Abstract
A model system of a single-stranded trisegment Brome mosaic bromovirus (BMV) was used to analyze the mechanism of homologous RNA recombination. Elements capable of forming strand-specific stem-loop structures were inserted at the modified 3′ noncoding regions of BMV RNA3 and RNA2 in either positive or negative orientations, and various combinations of parental RNAs were tested for patterns of the accumulating recombinant RNA3 components. The structured negative-strand stem-loops that were inserted in both RNA3 and RNA2 reduced the accumulation of RNA3-RNA2 recombinants to a much higher extent than those in positive strands or the unstructured stem-loop inserts in either positive or negative strands. The use of only one parental RNA carrying the stem-loop insert reduced the accumulation of RNA3-RNA2 recombinants even further, but only when the stem-loops were in negative strands of RNA2. We assume that the presence of a stable stem-loop downstream of the landing site on the acceptor strand (negative RNA2) hampers the reattachment and reinitiation processes. Besides RNA3-RNA2 recombinants, the accumulation of nontargeted RNA3-RNA1 and RNA3-RNA3 recombinants were observed. Our results provide experimental evidence that homologous recombination between BMV RNAs more likely occurs during positive- rather than negative-strand synthesis.
RNA recombination is a general phenomenon in animal, plant, and bacterial RNA viruses, and it plays an important role in the fitness and evolution of virus genomes (24, 28, 36). Model systems to study RNA recombination have been developed for the Brome mosaic bromovirus (BMV), coronaviruses, poliovirus, Turnip crinkle carmovirus (TCV), tombusviruses, and for RNA phage Qβ (5, 6). Most of the RNA recombination events are thought to occur via a copy-choice mechanism. The evidence for participation of the replicase (RdRp) proteins in recombination has been provided for BMV (15-17, 35). Other proposed mechanisms are cleavage/religation (25, 28, 45) and transesterification (10).
According to a copy-choice model, prior to the actual switch the replicase stalls or dislodges from its template (21, 28, 36), which may occur at the 5′ end of the template (47, 49), at a template break (39), or at the nascent strand stem-loop structure. The latter is analogous to rho-independent transcription termination (46, 50), although it may be distinct from a dislodging RdRp in a copy-choice model. Polymerase stalling may be induced by RNA secondary and/or tertiary structures, by homopolymer runs, or by the misincorporation of nucleotides (12, 20, 21, 48, 50).
The success of the switch to the acceptor template can be influenced by a number of factors (5, 6, 23, 26, 28), such as (i) intermolecular RNA-RNA interactions (18, 31), (ii) intramolecular RNA-RNA interactions, or (iii) RNA-protein interactions (9, 36, 38). Altogether, RNA recombination appears to be a multistep process that involves primary and secondary structures, and the progeny recombinants are further selected on the basis of their fitness (43).
The intersegmental RNA-RNA recombination was extensively studied in BMV, a tripartite positive-strand RNA virus (5, 17). The 3′ noncoding regions (3′ NCR) of the three BMV RNAs share high sequence identity, and homologous recombination occurs frequently among these regions (7, 8). In particular, the so-called region R sequence, which is part of the wild-type (wt) 3′ NCRs of RNA2 and -3, can support homologous crossovers between RNA2 and RNA3 (32). A correlation between the length of the homologous sequence and recombination frequency as well as the effects of AU-GC contents were observed (32-34, 37).
Several authors have suggested the significance of RNA secondary structure in recombination. Crossovers between intertypic strains of poliovirus have been found within regions predicted to be single stranded (44). Recombination between TCV RNA and one of its satellites, RNA-D, frequently occurs at the base of a stem-loop structure (9, 36). A role of template RNA folding in the formation of defective RNAs was suggested for bromoviruses (41), coronaviruses (30), nodaviruses (29), and tombusviruses (49). However, these experiments could not rule out that selection for replicable molecules was responsible for the observed outcome.
The effect of RNA secondary structure on homologous recombination was not observed in BMV, and previous mutations had unpredictable effects on RNA folding (32). Here we show that stable stem-and-loop structures in negative strands of BMV RNA3 and RNA2 can significantly affect the distribution of RNA recombinants compared to unstable structures in negative strands or to either stable or unstable structures in positive strands. We conclude that homologous RNA3-RNA2 recombination more likely can occur during positive-strand than during negative-strand synthesis. The mechanistic implications of these results are discussed.
MATERIALS AND METHODS
Engineering of plasmid constructs and synthesis of transcripts.
Restriction enzymes, T4 DNA polymerase, T7 RNA polymerase, and Moloney murine leukemia virus (MMLV) reverse transcriptase were from Promega Corporation (Madison, Wis.) and from Fisher Scientific. In order to make insertions in the HindIII site within viral RNA3 and RNA2 sequences (nucleotide positions 1911 and 2665, respectively), a vector HindIII site was inactivated in PN-H66 (32) by filling it in with T4 DNA polymerase and deoxynucleoside triphosphates. Then the PstI-EcoRI insert was replaced in the resulting PN-H66H plasmid by the corresponding fragment of PN-H39 (32), and the resulting pB3H plasmid was used to introduce various HindIII fragments into RNA3. A similar clone for RNA2 was obtained by ligating the PstI-EcoRI fragment (the entire RNA2 sequence) of pB2TP5 (22) into the corresponding sites of PN-H66H. This yielded pB2H. Complementary (with overhangs) oligonucleotides 195 (5′-AGCTTATTAGCGCTCGAAAGAGTGTTTATTTA-3′) and 197 (5′-AGCTTAAATAAACACTCTTTCGAGCGCTAATA-3′) were annealed and then ligated in either orientation into HindIII-digested pB2H and pB3H plasmids in order to generate the stable stem-loop insertion mutants (Fig. 1). The resulting p2hp+ and p3hp+ constructs carried the stem-loop structure that was predicted to form only in positive strands of RNA2 and RNA3, respectively, while p2hp− and p3hp− constructs carried the stem-loop structure only in negative strands. Likewise, constructs p2ss+, p3ss+, p2ss−, and p3ss− were obtained by the ligation of another pair of oligonucleotides 24 (5′-AGCTTATTAGCGCCCGAAAGAATGTTTATTTA-3′) and 25 (5′-AGCTTAAATAAACATTCTTTCGGGCGCTAATA-3′) in either orientation into the HindIII sites of pB2H and pB3H.
