Abstract
Force-induced changes in genome expression as well as remodeling of nuclear architecture in development and disease motivate a deeper understanding of nuclear mechanics. Chromatin and green fluorescent protein-lamin B dynamics were visualized in a micropipette aspiration of isolated nuclei, and both were shown to contribute to viscoelastic properties of the somatic cell nucleus. Reversible swelling by almost 200% in volume, with changes in salt, demonstrates the resilience and large dilational capacity of the nuclear envelope, nucleoli, and chromatin. Swelling also proves an effective way to separate the mechanical contributions of nuclear elements. In unswollen nuclei, chromatin is a primary force-bearing element, whereas swollen nuclei are an order of magnitude softer, with the lamina sustaining much of the load. In both cases, nuclear deformability increases with time, scaling as a power law—thus lacking any characteristic timescale—when nuclei are either aspirated or indented by atomic force microscopy. The nucleus is stiff and resists distortion at short times, but it softens and deforms more readily at longer times. Such results indicate an essentially infinite spectrum of timescales for structural reorganization, with implications for regulating genome expression kinetics.
INTRODUCTION
Genome expression in eukaryotic cells is influenced by higher order chromatin structure, nuclear compartmentalization, and other regulatory factors that are all physically contained by the nuclear envelope. The nuclear envelope separates the chromatin from the cytoplasm and consists of a double lipid bilayer plus a cortical protein lamina (1) attached by nuclear membrane proteins that stabilize the envelope and provide sites for chromatin binding and organization (2–5). The lamina comprises intermediate lamin filaments (6) as well as putative accessory proteins (7) and is a stiff but expandable network when compared to other cell membrane networks (8). At least 10 diseases, known as laminopathies, have been linked to mutations in lamins or lamin-binding proteins (4). Proposed mechanisms include mechanical weakness of the nuclear envelope coupled to defects in tissue-specific gene expression (9–11). Separating the roles of lamins and chromatin is vital to understanding nuclear mechanics and its implications for function.
Cells respond to mechanical cues from their environment by altering genome expression (12,13). Although the exact mechanisms governing force transduction are unknown, the relative stiffness of the nucleus and lamins should regulate the effects of cellular forces on chromatin or genome expression. Previously measured mechanical properties have not separated the relative contributions of the load-bearing structures of the somatic nucleus. Force transmission to nuclear subdomains such as nucleoli and the relative involvement of the lamina, chromatin, etc. are critical for functional insights. Measurements of forces on nuclei within cells are likely to be skewed due to force redistribution by the cytoskeleton and force dissipation by other cytoplasmic factors.
Here, we begin to map out the basic mechanics of isolated nuclei over a large range of strains and timescales. The goal is to separately characterize the contributions of the envelope, chromatin, and subnuclear compartments to nuclear viscoelasticity. The general result that emerges here from both micropipette aspiration (MPA) and atomic force microscopy (AFM) is that the nucleus and its components exhibit power-law rheology, whether the deformations are small or large, tensile or compressive. Reversible nuclear swelling with divalent salt alters the effective chromatin contribution and shows that intranuclear domains are also strained. Swelling provides a means of shifting the load from chromatin dominated to lamina dominated, and swollen nuclei are more compliant compared to normal isolated nuclei. Overall, the deformation behavior and power-law rheology indicate an extremely broad range of timescales that underlie reversible changes in nuclear structure.
MATERIALS AND METHODS
Cell culture
TC7 cells (14), a subline of African green monkey kidney epithelium, were a kind gift of M. Goulian (University of Pennsylvania, Philadelphia, PA). They were cultured under standard conditions in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum, 1% L-glutamine, and 0.5% penicillin-streptomycin (Invitrogen, Carlsbad, CA) at 37°C in 5% CO2.
Transfections
Cells were passaged to 50% confluence 24 h before transfection. A vector containing a green fluorescent protein (GFP) construct of Xenopus lamin B1 (15), the kind gift of D. M. Gilbert (State University of New York, Syracuse, NY), was transfected into TC7 cells using 5 μg DNA and 10 μL Lipofectoamine 2000 reagent (Life Technologies, Gaithersburg, MD) for 6 h, per manufacturers instructions. Cells were grown 1–2 days posttransfection to confluence, giving a high density of cells with a relatively low transfection efficiency (<20% transfected cells) for facile nuclear isolation.
Nuclear isolation
TC7 nuclei were isolated by a modification of a previously described protocol (14). Briefly, cells were cultured to confluence in T75 flasks. Cells were washed twice with PBS (Invitrogen) and once with 10 mM HEPES, 1 mM DTT (Sigma-Aldrich, St. Louis, MO) pH 7.5. Cells were scraped into 1 ml of the HEPES/DTT solution and allowed to equilibrate for 10 min on ice. After 25 strokes in a Dounce homogenizer, 0.9 ml of the cell lysate was added to a one-fifth volume of 5× STKMC (1.25 M sucrose, 250 mM Tris, pH 7.6, 125 mM KCl, 15 mM MgCl2, 15 mM CaCl2, 10 μg/ml each of pepstatin and leupeptin; Sigma-Aldrich) and incubated for 10 min on ice. A total of 2.3 M sucrose in TKMC (50 mM Tris, pH 7.6, 25 mM KCl, 3 mM MgCl2, 3 mM CaCl2, 2 μg/ml each leupeptin and pepstatin) was added to the cell lysate suspension. The 1.6 M sucrose or STKMC suspension was layered onto a 150 μL cushion of 2.3 M STKMC and spun at 166,000 × g in an Optima TLX ultracentrifuge (Beckman Coulter, Palo Alto, CA). Nuclei were resuspended in 200 μL TKMC or 200 μL Tris (10 mM Tris-HCl, pH 7.6, 2 μg/m each of pepstatin and leupeptin and 1% bovine serum albumin (BSA); Sigma-Aldrich).
Nuclear swelling
Varying concentrations of a 3 mM solution of EDTA were added to swell unswollen nuclei, and varying concentrations of 5× TKMC were added to shrink swollen nuclei. In some cases, DNase and RNase (Sigma-Aldrich) were added to both swollen and unswollen nuclei at a final concentration of 0.01 mg/ml each and incubated for 30 min.
DNA labeling
For DNA visualization, isolated nuclei were labeled with Hoechst 33342 or YO-PRO (both from Molecular Probes, Eugene, OR). For more quantitative DNA measures, nuclei were labeled with PICO-Green (Molecular Probes). Nucleoli were identified in cells fixed with methanol at 4°C by labeling nucleolin with tri-rhodamine isothiocyanate (TRITC) conjugated murine antibody C23 (MS-3; IgG1; Santa Cruz Biotechnology, Santa Cruz, CA).
