DE3)pLysS. The cells were grown at 25°C in TB medium (1.2% bactotryptone, 2.4% yeast extract, 0.4% glycerol, 72 mM potassium phosphate buffer, pH 7.5) containing 200 μg/ml ampicillin, 30 μg/ml chloramphenicol, and 2.5 mM δ-aminolevulinic acid. The expression was induced, when the culture had an absorption A600 = 1.7, by addition of isopropyl-1-thio-β-d-galactopyranoside to a final concentration of 0.4 mM, and growth was continued overnight. Cells were harvested, washed, and resuspended in lysis buffer (50 mM Tris-HCl, pH 8; 1 mM EDTA, 0.5 mM dithiothreitol). The cells were then exposed to two freeze-thaw cycles and were sonicated until completely lysed (19).
The extract was clarified by low (10 min at 10,000 × g) and high (60 min at 105,000 × g) speed centrifugation at 4°C and fractioned by ammonium sulfate precipitation (40–90%). The 90% ammonium sulfate pellet containing the crude SOLly GLB1 was dissolved in 5 mM Tris-HCl, pH 8.5, dialysed, and loaded onto a CM-Sepharose Fast Flow column equilibrated in the same buffer. After washing of the unbound material, SOLly GLB1 was eluted with 200 mM NaCl. The hemoglobin was then concentrated by Amicon filtration (PM10) and passed through a Sephacryl S200 column (19). The fractions containing SOLly GLB1 were pooled, concentrated, and stored at −20°C.
Measurement of the autooxidation rate
The spectral measurements were done with a SLM DW2000 spectrophotometer (Urbana, IL). To study the autooxidation, a stock solution of ∼100 μM was first reduced with a slight excess of sodium dithionite to obtain the deoxy ferrous form of the protein. An aliquot was then diluted 20-fold by injection into a cuvette containing 100 mM potassium phosphate buffer at pH 7, 37°C, and equilibrated under 1 atmosphere of oxygen; the absorption spectra were then recorded versus time.
Kinetics of ligand binding
All ligand-binding experiments were performed in 50 mM potassium phosphate buffer (at pH 7 or 8.5) at 25°C. The bimolecular rebinding kinetics of the protein with either O2, or CO were measured after flash photolysis with 10 ns YAG laser pulse delivering 160 mJ at 532 nm (Quantel, Les Ulis, France) with at least 4 s between photolysis pulses to allow sample recovery. A mixed atmosphere of O2 and CO was also used, which allows a measurement of the O2 to CO replacement reaction to determine the oxygen dissociation rate. Different detection wavelengths (such as 436 nm for CO rebinding or 419 or 425 nm for the replacement of O2 or the internal ligand, respectively, by CO) and different ligand concentrations were used to observe separately the binding of the external and the protein ligands. The dissociation rates of the protein (E7-His) and the O2 ligands were determined from the flash photolysis kinetics by numerical integration as described previously (19,20). CO dissociation from SOLly GLB1 was detected spectrally in the Soret region using a diode array spectrophotometer HP8453 by replacement with O2 by dilution into a buffered solution equilibrated under 1 atm oxygen (so that the final [O2] was 1.4 mM, over 100 times more than the remaining [CO] after the mixing).
Gel filtration chromatography
Gel filtration chromatography analysis was done according to the previous method (21) with minor modifications. Protein samples were loaded onto a Superose 12 HR 10/30 column (Amersham Biosciences, Uppsala, Sweden) equilibrated at 25°C with 150 mM tris-acetate buffer at pH 7.5 and linked to a Gilson HPLC system (Middleton, WI). Elution profiles were monitored at 415 and 280 nm. The flow rate was constant and equal to 0.04 ml/mn. The dilution factor for each elution profile was estimated by the ratio between the width at half-height (converted into milliliters) and the sample volume. From an average of several experiments the dilution factor was found to be 60. Included (Vi) and excluded (V0) volumes were measured by using a 10% acetone solution and blue dextran, respectively. The elution volume of a protein sample (Ve) corresponds to the elution profile peak (similar to the value obtained by comparison of elution peaks at peak half-width and peak half-height).
From the column characteristics, the weight-averaged partition coefficient, 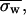 is given by the relationship:
is given by the relationship:
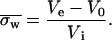 |
For a dimer this coefficient depends upon the fractions of monomers (fM) and dimers (fD) and their respective partition coefficients, σM and σD, following the relationship:
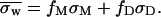 |
Finally dimer and tetramer fractions are related to the protein concentration (P) and the equilibrium constant (KMD) of the monomer-dimer autoassociation by the following relationship:
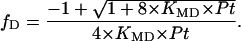 |
The elution volume of a globin tetramer was determined with a solution of diaspirin cross-linked hemoglobin from Baxter (Deerfield, IL) and the elution volume of a monomeric globin with a solution of lyophilized sperm whale Mb from Sigma (St. Louis, MO). Data simulations and standard errors were carried out using the nonlinear least-squares procedure of Scientist (MicroMath, Salt Lake City, UT).
Optical and resonance Raman spectra
Optical measurements were done with a CARY-5 UV-Vis-NIR spectrophotometer (Varian, Palo Alto, CA). All optical spectra were measured in a range from 350 to 700 nm. Resonance Raman measurements were carried out on an 80-cm DILOR XY-800 Raman scattering spectrometer (Dilor, Lille, France) equipped with a triple monochromator allowing for multichannel liquid-nitrogen cooled charge-coupled device detection. The excitation source was a Kr-ion laser (Spectra Physics 2000, Mountain View, CA) (413.1 nm). The protein solution was stirred at 6000 rpm to avoid local heating. Five spectra (with individual recording times of 120 s) were recorded and averaged after the removal of cosmic ray spikes by an in-house-developed program. Frequency shifts in the Raman spectra were calibrated using acetone-CCl4 as a reference. The laser power was maintained at 17 mW.
The ferric state of the protein was formed after exposure of the protein to air for >1 h. The deoxy ferrous form of the sample was obtained by equilibration under nitrogen and adding an excess of sodium dithionite. The concentration of the protein samples used for optical and RR measurements was typically ∼60 μM in Tris buffer, pH 8.5.
Continuous-wave electron paramagnetic resonance
CW-EPR spectra were recorded on a Bruker ESP300E spectrometer (Bruker BioSpin, Karlsruhe, Germany) (microwave frequency 9.43 GHz) equipped with a gas-flow cryogenic system (Oxford Instruments, Oxon, UK), allowing operation from room temperature down to 2.5 K. All the spectra were recorded with a microwave power of 10 mW, a modulation frequency of 100 kHz, and modulation amplitude of 0.5 mT. The calibration of the magnetic field was done using a sample of diphenylpicrylhydrazyl and an NMR Gaussmeter (Bruker ER 035 M). All the EPR spectra were simulated with the EasySpin program (www.esr.ethz.ch).
Pulsed electron paramagnetic resonance
All pulse EPR experiments were carried out on a Bruker ESP 380E spectrometer (Bruker BioSpin) (microwave frequency 9.76 GHz) equipped with a liquid-helium cryostat from Oxford. The experiments were performed at a temperature of 6 K with a repetition rate of 1 kHz. The magnetic field was measured with a Bruker ER035M NMR Gaussmeter. The following pulse EPR techniques were used.
Three-pulse electron spin echo envelope modulation (ESEEM) (11). The pulse sequence used was π/2-τ-π/2-T-π/2-τ-echo with pulse lengths tπ/2 = 16 ns. The time interval T was varied from 96 to 5696 ns in steps of 16 ns, whereas the time interval τ was varied from 96 to 336 ns in steps of 8 ns. A four-step phase cycle was used to eliminate the unwanted echoes. The individual time traces were baseline corrected with a third-order polynomial, apodized with a Hamming window, and zero-filled. After 1D Fourier transformation the absolute value spectrum was calculated. To get a blind-spot free 1D-CP spectrum the τ-traces were summed after 1D Fourier transformation.