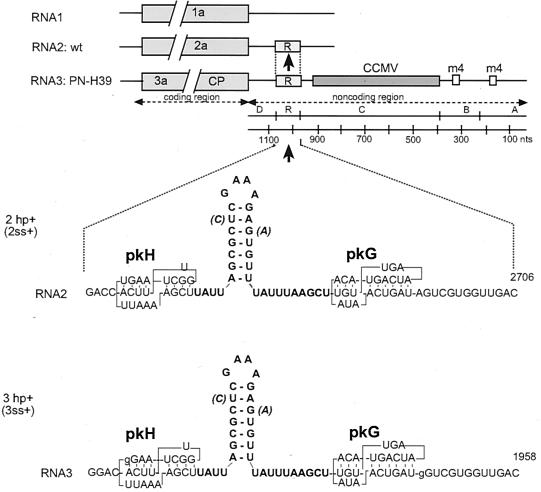
FIG. 1.
Schematic representation of BMV RNA recombination constructs and the stem-loop inserts. The top portion shows the 3′ regions of wt BMV RNA1 and -2 and the modified 3′ region of PN-H39 RNA3. The latter consists of a duplication of the tRNA-like structure sequence (regions A and B), each carrying a viable m4 deletion (represented by small rectangular boxes) and an insertion of a 765-nt 3′ fragment of CCMV RNA3 (region C; see reference 32). The recombinationally active 60-nt region R, which is homologous between RNA3 and -2, is represented by a box with the letter R, and the arrow points to the sites of the stem-loop inserts. Region D comprises the rest of the 3′ NCR (32). The large gray boxes to the left represent the open reading frames (not to scale) of protein 1a, 2a, or protein 3a and the coat protein (CP), which are encoded by RNA1, -2, and -3, respectively. The bottom portion of the figure shows computer-predicted secondary structures of the positive-strand stem-loop inserts (in bold face) that were introduced between pseudoknots pkG and pkH (27) of region R. Nucleotides in brackets represent the helix-destabilizing substitutions (in constructs 2ss+ and 3ss+). The negative-strand stem-loop inserts are not shown.
EcoRI-linearized plasmids pB1TP3, pB2TP5, and pB3TP7 (22) were transcribed in vitro (by using a MEGAscript T7 kit; Ambion, Austin, Tex.) to synthesize the infectious wild-type (wt) BMV RNA1, -2, and -3, respectively. The same kit was used to synthesize the infectious RNAs for all modified RNA constructs.
In vivo recombination assays and analysis of recombinants.
Mixtures of wt RNA1, wt or mutant RNA2, and RNA3 were inoculated on Chenopodium quinoa leaves, as previously described (31). Briefly, a mixture of 1 μg of each transcript in 15 μl of the inoculation buffer (10 mM Tris [pH 8.0], 1 mM EDTA, 0.1% Celite) was rubbed on a fully expanded leaf. Four separate leaves per plant (two to three plants) were inoculated per experiment. The experiment was repeated two to three times. The inoculated C. quinoa plants were maintained in a standard greenhouse at 25°C, with 16 h of light. Local lesions were counted, collected 14 days postinoculation, and stored frozen (at −80°C).
In this work we monitored only one subset of putative recombinants among the BMV RNAs, i.e., the positive-strand recombinants originating from RNA3. For each combination of parental RNA1, -2, and -3, total RNA was isolated from separate local lesions as described previously (4, 31, 40) and the progeny RNA3 sequences were amplified by reverse transcription-PCR (RT-PCR) (31). Primer 75 (5′-CAGTGAATTCTGGTCTCTTTTAGAGATTTACAGTG-3′), which was complementary to the common 3′-terminal sequence of all three BMV RNAs (nucleotides 2093 to 2117 for RNA3), was used for the first-strand cDNA synthesis with MMLV-RT. The resulting cDNA was amplified by PCR with TaqI DNA polymerase, primer 75, and RNA3-specific 5′ primer 74 (5′-CTGAAGCAGTGCCTGCTAAGGC-3′), representing nucleotides 1726 to 1747. The double-stranded DNA products were separated on a 1.5% agarose gel, purified by using a QIAquick gel extraction kit (QIAGEN Inc.), and sequenced in both directions.
RESULTS
Generation and infectivity of BMV RNA3 and RNA2 constructs.
The system that we used in this work relies on the repair of debilitated RNA3 constructs by homologous crossovers with RNA2. The crossovers are mediated by recombinationally active region R (32). The resulting recombinant RNA3s easily accumulate because of a selection advantage over the unrepaired parental RNA3s (31). PN-H39 is a BMV RNA3 cDNA construct that carries an extended 3′ NCR composed of a duplicated 3′ terminal sequence (Fig. 1), each carrying a 20-nucleotide (nt) deletion (designated m4). In addition, PN-H39 has an upstream 765-nt-long heterologous sequence of the cowpea chlorotic mottle virus (CCMV) RNA3 (32). The homologous crossovers occur within a further-upstream 60-nt sequence of region R that is also present in the wt 3′ NCR of RNA2. Previously it was shown that PN-H39 RNA3 supported 89% of homologous recombinants between RNA3 and RNA2 and that PN-H39 RNA3 could tolerate additional sequence inserts without measurable effects on the infectivity (34, 37).
Region R consists of two pseudoknot structures, pkH and pkG (Fig. 1). To examine the role of RNA structure on homologous recombination between RNA3 and RNA2, a synthetic stem-loop structure was inserted between pkG and pkH in both PN-H39 RNA3 and wt RNA2 (Fig. 1, constructs 3hp+ and 2hp+, respectively). The top part of the structure (CUCGAAAGAG) is identical to a sequence in satRNA-D of TCV that appeared to be a hotspot for recombination with genomic TCV RNA (9), whereas the remaining 4 bp were introduced to achieve the strand-specific effect. The stability of the inserted stem-loop conformation was not determined experimentally. However, both the computer-run analyses with RNA folding algorithms (RNAdraw and MFOLD) and the stabilizing effects of the GNRA loop motif present in the insert (where N is any nucleotide and R corresponds to G or A [19]) strongly suggested the existence of the predicted structure. While the structure was stable in positive strands (−41.2 kJ/mol at 25°C), in the complementary negative strand it was nearly single-stranded (−0.8 kJ/mol) due to G-U to A:C replacements (Fig. 1). In contrast, two other RNA3 variants (designated 2hp− and 3hp−) carried the insert in a reverse orientation, so the stable stem-loops were formed in negative strands. Four other RNA constructs (designated 2ss+, 3ss+, 2ss−, and 3ss−) carried the additional helix-destabilizing U:C or G:A mismatches (shown in parentheses in Fig. 1), so the stem-loop inserts were unstable in either strand (−3.7 and −4.5 kJ/mol at 25°C).