Micropipette aspiration
Capillary tubes of 1.0 mm inner diameter (World Precision Instruments, Sarasota, FL) were pulled into micropipettes using a Flaming-Brown Micropipette Puller (Sutter Instrument, Novato, CA) and cut to various diameters using a deFonbrune-type microforge (Vibratome, St. Louis, MO). Micropipette diameters ranged from Rp = 1.86 to 3.53 μm for unswollen and shrunken nuclei and 3.40–4.71 μm for swollen nuclei. Micropipettes were attached to a dual-stage water manometer with reservoirs of adjustable height. Suction was applied by a syringe, and the corresponding pressure was measured by pressure transducer (Validyne, Northridge, CA) calibrated by a mercury U-tube manometer. Pressures for different experiments ranged from 1.9 to 20 kPa for unswollen and shrunken nuclei and 0.66 to 3.6 kPa for swollen nuclei. Before nuclear aspiration, large volumes (50–100 μL) of the appropriate buffer were aspirated into the micropipette.
Image acquisition
Swelling and MPA images were acquired on a Nikon TE300 inverted microscope with a 60× (oil, 1.4 numerical aperture) objective and recorded on videotape with a video camera, JE8242 (Javelin Systems, Torrance, CA). Images were captured from video, and sizes were measured using Scion Image (Scion Corporation, Frederick, MD).
AFM and sample preparation
Nuclei were allowed to adhere to a glass slide using a method established by Hategan and co-workers (16). A small quantity (100 μl) of isolated normal, swollen, and shrunken nuclei was washed in their corresponding buffer solutions and allowed to adhere to each poly-L-lysine-coated glass slide for 10 min. The force-distances curves of adherent, spread nuclei were acquired at room temperature with an Asylum 1-D AFM (Asylum Research, Santa Barbara, CA). The nuclei were tested in the appropriate buffer with borosilicate sphere tipped cantilevers (R = 2.5 μm; Bioforce Nanoscience, Ames, IA) having spring constants of ∼60 pN/nm, which were confirmed by a thermal fluctuations method. Indentation velocity of the cantilever was varied between 0.1 and 100 μm/s, which changed the corresponding sample contact time over a similar range. Indentation (δ) is obtained by subtracting the deflection (d) to the movement of the piezoelectric ceramic (Δz = z − z0) in the z direction, where z0 is the contact point. The contact point (z0) was determined following the modified method proposed by Domke and Radmacher (17), using an automatic least-square fitting procedure in the deflection range 10 nm–100 nm.
AFM analysis
For each force-indentation profile, the apparent Young's modulus was determined by fitting the indentation curve using the finite thickness corrected Hertz sphere model (18). Nuclear height was estimated by assuming an adherent hemisphere of the same average volume as nuclei suspended under the same solution condition. Nuclear volume was approximated from the projected area determined by microscopy (see Fig. 2 C), assuming the nucleus to be a sphere. These geometrical assumptions yield estimated heights without the use of AFM imaging, which would tend to distort the nucleus and alter mechanical results. Indentations up to 10% of the estimated sample height were fit (e.g., effective strains <10%). Since the samples were highly hydrated, ν is taken equal to 0.5. A mean value of E was calculated by averaging over numerous locations and nuclei within a given sample. Contact time was calculated from the contact distance and multiplying by the indentation velocity.
FIGURE 2.
Isolated nuclei swell with changes of divalent salts. (A) Isolated nuclei attached by polylysine to a coverslip show dilation and recovery of the nucleus prelabeled with YO-PRO, with a heterogeneous label. (B) Nuclei free in solution swell to 170% of their original projected area with the chelation of salt from the media with EDTA. Adding back divalent salt returns nuclei to their original size. (C) The projected area of native isolated nuclei (▪) increases linearly with the addition of EDTA (□). Conversely, nuclei isolated into media with no salt (•) shrink linearly with addition of concentrated salts (○). DNase increases the projected area of both swollen and unswollen nuclei, indicating that the chromatin network contributes to regulation of nuclear size.
RESULTS
MPA and the creep compliance of isolated nuclei
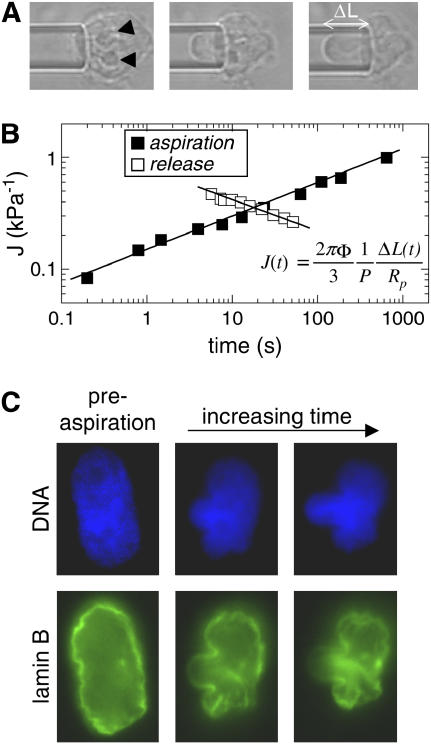
Nuclei were isolated from cultured TC7 cells into TKMC buffer (Tris buffer, potassium, magnesium, and calcium; see Materials and Methods) developed previously for efficient isolation of viable, active nuclei (14). The creep compliance—a ratio of strain to stress as a function of time—of an aspirated nucleus was measured by applying a step pressure and following the projection of the nucleus into the micropipette as a function of time (Fig. 1 A). The portion of the nucleus outside of the micropipette could not be reliably measured by the two-dimensional (2D) projection due to surface roughness, so it was treated as an infinite half space. The creep compliance, J, was determined from a variation of the equation developed by Theret and co-workers as a solution for aspiration of an infinite half space (19):
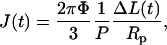 |
(1) |
where Φ depends on micropipette wall thickness (here, Φ = 2.1 as per Theret et al. (19)), P is the constant pressure applied, and ΔL is the aspirated length normalized to the pipette radius, Rp. A similar equation to Eq. 1 has been used for viscoelastic property determinations (20), but the simple modification here gives a general form of creep as a function of time while making no a priori assumption of springs or dashpots. On a log-log plot, J(t) shows a power law over several decades (Fig. 1 B).
FIGURE 1.