Hyperfine sublevel correlation (HYSCORE) (11,22) experiments were carried out with the pulse sequence π/2-τ-π/2-t1-π/2-t2-π-τ-echo. An eight-step phase cycle was performed to eliminate unwanted echo contributions. The following parameters were used: pulse lengths of tπ/2 = 16 or 24 ns and tπ = 16 ns, starting times of 96 ns for t1 and t2 and time increments Δ = 16 ns (data matrix 300 × 300). Different τ-values were taken to reduce the blind spots. The individual time traces were baseline corrected with a third-order polynomial, apodized with a Hamming window and zero-filled. After 2D Fourier transformation the absolute value spectrum was calculated. The HYSCORE spectra were simulated using programs developed at the Swiss Federal Institute of Technology Zurich (23). The same sets of τ-values as in the experiments were taken. For the simulation, nonideal pulses were used with the same pulse lengths as in the experiment. The Euler angles α, β, and γ define an active rotation (right-hand) of the matrices and tensors with respect to the g principal axes system. To optimize the proton signal intensity, the second and third π/2 pulses were experimentally replaced by matched pulses of 48 ns length (24).
Combination-peak experiments (11). The experiments were performed with the pulse sequence: π/2-τ-π/2-T-π-T-π/2-τ-echo, with pulse lengths of tπ/2 = 16 ns and tπ = 16 ns. An eight-step phase cycle was used. The interpulse time T was varied from 96 to 2888 ns in steps of 8 ns; τ was varied from 96 to 328 ns in steps of 8 ns. An eight-step phase cycle was performed to eliminate unwanted echoes. The data were manipulated similarly to the three-pulse ESEEM data. The proton combination frequencies in the 1D-CP spectra were analyzed using the procedure outlined in Astashkin et al. (14).
Parameters determined by EPR experiments
From the CW-EPR spectra the g tensor of the ferric form can be determined. The principal values of this tensor give direct information on the axial ligands of the heme complex (9). The pulsed EPR experiments reveal the hyperfine interactions (A tensor) between the unpaired electron(s) and surrounding nuclei with a nuclear spin (I > 0) and give information on the nuclear-quadrupole-interaction tensors (P tensors) for the surrounding nuclei with I ≥ 1. The principal values Px, Py, and Pz of the traceless P tensor are usually expressed by the quadrupole-coupling constant K = e2qQ/(4h) and the asymmetry parameter η, with Px = −(e2qQ/(4h))(1 − η), Py = −(e2qQ/(4h))(1 + η), and Pz = e2qQ/(2h). For a more detailed description of the fundaments of the EPR experiments we refer to the Supplementary Material and to the basic literature (11).
RESULTS AND DISCUSSION
Amino acid sequence
Fig. 1 shows the alignment of SOLly GLB1 and other known plant and vertebrate globins. Based on the sequence similarity it is clear that this tomato globin belongs to group I of the plant globins. The SOLly GLB1 protein shows 74% similarity with Arabidopsis thaliana GLB1 and 71% with rice (Oryza sativa) GLB1. It contains the proximal histidine F8, the distal histidine E7, the phenylalanine CD1, and the proline C2. These amino acid residues are conserved among all plant hemoglobins, with the exception of the new family of the truncated hemoglobins (25). There are a few remarkable amino acid substitutions in the SOLly GLB1 compared with other (group I) plant globins. At position B5 there is an unprecedented tryptophan, a lysine is present at position E9 where in other plants a hydrophobic residue is present. A lysine at this position is, however, not unique as this exists in vertebrate globins as well.
Analytical gel filtration chromatography
Analytical gel chromatography of the oxidized hexa-coordinated tomato globin showed a transition from a monomer species to a species with higher molecular weight, which was reasonably attributed to a homodimer species (see Supplementary Material). Because of the elevated dilution factor of our chromatography equipment, we were not able to monitor elution profiles for final protein concentrations higher than 10 μM, where higher molecular weight species might have been observed. Any high molecular weight forms, such as a tetrameric state, are apparently not very stable and probably not relevant for the in vivo concentrations. The monomer-dimer equilibrium binding coefficient was 2 × 107/M (with a precision of 25%), being equivalent to a monomer-dimer assembly free energy of 10 kcal/mol heme.
Optical experiments
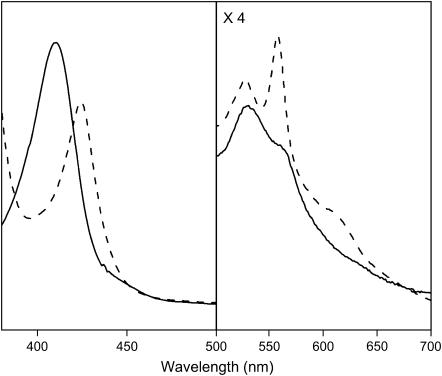
The deoxy ferrous form of tomato nonsymbiotic hemoglobin, SOLly GLB1, reveals a Soret band at 425 nm and the α- and β-bands at 527 and 558 nm, respectively (Fig. 2). Similar values were reported for nonsymbiotic barley Hb (26), nonsymbiotic rice Hb1 (7), neuroglobin (27), and cytoglobin (28). They are characteristic of a hexacoordinated heme with the iron in the low-spin FeII state. The sequence alignment identifies the proximal (F8) and distal (E7) residues of SOLly GLB1 as histidines, suggesting a His-Fe2+-His conformation as the most likely binding scheme. The α-absorption band was not as strong as for Ngb (27), suggesting only a partial hexacoordination, as observed for rice Hb1 (29). A small increase in the contribution of the pentacoordinated form was found when going from pH 7 to pH 8.5 (see Supplementary Material).
FIGURE 2.
Absorption spectra of the ferric (solid line) and the deoxy ferrous (dashed line) form of nonsymbiotic tomato hemoglobin, SOLly GLB1. The proteins (∼60 μM) were dissolved in 5 mM Tris-HCl buffer at pH 8.5.
The ferric form of the protein displays absorptions at 410, 531, and 565 nm (Fig. 2), typical for a hexacoordinated low-spin FeIII heme, suggesting predominant bis-histidine coordination. Also, the absorption spectrum shows a tail extending in the 630-nm region, which suggests a smaller contribution of an aquomet form (typical absorption maxima for aquomet globins are 408, 503, and 633 nm (30)). The presence of a minor contribution of an aquomet form is more clearly visible in the CW-EPR and RR spectra (see following sections). No significant pH effect from 7.0 to 9.5 was observed, in agreement with the presence of a small fraction of the aquomet form.
Resonance Raman experiments
It is well established that the high-frequency region (900–1700 cm−1) of the RR spectra of heme proteins is comprised of porphyrin in-plane vibrational modes, which are markers of the oxidation state, coordination state, and the spin state of the central iron atom (10). The low-frequency region of the RR spectra (100–900 cm−1) shows several in-plane and out-of-plane vibrational modes of the heme, including heme-propionate modes and ligand vibrational modes.
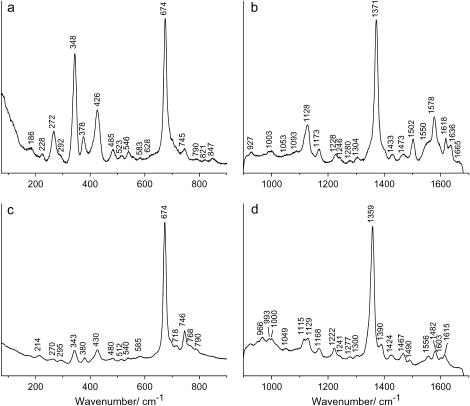
The RR spectrum of the ferric form of SOLly GLB1 at pH 8.5 displays the electron-density marker ν4 at 1371 cm−1, which is characteristic of the Fe(III) state (Fig. 3 b). The values of ν3 (1502 cm−1) and ν10 (1636 cm−1) are typical for a hexacoordinated low-spin FeIII heme form. The appearance of a weak line at 1473 cm−1 suggests that a minor population of a hexacoordinated high-spin FeIII complex is present (18). This indicates the presence of an aquomet form, as will be confirmed by the CW-EPR results. The line at 1473 cm−1 disappears almost completely at pH 7 (see Supplementary Materials), indicating a lower amount of the aquomet form at lower pH. The above observation thus suggests that the bis-hisitidine coordination becomes less favorable at higher pH, although it remains the dominant coordination form for ferric SOLly GLB1 (see EPR data later in the article). The porphyrin core-size markers, ν3, ν38, ν2, and ν10, of the ferric low-spin form are similar to those observed for other hexacoordinated globins (28,31,18) and predict a porphyrin center to pyrrole nitrogen distance of 0.1995 nm (see Supplementary Material; (32)).
