Abstract
Protein conformational changes may be associated with particular properties such as its function, transportation, assembly, tendency to aggregate, and potential cytotoxicity. In this research, the conformational change that is responsible for the fast destabilization and aggregation of rabbit muscle creatine kinase (EC 2.7.3.2) induced by heat was studied by intrinsic fluorescence and infrared spectroscopy. A pretransitional change of the tryptophan microenvironments was found from the intrinsic fluorescence spectra. A further analysis of the infrared spectra using quantitative second-derivative and two-dimensional correlation analysis indicated that the changes of the β-sheet structures in the C-terminal domain and the loops occurred before the formation of intermolecular cross-β-sheet structures and the unfolding of α-helices. These results suggested that the pretransitional conformational changes in the active site and the C-terminal domain might result in the modification of the domain-domain interactions and the formation of an inactive dimeric form that was prone to aggregate. Our results highlighted the fact that some minor conformational changes, which were usually negligible or undetectable by normal methods, might play a crucial role in protein stability and aggregation. Our results also suggested that the changes in domain-domain interactions, but not the dissociation of the dimer, might play a crucial role in the thermal denaturation and aggregation of this dimeric two-domain protein.
INTRODUCTION
It has been widely accepted that many large proteins contain a number of individual domains that can be considered as the units of structure, folding, evolution, and function (1–3). In many cases, an isolated single domain can be refolded independently and domains can be taken as the folding units with differing stabilities in multidomain proteins (1,4). Accordingly, the overall stability of these proteins can be regarded as the sum of their modules (5). However, individual properties of the domains are not a general aspect, and domain-domain interactions are also found to play a crucial role in the stability and folding of some multidomain proteins (6–8) as well as their in vivo functions (9). Particularly, for a two-domain protein, the folding/unfolding transitions may follow a two-state, an independent folding/unfolding, or a cooperative model, which are determined by the properties of domains as well as domain-domain interactions (2).
Creatine kinase (CK, EC 2.7.3.2), an oligomeric two-domain protein, catalyzes the reversible transfer of the phosphoryl group from MgATP to creatine in the cells of excitable tissues (10). CK, a key enzyme in cellular energy metabolism found primarily in vertebrates, belongs to a large family of phosphagen kinases. All phosphagen kinases show a high level of sequence homology (11) and a high degree of structural similarity. Cytosolic CK isozymes are exclusively dimeric and are formed by homo- or heterosubunits, whereas mitochondrial CK forms both octameric and dimeric structures. The structures of the various CK isozymes have the same subunit topology (12–15), and each monomer consists of two domains: a small N-terminal domain containing only α-helices and a large C-terminal domain containing an antiparallel β-sheet surrounded by several long α-helices (Fig. 1). The two domains are connected by a long linker and the active site is located in the cleft between the two domains. CK has been taken as a model protein, and numerous studies have been carried out in enzymology, structure, and folding mechanisms for more than five decades since the first purification of a homodimeric CK from rabbit muscle (RMCK) (16). The roles of the two domains as well as the subunit interactions in RMCK's catalytic and folding mechanisms have been studied by various probes. The folding and unfolding of RMCK by denaturants show rather complex pathways involving several intermediates (17–23). Particularly, a partly folded monomeric intermediate is found at low concentrations of guanidinium chloride. This stable intermediate is characterized by a mainly unstructured N-terminal domain and a rather compact C-terminal domain (22). The concept that domain-domain interactions may play a crucial role in the structural stability of RMCK was also confirmed by an increase in stability by cross-linking of the subunits (24) and an unstructured N-terminal fragment when expressed separately (25).
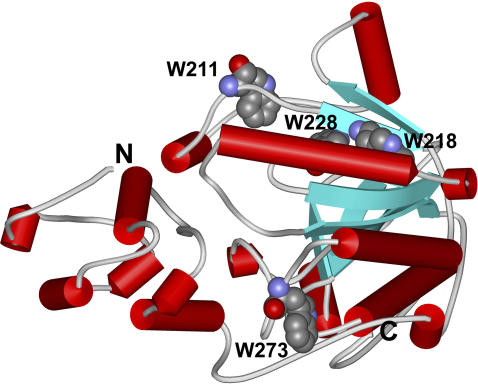
FIGURE 1.
Schematic 3D structure of the RMCK monomer showing the N- and C- terminal domains and the position of the four Trp residues (Protein Data Bank entry 2CRK).
In contrast to the complex multistate folding/unfolding mechanism of RMCK induced by denaturants, the thermal denaturation of RMCK was found to follow a simple two-state model (26). However, a fast loss of RMCK activity was found at a temperature far below the Tm of the enzyme (27,28). Similar to the phenomena observed when induced by chemical denaturants (18,29,30), the changes in the active site were found to occur far before the overall denaturation of the protein (28). These results suggested that pretransitional conformational changes might occur during the thermal denaturation of RMCK. Moreover, the protein was prone to aggregate when denatured by heat and the aggregation also occurred at a temperature (∼47°C) far below the Tm (∼56°C) of the protein. However, the mechanism of the fast destabilization and aggregation of RMCK induced by heat is not clear yet. In this research, the sequential events of RMCK thermal aggregation were studied by spectroscopic methods. A combination of quantitative second-derivative infrared analysis (QSD-IR; 31) and two-dimensional infrared correlation spectroscopy (2D IR; 32–34) was used to characterize the events that are responsible for inactivation and aggregation of RMCK induced by heat. The results indicated that the pretransitional changes of the Trp residue in the active site and the β-sheet structures in the C-terminal domain occurred before the formation of intermolecular cross-β-sheet structures and the unfolding of α-helices. These results suggested that the hierarchy in structural building blocks existed in RMCK thermal denaturation, though the differential scanning calorimetry (DSC) and circular dichroism (CD) results could be interpreted by an irreversible two-state approximation. Of particular importance is that the minor pretransitional changes in the active site and the C-terminal domain might weaken the domain-domain interactions and induce the formation of an inactive dimeric form that is prone to aggregate. These results highlighted the fact that those minor noncooperative events, which are usually negligible by normal probes, may play a crucial role in protein stability, activity, and aggregation.