MPA of isolated nuclei shows power-law creep behavior dominated by the DNA. (A) An isolated nucleus aspirated into a micropipette of 6 μm inner diameter at a constant applied pressure of 15 kPa shows deformation of the nucleus as well as the nucleoli (arrowheads). (B) Creep compliance, J, as a function of time shows power-law behavior over four decades with an exponent α ≈ 0.3. When the pressure is released, the projection length decreases with the same exponent. (C) Aspirated nuclei, with GFP-lamin B and Hoechst 33342-labeled DNA, show the relative contributions under strain. The DNA appears to percolate throughout the nucleus and fill the micropipette, and lamins appear to be dominated by an internal network which causes buckling at sites exterior to the micropipette.
When the aspiration pressure was released, the nuclei also showed creep behavior during relaxation out of the pipette. The initial reverse creep showed a similar power-law exponent as during aspiration. Although nuclei did not expel completely from the micropipette in every case, any irreversibility observed seems due more to adhesion of the nuclei to the inner wall of the micropipette than nuclear plasticity. AFM data, described below, also did not show any obvious plastic deformation after repeated indentation.
Limitations to MPA and analysis
To limit adhesion to glass during MPA, BSA was added to the suspending solutions—per work with cells (e.g., Discher et al. (21))—and the solution was aspirated into the micropipettes before nuclear aspiration. Adhesion during aspiration would tend to give, of course, an overestimate of nuclear stiffness since adhesion would limit stretching inside the pipette. As evidence that adhesion was minimal, decreasing the aspiration pressure to zero after aspiration showed that the nuclei crept most of the way out of the micropipette.
Although Eq. 1 assumes an infinite half space outside of the micropipette, the ratio of nuclear size to pipette size (2 > Dnucleus/Dpipette > 5) ensures that a significant amount of the nucleus is at least initially excluded from the pipette during aspiration. The continuity of J(t) from early to late times and an apparent lack of (strong) dependence of J(t) on micropipette diameter suggest that stretching of the nucleus provides the majority of the resistance to aspiration. In addition, fluorescence visualization of the lamina below clearly shows nuclear stretching in the pipette.
Equation 1 was developed for small strains, but experiments with large deformations (ΔL) into large radii pipettes (Rp) yield similar J(t) to small deformation into smaller radii pipettes. Likewise, similar results for different aspiration pressures suggest that Eq. 1 is adequate for a first analysis of these types of experiments. Variations among experiments on all nuclei fit within a coefficient of variation of 20%–40%.
Chromatin provides a majority of the resistance deformation in unswollen nuclei
Nuclei isolated from GFP-lamin B transfected cells were labeled with Hoechst 33342 for dual visualization of DNA and lamins during aspiration (Fig. 1 C). The aspirated lamina, shown in green and primarily located at the periphery of the nucleus, appeared to deform together with the DNA. Increased aspiration pressure (or increased time) caused greater deformation and induced the lamina external to the pipette to roughen and buckle. This suggests a strong mechanical coupling of the lamina throughout the nucleoplasm via distinct internal foci rather than via cortical tension alone; the latter would otherwise cause the lamina to become smoother as seen in the swollen case (below). The internal foci likely reflect chromatin tethering elements or chromatin entanglements, and they appear to be easily disrupted by nuclear swelling.
Nuclear swelling reduces chromatin density
To determine if chromatin dominates the viscoelastic resistance to aspiration, nuclei were reversibly swollen through salt changes that increase the volume of the nucleus and thus dilute the DNA. Nuclei were isolated into TKMC buffer, and EDTA was added to chelate divalent salt in two different ways (Fig. 2, A and B). Nuclei were labeled with a fluorescent dye YO-PRO, which showed spotty fluorescence within the nucleus, highlighting smaller, subnuclear domains, which colocalized with nucleolar antibody staining in fixed cells (Supplemental Fig. 1). These nuclei were adhered to a polylysine-coated coverslip to prevent convection away from the surface during addition of EDTA solution. Upon addition, nuclear and nucleolar swelling was followed over time (Fig. 2 A). Adhered nuclei swelled 87(±19)% of their original projected area, and the YO-PRO labeled nucleoli swelled 235(±44)%. Nuclear and nucleolar swelling could be mostly reversed by adding back divalent salts, with slight deviations from the original projected area that are likely due to adhesion affects.
To eliminate possible constraints on size due to adhesion, large numbers of nuclei were swollen in solution and their projected area was measured (Fig. 2 B, quantified in Fig. 2 C). Again, the removal of salt swelled nuclei, but with an average increase of 170% in projected area. For a simple sphere, this translates to a relative change in volume of (170%)3/2 = 222%, which corresponds to a >2-fold dilution of chromatin. Conversely, “swollen” nuclei isolated into Tris buffer alone could be incrementally shrunken by adding small volumes of concentrated salts to return the buffer to the normal “unswollen” salt concentration (Fig. 2, B and C). Adding more salt to unswollen nuclei (to twice the original salt concentrations) did not change their size (p-value = 0.72). Also, in the presence of the appropriate salts, drastically changing the Tris buffer by up to fivefold had no effect on nuclear size (data not shown, p-value = 0.51), so swelling does not appear to be a simple osmotic effect. Both unswollen and swollen nuclei could be further enlarged with the addition of DNase and RNase (Fig. 2 C). There are numerous direct and indirect connections of chromatin to the lamina (4), and the increased distension here in the DNase-treated nuclei suggests that severing DNA allows envelope expansion. Internal DNA structures, even when decondensed (in the swollen state), impose an internal tension on the lamina since the swollen nucleus can dilate even further when these structures are disrupted.
DNA in the swollen nuclei appeared diffuse as visualized by Hoechst 33342 staining (Fig. 3 A). Mean intensity of DNA labeled by PICO Green, an extremely sensitive probe for double stranded DNA, normalized by projected area remains statistically similar (Fig. 3 B, p-value > 0.15) despite the large changes in projected area (Fig. 2 C). DNA fluorescence (related directly to concentration) plotted against nuclear area shows a linear, inverse trend for several different conditions of nuclear swelling.
FIGURE 3.
With swelling, the lamina smoothes and the DNA distributes homogeneously. (A) When nuclei swell, the lamina remains intact and smoothes; lamina wrinkles reappear when nuclei shrink. (B) The DNA disperses when nuclei swell such that integrated DNA fluorescence (measured by PICO-green probe) remains constant during nuclear swelling.