FIGURE 3.
Resonance Raman spectra of tomato nonsymbiotic hemoglobin, SOLly GLB1. Low-frequency (a,c) and high-frequency (b,d) region of ferric SOLly GLB1 (a,b) and deoxy ferrous SOLly GLB1 (c,d). The laser-excitation wavelength was 413.1 nm at 17 mW power. The proteins were dissolved in 5 mM Tris-HCl buffer at pH 8.5.
The high-frequency region of the RR spectrum of deoxy ferrous SOLly GLB1 (Fig. 3 d) is dominated by a hexacoordinated low-spin FeII heme form (ν4 = 1359 cm−1, ν3 = 1490 cm−1). In accordance with the absorption data (Fig. 2), a peak at ν3 = 1467 cm−1 indicates the presence of a pentacoordinated high-spin ferrous heme form. The intrinsic intensity of ν3 is very high for pentacoordinated species compared to hexacoordinated species, making a direct assessment of the relative populations by inspection of the RR spectra difficult (18). Comparison of the RR spectra of deoxy ferrous SOLly GLB1 at pH 7 and pH 8.5 (Supplementary Material) shows a slight increase in the 1467 cm−1 signal (thus pentacoordination) with pH, confirming the absorption data. We thus observe also for the ferrous form of the protein a decrease in the bis-histidine coordination upon increase of pH.
For both the ferric and ferrous form of SOLly GLB1, the out-of-plane modes γ6, γ7, γ12, and γ21 are hardly visible indicating, as is typical for a bis-histidine coordination, a relaxed state of the heme group with the iron almost totally in the porphyrin plane (Fig. 3, a and c).
In accordance with the findings for barley Hb (18), the peak at 214 cm−1 is assigned to the Fe-His(proximal) stretching mode, νFe-His (Fig. 3 c). This stretching mode appears for pentacoordinated ferrous heme proteins, confirming the earlier findings that a considerable percentage of the ferrous SOLly GLB1 is in a pentacoordinated form. In peroxidases, the νFe-His mode is detected at frequencies higher than 240 cm−1 (33,34). This high frequency is ascribed to the imidazolate character of the proximal histidine (34). The low νFe-His mode of SOLly GLB1 (similar to the one of mammalian Hbs and Mbs (35) and Barley Hb (18)) suggests the presence of an uncharged proximal imidazole.
Finally, it should be remarked that the RR spectra of ferric/ferrous SOLly GLB1 and Barley Hb (18) are highly similar.
CW-EPR experiments on the ferric form of SOLly GLB1
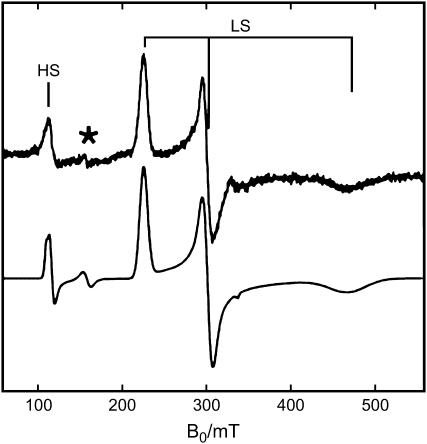
Fig. 4 shows the EPR spectrum of the nonsymbiotic tomato Hb, SOLly GLB1. It is dominated by a rhombic signal, characteristic of a low-spin Fe(III) heme. A smaller contribution of a high-spin ferric form is also discerned in the spectrum. The observation of the latter signal corroborates the optical absorption and RR data, indicating the presence of a minor population of an aquomet form.
FIGURE 4.
CW-EPR spectrum of ferric tomato nonsymbiotic hemoglobin, SOLly GLB1. Experimental (top) and simulated (bottom) spectrum. HS, high-spin, LS, low-spin. *Indicates the signals from nonheme iron. The protein was dissolved in 5 mM Tris-HCl buffer at pH 8.5. The measurement was performed at 10 K.
The g-values of the low-spin ferric form of SOLly GLB1 are given in Table 1 in comparison with those of other bis-histidine coordinated heme proteins. The g-values can be related to the ligand-field parameters V/Δ and Δ/λ (V = rhombic splitting parameter; Δ = tetragonal splitting parameter; λ = spin-orbit coupling) using the formulae of Taylor (36) (Table 1). According to Blumberg-Peisach's truth tables (9), the EPR parameters of SOLly GLB1 agree with a bis-imidazole ligation, confirming our earlier assumptions that the ferric protein is in a F8His-Fe3+-E7His ligation form. The ligand-field parameters can reveal also valuable information on the histidine-plane orientations in the heme pocket. When V/λ ≈ 2, the histidine planes are coplanar (37,38). As the histidine planes get twisted and tilted, V/λ will decrease and the gz-value will become larger than 3 (37,38). Furthermore, if the histidines are quasiparallel, the V/Δ value gives an indication on whether the histidine planes are eclipsing or bisecting a Nheme-Fe-Nheme axis (37). V/Δ = 2/3 is the expected value for a full eclipsing of two coplanar histidine planes with a Nheme-Fe-Nheme axis (37). This is in accordance with the data for MetMbIm, where the x-ray structure shows only a deviation of 16° from coplanarity of the imidazoles and a full eclipsing case for one of the imidazoles (39). For ferric neuroglobin x-ray studies have proven that the histidine planes are perpendicular to each other and in a staggered position with respect to the pyrroles (pointing versus the meso-carbons of the heme group) (40,41), fully in accordance with the low V/λ value and even the low V/Δ value (despite the fact that the His ligands are no longer parallel) (Table 1). The ligand-field parameters for ferric SOLly GLB1 lie in between the two extreme cases. From the V/λ value we derive that the angle between the two histidine planes is considerably lower than 90°. Furthermore, the V/Δ value of ferric SOLly GLB1 suggests that the histidines are not eclipsing with a Nheme-Fe-Nheme axis. Based on Quinn et al. (37) this value agrees with a turning angle of the histidine planes versus the Nheme-Fe-Nheme axis of ∼28° if coplanarity of the histidines is assumed. The latter assumption is not fully correct (see V/λ value), implying that more advanced EPR techniques are needed to determine the histidine orientation in SOLly GLB1.
TABLE 1.
The g-values of the dominant low-spin component in ferric SOLly GLB1in comparison with other hexacoordinated heme proteins
| gx (±0.02) | gy (±0.01) | gz (±0.01) | V/λ | V/Δ | Reference | |
|---|---|---|---|---|---|---|
| Ferric SOLly GLB1 | 1.44 | 2.23 | 2.98 | 1.72 | 0.54 | This work |
| Ferric Barley Hb | 1.48 | 2.22 | 3.02 | 1.72 | 0.49 | 18 |
| Cytochrome b5 (house fly) | 1.35 | 2.22 | 3.07 | 1.54 | 0.51 | 55 |
| Cytochrome b5 (bovine liver) | 1.43 | 2.23 | 3.03 | 1.66 | 0.52 | 56 |
| Cytochrome b559 | 1.53 | 2.25 | 2.98 | 1.84 | 0.53 | 15 |
| Flavocytochrome b2 | 1.47 | 2.22 | 2.99 | 1.74 | 0.51 | 42 |
| Ferric MbIm | 1.53 | 2.26 | 2.91 | 1.93 | 0.58 | 57 |
| Fe(III)PPIX(im)2 | 1.52 | 2.25 | 2.98 | 1.83 | 0.53 | 15 |
| Ferric mNgb | 1.29 | 2.15 | 3.12 | 1.41 | 0.44 | 58 |
| Ferric hCygb | 1.20 | 2.08 | 3.20 | 1.26 | 0.39 | 58 |
Note also that the EPR parameters of ferric SOLly GLB1 are close to those of flavocytochrome b2 (flavocyt b2) (Table 1; (42)), for which x-ray studies predict a deviation from coplanarity of 13–18° and the histidine planes are oriented in positions in between the eclipsed and staggered extrema (43,44). Analogously, an x-ray study of oxidized bovine cytochrome b5 (cyt b5) predicts an angle of ∼21° between the histidine planes, with one histidine exactly bisecting and the other histidine tilted 26° away from a Nheme-Fe-Nheme axis (45). Finally, the close resemblance of Barley Hb and SOLly GLB1 is again reflected in the EPR parameters (Table 1), in accordance with the analogies in the RR spectra noted earlier.