MATERIALS AND METHODS
Materials
Purification and identification of RMCK was as described previously (17). The enzyme concentration was determined by measuring the absorbance at 280 nm with 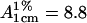 (17). Deuterated solvents were from Cambridge Isotope Laboratory (Andover, MA). Samples for spectroscopic experiments were prepared by using a 50-mM Tris-HCl buffer, pH 8.0.
(17). Deuterated solvents were from Cambridge Isotope Laboratory (Andover, MA). Samples for spectroscopic experiments were prepared by using a 50-mM Tris-HCl buffer, pH 8.0.
Fluorescence spectroscopy
The intrinsic fluorescence emission spectra of RMCK were measured using a Hitachi F-2500 spectrofluorometer (Tokyo, Japan) using 1-cm pathlength cuvettes. An excitation wavelength of 295 nm was used to avoid the contribution of the emission of residues other than tryptophan (Trp). The final spectrum was the average of three scans, and every spectrum was corrected by subtraction of the corresponding blank sample without protein. The fluorescence was measured every 0.5 nm in the range of 300–400 nm after 2 min equilibrium at the desired temperature, which was controlled with a circulating water bath. The spectra were collected from 30°C up to 80°C at increments of 2°C. The thermal unfolding of RMCK was obtained by monitoring the ratio of the intensity at 320 nm to that at 365 nm. The fitting of the fluorescence spectra was carried out using the discrete states model of Trp residues in proteins and was calculated by program developed in-house based on the SIMS algorithms of decomposition (35). In brief, all the fluorescence spectra were normalized before calculation. All three classes of Trp residues—that is, classes I, II, and III (36,37)—were assumed to be present in the protein and were included in the curve-fitting process. A biparametic long-normal fitting function was used to describe the three possible components of a spectrum (38). All possible fittings were obtained by varying the maximum intensity (Im) and wavelength (Vm) of each component independently within appropriate limits, according to the previous researches (35–37), and the best fitting results were obtained according to the least root mean-square criterion.
CD spectroscopy
CD spectra were recorded with a Jasco J-715 spectrophotometer (Tokyo, Japan) equipped with a thermoelectrically controlled cell holder over a wavelength range of 190–250 nm with a 1-mm pathlength cell. Each spectrum was the result of three scans obtained by collecting data with a resolution of 0.5 nm and a bandwidth of 2 nm. The spectra were measured at intervals of 2°C in the temperature range from 30°C to 80°C. The thermal unfolding of RMCK was obtained by monitoring the change of the mean residue ellipticity at 222 nm.
Fourier transform infrared spectroscopy
Details with regard to the IR measurements were the same as those described before (39). In brief, IR spectra were measured with a Perkin-Elmer Spectrum 2000 spectrometer (Wellesley, MA) equipped with a dTGS detector. IR samples were prepared by dissolving 50 mg protein in 50 mM Tris-HCl buffer prepared using D2O instead of H2O, pD 8.45. The samples were stored overnight at room temperature and lyophilized. The deuterated protein was dissolved in D2O and centrifuged at 6000 × g for 10 min before use; ∼30 μl sample was placed between a pair of CaF2 windows separated by a 50-μm Teflon spacer. Either 128 or 256 scans were recorded with a spectral resolution of 4 cm−1 in single-beam mode. For thermal transition studies, spectra were recorded from 30°C up to 80°C at increments of 2°C. Second derivative spectra were obtained using the algorithm in the software Spectrum v3.02 provided by Perkin-Elmer (Wellesley, MA) with a 9- or 13-point Savitzky-Golay smoothing. The changes of various secondary structures were evaluated using the QSD-IR method (31). Fourier self-deconvolution (FSD) was performed using the Spectrum v3.02 software program with a gamma factor of 2.5 and a Bessel smoothing of 70%.
2D IR correlation analysis
To minimize the artifacts that may be caused by baseline offsets, a linear baseline was subtracted from each 1D IR spectrum before further analysis. To generate the 2D IR plots, heating was used as the perturbation to induce time-dependent spectral fluctuations and to detect the dynamic spectral variations of the secondary structure of RMCK. Synchronous and asynchronous correlation plots were calculated from the FSD spectra with a spectral region of 1700–1600 cm−1 using SDIAPP software developed in-house (39) according to the generalized 2D correlation algorithm based on the Hilbert transform (34). In light of recent publications concerning the artifacts that might be introduced into the 2D correlation plots due to baseline offsets, band overlapping, noise, and any distortions in the spectra used for the 2D correlation analysis (40–44), various methods including normalization, static spectrum removal, and low-pass Fourier filtering procedures have been tested in addition to using the FSD spectra directly. We found that the best results were obtained using nonnormalized spectra and using time-averaged spectrum as a reference. Since the 2D correlation spectra with time-average method are more sensitive to noise than those without the reference spectra (41,43), the noise was evaluated by comparing results obtained with or without reference spectrum. Moreover, the deviation from spherical peak shape was also used as an indicator of noise (41). Finally, the absence of any misleading results was achieved by confirming that the events from the 2D IR analysis matched those obtained from other biophysical techniques. The 2D IR correlation plots were presented as contour maps, constructed by drawing the contour lines every 10% off from the maximum intensity of the corresponding map. The order of the events was characterized by analyzing the sign of the peaks in the 2D IR correlation plots using rules proposed by Noda (34).