Each construct generated a similar number of lesions in C. quinoa plants, a BMV local lesion host, compared to that generated by wt BMV RNA (not shown).
Effects of stem-loop structures in both RNA3 and RNA2 on homologous RNA3-RNA2 recombination.
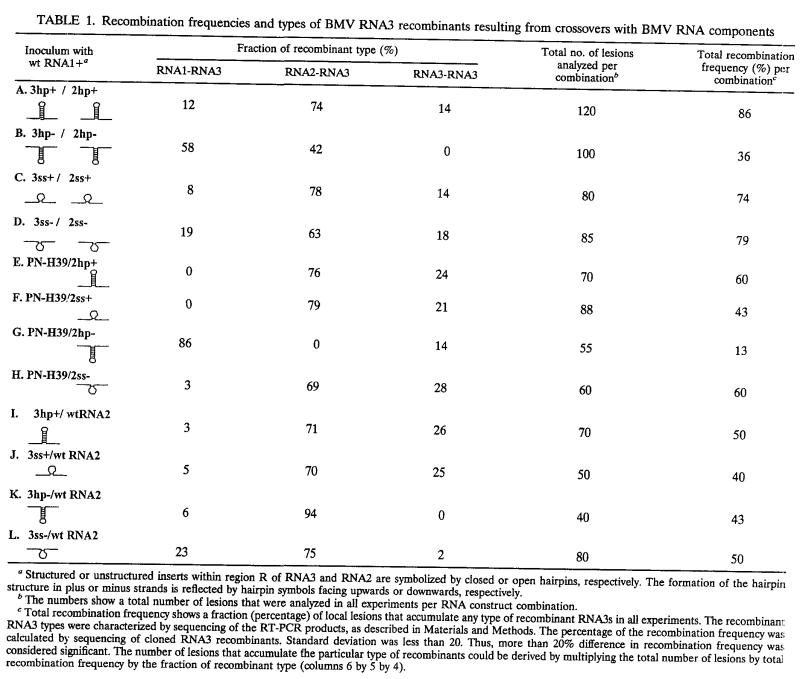
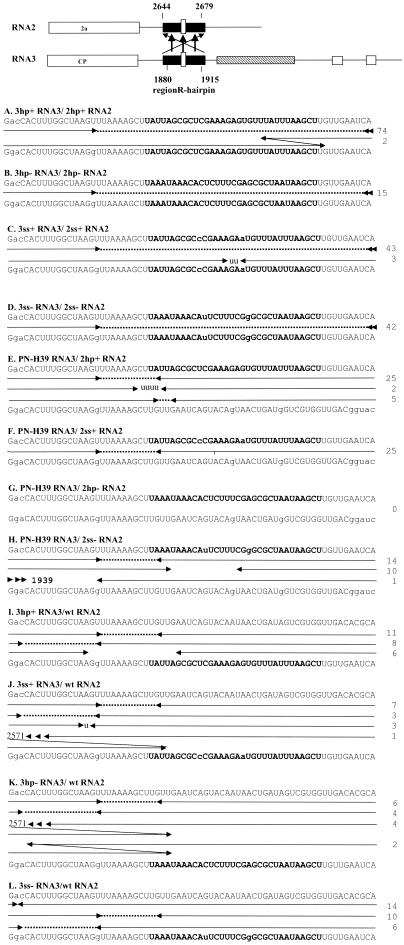
C. quinoa plants were coinoculated with wt RNA1, 3hp+ RNA3, and 2hp+ RNA2, following a methodology originally described by Rao et al. (40) and modified by Nagy and Bujarski (31). Since all further inoculations involved wt RNA1, for clarity and brevity it will not be mentioned throughout the rest of the text. The accumulation of recombinant RNA3s was monitored by RT-PCR with primers 74 and 75, which amplified the sequences carrying the RNA3 portion at the upstream side. This confirmed the presence of recombinants in 86% of local lesions (Table 1, row [experiment] A), which was similar to the frequency observed with the original PN-H39 RNA3 and wt RNA2 (32). Sequence analysis of 120 local lesions revealed that 74% (76 lesions) of the recombinants were derived by crossovers between RNA3 and RNA2, which again was comparable to the results from PN-H39 RNA3/wt RNA2 infection. Except for 2 recombinant RNA3s, which carried a 12-nt duplication within the insert, 74 characterized recombinants were precise and retained the parental stem-loop insert (Fig. 2A). The observed level of imprecise recombinants is consistent with the results on the unmodified PN-H39 RNA3 and wt RNA2 infection (32).
TABLE 1.
Recombination frequencies and types of BMV RNA3 recombinants resulting from crossovers with BMV RNA components
Structured or unstructured inserts within region R of RNA3 and RNA2 are symbolized by closed or open hairpins, respectively. The formation of the hairpin structure in plus or minus strands is reflected by hairpin symbols facing upwards or downwards, respectively.
The numbers show a total number of lesions that were analyzed in all experiments per RNA construct combination.
Total recombination frequency shows a fraction (percentage) of local lesions that accumulate any type of recombinant RNA3s in all experiments. The recombinant RNA3 types were characterized by sequencing of the RT-PCR products, as described in Materials and Methods. The percentage of the recombination frequency was calculated by sequencing of cloned RNA3 recombinants. Standard deviation was less than 20. Thus, more than 20% difference in recombination frequency was considered significant. The number of lesions that accumulate the particular type of recombinants could be derived by multiplying the total number of lesions by total recombination frequency by the fraction of recombinant type (columns 6 by 5 by 4).