Pulsed EPR experiments
To get more detailed information on the heme-pocket electronic and geometric structure, pulsed EPR experiments were undertaken. These experiments reveal information on the interaction between the unpaired electron on the iron and the surrounding nitrogens and protons. We recently outlined a general strategy for the study of low-spin ferric porphyrin complexes with pulsed EPR (16). Here, we apply this strategy to study ferric SOLly GLB1. In the following paragraphs, we report on the major steps and results of the procedure. We refer the reader to the Supplementary Materials for a more detailed and technical description of the analysis.
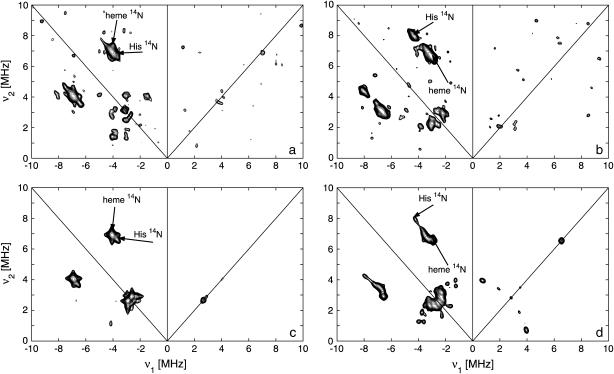
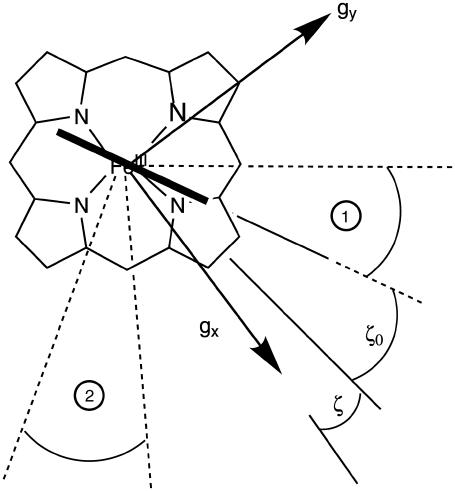
The first step in the analysis involves the recording of the two-dimensional HYSCORE spectra at different observer positions. Fig. 5, a and b, shows two such nitrogen HYSCORE spectra. Further spectra are shown in the Supplementary Materials with complementary three-pulse ESEEM spectra. Based on the previous isotope-labeling work of a heme-model system by García-Rubio et al. (15), the double-quantum crosspeaks in the HYSCORE spectra could be attributed to contributions of the heme and nearest histidine nitrogens (Fig. 5, a and b; Supplementary Material). Using a recently developed simulation program (23), the HYSCORE spectra could be simulated using the hyperfine and nuclear-quadrupole parameters given in Tables 2 and 3 (Fig. 5, c and d; Supplementary Material). This procedure reveals the orientation of the A and P tensors in the g axes frame. However, to relate these parameters to structural information, we need to determine the position of the g-tensor axes in the molecular frame. From the g-values, it can be predicted that the gz axis is approximately along the heme normal (46). The orientation of the gx and gy axes in the porphyrin plane is less trivial. The HYSCORE spectra of the porphyrin nitrogens are sensitive to the in-plane rotation of the g axes versus the hyperfine and nuclear-quadrupole tensors of the porphyrin nitrogens. Earlier single-crystal measurements on different metallo-porphyrin systems showed that the largest nuclear-quadrupole principal value lies in the porphyrin plane perpendicular to the Fe-Np bond (47,48). Using this information the g axes can be oriented in the molecular frame. Best agreements were found between the experimental and simulated HYSCORE spectra when the g-tensor axes are ∼10° (±10°) tilted away from the Nheme-Fe-Nheme axes (Fig. 6, angle ζ; Table 2).
FIGURE 5.
Nitrogen HYSCORE spectra of ferric SOLly GLB1. (a,b) Experimental spectra at observer position g = gz (a) and g = gy (b). The experimental settings are given in the text. The double-quantum crosspeaks are indicated with arrows and ascribed to the different nitrogen types. (c,d) Simulations of the spectra (a,b) using the parameters in Tables 2 and 3.
TABLE 2.
Hyperfine and nuclear-quadrupole parameters for the porphyrin nitrogens of ferric heme complexes and heme proteins
| Ax (MHz) ±0.2 | Ay (MHz) ±0.2 | Az (MHz) ±0.2 | α,β,γ (°) ±10° | e2qQ (MHz) ±0.1 | η ±0.05 | α,β,γ (°) ±10° | Reference | |
|---|---|---|---|---|---|---|---|---|
| SOLly GLB1 | −4.5 | −3.6 | −5.4 | 0,10,10 | 1.75 | 0.05 | 0,90,100 | This work |
| MbOH | −4.9 | −5.1 | −5.3 | 0,0,0 | 2.2 | 0.1 | 0,45,0 | 12 |
| AquometMb | 9.86 | 6.89 | 7.11 | 0,0,0 | 2.08 | 0.48 | 90,90,0 | 47 |
| FeTPP(4-MeIm)2* | −4.8 | −4.8 | −5.8 | 0,0,0 | 1.85 | 0.1 | 90,90,0 | 16 |
| Cytochrome b559 | −4.9† | −4.7† | −5.8 | 0,20–38,0 | 1.6–2.2 | 0–0.27 | 0,90,0 | 15 |
TPP, tetraphenylporphyrin; 4-MeIm, 4-methyl imidazole.
These are the Axx and Ayy values in the g axis frame with 0.4 < Axy < 1.3 MHz. The real Ax and Ay values will thus deviate from these values.
TABLE 3.
Hyperfine and nuclear-quadrupole parameters for the nearest imidazole nitrogens of ferric heme complexes and heme proteins
| Ax (MHz) ±0.3 | Ay (MHz) ±0.3 | Az (MHz) ±0.3 | α,β,γ (°) ±15° | e2qQ (MHz) ±0.2 | η ± 0.1 | α,β,γ (°) ±10° | Reference | |
|---|---|---|---|---|---|---|---|---|
| SOLly GLB1 | −5.1 | −6.4 | −4.9 | 0,5,30 | −1.6 | 0.9 | 0,5,30 | This work |
| −5.4 | −6.7 | −4.9 | 0,5,55 | −1.6 | 0.9 | 0,5,55 | ||
| MbOH | −5.5 | −5.5 | −4.2 | 0,7,0 | −2.3 | 0.1 | 0,13,0 | 12 |
| MbCN | −2.6 | −2.6 | −1.5 | 0,15,0 | −2.5 | 0.3 | 0,5,0 | 13 |
| MbN3 | −4.6 | −4.6 | −3.0 | 0,10,0 | −3.2 | 0.1 | 0,23,0 | 13 |
| Mb(β-mercaptoethanol) | −2.6 | −2.6 | −1.4 | 0,7,0 | −2.5 | 0.3 | 0,18,0 | 13 |
| AquometMb | 8.33 | 8.08 | 11.55 | 0,0,0 | −2.24 | 0.53 | 0,0,0 | 47 |
| FeTPP(4-MeIm)2* | −5.7 | −6.2 | −5.3 | 0,0,0 | −1.7 | 0.2 | 0,0,0 | 16 |
| Cyt b559 | −5.6† | −6.2† | −5.1 | 0,65,0 | −1.6 | 1 | – | 15 |
TPP, tetraphenylporphyrin; 4-MeIm, 4-methyl imidazole.