RESULTS
Fluorescence spectroscopy
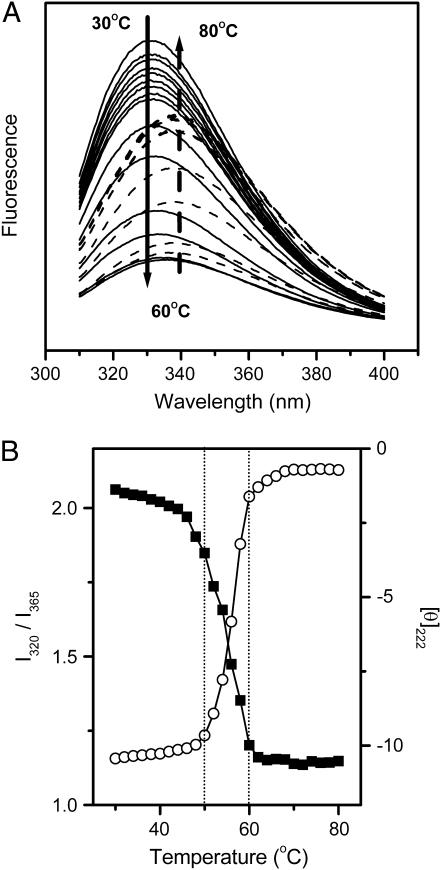
The crystal structure of RMCK (13) reveals that the enzyme contains two domains in each subunit (Fig. 1). The unequal distribution of the secondary structures and the aromatic residues provides a useful tool in unraveling the different contributions of the two domains to the stability of RMCK. Particularly, the β-sheet structures and all of the four Trp residues are located in the C-terminal domain. As presented in Fig. 2 A, the fluorescence spectrum of the native protein showed a maximum intensity at ∼330 nm. A red shift of the emission maximum from 330 nm to 340 nm was found when the sample was heated from 30°C to 80°C. Since the intrinsic fluorescence is gradually affected by temperature, the ratio of the intensity at 320 nm to that at 365 nm (I320/I365), which is characteristic of the shape and position of the fluorescence spectrum (45), was used to monitor the conformational changes of RMCK during thermal denaturation. Fig. 2 B shows that the data from CD experiments revealed a typical two-state process, but the data from intrinsic fluorescence could not be fitted into a simple two-state model. The value of I320/I365 decreased slightly with the increase of temperature from 30°C to 44°C, greatly from 44°C to 54°C, and acutely from 54°C to 60°C. The temperature at which the protein had significant conformational changes was much lower than that from the CD (Fig. 2 B, circles), DSC (26), or IR method (see Fig. 4, A and D). The deviations of the fluorescence data from the CD data suggested that the microenvironments around the Trp residues might change earlier than the overall structural disruption.
FIGURE 2.
Thermal dependence of the intrinsic fluorescence spectra (A) and thermal melting curves (B) of RMCK. In A, the spectra recorded at temperatures between 30 and 60°C are represented by solid lines, whereas those recorded at temperatures between 60 and 80°C are represented by dashed lines. The arrows show the direction of the intensity change with increasing temperature. In B, thermal melting curves were obtained by monitoring the ratio of the intensity of the intrinsic fluorescence at 320 nm to that at 365 nm (I320/I365, ▪) and the mean residue ellipticity at 222 nm ([θ]222, ○). [θ]222 was expressed in [103 × deg × cm2 × dmol−1].
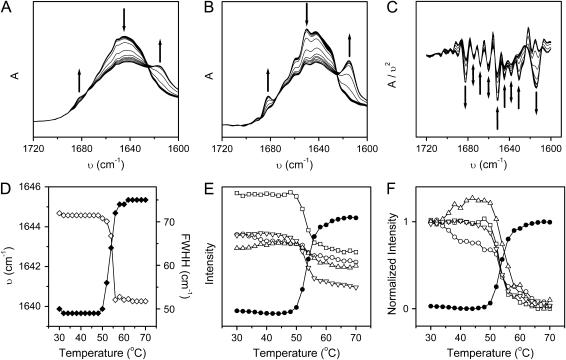
FIGURE 4.
Thermal dependence of the original (A), Fourier self-deconvolution (B), and second derivative (C) infrared spectra and the thermal melting curves (D, E, and F) of RMCK in 50 mM Tris-HCl buffer, pD 8.45. In A, B, and C, the arrows show the direction of the intensity change of characteristic bands with increasing temperature from 30 to 70°C. The arrows in A and B indicate the position of bands around 1682, 1652, and 1614 cm−1, from left to right, respectively, whereas those in C indicate the position of bands at 1682, 1674, 1668, 1660, 1652, 1645, 1638, 1631, and 1614 cm−1, from left to right. (D) Thermal dependence of the band shift (⋄) and band width at half height (♦) were calculated from the original IR spectra in Fig. 4 A. (E) Thermal dependence of the change of the intensity of the bands at 1652 (□), 1645 (○), 1638 (▵), 1631 (▿), and 1614 cm−1 (•), respectively. The data were calculated from the spectra in Fig. 4 C using the QSD-IR method (31). (F) Thermal dependence of the normalized intensity calculated from the data is presented in B.
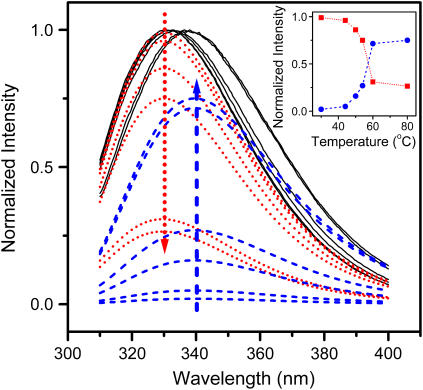
The experimental fluorescence spectra were further analyzed by the theoretical model of discrete states of Trp residues in proteins (35–37). As presented in Fig. 3, the fluorescence spectrum of the native enzyme had only one dominant class I component centered at 330 nm, which suggested that all of the four Trp residues in RMCK were buried in a hydrophobic environment. As the temperature increased, the intensity of the class I component decreased, whereas the class II component centered at 339.5 nm, which represents the fluorophores exposed to bonded waters, increased. The class III component, which comes from fully water-exposed fluorophores, was very small (not more than 1%), even for the fluorescence spectra recorded at high temperatures. Meanwhile, the class I component contributed ∼30% to the emission of the protein at temperatures >60°C. These results suggested that the protein was not fully unfolded upon heating.
FIGURE 3.
Fitting of the experimental fluorescence spectra recorded at 30, 44, 50, 54, 60, and 80°C to the theoretical model of discrete states of Trp residues in proteins. The fitted spectra (black solid lines) are the sum of the spectral components, class I (dotted red lines), and class II (dashed blue lines) fluorophores. The fitting error was estimated to be ∼5%. The other components (class S and class III) were so small that their presence was negligible. The arrows show the direction of the intensity changes of class I (dotted red line) and class II (dashed blue line) fluorophores with increasing temperature from 30 to 80°C. The inset shows the temperature dependence of the maximum intensity of components I (red squares) and II (blue circles), respectively.