Chromatin and lamina network in the swollen nuclei
The lamina network in swollen nuclei was again visualized with GFP-lamin B. In unswollen nuclei the lamina network showed a roughened or wrinkled peripheral fluorescence in Fig. 3 A. The lamina remained intact as the nuclei swelled and appeared much smoother after swelling. When swollen nuclei were shrunken, the lamina network reverted to its original, roughened state. Lamin overexpression did not significantly affect the size of the isolated nuclei (data not shown, p-value = 0.85). It is intriguing that the smoothed lamina in the swollen nucleus has an appearance more typical of that seen in intact cells. This morphological difference between an isolated nucleus and a nucleus in the cell is addressed in the discussion.
Fluorescence of DNA and the lamina network in aspirated swollen nuclei revealed considerably different behavior compared to unswollen nuclei (shown in Fig. 1 C). The nuclear lamina external to the micropipette smoothes upon aspiration (Fig. 4 A), indicating that the tension imposed by the micropipette distributes along the cortex. Further, the nuclear lamina shows an increased fluorescence at the entrance to the pipette and a reduced but finite fluorescence at the distended cap of the projection (Fig. 4 A, arrows). This fluorescence gradient from the entrance to the cap increases upon further aspiration and is reminiscent of the dilation in the red cell membrane skeleton, which shows similar fluorescent gradients upon aspiration (21).
FIGURE 4.
Swollen, aspirated nuclei are dominated by the lamina and are much less stiff than unswollen nuclei. (A) MPA of swollen nuclei show a smoothing of the lamina external to the pipette, indicating a 2D cortical network. Increased aspiration shows a nonuniform projection of lamina fluorescence distended from the micropipette entrance to the tip of the projection (arrow), indicating a stretched network. (B) Swollen nuclei are much less stiff (higher J) and show more elastic character (shallower slope) than the unswollen nuclei. Filled versus unfilled or crosshatched symbols represent different experiments to show variation between individual nuclei. The J of unswollen and shrunken nuclei are mostly indistinguishable. Power-law fits are summarized in Table 1.
Mechanics depend on swollen state of nuclei
Using the MPA techniques described earlier, unswollen nuclei, swollen nuclei, and shrunken nuclei were measured for their creep compliance J using Eq. 1 (Fig. 4 B). The creep compliance data, representing a variety of pipette diameters and pressures, proved to be best described by the power-law form:
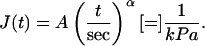 |
(2) |
Unswollen and shrunken nuclei showed similar power-law behaviors and statistically similar values for both prefactor A (p-value = 0.46) and exponent α (p-value = 0.17) (Table 1). Swollen nuclei also exhibited power-law behavior (Fig. 4 B) but with a higher A, suggesting softer nuclei, and a lower α, suggesting a more elastic than viscous response. For convenience and comparison to data obtained from AFM, the creep compliance was inverted to a time-dependent elastic stiffness (22), E:
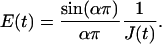 |
(3) |
This apparent elasticity measured by both MPA and AFM is reported in the form
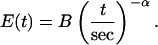 |
(4) |
The prefactor B (in units of kPa) and exponent α are listed in Table 1 for MPA and AFM measurements. Note that differences between methods are less than a factor of two.
TABLE 1.
Creep prefactor (A), complex elasticity prefactor (B), and exponent (α) of the power-law form of the creep compliance of unswollen (native), swollen, and shrunken nuclei as measured by MPA compared with AFM measurements (mean ± SD)
| Unswollen | Swollen | Shrunken | ||
|---|---|---|---|---|
| MPA | A (kPa−1) | 0.16 ± 0.06 | 1.11 ± 0.33 | 0.14 ± 0.06 |
| B (kPa) | 5.72 ± 2.15 | 0.90 ± 0.26 | 6.99 ± 2.81 | |
| α | 0.32 ± 0.07 | 0.21 ± 0.05 | 0.28 ± 0.05 | |
| AFM | B (kPa) | 2.80 ± 0.10 | N/A | 3.09 ± 0.06 |
| α | 0.20 ± 0.02 | N/A | 0.19 ± 0.02 |
AFM measurements show similar mechanical trends
Mechanical properties of nuclei at various levels of swelling were also measured by AFM (Supplemental Fig. 2). AFM, which has long been accepted for measurement of molecular forces, also continues to emerge as a method for determining elastic properties of cells (17,18,23) and macroscopic (24) samples. With the isolated nuclei here, more than 10 locations were typically probed per nucleus, with several nuclei tested per sample (n = 3). The reproducibility of nuclear indentations both with increasing and decreasing indentation velocity suggests no rate-dependent plastic deformations.
Data were fit to a modified Hertz sphere model that was corrected for sample height (17,18,23). We assumed an adherent hemisphere (6.8 μm measured for unswollen and shrunken nuclei, 8.4 μm for swollen nuclei, see Materials and Methods). Moduli from each sample were averaged and plotted against the AFM tip's contact time. The data fit well to a power-law model (r2 = 0.978, unswollen and r2 = 0.916, shrunken) similar to the MPA experiments as shown in Eq. 4. Table 1 summarizes the prefactor B and exponent α for the AFM-indented unswollen and shrunken nuclei.
Swollen nuclei were also measured by AFM, but the results appeared inconsistent with the three-dimensional (3D) analyses above. We hypothesize that the combination of nuclear swelling, due to disruption of internal cohesion, and strong membrane adhesion to the polylysine surface produces a highly tensed nuclear envelope. Analogous to red cells bound and tensed in adhesion to polylysine (16), AFM probing of the nuclear envelope appears resisted as much or more by this cortical tension than by the interior chromatin. It also shows no time dependence, seeming completely elastic (i.e., α = 0). By fitting the small indentation measurements on the tensed, swollen nucleus to a recently introduced membrane elasticity model (16), the membrane elastic modulus was determined to be K = 325 ± 150 mN/m. This 2D-dilational modulus is similar to the K = 390 mN/m determined recently for Xenopus oocyte nuclear envelopes by osmotic swelling (8) as well as to the elasticity expected for double bilayer lipid membranes (25).
It is possible that adhesion to the surface and subsequent tensing of the nuclear membrane could lead to overestimates of the elastic modulus of the unswollen nucleus compared to a freely suspended nucleus. However, since the AFM-imposed deformations yield a somewhat similar scaling of E with time as the MPA deformations, it seems that cortical membrane adhesion does not contribute significantly to mechanical properties. Since unswollen nuclei show some membrane and lamina buckling (Fig. 3 A) which smoothes and stretches upon swelling, this excess membrane may allow adhesion and spreading of the nuclei on the polylysine substrate without a pretensing of the membrane.