These are the Axx- and Ayy-values in the g axis frame with Axy < 0.7 MHz. The real Ax- and Ay-values will thus deviate from these values.
FIGURE 6.
Schematic drawing of the orientation of the g-tensor axes and the histidine planes in the molecular frame as derived from the pulsed EPR experiments. The angles ζ and ζ0 and the explanations of the regions 1 and 2 are given in the text.
In a second step, the results from the CP experiments and proton HYSCORE experiments in combination with the counterrotation principle are used to determine the orientation of the histidine planes in the g axes frame (14,16,46) (see details in Supplementary Material). The analysis shows that one of the histidine planes is turned over an angle ζ0 = −20° (±10°) in the counterdirection of the tilt of the gx axis (Fig. 6). The orientation of the second histidine plane is less well determined and may lie in the regions marked by 1 and 2 in Fig. 6. However, our earlier CW-EPR analysis predicted a small deviation from coplanarity between the two histidine planes (10–25° based on the spectral analogies with bovine cyt b5 and flavocyt b2 (43–45)). This suggests that the second histidine plane lies within region 1 rather than within region 2 (Fig. 6).
The distance between the iron atom and the nearest protons on the axially ligated histidines is found to be 0.325 (±0.005) nm whereby the Fe-H axis is found to be tilted over an angle 32° (±5°) with respect to the heme normal. This leads to an estimation of the Fe-NHis distance of 0.224 (±0.020) nm, which is normal for hexacoordinated globins (49,17).
Table 2 shows that the hyperfine and nuclear-quadrupole parameters of the heme nitrogens of ferric SOLly GLB1 are typical values for low-spin heme proteins. The hyperfine values are negative due to an excess of β-spin in the nitrogen 2s-orbital (16,50). This is in contrast to the hyperfine couplings of the heme nitrogens of high-spin ferric globins, characterized by a positive sign and larger average value (47). Analogous trends are observed for the imidazole nitrogens (Table 3). From earlier studies, it is known that the e2Qq value of the coordinated nitrogen of the proximal histidine in Mb is very sensitive to axial bond changes, such as differences in the Fe-N distance and the iron-nitrogen π bonds (13). Inspection of Table 3 suggests that the |e2Qq| value of the heme nitrogen(s) is considerably lower in the case of a bis-imidazole coordination than in the case where one of the two axial ligands is not an imidazole. Pulsed EPR measurements on neuroglobin and cytoglobin are in progress to determine whether this is a feature general for all bis-histidine ligated heme proteins or whether it depends on the relative orientation of the histidines.
Kinetics study
It is clear that the equilibrium and kinetic proprieties of the external ligand binding will be closely related to the heme-pocket structure. Kinetic studies of O2 and CO binding to SOLly GLB1 are presented in Table 4 and compared with other known hexacoordinated proteins. Ferrous SOLly GLB1 shows a very high affinity for oxygen. Its O2-association constant (kon) is similar to other O2 transport proteins, such as human Hb, however, the high affinity of SOLly GLB1 for O2 results from a low dissociation constant (koff) indicating a favorable orientation of the distal histidine after O2 binding for the stabilization of the dipolar Feδ+-O2δ− bond via a hydrogen bond. Similar observations were made for Arabidopsis GLB1 (3), Barley Hb1 (26), and rice Hb1 (7).
TABLE 4.
Ligand binding parameters for ferrous SOLly GLB1 and other globins
| Protein |
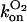 (μM−1s−1) (μM−1s−1) |
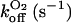 |
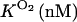 |
KHis |
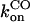 (μM−1s−1) (μM−1s−1) |
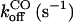 |
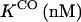 |
Reference |
|---|---|---|---|---|---|---|---|---|
| SOLly GLB1 | 30 | 0.5 | 15 | 0.7 | 1 | 0.02 | 20 | This study |
| Oryza sativa Hb1 (rice) | 68 | 0.038 | 0.56 | 0.3 | 7.2 | 0.001 | 0.138 | 29 |
| Arabidopsis Hb | 150 | 2 | 14 | 50 | – | – | – | 20 |
| Soybean Lb | 120 | 5.6 | 47 | – | 13 | 0.0078 | 0.62 | 59 |
| Barley Hb | 9.5 | 0.0272 | 2.86 | – | 0.57 | 0.0011 | 1.93 | 26 |
| Parasponia Hb | 165 | 15 | 90 | – | 14 | 0.019 | 1.4 | 59 |
| Sperm whale Mb | 14 | 12 | 857 | – | 0.51 | 0.019 | 37 | 60 |
| Human Hb | 40 | 50 | 1250 | – | 6 | – | – | 30 |
| Ascaris Hb | 1.5 | 0.0041 | 2.7 | – | 17 | 0.018 | 1.1 | 61 |
| mNgb | 300 | 0.4 | 1.3 | ∼1650 | 72 | 0.013 | 0.18 | 27 |
| HCygb | 27 | 0.9 | 33 | 100 | 5.6 | 0.003 | 21.7 | 49,62 |
On/off-rates were measured at 25°C; Kd= koff/kon (nM) for O2 or CO is the dissociation coefficient. The observed affinity for binding to exogenous ligands for hexacoordinated globins is then Kobs = Ka/(1 + KHis) with Ka = 1 / Kd and KHis the association coefficients.
Furthermore, the association and dissociation rate constants of the distal histidine have been determined: 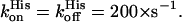 The values are different from those reported for Cygb (
The values are different from those reported for Cygb (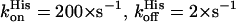 ) (49,51) or mNgb (
) (49,51) or mNgb (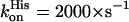 and
and 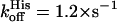 ) (27), for which the observed O2 and CO affinities are greatly reduced by the competition with the distal histidine (Kobs = Kpenta/(1 + KHis)). Indeed, the histidine on-and off-rates are similar resulting in a weak affinity of the distal histidine. There is a large pH dependence in these rates (
) (27), for which the observed O2 and CO affinities are greatly reduced by the competition with the distal histidine (Kobs = Kpenta/(1 + KHis)). Indeed, the histidine on-and off-rates are similar resulting in a weak affinity of the distal histidine. There is a large pH dependence in these rates (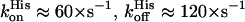 at pH 8.2); however, the ratio is little changed, leading to a small decrease of the O2 affinity (0.02 vs. 0.015 mmHg). Note that large pH effects on the O2 affinity are rarely observed for nonallosteric hemoglobins. These differences between Ngb and SOLly GLB1 in the histidine binding rates clearly reflect the differences in the electronic and geometric structure of the heme as was already clear from comparison of the spectroscopic data.
at pH 8.2); however, the ratio is little changed, leading to a small decrease of the O2 affinity (0.02 vs. 0.015 mmHg). Note that large pH effects on the O2 affinity are rarely observed for nonallosteric hemoglobins. These differences between Ngb and SOLly GLB1 in the histidine binding rates clearly reflect the differences in the electronic and geometric structure of the heme as was already clear from comparison of the spectroscopic data.
A different heme-pocket environment is also suggested by the autooxidation rate of SOLly GLB1 (koxid = 0.7/h). This value is much lower than the reported values for mNgb (19/h), but denotes a faster autooxidation relative to Hbs and Mbs involved in O2 delivery.