FTIR spectroscopy
As mentioned above, all of the β-sheet structures are located in the C-terminal domain of RMCK, whereas α-helices are located in both the N- and C-terminal domains (Fig. 1). Since CD spectroscopy is not as sensitive to β-sheets as to α-helices and little change was found in the pretransitional stage during RMCK thermal denaturation (Fig. 2 B), IR spectroscopy was used to verify the results above from the intrinsic fluorescence spectra. Consistent with the crystal structure of RMCK (13) and previous IR studies (46,47), the amide I IR region of the native protein (Fig. 4) was dominated by bands from α-helices (1650–1658 cm−1), β-sheet (1630–1640 cm−1), and β-turns (1668 and 1674 cm−1). Upon heating, the intensity of bands from native structures decreased and two new bands, located at 1614 and 1682 cm−1, appeared at a temperature of ∼52°C. These two bands have been assigned to the sign of the formation of intermolecular β-sheet in aggregates (39,46,47). The correlation between the dominant secondary structure and the amide I frequencies of RMCK at low and high temperatures are summarized in Table 1. The change of the band position and bandwidth at half-height, which are characteristic of the IR spectra, shows a typical two-state thermal melting curve (Fig. 4 D). An abrupt change was found from 50°C to 56°C.
TABLE 1.
Correlation between the dominant secondary structure and the amide I frequencies of RMCK at low and high temperatures
| Band frequency (cm−1)
|
Assignment
|
|
|---|---|---|
| 30–48°C | 50–70°C | |
| 1611–1618 | Side chain | Cross-β-structures in aggregates (low wavenumber component) |
| 1626 | Extended chains or β-sheet | Extended chains or β-sheet |
| 1631–1640 | β-sheet | β-sheet |
| 1644–1649 | Random coil | Random coil |
| 1650–1657 | α-Helix | α-Helix |
| 1660, 1668, 1674 | β-Turns | β-Turns |
| 1682 | β-Sheet | Cross-β-structures in aggregates (high wavenumber component) |
For decomposition of the changes of different IR bands, the QSD-IR method (31) was used to obtain the melting curves of different secondary structures. As presented in Fig. 4 E, the changes of the bands at 1652 (α-helices) and 1614 cm−1 (intermolecular cross-β-sheet in aggregates) were similar to the overall change of the molecular structure. However, the changes of the bands at 1645 (random coils), 1638, and 1631 cm−1 (β-sheet) were quite different. A significant intensity decrease was found before the overall melting of the molecules for the band at 1645 cm−1; a slight decrease was found for the band at 1631 cm−1, whereas an increase was found for the band at 1638 cm−1. A clearer demonstration was achieved by normalizing the changes, as presented in Fig. 4 F. These results suggested that a pretransitional conformational change occurred before the overall disruption of the RMCK tertiary structure induced by heat, though the change was minor and was negligible when normal biophysical methods were used.
2D IR correlation analysis
2D IR correlation spectroscopy, which has been shown to be a powerful tool in obtaining the order of events upon perturbation (32–34), was used to further characterize the events that directly related to RMCK aggregation. The methodology of the generalized 2D IR correlation spectroscopy used in this work was the same as that described by Noda (34). Synchronous and asynchronous plots were constructed using temperature as the perturbation. In general, the synchronous plot has two types of peaks: autopeaks, which appear on the diagonal and indicate that the corresponding bands are changing with the perturbation, and crosspeaks, which reflect that the two correlated bands are changing in the same (positive) or opposite (negative) direction. The asynchronous plot is symmetrical and has only crosspeaks, and the appearance of a crosspeak indicates that the changes of the two correlated bands are out of phase. The sequence of events can be obtained by comparing the signs of the corresponding crosspeaks in the synchronous and asynchronous plots.
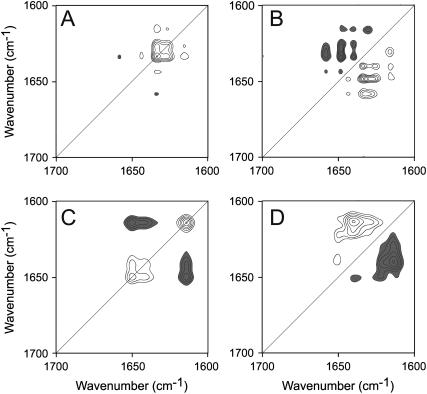
To get a better insight into the thermal denaturation processes of RMCK, 2D IR plots were constructed in the range of 30–48°C, in which the pretransitional conformational change of RMCK was observed, and in the range of 50–70°C, in which significant unfolding and aggregation was observed. In the pretransitional stage, two autopeaks located at 1626 and 1633 cm−1 were evident in Fig. 5 A, which suggested that the changes of the β-sheet structures and the extended chains were the major events in this stage. Positive crosspeaks were found for the band pairs 1615/1626, 1615/1633, 1626/1633, and 1633/1645 cm−1, which indicated that these peaks were changing in the same direction. A negative crosspeak was found at 1633/1660 cm−1, which indicated that the change of the band at 1660 cm−1 (β-turns) was increasing along with the decreasing of the band at 1633 cm−1. Besides the bands characterized in the synchronous plot, two additional bands (1640 and 1648 cm−1) were found to be correlated with the bands at 1615, 1626, and 1633 cm−1 in the asynchronous plot (Fig. 5 B). The sequence of the events could be identified by the signs of the crosspeaks as 1640 and 1649 cm−1 > 1615, 1626, and 1633 cm−1 > 1645 and 1660 cm−1. An interesting finding was that both β-sheet structures and random coils had two corresponding bands in the 2D IR plots, which might reflect that these bands had a different response to the perturbation (36,48). These results were quite consistent with those obtained by QSD-IR spectroscopy.
FIGURE 5.
2D correlation analysis of the IR spectra of RMCK thermal denaturation. The synchronous (A and C) and asynchronous (B and D) plots were constructed from the Fourier self-deconvolution spectra in Fig. 4 B in the range of 30–48°C (A and B) and 50–70°C (C and D). The plots are presented as contour maps, constructed by drawing the contour lines every 10% off from the maximum intensity of the corresponding map. Clear peaks are positive and dark peaks are negative.