DISCUSSION
Physiological forces felt by cells and tissues undoubtedly span a broad range of timescales as do transcriptional processes and chromatin reorganization (26). Stress and strain fields within different tissues also vary greatly. The focus here has been on mechanical properties of isolated somatic nuclei probed over a wide range of times and strains. Swelling, distension in aspiration, and in situ fluorescent labeling were combined to develop an understanding of which structural elements—the lamina network or the chromatin—contribute to nuclear mechanics and integrity. At different salt concentrations and resulting nuclear dilation, the relevant mechanical elements shift and so does the measured stiffness (Fig. 5). The relative contributions provide novel and fundamental information about mechanical force transduction in the nucleus as well as needed insight into pathological processes associated with the lamina network.
FIGURE 5.
Changes in nuclear area, shape, or volume alter dominant mechanical elements and possibly influence genome access. As nuclei deform, changes in chromatin organization could expose new sites for transcriptional regulation. The reversibility of nuclear deformation, both by swelling and applied force, suggests a controlled mechanism to alter chromatin states.
Changes in genome expression and protein synthesis correlate with alterations in nuclear shape and architecture (27). Morphological changes in chromatin structure have also been observed during expression of individual genes (28). The ability of the nucleus to reversibly undergo large morphological changes may thus prove functionally important. A quantification of distortion and dilation of nuclear elements such as the lamina, chromatin, and nucleoli is an important step in connecting subnuclear forces to genome expression. Nuclei distort as cells are compressed or spread, changing interchromosomal interactions and possibly removing steric inhibitions to transcription factors (Fig. 5), thus leading to mechanically induced changes in genome expression.
Power-law behavior of isolated nuclei
Previous micromechanical studies of isolated nuclei have reported that nuclei are significantly stiffer than the extranuclear, cytoplasmic space of the cell. The effective elasticity of endothelial nuclei measured by parallel plate compression was reported, for example, to be 8 kPa compared to 0.5 kPa for the cytoplasm (29). MPA of chondrocyte nuclei gave static elastic moduli from 1 to 5 kPa, with data fit best by a three-parameter viscoelastic model (30). Our prefactor B ranges from 0.9 to 7 kPa (Table 1) and is similar to these elastic moduli. However, the viscous term in spring-dashpot models assumes a single time constant. The power-law rheology seen here, as in a number of cellular systems (31,32), rules out any single timescale.
Systems with power-law rheology include microgels and are metastable rather than equilibrated (33), possessing an essentially infinite number of intermediate conformations corresponding to an infinite number of relaxation modes or timescales. Nuclear elements at different length scales such as nucleosomes (34), chromatin fibers (35), and chromosomes (36) exhibit diverse populations of organization, forces, and energies within the nucleus. At each level, these nuclear elements seem likely to show intermediate conformations of mechanical relaxation, consistent with many modes or mechanisms of metastability that could, in turn, regulate genome expression kinetics.
Length scales and nuclear rheology
Here we have presented mechanics of whole nuclei under tension and compression on the approximate length scale of the entire nucleus. Previous studies have used particle tracking inside the nucleus to determine diffusive properties and also rheological properties. Although these studies have an obvious advantage in that they measure properties inside living cells, factors such as tracer size, tracer chemistry, cell type, and detection method influence the results. Diffusivities range from 1.1 μm2/s (37) to 40 μm2/s (38) with many reports in between (39,40). The elastic modulus of the nuclear interior measured by nanoparticle tracking has been reported to be 18 Pa (41), which is much lower than the numbers reported here even at the lowest salt conditions. The same study suggested the presence of microdomains that are ∼300 nm and are mostly absent outside the nucleus. Similarly, a broad distribution of movements reported by single molecule studies in the nucleus is not suggestive of simple diffusion (37) and may hint at microdomains in the nucleus. It is thus possible that studying the nucleus at the nanometer length scale shows considerably different behavior than at the whole-nucleus level. Further studies on both length scales using similar cell types and experimental conditions would provide interesting insight to the obvious importance of length scale in rheological measures in the nucleus.
Salt conditions affect nuclear size and shape
By changing external salt concentration, the size and shape of the isolated nuclei changed dramatically. With higher salt concentrations the nucleus was in an unswollen state and proved to be phase dense with a “wrinkled” or compressed peripheral lamina network. This morphology could be due to chromatin condensation by the salt that, in turn, buckles the lamina. The phenomenon could be similar to buckling of colloidal particles during isotropic shrinkage (42). It is also possible that the wrinkling results from the removal of stresses exerted by the cytoskeleton inside the cells. Cells lacking intermediate filament networks have large nuclear folds and invaginations similar to the buckling we observe here (43).
Isolating nuclei into media without salt or removing the salt with EDTA swelled the nuclei to 170% of their original size, reducing chromatin density and smoothing the lamina network. Nucleoli were similarly dilated by the changes in salt concentration, and dilation kinetics of nuclear and nucleolar swelling were similar. Previous work has shown the importance of cation concentration on nuclear turbidity and chromatin organization by electron microscopy (44,45), which is consistent with the loss of phase density and DNA concentration observed here in swollen nuclei.
Depleting the extranuclear media of salt, at least divalent cations, causes a concentration gradient of salt across the nuclear envelope and drives an efflux of salt. There is no evidence for simple osmotic effects since dramatic changes in Tris buffer concentration did not change nuclear size. The change in salt concentration inside the nucleus most likely “remodels” the chromatin. Although DNA can unwind from histones at differing salt concentrations (46), the completely reversible nature of the nuclear swelling does not fully support this theory as unbound histones could be lost or mislocalized. A more plausible explanation is that changes in chromatin organization are induced by chromatin tethering elements, possibly internucleosomal interactions, recognized in studies with individual chromatin fibers (35,36). Like the reversibility of mechanically (36) and chemically (47) extended chromosomes, the nucleus is seen here to return to its original unswollen state, suggesting a high degree of quaternary organization. The return (or not) of functional organization in reversibly swollen nuclei requires further investigation.
Nuclear swelling and applied force
Fluorescence of the nuclear lamina network and chromatin in unswollen nuclei during aspiration indeed shows a dominance of the chromatin. Buckling of the distal lamina also reveals significant 3D interior attachments throughout the nucleus. These interior attachments may represent a nuclear matrix (48) or similar 3D network of proteins (49), but they could also be explained by chromatin attachments (50).