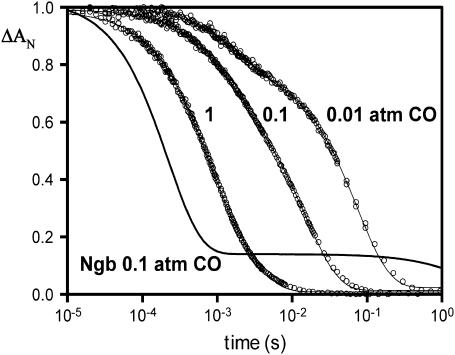
The CO rebinding kinetics after photodissociation are shown in Fig. 7. The initial kinetics shows the usual dependence on the CO concentration, as expected for a bimolecular reaction. The slower phase is less distinct for SOLly GLB1, as compared to Ngb, indicating a much higher rate of dissociation for the internal ligand. As observed for the absorption spectra, the kinetics indicate only a weak pH dependence in the fraction hexacoordinated form for the ferrous deoxy protein.
FIGURE 7.
Rebinding kinetics after photodissociation of CO (detected at 436 nm) at pH 7, 25°C. Data are shown for SOLly GLB1 (tomato) Hb equilibrated under 1, 0.1, and 0.01 atm CO, and compared to neuroglobin under 0.1 atm CO. In general the hexacoordinated globins display two kinetic phases, because CO and histidine will compete for binding sites; histidine binding becomes more competitive at the lower [CO]; the fraction that bind histidine may display a very slow replacement by CO depending on the histidine dissociation rate. For Ngb the fraction that binds histidine requires >1 s for full recovery of the CO form indicating a high histidine affinity, whereas SOLly GLB1 shows a much faster return to the preflash (CO bound) condition. The slow phase is less distinct for SOLly GLB1, because the histidine on- and off-rates are nearly equal at 200/s.
There was no evidence of cooperativity of ligand binding in the CO binding kinetics of the tomato globin for which the short-lived pentacoordinated state does not exceed a few milliseconds even at low [CO] due the histidine binding rate. Indeed, we did not observe a change of the kinetic features at different levels of CO photodissociation. A cooperativity has been reported for dimers of Scapharca homodimer Hb (52), which dimerize in the same concentration range. Note that the formation of homodimers occurs in Barley Hb (26), Parasponia Hb (53), and at a much higher concentration for rice Hb1 (29). Further studies on the oxygenated globin aggregation state could also confirm if a change of stability of the dimer interface occurs upon ligation of an external ligand as opposed to the distal histidine in the oxidized form.
Comparison with other nonsymbiotic plant hemoglobins
Because of the 71% sequence similarity between SOLly GLB1 and rice Hb1, one would expect a strong analogy between the two hemoglobins on the level of the heme-pocket structure and their O2 and CO binding capacities. Inspection of Table 4 shows us that there is, however, a large difference in the O2 and CO affinities of both proteins, which strongly indicates differences in the heme-pocket area.
An x-ray study on rice Hb1 revealed that this protein is a homodimer with an unusual bis-histidyl heme coordination (17). Our studies confirm both characteristics for SOLly GLB1. Furthermore, the x-ray study of ferric rice Hb1 shows an angle of ∼28° between the proximal histidine and the nearest Nheme-Fe-Nheme axis, and places the distal histidine in a staggered position with respect to the pyrrole nitrogens with an angle of ∼65° between the two histidine planes ((17); Brookhaven Protein Data Bank). The pulsed EPR data of ferric SOLly GLB1 reveal that one of the axial histidine planes is rotated 20° (±10°) away from a Nheme-Fe-Nheme axis. This agrees with the orientation of the proximal histidine in rice Hb1. Two orientations of the second histidine are compatible with the pulsed EPR data, namely an orientation in region 1 or 2 as indicated schematically in Fig. 6. Based on the g-values and the many spectral analogies with cyt b5 and flavocyt b2, for which x-ray structures are known, an orientation of the distal histidine in region 1 is found to be more likely. This assignment deviates from the structural arrangement found in rice Hb1. This can explain the significant differences in the exogenous ligand affinities. Indeed, the extreme slow oxygen and CO dissociation, observed both for rice Hb1 and Barley Hb (Table 4), implies stabilization of the bound ligand, commonly by hydrogen-bond formation from a distal residue to the bound ligand as in whale Mb (54). For CO-ligated ferrous Barley Hb, there is strong hydrogen bonding between the CO molecule and the distal histidine (18). The increase in the O2 and CO dissociation rates by an order of magnitude upon going from rice and Barley Hb to SOLly GLB1 implies a less strong stabilization of the exogenous ligand by the distal histidine, which indicates a different orientation of the histidine in the heme pocket. Note that the SOLly GLB1 has a Lys on position E9 instead of the hydrophobic amino acids found in the other nsHbs. Although this amino acid is not in the heme pocket, the change in polarity may well influence the overall position of the E-helix and thus cause the change in the distal histidine orientation. X-ray studies are in progress to clarify these analyses.
The RR spectra of both the ferrous deoxy and ferric forms of SOLly GLB1 and Barley Hb1 are very similar, despite a clear difference in the ligand affinities (Table 4). This is not surprising, because the RR spectra are not sensitive on the orientation of the distal histidine, as long as this conformational difference does not cause major out-of-plane distortions in the heme.
The high O2 affinity for all nonsymbiotic Hbs in comparison to mammalian Hbs clearly excludes the earlier hypothesis that nsHbs can function as O2 transporters that sequester O2 under hypoxic conditions and that facilitate O2 diffusion to cells needing aerobic mitochondrial respiration (7). The hypothesis that nsHbs may act as O2 sensors that undergo significant conformational change in response to ligand binding and release whereby other proteins or enzymes are activated (6,7) can agree with the bis-histidine coordination observed in all investigated nsHbs. Indeed, this coordination implies that large tertiary conformational changes will occur in response to ligand binding.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
We gratefully acknowledge M. L. Van Hauwaert (University of Antwerp) for technical assistance.
This work was supported by the Institut National de la Santé et de la Recherche Médicale (France), the University of Paris XI. S.D. is a postdoctoral fellow of the Fund for Scientific Research-Flanders (FWO). I.I. and S.V.D. thank the FWO for financial support (grant G. 0468.03).
Abbreviations used: Hbs, hemoglobins; nsHb, nonsymbiotic hemoglobin; Mb, myoglobin; SOLly GLB1, nonsymbiotic tomato hemoglobin of group I; RR, resonance Raman; CW, continuous-wave; EPR, electron paramagnetic resonance; ESEEM, electron spin echo envelope modulation; HYSCORE, hyperfine sublevel correlation; CP, combination peak.
References
- 1.Hardison, R. C. 1996. A brief history of hemoglobins: plant, animal, protist, and bacteria. Proc. Natl. Acad. Sci. USA. 93:5675–5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Appleby, C. A. 1992. The origin and functions of hemoglobin in plants. Sci. Prog. 76:365–398. [Google Scholar]
- 3.Trevaskis, B., R. A. Watts, C. R. Andersson, D. J. Llewellyn, M. S. Hargrove, J. S. Olson, E. S. Dennis, and W. J. Peacock. 1997. Two hemoglobin genes in Arabidopsis thaliana: the evolutionary origins of leghemoglobins. Proc. Natl. Acad. Sci. USA. 94:12230–12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arredondo-Peter, R., M. S. Hargrove, J. F. Moran, G. Sarath, and R. V. Klucas. 1997. Plant hemoglobins. Plant Physiol. 118:1121–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunt, P. W., R. A. Watts, B. Trevaskis, D. J. Llewelyn, J. Burnell, E. S. Dennis, and W. J. Peacock. 2001. Expression and evolution of functionally distinct haemoglobin genes in plants. Plant Mol. Biol. 47:677–692. [DOI] [PubMed] [Google Scholar]
- 6.Sowa, A. W., S. M. G. Duff, M. G. Guy, and R. D. Hill. 1998. Altering hemoglobin levels changes energy status in maize cells under hypoxia. Proc. Natl. Acad. Sci. USA. 95:10317–10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arredondo-Peter, R., M. S. Hargrove, G. Sarath, J. F. Moran, J. Lohrman, J. S. Olson, and R. V. Klucas. 1997. Rice hemoglobins (gene cloning, analysis, and O2-binding kinetics of a recombinant protein synthesized in Escherichia coli). Plant Physiol. 115:1259–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang, Y.-H., L. V. Kochian, J. J. Doyle, and D. F. Garvin. 2003. Two tomato non-symbiotic haemoglobin genes are differentially expressed in response to diverse changes in mineral nutrient status. Plant Cell Environ. 26:673–680. [Google Scholar]
- 9.Peisach, J. 1998. Foundations of Modern EPR, 1st Ed. G. R. Eaton, S. S. Eaton, and K. Salikhov, editors. World Scientific Publishing, Singapore.