FIG. 2.
Diagram summarizing the recombination frequency and the distribution of crossover sites in BMV RNA3 recombinants that were derived by RNA3-RNA2 crossovers after infection with the indicated mixtures of parental RNA3 and RNA2 constructs. The region of RNA3-RNA2 crossovers is shown schematically on the top, with region R represented by a filled black box and a small open rectangle inside that symbolizes the stem-loop insert. The arrows mark the crossovers between RNA3 and RNA2. Each combination of the RNA2 and RNA3 positive sequences are shown in the top and the bottomlines, respectively. Capital letters depict the homologous regions, the stem-loop inserts are in bold letters, and the helix-destabilizing substitutions within these inserts are in lower case. Each recombinant contains a 3′-terminal sequence derived from RNA2 on the right side and a 5′ sequence from RNA3 on the left side. Consequently, each rightward-pointing arrowhead denotes the last nucleotide derived from RNA3 and each leftward-pointing arrowhead denotes the first nucleotide derived from RNA2. Multiple arrowheads symbolize the occurrence of crossovers outside the range of the indicated sequences. Dashed lines show ambiguous (identical) regions (derived from either RNA2 or RNA3) in the precise homologous recombinants. Gaps between opposing arrowheads show deleted nucleotides, whereas nontemplated nucleotides generated during the crossover events are shown by lowercase letters between the arrows. The nucleotide sequences in the imprecise recombinants with ambiguous crossovers were arbitrarily placed with the upstream junction. Each entry represents a different RNA2/RNA3 recombinant type isolated from separate local lesions. Numbers on the right show the incidence of each type of RNA3 recombinant.
A different pattern arose after coinoculation with 3hp− RNA3 and 2hp− RNA2, the constructs that carried the stable stem-loop in their negative strands. The recombination frequency was reduced to 36% and the fraction of RNA3-RNA2 recombinants was reduced to 42% (15 lesions) (Table 1, row B). The recombinants were precise and mapped only within the flanking markers (Fig. 2B), confirming the importance of the region R sequence for recombination.
To check whether the observed change in the original pattern of RNA3-RNA2 recombinants was due to stable stem-loops, plants were coinfected with 3ss+ RNA3 and 2ss+ RNA2 (Fig. 1) and the accumulating RNA3 molecules were analyzed. As shown in Table 1, row C, the recombination frequency reached 74% and the fraction of RNA3-RNA2 recombinants was 78% (46 lesions; Fig. 2C). A similar trend was discernible for 3ss− RNA3/2ss− RNA2 infection (Table 1, row D). These data revealed that the unstable stem-loop structure in either strand did not affect markedly the accumulation of RNA3-RNA2 recombinants compared to that of the structured minus-strand inserts.
Effects of stem-loop inserts in either RNA3 or RNA2 on RNA3-RNA2 recombination.
To further study the function of RNA3 and RNA2 during homologous crossovers, we used only one parental RNA component (RNA3 or RNA2) that carried the stem-loop insert. Coinfection of PN-H39 RNA3 and 2hp+ RNA2 generated 76% (32 lesions) of recombinant RNA3s that were derived from RNA3-RNA2 crosses (Table 1, row E). This proportion was similar to those found for 3hp+ RNA3/2hp+ RNA2 or 3ss+ RNA3/2ss+ RNA2 infections. The stem-loop inserts were retained in the recombinants (Fig. 2E), suggesting that the stem-loop structure did not interfere per se with the formation and accumulation of recombinants. Similar homologous recombinant RNA3s were identified during PN-H39 RNA3/2ss+ RNA2 or PHH39H RNA3/2ss− RNA2 infections (Fig. 2F and H), indicating that the unstable structure in either strand of RNA2 was not critical. In contrast, the presence of the stable structure in the negative strand of RNA2 (infection PN-H39 RNA3/2hp− RNA2) inhibited the formation of RNA3-RNA2 recombinants (Table 1, row G), and the overall recombination frequency was reduced to 13%. This showed that negative strands of RNA2 were critical during the accumulation of the recombinant RNA3 variants.
There were relatively small effects when using the reciprocal combinations of parental RNA3 and RNA2 constructs. The infections with 3hp+ RNA3/wt RNA2, 3ss+ RNA3/wt RNA2, 3hp− RNA3/wt RNA2, or 3ss− RNA3/wt RNA2 produced 71, 70, 94, and 75% (25, 14, 16, and 30 lesions), respectively, of RNA3-RNA2 recombinants (Table 1, rows I, J, K, and L, respectively). None of the recombinants retained the stem-loop inserts (Fig. 2I through L), revealing that the crossovers occurred at upstream homologous positions.
RNA3-RNA1 recombinants.
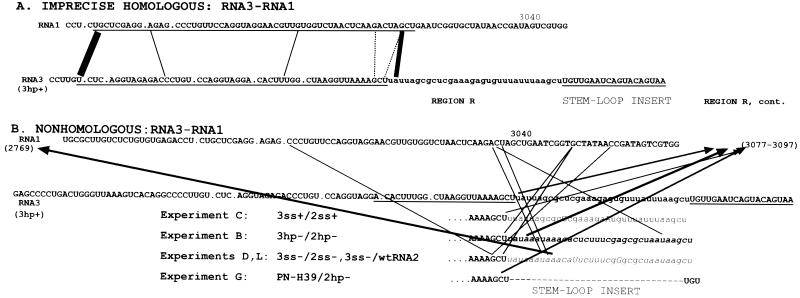
Besides homologous RNA3-RNA2 recombinants, nontargeted RNA3-RNA1 recombinants accumulated with variable frequency (Table 1). Two groups of recombinants were identified. One group (Fig. 3A) involved imprecise homologous recombinants with the majority of junction sites between region R of RNA3 and a homologous sequence of RNA1 (nucleotide positions 2986 to 3040 from the 5′ end, with 71% similarity to the RNA3 sequence). Six such recombinants were identified for 3hp+ RNA3/2hp+ RNA2, three for 3hp− RNA3/2hp− RNA2, and three for 3ss− RNA3/2ss− RNA2 infections (experiments A, B, D, respectively). Similar recombinants have been characterized previously (32, 37).
FIG. 3.