The swollen nucleus, in contrast, shows a more cortical or peripheral 2D strain as seen in Fig. 4 and as taken to a limit in the AFM analyses of swollen and tensed nuclei. This apparent shift from entirely 3D to a more substantial 2D contribution suggests that when higher order chromatin organization is disrupted by dilution, the lamina network is able to provide significant, if reduced, resistance to deformation. These results with swollen nuclei thus suggest a stiff lamina network that provides significant, if reduced, resistance to distortion when the chromatin is not compacted. The possibility of multiple load-bearing elements suggests that the nucleus with chromatin and lamina is a redundant and robust system. Quantitative MPA measurements of swollen nuclei show a slightly decreased α, suggesting a shift toward simple elasticity (i.e., more solid-like). This result is consistent with the model of swelling proposed here (Fig. 5), since swelling allows for easier chromatin reorganization and a tendency toward elastic stretching. Recent studies on the nuclear envelope of Xenopus oocyte nuclei in which chromatin is dilute and not a structural factor also show the lamina to be a stiff, purely elastic network (8). Unlike the Xenopus system, however, the somatic nuclei here always show a significant viscoelastic character that undoubtedly reflects the higher density of chromatin.
Further, the fluorescence maps of the lamina network in aspirated swollen nuclei are reminiscent of studies of network reorganization and dilation of the red cell's spectrin cytoskeletal network (21,51). Fluorescence imaged microdeformation of ordered 2D cortical networks shows increased fluorescence intensity in regions of network compression, at the entrance to the pipette, and decreased fluorescent intensity of the cap, which is dilated due to curvature effects (51). Recent work by Rowat et al. highlights microdeformation of the lamina (52).
Importantly, mechanical properties change with nuclear swelling. Earlier measurements of isolated nuclei were made under low concentrations of divalent ions (30), suggesting that the “swollen” regime of the isolated nuclei was measured. Although a difference between cell types prevents direct comparison, swollen nuclei here tend to show a lower power-law exponent that, for smaller ranges of time, could appear to be modeled with a spring-dashpot. The choice of appropriate salt concentrations for use with isolated nuclei is not obvious. In this case, we chose previously suggested values for potassium, magnesium, and calcium (14). Physiological levels of salts such as calcium are uncertain, since free intracellular levels are extremely low, whereas bound levels in the cytoskeleton, the nucleus, and other organelles are millimolar (53) and vary with cell type (54,55). Given the uncertainty, we study the range of conditions (unswollen at high divalent salt concentration to swollen at essentially zero divalent salt) that effectively bracket the correct mechanical properties of nuclei inside the cell. Moreover, these results may hint at the effects or importance of altered divalent salt concentrations in cancerous nuclei (54).
MPA results here are given for times from one to hundreds of seconds; during which times tensile stresses are applied to the nucleus and significant strains result. In contrast, AFM compresses nuclei locally (∼1 μm) with contact times under 2 s. Despite the differences, results from both methods fit to a power-law form for complex elasticity as in Eq. 4. Prefactors, B, and exponents, α, for MPA and AFM experiments are presented in Table 1. The quantitative differences in the effective elastic moduli between these two methods are not large but most likely reflect differences in the mode of applied force (compression versus tension) as well as timescales and strain levels.
Implications of nuclear mechanics
The ability of the nucleus to undergo large changes in surface area and volume is extremely important, if uncertain, in physiological significance. During cellular compression similar to loading of a joint, for example, the nuclear volume changes by 10% and nuclei change shape (12). The nuclear swelling seen here illustrates that a nucleus can maximally increase its projected surface area by at least 70%. Although some of this change in area at the membrane and lamina may be due to unbuckling or unfolding, the reproducibility of the nuclear swelling and shrinking suggests a reversible reorganization within the nucleus. Similar large, reversible changes in nuclear envelope surface area have been seen in other model systems (8).
The power-law rheology observed here relates to hypothetical mechanotransduced changes in genome expression. At short times the complex stiffness, E(t), is very high and would effectively protect the nuclear interior from brief changes in applied stress: gene expression would not be affected by sudden, transient changes in forces. However, at longer times stiffness is reduced, allowing for easier nuclear deformation and possible alterations in genetic access, which is important for spreading and crawling cells or for cells strained under constant shear or compression. Over these longer timescales, cells would be able to adapt their gene expression to the environment.
Spatial positioning and chromatin movements have been shown to affect genome expression (56), and remodeling within the nucleus is governed by the overall nuclear stiffness, which is shown here to exhibit complex dynamics. Likewise, diffusion of transcription factors and transcriptional machinery is affected by the fluid-like nature of the nuclear interior. The timescales of genome expression vary at many levels of nuclear organization from polymerase activity (tens of basepairs per second (57)), DNA exposure from the nucleosome (tens of seconds to minutes (58)), and chromatin movements to transcriptional centers (seconds to hours (59)). The time-invariant nature of nuclear and subnuclear movements described by power-law rheology suggests a possible mechanical regulation genome expression kinetics. Further studies are underway to study rheology of nuclei from other cell types (K. N. Dahl) as well as nuclear deformation inside of intact cells (J. D. Pajerowski).
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
The authors acknowledge the technical assistance of M. Tewari. We sincerely appreciate K. L. Wilson (Johns Hopkins, Baltimore, MD), B. D. Hoffman, J. C. Crocker, and S. Sen (University of Pennsylvania) for helpful discussions. We thank D. M. Gilbert (State University of New York, Syracuse, NY) for the GFP-lamin-B constructs, and M. Goulian (University of Pennsylvania) for the TC7 cells.
Funding from the Whitaker Foundation (K.N.D.) and the Muscular Dystrophy Association, National Science Foundation, and National Institutes of Health (D.E.D.) is gratefully acknowledged.