In the range 50–70°C, the appearance of an autopeak at 1614 cm−1 in the synchronous plot (Fig. 5 C) indicated that aggregates were formed in this stage. This autopeak was found to negatively correlate to two other autopeaks at 1640 and 1651 cm−1, which indicated that the intensity of the corresponding bands changed in the opposite direction. That is, the unfolding of the native structures was accompanied by the formation of nonnative structures in aggregates. A comparison of the signs in Fig. 5, C and D, indicated that the change of the band at 1640 cm−1 was earlier than the bands at 1614 and 1650 cm−1, which suggested that the unfolding of β-sheet structures occurred before the unfolding of the helical structures and the formation of aggregates. No crosspeak could be identified between the bands at 1614 and 1651 cm−1, which suggested that the changes of the helical structures and the formation of aggregates were synchronous events.
DISCUSSION
Pretransitional conformational changes of RMCK
A previous DSC study suggested that the thermal denaturation of RMCK was consistent with an irreversible two-state model (26,49). However, according to the criteria of a two-state transition proposed by Jackson and Fersht (50), a two-state transition must be independent of the probe used for observation. The loss of the enzyme activity occurred at a temperature far below the Tm of the enzyme (27) and this fast inactivation had been attributed to the conformational changes of the flexible active site reflected by the change of C283, which unfolded before the disruption of the protein structure (28). The results in this research also suggested that noncooperative events, which were termed pretransitional changes, occurred before the thermal denaturation of RMCK. Similar pretransitional stages have also been observed during the thermal unfolding of other proteins, such as ribonucleases (51,52). Furthermore, the usage of a combination of newly developed analytical methods provided detailed information about the minor events that might not have been available by normal observation.
In the IR analysis, two dominant events, the changes of the β-sheet and the loops, were found to be evident in the pretransitional stage of RMCK thermal transitions. No significant change was observed for the α-helices by either IR (Figs. 4 and 5) or CD spectroscopy (Fig. 2 B), which suggested that the α-helices in the N- and C-terminal domain changed little in this stage. According to the results in literature (28), the intensity decrease of the 1645 cm−1 band could be attributed to the disruption of the loops in the active site. The change of the β-sheet was composed of two distinct events in the QSD-IR analysis: the intensity decrease of the 1631 cm−1 band and the intensity increase of the 1638 cm−1 band. The change of the 1631 cm−1 band could be attributed to the partial unfolding of the native β-sheet. But the change of the 1638 cm−1 band is difficult to interpret. Two possible explanations are considered. One is that the conformational change in the pretransitional stage resulted in a partial rearrangement of the C-terminal domain and this rearrangement might have induced a further increase in the β-sheet content. The other is that the intensity increase of the 1638 cm−1 band might be caused by the formation of nonnative oligomers through the cross-β-sheet structures. At present, it is difficult to say which of the two explanations is correct. However, the second seems more likely to be true. A similar increase of the β-sheet content was observed for the oligomer formation of the interleukin-1 receptor antagonist (53) and bovine pancreatic ribonuclease A (Y.-B. Yan, J. Zhang, H.-W. He, and H.-M. Zhou, unpublished data).
The results above from IR methods were also quite consistent with the change of the microenvironments of the Trp residues revealed by intrinsic fluorescence. There are four Trp residues in each of the RMCK subunits, and the side chains of the four Trp residues are mainly buried in a hydrophobic microenvironment (Fig. 1), which is confirmed by the fact that the emission spectrum of the native protein was dominated by only the class I component centered at 330 nm (Fig. 3). Upon thermal unfolding, the contribution of the class I component decreased, whereas that of the class II component increased to the emission of the heated protein. Similar to the results from IR spectra, a pretransitional change of the state of the Trp residues could be identified (Figs. 2 and 3). A close inspection of the structure of RMCK indicates that the four Trp residues in each RMCK subunit have distinct positions: W228 is buried in the interior of the hydrophobic core of the C-terminal domain, W211 is located on the dimer interface, W218 is located near the active site, and W273 is located on the surface of the molecule. It is worth noting that although W273 is located on the surface, the side chain of W273 is buried in the molecule (Fig. 1) and the solvent-accessible surface is ∼30% (calculated by MOLMOL). It has been shown that the loss of RMCK activity induced by heat was not due to the disassociation of the subunits and thus the changes of two Trp residues, W218 and W273, were more likely to contribute to this pretransitional change. A previous study using fluorescence labeling techniques also indicated that the change of the Cys residue in the active site was responsible for the inactivation of the enzyme (28). Since the active site is located between the N- and C-terminal domains, these results suggested that in the pretransitional stage, the change of the active site accompanied by some structural rearrangement of the C-terminal domain might weaken the domain-domain interactions.
Thermal aggregation of RMCK
Protein aggregation is an off-pathway product in biotechnology and has been found to be associated with many serious diseases (54). Despite its scientific significance, the mechanism of protein aggregation is not fully understood yet (55). Particularly, the lack of any high-resolution structure of the aggregates limits our knowledge of how the aggregates form and what is responsible for the initiation of aggregation. In general, the partially folded intermediate, which contains a rather large amount of hydrophobic exposure, is thought to be responsible for protein aggregation. As for RMCK, a monomeric intermediate induced by denaturants has been well characterized (18–23,46,56). This partially folded intermediate is characterized by a mainly unstructured N-terminal domain and a rather compact C-terminal domain (22), which suggests that the N-terminal is more unstable and the domain-domain interactions help to maintain its native structure. In contrast, the C-terminal domain is regarded as an autonomous folding unit (22).