Diagram summarizing the location of junction sites in BMV RNA3 recombinants that were derived by RNA3-RNA1 crossovers. Panel A shows the imprecise homologous recombinants, and panel B shows the nonhomologous recombinants. The lines in each panel represent RNA1 sequence (top line) or RNA3 sequence (bottom line). Lines between the two sequences connect the last nucleotide of RNA3 and the first nucleotide of RNA1. The dashed and thin lines represent a low incidence (one or two, respectively) of the identified recombinants, while the thick lines represent those found at least three times. The positions of the crossovers (of panel B) that occurred outside the shown sequence range are depicted by arrowheads and by the nucleotide positions numbered in brackets. The underlined nucleotides represent the most similar sequence (A) or region R in the RNA3 sequence (B).
The second and largest group of recombinants had the crossover sites at heterologous (noncorresponding) positions between the two RNAs (Fig. 3B). In several cases, the crossover sites on RNA3 were located at the junction between region R and the stem-loop insert, especially for 3hp− RNA3/2hp− RNA2 (eight), 3hp+ RNA3/2hp+ RNA2 (six), 3ss+ RNA3/2ss+ RNA2 (one), 3ss− RNA3/2ss− RNA2 (one), and 3ss− RNA3/wt RNA2 (three) combinations (experiments B, C, D, L, respectively). The unmodified PN-H39 RNA3 (with 2hp− RNA2; experiment G) also supported such recombinants (three) at this location, suggesting the existence of a recombination hot spot that was not related to the presence of the stem-loop insert.
Other RNA3-RNA1 recombinants derived their RNA3 junctions from sequences within the stem-loop insert, i.e., of 3hp− RNA3/2hp− RNA2 (10), 3ss− RNA3/2ss− RNA2 (10), or 3ss− RNA3/wt RNA2 (6) infection (experiments B, D, and L, respectively). The corresponding junction sites on BMV RNA1 varied, either at upstream or downstream (the majority of recombinants) locations in the 3′ NCR. Overall, the isolation of the nontargeted RNA3-RNA1 recombinants revealed their high variability in sequence and size. Moreover, the majority of recombinants had the stem-loop inserts removed, irrespective of stem-loop stability or strandedness.
RNA3-RNA3 recombinants.
We also identified recombinants that occurred between parental RNA3 molecules (Fig. 4). One type of these recombinants carried only the terminal 3′ NCR, including the m4 marker deletion (Fig. 1, region A). This suggested that they arose by single RNA3-RNA3 crossovers. The highest fraction (10) of such recombinants were found during 3hp+ RNA3/2hp+ RNA2 infection (experiment A), and less (between 1 and 9) were found for other infections, with RNAs carrying either stable or unstable stem-loop structures (experiments C, D, I, J, and L). The unmodified PN-H39 RNA3 sequences also supported these crossovers (experiments E, F, and H).
FIG. 4.
Diagram summarizing the characteristics of BMV RNA3 recombinants that were derived by RNA3-RNA3 crossovers. The top panel shows the location of crossovers (either single or double crossovers) that occurred within the 3′ NCR between two parental RNA3 molecules of the same type. The numbers represent the nucleotide coordinates at the borders of the individual sequence elements (symbolized by letters A, B, C, R, and D on the top; for a description of individual elements, see the Fig. 1 legend). The connecting lines represent the crossover events (for double crossovers the dashed and undashed lines match the corresponding departing and returning positions). The location of the stem-loop insert is represented by a hairpin symbol inside region R. The bottom table shows the number of the types (single crossed versus double crossed) of RNA3-RNA3 recombinants as well as the fraction (in percentages) of recombinants carrying the parental stem-loop insert. Letters A to L on the left denote the particular experiments, as described in Table 1.
The second type of RNA3-RNA3 recombinants carried the additional CCMV sequence (25 to 35 nt long) that derived from the parental RNA3 constructs, indicating that these recombinants arose by double-crossovers between RNA3 molecules (see the top panel of Fig. 4). The highest proportion of the type 2 RNA3-RNA3 recombinants were identified during 3ss+ RNA3/2ss+ RNA2 and 3ss− RNA3/2ss− RNA2 infections but also during 3hp+ RNA3/2hp+ RNA2 and PN-H39 RNA3/2hp+ RNA2 or PN-H39 RNA3/2ss+ RNA2 infections (experiments C, D, A, E, and F, respectively). The majority of RNA3-RNA3 recombinants of either type did maintain the parental stem-loop inserts.
Interestingly, no RNA3-RNA3 recombinants were observed for combinations that involved 3hp− RNA3 (Table 1, rows B and K). This suggested that a stable structure in the negative strand of RNA3 also influences RNA3-RNA3 recombination (see Discussion).
DISCUSSION
The bulk of evidence from viral RNA recombination systems points toward template switching as the preferred mechanism of RNA recombination (18, 36). Our previous studies revealed that homologous crossovers between BMV RNA3 and RNA2 depended on the length and the extent of sequence identity, but the role of secondary structures was not apparent (32). Data obtained by mutation analysis of recombinationally active region R suggested that template switching predominantly occurred during positive-strand synthesis (32, 33, 34, 37).
In the experiments described here we provided more direct evidence for the template switching mechanism and for the role of secondary structure. We established that structured negative-strand stem-loops that were inserted in both RNAs reduced the accumulation of RNA3-RNA2 recombinants (experiment B) to a much higher extent than the same stem-loop inserts in positive strands (experiment A) or the unstructured inserts in either positive or negative strands (experiments C and D). The infections with only one RNA carrying the structured insert reduced the fraction of RNA3-RNA2 recombinants even further, but only when the stem-loops were in negative strands of RNA2 (compare experiments G and K). The role of RNA secondary structure was especially clear when comparing experiments G (2hp− RNA2) and H (2ss− RNA2). Just two nucleotide substitutions in 2ss− RNA2 were sufficient to prevent this RNA from taking part in recombination events with RNA3. A similar recombination silencing effect has been observed by using GC-rich inserts in BMV RNAs (34). Also, here the effect was only visible when the GC-rich insert was in region R of RNA2 but not of RNA3. However, there was a relation neither with the secondary structure of the insert nor with the polarity of the insert, since identical structures could form in either strand of the RNA.