References
- 1.Gruenbaum, Y., R. D. Goldman, R. Meyuhas, E. Mills, A. Margalit, A. Fridkin, Y. Dayani, M. Prokocimer, and A. Enosh. 2003. The nuclear lamina and its functions in the nucleus. Int. Rev. Cytol. 226:1–62. [DOI] [PubMed] [Google Scholar]
- 2.Moir, R. D., T. P. Spann, and R. D. Goldman. 1995. The dynamic properties and possible functions of nuclear lamins. Int. Rev. Cytol. 162B:141–182. [DOI] [PubMed] [Google Scholar]
- 3.Stuurman, N., S. Heins, and U. Aebi. 1998. Nuclear lamins: their structure, assembly, and interactions. J. Struct. Biol. 122:42–66. [DOI] [PubMed] [Google Scholar]
- 4.Burke, B., and C. L. Stewart. 2002. Life at the edge: the nuclear envelope and human disease. Nat. Rev. Mol. Cell Biol. 3:575–585. [DOI] [PubMed] [Google Scholar]
- 5.Gant, T. M., and K. L. Wilson. 1997. Nuclear assembly. Annu. Rev. Cell Dev. Biol. 13:669–695. [DOI] [PubMed] [Google Scholar]
- 6.Aebi, U., J. Cohn, L. Buhle, and L. Gerace. 1986. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 323:560–564. [DOI] [PubMed] [Google Scholar]
- 7.Sasseville, A. M., and Y. Langelier. 1998. In vitro interaction of the carboxy-terminal domain of lamin A with actin. FEBS Lett. 425:485–489. [DOI] [PubMed] [Google Scholar]
- 8.Dahl, K. N., S. M. Kahn, K. L. Wilson, and D. E. Discher. 2004. The nuclear envelope lamina network has elasticity and a compressibility limit suggestive of a molecular shock absorber. J. Cell Sci. 117:4779–4786. [DOI] [PubMed] [Google Scholar]
- 9.Morris, G. E. 2001. The role of the nuclear envelope in Emery-Dreifuss muscular dystrophy. Trends Mol. Med. 7:572–577. [DOI] [PubMed] [Google Scholar]
- 10.Wilson, K. L. 2000. The nuclear envelope, muscular dystrophy and gene expression. Trends Cell Biol. 10:125–129. [DOI] [PubMed] [Google Scholar]
- 11.Zastrow, M. S., S. Vlcek, and K. L. Wilson. 2004. Proteins that bind A-type lamins: integrating isolated clues. J. Cell Sci. 117:979–987. [DOI] [PubMed] [Google Scholar]
- 12.Guilak, F. 1995. Compression-induced changes in the shape and volume of the chondrocyte nucleus. J. Biomech. 28:1529–1541. [DOI] [PubMed] [Google Scholar]
- 13.Papadaki, M., and S. G. Eskin. 1997. Effects of fluid shear stress on gene regulation of vascular cells. Biotechnol. Prog. 13:209–221. [DOI] [PubMed] [Google Scholar]
- 14.Dean, D. A., and H. Kasamatsu. 1994. Signal- and energy-dependent nuclear transport of SV40 Vp3 by isolated nuclei. Establishment of a filtration assay for nuclear protein import. J. Biol. Chem. 269:4910–4916. [PubMed] [Google Scholar]
- 15.Izumi, M., O. A. Vaughan, C. J. Hutchison, and D. M. Gilbert. 2000. Head and/or CaaX domain deletions of lamin proteins disrupt preformed lamin A and C but not lamin B structure in mammalian cells. Mol. Biol. Cell. 11:4323–4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hategan, A., R. Law, S. Kahn, and D. E. Discher. 2003. Adhesively-tensed cell membranes: lysis kinetics and atomic force microscopy probing. Biophys. J. 85:2746–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domke, J., S. Dannohl, W. J. Parak, O. Muller, W. K. Aicher, and M. Radmacher. 2000. Substrate dependent differences in morphology and elasticity of living osteoblasts investigated by atomic force microscopy. Colloids Surf. B Biointerfaces. 19:367–379. [DOI] [PubMed] [Google Scholar]
- 18.Dimitriadis, E. K., F. Horkay, J. Maresca, B. Kachar, and R. S. Chadwick. 2002. Determination of elastic moduli of thin layers of soft material using the atomic force microscope. Biophys. J. 82:2798–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Theret, D. P., M. J. Levesque, M. Sato, R. M. Nerem, and L. T. Wheeler. 1988. The application of a homogeneous half-space model in the analysis of endothelial cell micropipette measurements. J. Biomech. Eng. 110:190–199. [DOI] [PubMed] [Google Scholar]
- 20.Sato, M., D. P. Theret, L. T. Wheeler, N. Ohshima, and R. M. Nerem. 1990. Application of the micropipette technique to the measurement of cultured porcine aortic endothelial cell viscoelastic properties. J. Biomech. Eng. 112:263–268. [DOI] [PubMed] [Google Scholar]
- 21.Discher, D. E., N. Mohandas, and E. A. Evans. 1994. Molecular maps of red cell deformation: hidden elasticity and in situ connectivity. Science. 266:1032–1035. [DOI] [PubMed] [Google Scholar]
- 22.Lakes, R. S. 1999. Viscoelastic Solids. CRC Mechanical Engineering Series. CRC Press, Boca Raton, FL.
- 23.Heinz, W. F., and J. H. Hoh. 1999. Spatially resolved force spectroscopy of biological surfaces using the atomic force microscope. Trends Biotechnol. 17:143–150. [DOI] [PubMed] [Google Scholar]
- 24.Engler, A., L. Bacakova, C. Newman, A. Hategan, M. Griffin, and D. Discher. 2004. Substrate compliance versus ligand density in cell on gel responses. Biophys. J. 86:617–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rawicz, W., K. C. Olbrich, T. McIntosh, D. Needham, and E. Evans. 2000. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys. J. 79:328–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hager, G. L., A. K. Nagaich, T. A. Johnson, D. A. Walker, and S. John. 2004. Dynamics of nuclear receptor movement and transcription. Biochim. Biophys. Acta. 1677:46–51. [DOI] [PubMed] [Google Scholar]
- 27.Thomas, C. H., J. H. Collier, C. S. Sfeir, and K. E. Healy. 2002. Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc. Natl. Acad. Sci. USA. 99:1972–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsukamoto, T., N. Hashiguchi, S. M. Janicki, T. Tumbar, A. S. Belmont, and D. L. Spector. 2000. Visualization of gene activity in living cells. Nat. Cell Biol. 2:871–878. [DOI] [PubMed] [Google Scholar]
- 29.Caille, N., O. Thoumine, Y. Tardy, and J. J. Meister. 2002. Contribution of the nucleus to the mechanical properties of endothelial cells. J. Biomech. 35:177–187. [DOI] [PubMed] [Google Scholar]
- 30.Guilak, F., J. R. Tedrow, and R. Burgkart. 2000. Viscoelastic properties of the cell nucleus. Biochem. Biophys. Res. Commun. 269:781–786. [DOI] [PubMed] [Google Scholar]
- 31.Fabry, B., G. N. Maksym, J. P. Butler, M. Glogauer, D. Navajas, and J. J. Fredberg. 2001. Scaling the microrheology of living cells. Phys. Rev. Lett. 87:148102–148102-4. [DOI] [PubMed] [Google Scholar]