- 10.Kincaid, J. R. 2000. Theoretical and physical characterization. In The Porphyrin Handbook, Vol. 7. K. M. Kadish, K. M. Smith, and R. Guilard, editors. Academic Press, New York, NY. 225–289.
- 11.Schweiger, A., and G. Jeschke. 2001. Principles of Pulse Electron Paramagnetic Resonance. Oxford University Press, Oxford, UK.
- 12.Magliozzo, R. S., and J. Peisach. 1992. Electron spin echo envelope modulation spectroscopic study of iron-nitrogen interactions in myoglobin hydroxide and iron(III) tetraphenylporphyrin models. Biochemistry. 31:189–199. [DOI] [PubMed] [Google Scholar]
- 13.Magliozzo, R. S., and J. Peisach. 1993. Evaluation of nitrogen nuclear hyperfine and quadrupole coupling parameters for the proximal imidazole in myoglobin-azide, -cyanide, and -mercaptoethanol complexes by electron spin echo envelope modulation spectroscopy. Biochemistry. 32:8446–8456. [DOI] [PubMed] [Google Scholar]
- 14.Astashkin, A. V., A. M. Raitsimring, and F. A. Walker. 1999. Two- and four-pulse ESEEM studies of the heme binding center of a low-spin ferriheme protein: the importance of a multi-frequency approach. Chem. Phys. Lett. 306:9–17. [Google Scholar]
- 15.García-Rubio, I., J. I. Martínez, R. Picorel, I. Yruela, and P. J. Alonso. 2003. HYSCORE spectroscopy in the cytochrome b559 of the photosystem II reaction center. J. Am. Chem. Soc. 125:15846–15854. [DOI] [PubMed] [Google Scholar]
- 16.Vinck, E., and S. Van Doorslaer. 2004. Analysing low-spin ferric complexes using pulse EPR techniques: a structure determination of bis (4-methylimidazole)(tetraphenylporphyrinato)iron(III). Phys. Chem. Chem. Phys. 6:5324–5330. [Google Scholar]
- 17.Hargrove, M. S., E. A. Brucker, S. Boguslaw, G. Sarath, R. Arredondo-Peter, R. V. Kucas, J. S. Olson, and G. N. Philips, Jr. 2000. Crystal structure of a non-symbiotic plant hemoglobin. Structure. 8:1005–1014. [DOI] [PubMed] [Google Scholar]
- 18.Das, T. K., C. H. Lee, S. Duff, R. D. Hill, J. Peisach, D. L. Rousseau, B. A. Wittenberg, and J. B. Wittenberg. 1999. The heme environment in barley hemoglobin. J. Biol. Chem. 274:4207–4212. [DOI] [PubMed] [Google Scholar]
- 19.Dewilde, S., M. Blaxter, M.-L. Van Hauwaert, K. Van Houte, A. Pesce, N. Griffon, L. Kiger, M. C. Marden, S. Vermeire, J. Vanfleteren, E. Esmans, and L. Moens. 1998. Structural, functional, and genetic characterization of Gastrophilus hemoglobin. J. Biol. Chem. 273:32467–32474. [DOI] [PubMed] [Google Scholar]
- 20.Uzan, J., S. Dewilde, T. Burmester, T. Hankeln, L. Moens, D. Hamdane, M. C. Marden, and L. Kiger. 2004. Neuroglobin and other hexacoordinated hemoglobins show a weak temperature dependence of oxygen binding. Biophys. J. 87:1196–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manning, L. R., W. T. Jenkins, J. R. Hess, K. Vandegriff, R. M. Winslow, and J. M. Manning. 1996. Subunit dissociations in natural and recombinant hemoglobins. Protein Sci. 5:775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Höfer, P., A. Grupp, H. Nebenfuehr, and M. Mehring. 1986. Hyperfine sublevel correlation (hyscore) spectroscopy: a 2D ESR investigation of the squaric acid radical. Chem. Phys. Lett. 132:279–284. [Google Scholar]
- 23.Madi, Z. L., S. Van Doorslaer, and A. Schweiger. 2002. Numerical simulation of one- and two-dimensional ESEEM experiments. J. Magn. Reson. 154:181–191. [DOI] [PubMed] [Google Scholar]
- 24.Jeschke, G., R. Rakhmatullin, and A. Schweiger. 1998. Sensitivity enhancement by matched microwave pulses in one- and two-dimensional electron spin echo envelope modulation spectroscopy. J. Magn. Reson. 131:261–271. [DOI] [PubMed] [Google Scholar]
- 25.Watts, R. A., P. W. Hunt, A. N. Hvitved, M. S. Hargrove, W. J. Peacock, and E. S. Dennis. 2001. A hemoglobin from plants homologous to truncated hemoglobins of microorganisms. Proc. Natl. Acad. Sci. USA. 98:10119–10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duff, S. M. G., J. B. Wittenberg, and R. D. Hill. 1997. Expression, purification, and properties of recombinant barley (Hordeum sp.) hemoglobin. Optical spectra and reactions with gaseous ligands. J. Biol. Chem. 272:16746–16752. [DOI] [PubMed] [Google Scholar]
- 27.Dewilde, S., L. Kiger, T. Burmester, T. Hankeln, V. Baudin-Creuza, T. Aerts, M. C. Marden, R. Caubergs, and L. Moens. 2001. Biochemical characterization and ligand binding properties of neuroglobin, a novel member of the globin family. J. Biol. Chem. 276:38949–38955. [DOI] [PubMed] [Google Scholar]
- 28.Sawai, H., N. Kawada, K. Yoshizato, H. Nakajima, S. Aono, and Y. Shiro. 2003. Characterization of the heme environmental structure of cytoglobin, a fourth globin in humans. Biochemistry. 42:5133–5142. [DOI] [PubMed] [Google Scholar]
- 29.Goodman, M. D., and M. S. Hargrove. 2001. Quaternary structure of rice non-symbiotic hemoglobin. J. Biol. Chem. 276:6834–6839. [DOI] [PubMed] [Google Scholar]
- 30.Antonioni, E., and M. Brunori. 1971. Hemoglobin and Myoglobin and Their Reactions with Ligands. North-Holland, Amsterdam, The Netherlands.
- 31.Couture, M., T. Burmester, T. Hankeln, and D. L. Rousseau. 2001. The heme environment of mouse neuroglobin. Evidence for the presence of two conformations of the heme pocket. J. Biol. Chem. 276:36377–36382. [DOI] [PubMed] [Google Scholar]
- 32.Choi, S., T. G. Spiro, K. C. Langry, K. M. Smith, D. L. Budd, and G. N. La Mar. 1982. Structural correlations and vinyl influences in resonance Raman spectra of protoheme complexes and proteins. J. Am. Chem. Soc. 104:4345–4351. [Google Scholar]
- 33.Dasgupta, S., D. L. Rousseau, H. Anni, and T. Yonetani. 1989. Structural characterization of cytochrome c peroxidase by resonance Raman scattering. J. Biol. Chem. 264:654–662. [PubMed] [Google Scholar]
- 34.Kitagawa, T. 1988. Resonance Raman spectra of heme and metalloproteins. In Biological Applications of Raman Spectroscopy, Vol. 3. T. G. Spiro, editor. John Wiley & Sons, New York, NY. 97–131.