To explain our data we propose a model in which the negative-strand stem-loops function as road blocks that interfere with the reattachment and reinitiation at the acceptor RNA2 negative strand (Fig. 5B). As a consequence, an increased fraction of the detached RdRps, including their nascent plus-strand RNA, will land on less-preferred RNA1 or RNA3 (Fig. 5B). Under normal circumstances, i.e., without the inserts, region R of RNA2 remains the favorite landing site for a detached RdRp (Fig. 5A).
FIG. 5.
Diagrammatic representation of a model that explains the shift of crossovers towards the nontargeted RNA3-RNA1 recombinants by stable stem-loop structures in negative strands of BMV RNA3 and -2. (A) The situation where the stable stem-loop insert is absent from the RNA3 and RNA2 parents. The RdRp complex starts the positive-strand synthesis on RNA3 and then switches onto the RNA2 acceptor negative strand at the homologous region R. In contrast, the presence of the structured element in negative strands of RNA2 (B) imposes difficulties for the RdRp to switch, and thus the RdRp complex jumps onto a less-homologous RNA1 template.
Stem-loops present in the donor RNA3 negative strand could possibly enhance RdRp pausing and detachment. Our data did not show a significant difference in the recombination activity of structured and unstructured inserts (Table 1, experiments K and L), although 3hp− RNA3 had the highest fraction of RNA2-RNA3 recombinants. Also, we did not find indications that the stem-loop might function as a terminator sequence as proposed in the Introduction. The formation of the stem-loop in the plus strand of RNA3 would then lead to increased detaching of RdRps from negative RNA3. Frequencies obtained with combinations involving 3hp+ RNA3 (experiments A and I), however, did not differ from those obtained with combinations involving 3ss+ or 3ss− RNA3 (experiments C, D, J, and L). We note that the lower frequencies of the single-insert combinations (experiments E through L) are probably due to the shorter stretch of (uninterrupted) homology between region R sequences of RNA2 and -3, a phenomenon described before (32).
Why does the inserted stem-loop interfere with recombination? We have good reason to believe that the insert folds as proposed (Fig. 1). The stem-loop resembles the well-studied GAAA-tetraloop hairpin, which has exceptional stability (19). Also, computer-assisted RNA folding (data not shown) did not reveal any significant interactions between the insert and the neighboring pseudoknots of region R in plus or minus strands. It should be noted that with the exception of one base pair, the G-U pair in pkH (Fig. 1), region R on the minus strand could form similar pseudoknots (data not shown). Thus, the stem-loop inserts likely function as autonomous structures and do not interfere with the recombination process by simply disturbing the structure of region R (although the inserts separate otherwise neighboring pseudoknots). Moreover, in previous experiments no clear correlation between the stability of region R and recombination activity could be found (32). We conclude that the stem-loop may interfere with the binding of the RdRp to RNA2. Secondly, the stem-loop may interfere with the rehybridization of the detached nascent strands onto the acceptor RNA2. The steric hindrance by the stem-loop itself may then be sufficient to prevent recombination from taking place.
Among other structural factors, one also needs to consider the postulated double-stranded rather than single-stranded forms of negative strands (3). Although this makes the existence of stem-loops in negative strands uncertain, the strand switching by the replicase may still occur within local single-stranded bubble structures, and the stem-loops can materialize during the unwinding of the replicative intermediates (34). One could envisage that the formation of a stem-loop in the plus strand of RNA2 could actually facilitate reattachment of the RdRp nascent-strand complex by locally opening up the double-stranded intermediate. To some extent this could to be true for BMV. In experiment A (3hp+/2hp+ RNAs), 76 out of 120 lesions analyzed contain RNA3-RNA2 recombinants (63%), whereas experiments C and D with unstructured inserts in both RNAs yield 58% (46 out of 80) and 49% (42 out of 85) RNA3-RNA2 recombinants. We emphasize that these numbers may not be significant and require a more systematic approach to verify such a model. In this respect it is of interest that stable tetraloop structures have been implicated in the separation of plus and minus strands during RNA phage replication (1). Also at the DNA level, small stem-loops have been shown to be capable of extruding from double-stranded structures (11).
In addition to precise homologous RNA3-RNA2 recombinants, we identified the imprecise variants that contained either single-nucleotide repeats, insertions, or deletions within the crossover region. The low incidence (in 3 to 7% of recombinants) of short repeats of the U residues was observed in experiments A and C (RNA2 and -3 bearing the structured or unstructured positive-strand inserts, respectively). This suggests that the positive strands participate in controlling the precision of homologous crossovers. However, the accuracy of homologous crosses also depended on other factors, because a higher fraction (22 to 40%) of imprecise recombinants accumulated while using single RNAs that carried the insert (experiments E, H, J, and K). This might be due to the reduced length of sequence identity between region R sequences. In these experiments the parental RNA2 carried either the structured positive or unstructured negative inserts, or conversely, the parental RNA3 carried the unstructured positive or structured negative inserts. Although a mechanistic explanation for these correlations remains unclear, it confirms that the precision depends on both donor and acceptor sequences.
Besides RNA3-RNA2 recombinants, the main topic of this work, the accumulation of nontargeted RNA3-RNA1 recombinants was observed (Fig. 3). Interestingly, the negative-strand stem-loop inserts either in both RNA3 and RNA2 or in only RNA2 increased the fraction of RNA3-RNA1 recombinants. This confirms the role of the RNA2 negative-strand stem-loops in the reattachment process, such that the released RdRps could jump on less-homologous RNA1 sequences. Previously, less dramatic shifts from RNA2 to the RNA1 acceptor have been reported (32). In these experiments decreasing sequence homology between RNA2 and -3 by the introduction of nucleotide substitutions in region R of RNA3 led to a higher fraction of RNA3-RNA1 recombinants (from 6 to 35%).
We also identified RNA3-RNA3 recombinants. They were nonhomologous in nature and arose either by single or double crossovers (Fig. 4), i.e., either by inter- or intramolecular recombination. These recombinants differed from previously described homologous RNA3-RNA3 recombinants within the intercistronic region (2, 13). Comparable to RNA3-RNA2 recombinants, structured negative stem-loops either in both RNA3 and RNA2 or in only RNA3 (experiments B and K, respectively) seemed to inhibit RNA3-RNA3 recombination. This suggests that RNA3-RNA3 recombinants also are generated during positive-strand synthesis, as negative-strand stem-loops and not positive-strand stem-loops in RNA3 inhibited the production of RNA3-RNA3 recombinants.