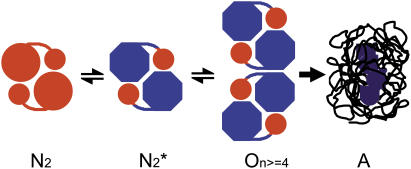
RMCK was found to have the tendency to aggregate when denatured by heat, and the aggregation occurred at a temperature (∼47°C) far below the Tm (∼56°C) of the protein (49). In our research, a combination of QSD-IR and 2D IR correlation analysis suggested that the minor conformational changes in the C-terminal domain occurred before the fast destabilization and aggregation of RMCK induced by heat. An analysis of the sequence of the events during RMCK thermal denaturation at high temperatures (Fig. 5 D) also indicated that the unfolding of β-sheet structures occurred before the formation of aggregates and the unfolding of α-helices. Since the β-sheet is located on the C-terminal domain (Fig. 1), these results suggested that the conformational change in the C-terminal domain of RMCK might be associated with its tendency to aggregate at a temperature far below the Tm of the protein. The aggregation of RMCK was not due to the disassociation of the two subunits since the enzyme did not dissociate when incubated at 50°C for 30 min (28). The intrinsic fluorescence spectra in Fig. 3 indicated that the enzyme still contained ∼86% class I fluorophores at 50°C and ∼75% at 54°C, which suggested that most of the four Trp residues were still buried in a hydrophobic environment at temperatures <54°C. As analyzed above, the microenvironments of W218 and W273 were more likely to change at the pretransitional stage. The change of W218 as well as other residues, such as C283 (28), reflected the disruption of the active site, which is located on the crevice of two domains. Thus the modification of the domain-domain interactions rather than the monomer-monomer interactions is more likely to contribute to the initiation of RMCK thermal aggregation. Our previous studies also indicated that the inactive dimeric intermediate was prone to aggregate in RMCK refolding from urea when a relatively high protein concentration was used (57,58). Moreover, it has also been suggested that the C-terminal domain plays a crucial role in the packing of monomer into native enzyme (25,56,59). Taking into account the pretransitional conformational changes discussed above, the aggregates of RMCK induced by heat might also form between similar inactive dimers. On the basis of the results herein and those in the literature (25,28,49,56–59), we propose a possible explanation for the mechanism of RMCK thermal inactivation and aggregation, as presented in Scheme 1. The minor changes in the active site and the C-terminal domain induced by heat resulted in the formation of an aggregation-prone species 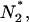 which acts as the building block of nonnative oligomers. Further unfolding and assembly can be obtained by increasing the temperature or prolonging the heating time. The thermally induced aggregates of RMCK contain a large amount of residual structures (∼30% from the intrinsic fluorescence spectra, Fig. 3).
which acts as the building block of nonnative oligomers. Further unfolding and assembly can be obtained by increasing the temperature or prolonging the heating time. The thermally induced aggregates of RMCK contain a large amount of residual structures (∼30% from the intrinsic fluorescence spectra, Fig. 3).
SCHEME 1.
A proposed model for RMCK thermal aggregation. N2 is the native dimeric enzyme, N2* is the inactive dimeric form induced by heat or other denaturants, O represents the nonnative oligomers formed from N2* as units, and A represents the nonnative disordered aggregates.
It is well known that protein conformational changes may be associated with the particular properties of a protein such as its activity, tendency to aggregate, cytotoxicity, binding to partners, and resistance to proteolysis. In general, partially folded intermediates are thought to be responsible for protein aggregation and have been verified in many proteins (54). However, some proteins were found to be prone to aggregate without any strong perturbations when stored at room temperature for days (60). Our results highlight the fact that some minor conformational changes, which may be undetectable or negligible by normal methods, also play a crucial role in protein stability and aggregation. Moreover, our results also suggested that the change in domain-domain interactions, but not the dissociation of the dimer, might play a crucial role in RMCK thermal denaturation and aggregation.
Acknowledgments
The authors thank Prof. Su-Qin Sun (Tsinghua University) for her help with the IR measurements and Mr. Jiang Zhu for his help with the fluorescence measurements. The authors also thank the anonymous reviewers for helpful suggestions.
This investigation was supported by funds from the National Key Basic Research Special Foundation (G1999075607), the National Natural Science Foundation of China (60401009), the Basic Research Fund (JC2003061), and the 985 Fund from Tsinghua University.
References
- 1.Wetlaufer, D. B. 1973. Nucleation, rapid folding, and globular intrachain regions in proteins. Proc. Natl. Acad. Sci. USA. 70:697–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jaenicke, R. 1999. Stability and folding of domain proteins. Prog. Biophys. Mol. Biol. 71:155–241. [DOI] [PubMed] [Google Scholar]
- 3.Vogel, C., M. Bashton, N. D. Kerrison, C. Chothia, and S. A. Teichmann. 2004. Structure, function and evolution of multidomain proteins. Curr. Opin. Struct. Biol. 14:208–216. [DOI] [PubMed] [Google Scholar]
- 4.Peng, Z. Y., and L. C. Wu. 2000. Autonomous protein folding units. Adv. Protein Chem. 53:1–47. [DOI] [PubMed] [Google Scholar]
- 5.Scott, K. A., A. Steward, S. B. Fowler, and J. Clarke. 2002. Titin: a multidomain protein that behaves as the sum of its parts. J. Mol. Biol. 315:819–829. [DOI] [PubMed] [Google Scholar]
- 6.Martin, A., and F. X. Schmid. 2003. Evolutionary stabilization of the gene-3-protein of phage fd reveals the principles that govern the thermodynamic stability of two-domain proteins. J. Mol. Biol. 328:863–875. [DOI] [PubMed] [Google Scholar]