- 35.Hu, S., K. M. Smith, and T. G. Spiro. 1996. Assignment of protoheme resonance Raman spectrum by heme labeling in myoglobin. J. Am. Chem. Soc. 118:12638–12646. [Google Scholar]
- 36.Taylor, C. P. S. 1977. The EPR of low spin heme complexes. Relation of the t2g hole model to the directional properties of the g tensor, and a new method for calculating the ligand field parameters. Biochim. Biophys. Acta. 491:137–149. [DOI] [PubMed] [Google Scholar]
- 37.Quinn, R., J. S. Valentine, M. P. Byrn, and C. E. Strouse. 1987. Electronic structure of low-spin ferric porphyrins: a single-crystal EPR and structural investigation of the influence of axial ligand orientation and the effects of pseudo-Jahn-Teller distortion. J. Am. Chem. Soc. 109:3301–3308. [Google Scholar]
- 38.Walker, F. A., B. H. Huynh, W. R. Scheidt, and S. R. Osvath. 1986. Models of the cytochromes b. Effect of axial ligand plane orientation on the EPR and Moessbauer spectra of low-spin ferrihemes. J. Am. Chem. Soc. 108:5288–5297. [Google Scholar]
- 39.Lionetti, C., M. G. Guanziroli, F. Frigerio, P. Ascenzi, and M. Bolognesi. 1991. X-ray crystal structure of the ferric sperm whale myoglobin: imidazole complex at 2.0 Å resolution. J. Mol. Biol. 217:409–412. [DOI] [PubMed] [Google Scholar]
- 40.Pesce, A., S. Dewilde, M. Nardini, L. Moens, P. Ascenzi, T. Hankeln, T. Burmester, and M. Bolognesi. 2003. Human brain neuroglobin structure reveals a distinct mode of controlling oxygen affinity. Structure. 11:1087–1095. [DOI] [PubMed] [Google Scholar]
- 41.Vallone, B., K. Nienhaus, M. Brunori, and G. U. Nienhaus. 2004. The structure of murine neuroglobin: novel pathways for ligand migration and binding. Proteins. 56:85–92. [DOI] [PubMed] [Google Scholar]
- 42.Capeillere-Blandin, C., R. C. Bray, M. Iwatsubo, and F. Labeyrie. 1975. Flavocytochrome b2: kinetic studies by absorbance and electron-paramagnetic-resonance spectroscopy of electron distribution among prosthetic groups. Eur. J. Biochem. 54:549–566. [DOI] [PubMed] [Google Scholar]
- 43.Xia, Z. X., and F. S. Mathews. 1990. Molecular structure of flavocytochrome b2 at 24 Å resolution. J. Mol. Biol. 212:837–863. [DOI] [PubMed] [Google Scholar]
- 44.Cunane, L. M., J. D. Barton, Z. W. Chen, F. E. Welsh, S. K. Chapman, G. A. Reid, and F. S. Mathews. 2002. Crystallographic study of the recombinant flavin-binding domain of baker's yeast flavocytochrome b2: comparison with the intact wild-type enzyme. Biochemistry. 41:4264–4272. [PubMed] [Google Scholar]
- 45.Durley, R. C. E., and F. S. Mathews. 1996. Refinement and structural analysis of bovine cytochrome b5 at 1.5 Å resolution. Acta Crystallogr. D Biol. Crystallogr. 52:65–76. [DOI] [PubMed] [Google Scholar]
- 46.Shokhirev, N., and F. A. Walker. 1998. Co- and counterrotation of magnetic axes and axial ligands in low-spin ferriheme systems. J. Am. Chem. Soc. 120:981–990. [Google Scholar]
- 47.Scholes, C. P., A. Lapidot, R. Mascarenhas, T. Inubushi, R. A. Isaacson, and G. Feher. 1982. Electron nuclear double resonance (ENDOR) from heme and histidine nitrogens in single crystals of aquometmyoglobin. J. Am. Chem. Soc. 104:2724–2735. [Google Scholar]
- 48.Brown, T. G., and B. M. Hoffman. 1980. 14N, 1H, and metal ENDOR of single crystal Ag(II)(TPP) and Cu(II)(TPP). Mol. Phys. 39:1073–1109. [Google Scholar]
- 49.Pesce, A., M. Bolognesi, A. Bocedi, P. Ascenzi, S. Dewilde, L. Moens, T. Hankeln, and T. Burmester. 2002. Neuroglobin and cytoglobin: fresh blood for the vertebrate globin family. EMBO Rep. 3:1146–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johansson, M. P., D. Sundholm, G. Gerfen, and M. Wikström. 2002. The spin distribution in low-spin iron porphyrins. J. Am. Chem. Soc. 124:11771–11780. [DOI] [PubMed] [Google Scholar]
- 51.Trent 3rd, J. T., and M. S. Hargrove. 2002. A ubiquitously expressed human hexacoordinate hemoglobin. J. Biol. Chem. 277:19538–19545. [DOI] [PubMed] [Google Scholar]
- 52.Royer, W. E., Jr., R. A. Fox, and F. R. Smith. 1997. Ligand linked assembly of Scapharca dimeric hemoglobin. J. Biol. Chem. 272:5689–5694. [DOI] [PubMed] [Google Scholar]
- 53.Wittenberg, J. B., B. A. Wittenberg, Q. H. Gibson, M. J. Trinick, and C. A. Appleby. 1986. The kinetics of the reactions of Parasponia andersonii hemoglobin with oxygen, carbon monoxide, and nitric oxide. J. Biol. Chem. 261:13624–13631. [PubMed] [Google Scholar]
- 54.Appleby, C. A., B. A. Wittenberg, and J. B. Wittenberg. 1973. Nicotinic acid as a ligand affecting leghemoglobin structure and oxygen reactivity. Proc. Natl. Acad. Sci. USA. 70:564–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guzov, V. M., H. L. Houston, M. B. Murataliev, F. A. Walker, and F. R. Feyereisen. 1996. Molecular cloning, overexpression in Escherichia coli, structural and functional characterization of house fly cytochrome b5. J. Biol. Chem. 271:26637–26645. [DOI] [PubMed] [Google Scholar]
- 56.Bois-Poltoratsky, R., and A. Ehrenberg. 1967. Magnetic and spectrophotometric investigations of cytochrome b5. Eur. J. Biochem. 2:361–365. [DOI] [PubMed] [Google Scholar]
- 57.Scholes, C. P., K. M. Falkowski, S. Chen, and J. Bank. 1986. Electron nuclear double resonance (ENDOR) of bis(imidazole) ligated low-spin ferric heme systems. J. Am. Chem. Soc. 108:1660–1671. [Google Scholar]
- 58.Vinck, E., S. Van Doorslaer, S. Dewilde, and L. Moens. 2004. Structural change of the heme pocket due to disulfide bridge formation is significantly larger for neuroglobin than for cytoglobin. J. Am. Chem. Soc. 126:4516–4517. [DOI] [PubMed] [Google Scholar]
- 59.Gibson, Q. H., J. B. Wittenberg, B. A. Wittenberg, D. Bogusz, and C. A. Appleby. 1989. The kinetics of ligand binding to plant hemoglobins. Structural implications. J. Biol. Chem. 264:100–107. [PubMed] [Google Scholar]
- 60.Springer, B. A., K. D. Egeberg, S. G. Sligar, R. J. Rohlfs, A. J. Mathews, and J. S. Olson. 1989. Discrimination between oxygen and carbon monoxide and inhibition of autooxidation by myoglobin. Site-directed mutagenesis of the distal histidine. J. Biol. Chem. 264:3057–3060. [PubMed] [Google Scholar]
- 61.Gibson, Q. H., and M. H. Smith. 1965. Rates of reaction of ascaris hemoglobins with ligands. Proc. R. Soc. Lond. B Biol. Sci. 163:206–214. [DOI] [PubMed] [Google Scholar]
- 62.Hamdane, D., L. Kiger, S. Dewilde, B. N. Green, A. Pesce, J. Uzan, T. Burmester, T. Hankeln, M. Bolognesi, L. Moens, and M. C. Marden. 2003. The redox state of the cell regulates the ligand binding affinity of human neuroglobin and cytoglobin. J. Biol. Chem. 278:51713–51721. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.