Major effects on BMV RNA3-RNA2 recombination were observed at the negative-strand level, despite earlier suggestions that RNA recombination occurred among positive strands (5, 6), allegedly because more positive strands are made during RNA virus life cycle (3). BMV RdRp was shown to switch between positive strands in vitro (14). In fact, for some viruses RNA recombination seems to prefer the positive strands, for others the negative strands, and for yet others it can occur between either strand (5, 6, 28, 29, 42). The final recombinant profiles likely depend on the combination of several factors, of which the switching properties of the RdRp replication complex and the properties of nucleotide sequences play an important role. Further experiments are required to determine whether different RNA sequences and/or different stem-loops can support the described patterns of the intersegmental BMV RNA recombination.
Acknowledgments
R.C.L.O. and A.B. contributed equally to this work.
This work was supported by a grant from the National Science Foundation (MCB-9983033) and by the Polish government through a grant (6 P04C 046 19) from the State Committee for Scientific Studies, awarded to J.J.B., and by the Plant Molecular Biology Center at Northern Illinois University. R.C.L.O. was financed by a TALENT stipend awarded by The Netherlands Organization for Scientific Research.
We thank R. Wierzchoslawski for valuable comments and discussions during the final stages of putting this paper together and L. Bujarski for editorial improvements.
REFERENCES
- 1.Beekwilder, M. J., R. Nieuwenhuizen, and J. van Duin. 1995. Secondary structure model for the last two domains of single-stranded RNA phage Q beta. J. Mol. Biol. 247:903-917. [DOI] [PubMed] [Google Scholar]
- 2.Bruyere, A., M. Wantroba, S. Flasinski, A. Dzianott, and J. J. Bujarski. 2000. Frequent homologous recombination events between molecules of one RNA component in a multipartite RNA virus. J. Virol. 74:4214-4219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buck, K. W. 1996. Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv. Virus Res. 47:159-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bujarski, J. J. 1998. Bromovirus isolation and RNA extraction. Methods Mol. Biol. 81:183-188. [DOI] [PubMed] [Google Scholar]
- 5.Bujarski, J. J. 1996. Experimental systems of genetic recombination and defective RNA formation in RNA viruses. Part I. Semin. Virol. 7:361-362. [Google Scholar]
- 6.Bujarski, J. J. 1997. Experimental systems of genetic recombination and defective RNA formation in RNA viruses. Part II. Semin. Virol. 8:75-76. [Google Scholar]
- 7.Bujarski, J. J., P. Nagy, and S. Flasinski. 1994. Molecular studies of genetic RNA-RNA recombination in brome mosaic virus. Adv. Virus Res. 43:275-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bujarski, J. J., and P. Kaesberg. 1986. Genetic recombination between RNA components of a multipartite plant virus. Nature 321:528-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cascone, P. J., C. D. Carpenter, X. H. Li, and A. E. Simon. 1990. Recombination between satellite RNAs of turnip crinkle virus. EMBO J. 9:1709-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chetverin, A. B., H. V. Chetverina, A. A. Demidenko, and V. I. Ugarov. 1997. Nonhomologous RNA recombination in a cell-free system: evidence for a transesterification mechanism guided by secondary structure. Cell 88:503-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai, X., M. Kloster, and L. B. Rothman-Denes. 1998. Sequence-dependent extrusion of a small DNA hairpin at the N4 virion RNA polymerase promoters. J. Mol. Biol. 283:43-58. [DOI] [PubMed] [Google Scholar]
- 12.Deng, L., and S. Shuman. 1997. Elongation properties of vaccinia virus RNA polymerase: pausing, slippage, 3′ end addition, and termination site choice. Biochemistry 36:15892-15899. [DOI] [PubMed] [Google Scholar]
- 13.Dzianott, A., S. Flasinski, S. Pratt, and J. J. Bujarski. 1995. Foreign complementary sequences facilitate genetic recombination in brome mosaic virus. Virology 208:370-375. [DOI] [PubMed] [Google Scholar]
- 14.Dzianott, A., N. Rauffer-Bruyere, and J. J. Bujarski. 2001. Studies on functional interaction between brome mosaic virus replicase proteins during RNA recombination, using combined mutants in vivo and in vitro. Virology 289:137-149. [DOI] [PubMed] [Google Scholar]
- 15.Figlerowicz, M., P. D. Nagy, and J. J. Bujarski. 1997. A mutation in the putative RNA polymerase gene inhibits nonhomologous, but not homologous, genetic recombination in an RNA virus. Proc. Natl. Acad. Sci. USA 94:2073-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figlerowicz, M., P. D. Nagy, N. Tang, C. C. Kao, and J. J. Bujarski. 1998. Mutations in the N terminus of the brome mosaic virus polymerase affect genetic RNA-RNA recombination. J. Virol. 72:9192-9200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Figlerowicz, M., and J. J. Bujarski. 1998. RNA recombination in brome mosaic virus, a model plus strand RNA virus. Acta Biochim. Pol. 45:847-868. [PubMed] [Google Scholar]
- 18.Figlerowicz, M., and A. Bibillo. 2000. RNA motifs mediating in vivo site-specific nonhomologous recombination in positive RNA virus enforce in vitro nonhomologous crossovers with HIV-1 reverse transcriptase. RNA 6:339-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heus, H. A., and A. Pardi. 1991. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science 253:191-194. [DOI] [PubMed] [Google Scholar]
- 20.Huang, J., L. G. Brieba, and R. Sousa. 2000. Misincorporation by wild type and mutant T7 RNA polymerases: identification of interactions that reduce misincorporation rates by stabilizing the catalytically incompetent open conformation. Biochemistry 39:11571-11580. [DOI] [PubMed] [Google Scholar]
- 21.Jager, J., and J. D. Pata. 1999. Getting a grip: polymerases and their substrate complexes. Curr. Opin. Struct. Biol. 9:21-28. [DOI] [PubMed] [Google Scholar]