- 32.Tsai, M. A., R. S. Frank, and R. E. Waugh. 1993. Passive mechanical behavior of human neutrophils: power-law fluid. Biophys. J. 65:2078–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sollich, P. 1998. Rheological constitutive equation for a model of soft glassy materials. Phys. Rev. E. 58:738–759. [Google Scholar]
- 34.Bennink, M. L., S. H. Leuba, G. H. Leno, J. Zlatanova, B. G. de Grooth, and J. Greve. 2001. Unfolding individual nucleosomes by stretching single chromatin fibers with optical tweezers. Nat. Struct. Biol. 8:606–610. [DOI] [PubMed] [Google Scholar]
- 35.Cui, Y., and C. Bustamante. 2000. Pulling a single chromatin fiber reveals the forces that maintain its higher-order structure. Proc. Natl. Acad. Sci. USA. 97:127–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poirier, M., S. Eroglu, D. Chatenay, and J. F. Marko. 2000. Reversible and irreversible unfolding of mitotic newt chromosomes by applied force. Mol. Biol. Cell. 11:269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goulian, M., and S. M. Simon. 2000. Tracking single proteins within cells. Biophys. J. 79:2188–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Politz, J. C., E. S. Browne, D. E. Wolf, and T. Pederson. 1998. Intranuclear diffusion and hybridization state of oligonucleotides measured by fluorescence correlation spectroscopy in living cells. Proc. Natl. Acad. Sci. USA. 95:6043–6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wachsmuth, M., W. Waldeck, and J. Langowski. 2000. Anomalous diffusion of fluorescent probes inside living cell nuclei investigated by spatially-resolved fluorescence correlation spectroscopy. J. Mol. Biol. 298:677–689. [DOI] [PubMed] [Google Scholar]
- 40.Seksek, O., J. Biwersi, and A. S. Verkman. 1997. Translational diffusion of macromolecule-sized solutes in cytoplasm and nucleus. J. Cell Biol. 138:131–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tseng, Y., J. S. Lee, T. P. Kole, I. Jiang, and D. Wirtz. 2004. Micro-organization and visco-elasticity of the interphase nucleus revealed by particle nanotracking. J. Cell Sci. 117:2159–2167. [DOI] [PubMed] [Google Scholar]
- 42.Tsapis, N., E. R. Dufresne, S. S. Sinha, C. S. Riera, J. W. Hutchinson, L. Mahadevan, and D. A. Weitz. 2005. Onset of buckling in drying droplets of colloidal suspensions. Phys. Rev. Lett. 94:018302-1–018302-4. [DOI] [PubMed] [Google Scholar]
- 43.Sarria, A. J., J. G. Lieber, S. K. Nordeen, and R. M. Evans. 1994. The presence or absence of a vimentin-type intermediate filament network affects the shape of the nucleus in human SW-13 cells. J. Cell Sci. 107:1593–1607. [DOI] [PubMed] [Google Scholar]
- 44.Aaronson, R. P., and E. Woo. 1981. Organization in the cell nucleus: divalent cations modulate the distribution of condensed and diffuse chromatin. J. Cell Biol. 90:181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Engelhardt, M. 2004. Condensation of chromatin in situ by cation-dependent charge shielding and aggregation. Biochem. Biophys. Res. Commun. 324:1210–1214. [DOI] [PubMed] [Google Scholar]
- 46.Yager, T. D., C. T. McMurray, and K. E. van Holde. 1989. Salt-induced release of DNA from nucleosome core particles. Biochemistry. 28:2271–2281. [DOI] [PubMed] [Google Scholar]
- 47.Poirier, M. G., T. Monhait, and J. F. Marko. 2002. Reversible hypercondensation and decondensation of mitotic chromosomes studied using combined chemical-micromechanical techniques. J. Cell. Biochem. 85:422–434. [DOI] [PubMed] [Google Scholar]
- 48.Hancock, R. 2000. A new look at the nuclear matrix. Chromosoma. 109:219–225. [DOI] [PubMed] [Google Scholar]
- 49.Shumaker, D. K., E. R. Kuczmarski, and R. D. Goldman. 2003. The nucleoskeleton: lamins and actin are major players in essential nuclear functions. Curr. Opin. Cell Biol. 15:358–366. [DOI] [PubMed] [Google Scholar]
- 50.Maniotis, A. J., K. Bojanowski, and D. E. Ingber. 1997. Mechanical continuity and reversible chromosome disassembly within intact genomes removed from living cells. J. Cell. Biochem. 65:114–130. [PubMed] [Google Scholar]
- 51.Discher, D. E., and N. Mohandas. 1996. Kinematics of red cell aspiration by fluorescence-imaged microdeformation. Biophys. J. 71:1680–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rowat, A. C., L. J. Foster, M. M. Neilsen, M. Weiss, and J. H. Ipsen. 2005. Characterization of the elastic properties of the nuclear envelope. Journal of The Royal Society Interface. 2:63–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chandra, S., C. Fewtrell, P. J. Millard, D. R. Sandison, W. W. Webb, and G. H. Morrison. 1994. Imaging of total intracellular calcium and calcium influx and efflux in individual resting and stimulated tumor mast cells using ion microscopy. J. Biol. Chem. 269:15186–15194. [PubMed] [Google Scholar]
- 54.Tvedt, K. E., G. Kopstad, O. A. Haugen, and J. Halgunset. 1987. Subcellular concentrations of calcium, zinc, and magnesium in benign nodular hyperplasia of the human prostate: x-ray microanalysis of freeze-dried cryosections. Cancer Res. 47:323–328. [PubMed] [Google Scholar]
- 55.Dobi, A., and D. v Agoston. 1998. Submillimolar levels of calcium regulates DNA structure at the dinucleotide repeat (TG/AC)n. Proc. Natl. Acad. Sci. USA. 95:5981–5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Misteli, T. 2004. Spatial positioning; a new dimension in genome function. Cell. 119:153–156. [DOI] [PubMed] [Google Scholar]
- 57.Tolic-Norrelykke, S. F., A. M. Engh, R. Landick, and J. Gelles. 2004. Diversity in the rates of transcript elongation by single RNA polymerase molecules. J. Biol. Chem. 279:3292–3299. [DOI] [PubMed] [Google Scholar]
- 58.Protacio, R. U., K. J. Polach, and J. Widom. 1997. Coupled-enzymatic assays for the rate and mechanism of DNA site exposure in a nucleosome. J. Mol. Biol. 274:708–721. [DOI] [PubMed] [Google Scholar]
- 59.Vazquez, J., A. S. Belmont, and J. W. Sedat. 2001. Multiple regimes of constrained chromosome motion are regulated in the interphase Drosophila nucleus. Curr. Biol. 11:1227–1239. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.