- 7.Martin, A., and F. X. Schmid. 2003. The folding mechanism of a two-domain protein: folding kinetics and domain docking of the gene-3 protein of phage fd. J. Mol. Biol. 329:599–610. [DOI] [PubMed] [Google Scholar]
- 8.Sanderová, H., M. Hulková, P. Malon, M. Kepková, and J. Jonák. 2004. Thermostability of multidomain proteins: elongation factors EF-Tu from Escherichia coli and Bacillus stearothermophilus and their chimeric forms. Protein Sci. 13:89–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hawkins, A. R., and H. K. Lamb. 1995. The molecular biology of multidomain proteins. Selected examples. Eur. J. Biochem. 232:7–18. [DOI] [PubMed] [Google Scholar]
- 10.Wallimann, T., M. Wyss, D. Brdiczka, K. Nicolay, and H. M. Eppenberger. 1992. Intracellular compartmentation, structure and function of creatine kinase isoenzymes in tissues with high and fluctuating energy demands: the ‘phosphocreatine circuit’ for cellular energy homeostasis. Biochem. J. 281:21–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki, T., and T. Furukohri. 1994. Evolution of phosphagen kinase. Primary structure of glycocyamine kinase and arginine kinase from invertebrates. J. Mol. Biol. 237:353–357. [DOI] [PubMed] [Google Scholar]
- 12.Fritz-Wolf, K., T. Schnyder, T. Wallimann, and W. Kabsch. 1996. Structure of mitochondrial creatine kinase. Nature. 381:341–345. [DOI] [PubMed] [Google Scholar]
- 13.Rao, J. K. M., G. Bujacz, and A. Wlodawer. 1998. Crystal structure of rabbit muscle creatine kinase. FEBS Lett. 439:133–137. [DOI] [PubMed] [Google Scholar]
- 14.Eder, M., U. Schlattner, A. Becker, T. Wallimann, W. Kabsch, and K. Fritz-Wolf. 1999. Crystal structure of brain-type creatine kinase at 1.41 Å resolution. Protein Sci. 8:2258–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen, Y.-Q., L. Tang, H.-M. Zhou, and Z.-J. Lin. 2001. Structure of human muscle creatine kinase. Acta Crystallogr. D. 57:1196–1200. [DOI] [PubMed] [Google Scholar]
- 16.Kuby, S. A., L. Noda, and H. A. Lardy. 1954. Adenosinetriphosphate-creatine transphosphorylase. I. Isolation of the crystalline enzyme from rabbit muscle. J. Biol. Chem. 209:191–201. [PubMed] [Google Scholar]
- 17.Yao, Q.-Z., L.-X. Hou, H.-M. Zhou, and C.-L. Tsou. 1982. Conformational changes of creatine kinase during guanidine denaturation. Sci. Sin. [B]. 25:1186–1193. [PubMed] [Google Scholar]
- 18.Zhou, H.-M., and C.-L. Tsou. 1986. Comparison of activity and conformation changes during refolding of urea-denatured creatine kinase. Biochim. Biophys. Acta. 869:69–74. [DOI] [PubMed] [Google Scholar]
- 19.Grossman, S. H. 1994. An equilibrium study of the dependence of secondary and tertiary structure of creatine kinase on subunit association. Biochim. Biophys. Acta. 1209:19–23. [DOI] [PubMed] [Google Scholar]
- 20.Gross, M., A. Lustig, T. Wallimann, and R. Furter. 1995. Multiple-state equilibrium unfolding of guanidino kinases. Biochemistry. 34:10350–10357. [DOI] [PubMed] [Google Scholar]
- 21.Zhu, L., Y.-X. Fan, and J.-M. Zhou. 2001. Identification of equilibrium and kinetic intermediates involved in folding of urea-denatured creatine kinase. Biochim. Biophys. Acta. 1544:320–332. [DOI] [PubMed] [Google Scholar]
- 22.Webb, T. I., and G. E. Morris. 2001. Structure of an intermediate in the unfolding of creatine kinase. Proteins Struct. Funct. Genet. 42:269–278. [PubMed] [Google Scholar]
- 23.Kuznetsova, I. M., O. V. Stepanenko, K. K. Turoverov, L. Zhu, J.-M. Zhou, A. L. Fink, and V. N. Uversky. 2002. Unraveling multistate unfolding of rabbit muscle creatine kinase. Biochim. Biophys. Acta. 1596:138–155. [DOI] [PubMed] [Google Scholar]
- 24.Guo, Z., Z. Wang, and X. C. Wang. 2002. Subunit interaction slows the unfolding of the N terminal domain of creatine kinase in urea. Biochemistry (Mosc.). 67:1388–1394. [DOI] [PubMed] [Google Scholar]
- 25.Gross, M., M. Wyss, E. M. Furter-Graves, T. Wallimann, and R. Furter. 1996. Reconstitution of active octameric mitochondrial creatine kinase from two genetically engineered fragments. Protein Sci. 5:320–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lyubarev, A. E., B. I. Kurganov, V. N. Orlov, and H.-M. Zhou. 1999. Two-state irreversible thermal denaturation of muscle creatine kinase. Biophys. Chem. 79:199–204. [DOI] [PubMed] [Google Scholar]
- 27.Dawson, D. M., H. M. Eppenberger, and N. O. Kaplan. 1967. The comparative enzymology of creatine kinase. II. Physical and chemical properties. J. Biol. Chem. 242:210–217. [PubMed] [Google Scholar]
- 28.Bai, J.-H., S.-Y. Zhang, and H.-M. Zhou. 1998. Inactivation of creatine kinase is due to the conformational changes of the active sites during thermal denaturation. Biochem. Mol. Biol. Int. 45:941–951. [DOI] [PubMed] [Google Scholar]
- 29.Yao, Q.-Z., M. Tian, and C.-L. Tsou. 1984. Comparison of the rates of inactivation and conformational changes of creatine kinase during urea denaturation. Biochemistry. 23:2740–2744. [DOI] [PubMed] [Google Scholar]
- 30.Zhou, H.-M., X.-H. Zhang, Y. Yin, and C.-L. Tsou. 1993. Conformational changes at the active site of creatine kinase at low concentrations of guanidinium chloride. Biochem. J. 291:103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang, J., and Y.-B. Yan. 2005. Probing conformational changes of proteins by quantitative second derivative infrared spectroscopy. Anal. Biochem. 340:89–98. [DOI] [PubMed] [Google Scholar]
- 32.Noda, I. 1989. Two-dimensional infrared spectroscopy. J. Am. Chem. Soc. 111:8116–8118. [Google Scholar]
- 33.Noda, I. 1990. Two-dimensional infrared (2D-IR) spectroscopy: theory and applications. Appl. Spectrosc. 44:550–561. [Google Scholar]
- 34.Noda, I. 1993. Generalized two-dimensional correlation method applications to infrared, Raman, and other types of spectroscopy. Appl. Spectrosc. 47:1329–1336. [Google Scholar]