- 22.Janda, M., R. French, and P. Ahlquist. 1987. High efficiency T7 polymerase synthesis of infectious RNA from cloned brome mosaic virus cDNA and effects of 5′ extensions of transcript infectivity. Virology 158:259-262. [DOI] [PubMed] [Google Scholar]
- 23.Jarvis, T. C., and K. Kirkegaard. 1991. The polymerase in its labyrinth: mechanisms and implications of RNA recombination. Trends Genet. 7:186-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirkegaard, K., and D. Baltimore. 1986. The mechanism of RNA recombination in poliovirus. Cell 47:433-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klovins, J., J. van Duin, and R. C. L. Olsthoorn. 1997. Rescue of the RNA phage genome from RNase III cleavage. Nucleic Acids Res. 25:4201-4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korzheva, N., A. Mustaev, M. Kozlov, A. Malhotra, V. Nikiforov, A. Goldfarb, and S. A. Darst. 2000. A structural model of transcription elongation. Science 289:619-625. [DOI] [PubMed] [Google Scholar]
- 27.Lahser, F. C., L. E. Marsh, and T. C. Hall. 1993. Contributions of the brome mosaic virus RNA-3 3′-nontranslated region to replication and translation. J. Virol. 67:3295-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lai, M. M. C. 1992. RNA recombination in animal and plant viruses. Microbiol. Rev. 56:1-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, Y., and L. A. Ball. 1993. Nonhomologous RNA recombination during negative-strand synthesis of flock house virus RNA. J. Virol. 67:3854-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Makino, S., C.-K. Shieh, L. H. Soe, S. C. Baker, and M. M. C. Lai. 1988. Primary structure and translation of a defective interfering RNA of murine coronavirus. Virology 166:550-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nagy, P. D., and J. J. Bujarski. 1993. Targeting the site of RNA-RNA recombination in brome mosaic virus with antisense sequences. Proc. Natl. Acad. Sci. USA 90:6390-6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagy, P. D., and J. J. Bujarski. 1995. Efficient system of homologous RNA recombination in brome mosaic virus: sequence and structure requirements and accuracy of crossovers. J. Virol. 69:131-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagy, P. D., and J. J. Bujarski. 1997. Engineering of homologous recombination hotspots with AU-rich sequences in brome mosaic virus. J. Virol. 71:3799-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagy, P. D., and J. J. Bujarski. 1998. Silencing homologous RNA recombination hot spots with GC-rich sequences in brome mosaic virus. J. Virol. 72:1122-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagy, P. D., A. Dzianott, P. Ahlquist, and J. J. Bujarski. 1994. Effect of mutations in the helicase-like domain of 1a protein on RNA-RNA recombination in brome mosaic virus. J. Virol. 69:2547-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nagy, P. D., and A. E. Simon. 1997. New insights into the mechanisms of RNA recombination. Virology 235:1-9. [DOI] [PubMed] [Google Scholar]
- 37.Nagy, P. D., C. Ogiela, and J. J. Bujarski. 1999. Mapping sequences active in homologous RNA recombination in brome mosaic virus: prediction of recombination hot spots. Virology 254:92-104. [DOI] [PubMed] [Google Scholar]
- 38.Nagy, P. D., C. Zhang, and A. E. Simon. 1998. Dissecting RNA recombination in vitro: role of RNA sequences and the viral replicase. EMBO J. 17:2392-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peliska, J. A., and S. J. Benkovic. 1992. Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Science 258:1112-1118. [DOI] [PubMed] [Google Scholar]
- 40.Rao, A. L., B. P. Sullivan, and T. C. Hall. 1990. Use of Chenopodium hybridum facilitates isolation of brome mosaic virus RNA recombinants. J. Gen. Virol. 71:1403-1407. [DOI] [PubMed] [Google Scholar]
- 41.Romero, J., Q. Huang, J. Pogany, and J. J. Bujarski. 1993. Characterizations of defective interfering RNA components that increase symptom severity of broad bean mottle virus infections. Virology 194:576-584. [DOI] [PubMed] [Google Scholar]
- 42.Rong, M., R. K. Durbin, and W. T. McAllister. 1998. Template strand switching by T7 RNA polymerase. J. Biol. Chem. 273:10253-10260. [DOI] [PubMed] [Google Scholar]
- 43.Roossinck, M. 1997. Mechanisms of plant-virus evolution. Annu. Rev. Phytopathol. 35:191-209. [DOI] [PubMed] [Google Scholar]
- 44.Tolskaya, E. A., L. I. Romanova, V. M. Blinov, E. G. Viktorova, A. N. Sinyakov, M. S. Kolesnikova, and V. I. Agol. 1987. Studies on the recombination between RNA genomes of poliovirus: the primary structure and nonrandom distribution of crossover regions in the genomes of intertypic poliovirus recombinants. Virology 161:54-61. [DOI] [PubMed] [Google Scholar]
- 45.Tsagris, M., M. Tabler, and H. L. Sanger. 1991. Ribonuclease T1 generates circular RNA molecules from viroid-specific RNA transcripts by cleavage and intramolecular ligation. Nucleic Acids Res. 19:1605-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uptain, S. M., and M. J. Chamberlain. 1997. Escherichia coli RNA polymerase terminates transcription efficiently at rho-independent terminators on single-stranded DNA templates. Proc. Natl. Acad. Sci. USA 94:13548-13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Meerten, D., H. Groeneveld, D. M. Miller, G. B.Marechal, N. V. Tsareva, R. C. L. Olsthoorn, M. de la Pena, and J. van Duin. 2002. In vivo generation of hybrids between different species of RNA phages. J. Gen. Virol. 83:1223-1235. [DOI] [PubMed] [Google Scholar]
- 48.von Hippel, P. H. 1998. An integrated model of the transcription complex in elongation, termination, and editing. Science 281:660-665. [DOI] [PubMed] [Google Scholar]
- 49.White, K. A., and T. J. Morris. 1995. RNA determinants of junction site selection in RNA virus recombinants and defective interfering RNAs. RNA 1:1029-1040. [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson, K. S., and P. H. von Hippel. 1995. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc. Natl. Acad. Sci. USA 92:8793-8797. [DOI] [PMC free article] [PubMed] [Google Scholar]