- 35.Burstein, E. A., S. M. Abornev, and Y. K. Reshetnyak. 2001. Decomposition of protein tryptophan fluorescence spectra into log-normal components. I. Decomposition algorithms. Biophys. J. 81:1699–1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reshetnyak, Y. K., and E. A. Burstein. 2001. Decomposition of protein tryptophan fluorescence spectra into log-normal components. II. The statistical proof of discreteness of tryptophan classes in proteins. Biophys. J. 81:1710–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reshetnyak, Y. K., Y. Koshevnik, and E. A. Burstein. 2001. Decomposition of protein tryptophan fluorescence spectra into log-normal components. III. Correlation between fluorescence and microenvironment parameters of individual tryptophan residues. Biophys. J. 81:1735–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burstein, E. A., and V. I. Emelyanenko. 1996. Log-normal description of fluorescence spectra of organic fluorophores. Photochem. Photobiol. 64:316–320. [Google Scholar]
- 39.Yan, Y.-B., Q. Wang, H.-W. He, X.-Y. Hu, R.-Q. Zhang, and H.-M. Zhou. 2003. Two-dimensional infrared correlation spectroscopy study of the heat-induced unfolding and aggregation process of myoglobin. Biophys. J. 85:1959–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Czarnecki, M. A. 1998. Interpretation of two-dimensional correlation spectra: science or art? Appl. Spectrosc. 52:1583–1590. [Google Scholar]
- 41.Tandler, P. J., P. B. Harrington, and H. Richardson. 1998. Effects of static spectrum removal and noise on 2D-correlation spectra of kinetic data. Anal. Chim. Acta. 368:45–57. [Google Scholar]
- 42.Czarnecki, M. A. 1999. Two-dimensional correlation spectroscopy: effect of normalization of the dynamic spectra. Appl. Spectrosc. 53:1392–1397. [DOI] [PubMed] [Google Scholar]
- 43.Czarnecki, M. A. 2003. Two-dimensional correlation spectroscopy: effect of reference spectrum on noise-free and noisy spectra. Appl. Spectrosc. 57:991–995. [DOI] [PubMed] [Google Scholar]
- 44.Yu, Z. W., Y. W. Wang, and J. Liu. 2005. Overlap may cause misleading results in two-dimensional correlation spectra. Appl. Spectrosc. 59:388–391. [DOI] [PubMed] [Google Scholar]
- 45.Turoverov, K. K., S. Y. Haitlina, and G. P. Pinaev. 1976. Ultra-violet fluorescence of actin. Determination of actin content in actin preparations. FEBS Lett. 62:4–6. [DOI] [PubMed] [Google Scholar]
- 46.Raimbault, C., F. Couthon, C. Vial, and R. Buchet. 1995. Effects of pH and KCl on the conformations of creatine kinase from rabbit muscle. Infrared, circular dichroic and fluorescence studies. Eur. J. Biochem. 234:570–578. [DOI] [PubMed] [Google Scholar]
- 47.Granjon, T., M.-J. Vacheron, C. Vial, and R. Buchet. 2001. Mitochondrial creatine kinase binding to phospholipids decreases fluidity of membranes and promotes new lipid-induced β structures as monitored by red edge excitation shift, Laurdan fluorescence, and FTIR. Biochemistry. 40:6016–6026. [DOI] [PubMed] [Google Scholar]
- 48.Yan, Y.-B., Q. Wang, H.-W. He, and H.-M. Zhou. 2004. Protein thermal aggregation involves distinct regions: sequential events in the heat-induced unfolding and aggregation of hemoglobin. Biophys. J. 86:1682–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meng, F.-G., Y.-K. Hong, H.-W. He, A. E. Lyubarev, B. I. Kurganov, Y.-B. Yan, and H.-M. Zhou. 2004. Osmophobic effect of glycerol on irreversible thermal denaturation of rabbit creatine kinase. Biophys. J. 87:2247–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jackson, S. E., and A. R. Fersht. 1991. Folding of chymotrypsin inhibitor 2. 1. Evidence for a two-state transition. Biochemistry. 30:10428–10435. [DOI] [PubMed] [Google Scholar]
- 51.Stelea, S. D., P. Pancoska, A. S. Benight, and T. A. Keiderling. 2001. Thermal unfolding of ribonuclease A in phosphate at neutral pH: deviations from two-state model. Protein Sci. 10:970–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stelea, S. D., and T. A. Keiderling. 2002. Pretransitional structural changes in the thermal denaturation of ribonuclease S and S protein. Biophys. J. 83:2259–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang, B. S., R. M. Beauvais, T. Arakawa, L. O. Narhi, A. C. Dong, D. I. Aparisio, and J. F. Carpenter. 1996. Formation of an active dimer during storage of interleukin-1 receptor antagonist in aqueous solution. Biophys. J. 71:3399–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dobson, C. M. 2003. Protein folding and misfolding. Nature. 426:884–890. [DOI] [PubMed] [Google Scholar]
- 55.Tycko, R. 2004. Progress towards a molecular-level structural understanding of amyloid fibrils. Curr. Opin. Struct. Biol. 14:96–103. [DOI] [PubMed] [Google Scholar]
- 56.Leydier, C., E. Clottes, F. Couthon, O. Marcillat, C. Ebel, and C. Vial. 1998. Evidence for kinetic intermediate states during the refolding of GdnHCl-denatured MM-creatine kinase. Characterization of a trapped monomeric species. Biochemistry. 37:17579–17589. [DOI] [PubMed] [Google Scholar]
- 57.Li, S., J.-H. Bai, Y.-D. Park, and H.-M. Zhou. 2001. Aggregation of creatine kinase during refolding and chaperonin-mediated folding of creatine kinase. Int. J. Biochem. Cell Biol. 33:279–286. [DOI] [PubMed] [Google Scholar]
- 58.Zhao, T.-J., W.-B. Ou, Q. Xie, Y. Liu, Y.-B. Yan, and H.-M. Zhou. 2005. Catalysis of creatine kinase refolding by protein disulphide isomerase involves disulphide cross-link and dimer to tetramer switch. J. Biol. Chem. 280:13470–13476. [DOI] [PubMed] [Google Scholar]
- 59.Mazon, H., O. Marcillat, C. Vial, and E. Clottes. 2002. Role of C-terminal sequences in the folding of muscle creatine kinase. Biochemistry. 41:9646–9653. [DOI] [PubMed] [Google Scholar]
- 60.Krishnan, S., E. Y. Chi, J. N. Webb, B. S. Chang, D. Shan, M. Goldenberg, M. C. Manning, T. W. Randolph, and J. F. Carpenter. 2002. Aggregation of granulocyte colony stimulating factor under physiological conditions: Characterization and thermodynamic inhibition. Biochemistry. 41:6422–6431. [DOI] [PubMed] [Google Scholar]