Abstract
The NS1 protein of influenza A/WSN/33 virus is a 230-amino-acid-long protein which functions as an interferon alpha/beta (IFN-α/β) antagonist by preventing the synthesis of IFN during viral infection. In tissue culture, the IFN inhibitory function of the NS1 protein has been mapped to the RNA binding domain, the first 73 amino acids. Nevertheless, influenza viruses expressing carboxy-terminally truncated NS1 proteins are attenuated in mice. Dimerization of the NS1 protein has previously been shown to be essential for its RNA binding activity. We have explored the ability of heterologous dimerization domains to functionally substitute in vivo for the carboxy-terminal domains of the NS1 protein. Recombinant influenza viruses were generated that expressed truncated NS1 proteins of 126 amino acids, fused to 28 or 24 amino acids derived from the dimerization domains of either the Saccharomyces cerevisiae PUT3 or the Drosophila melanogaster Ncd (DmNcd) proteins. These viruses regained virulence and lethality in mice. Moreover, a recombinant influenza virus expressing only the first 73 amino acids of the NS1 protein was able to replicate in mice lacking three IFN-regulated antiviral enzymes, PKR, RNaseL, and Mx, but not in wild-type (Mx-deficient) mice, suggesting that the attenuation was mainly due to an inability to inhibit the IFN system. Remarkably, a virus with an NS1 truncated at amino acid 73 but fused to the dimerization domain of DmNcd replicated and was also highly pathogenic in wild-type mice. These results suggest that the main biological function of the carboxy-terminal region of the NS1 protein of influenza A virus is the enhancement of its IFN antagonist properties by stabilizing the NS1 dimeric structure.
Folding of proteins into highly ordered structures is often essential for their functions. This is especially critical for proteins which perform multiple functions. In addition, multifunctional proteins usually show a modular organization, with different domains responsible for their different functions. RNA viruses typically encode a small number of proteins which need to take over the host cellular machinery in order to generate new infectious viruses. Therefore, most viral proteins perform multiple functions required for optimal viral replication. The NS1 protein of influenza A virus, a negative-strand RNA virus, appears to be one of those multifunctional proteins. At least three functional domains have been described in this 230-amino-acid protein: an RNA binding domain, an eIF4GI binding domain, and an effector domain.
In vitro studies showed that the core sequence of the RNA binding domain of the NS1 protein is amino acids 19 to 38 (37). NS1 has been reported to bind to heterogeneous RNAs, including poly(A) RNA (39), viral genomic RNA (18, 29), the 5′ untranslated region of viral mRNAs (36), U6 (40) and U6atac snRNA (51), and double-stranded RNA (dsRNA) (16, 26). The eIF4GI binding domain requires amino acids 81 to 113. Binding of eIF4GI by the NS1 protein has recently been shown to facilitate the preferential translation of viral mRNAs (2). Previous reports suggested that the NS1 effector domain, with a core sequence of amino acids 134 to 161 (37), blocks host mRNA splicing (24), polyadenylation (32, 42), and nuclear export (7, 10, 37). For these inhibitory functions on host mRNA processing, the RNA binding activity of NS1 seems not to be required (24, 32). However, the biological significance of these NS1 functions in the context of an infectious virus has not been fully elucidated.
It has been established that the induction of interferon alpha/beta (IFN-α/β) synthesis and secretion represents one of the first antiviral (innate) responses of the host (44). IFN-α/β induces the transcriptional activation of many genes, some of which play essential roles in the host antiviral defense. The importance of IFNs can be gleaned from the fact that most viruses encode one or more factors to combat the IFN system of the host in order to launch productive infections (1, 14, 23). The influenza A virus NS1 protein is one of these virally encoded IFN antagonists. A mutant influenza virus which has a deleted NS1 gene, delNS1, was generated and found to replicate efficiently in IFN-α/β-deficient systems (12). Previous studies also demonstrated that infection of different cell types with the delNS1 virus, but not with the wild-type PR8 virus, induces high levels of IFN-α/β (45, 53). Furthermore, expression of the NS1 protein blocks dsRNA-, delNS1-, and Sendai virus-mediated activation of the IFN-β promoter (53). It was also demonstrated that expression of the NS1 protein prevents the virus- and dsRNA-mediated activation of NF-κB (53) and IRF-3 (45), both of which are key transcription factors for the induction of IFN-β (54).
delNS1 virus is attenuated in wild-type mice but is virulent in STAT1−/− (12) or PKR−/− mice (3), which lack key components of the IFN system, indicating that the main function of the NS1 protein is the inhibition of the IFN response. Overexpression of the first 73 amino acids of NS1, which contains its RNA binding domain, inhibits the induction of IFN-α/β (53), suggesting that the RNA binding activity of the NS1 protein plays a critical role for its IFN antagonist function. However, the role of the RNA binding, eIF4GI binding, and effector domains of the NS1 protein in inhibiting the IFN system in vivo and in viral pathogenicity has not been elucidated. A 12-amino-acid deletion in the NS1 protein (amino acids 66 to 77) resulted in a host range temperature-sensitive influenza virus mutant, CR43-3 virus (5). Strikingly, a single amino acid substitution at position 23 of the NS1 protein led to the phenotypic reversion of CR43-3 virus (48). Although the carboxy-terminal NS1 effector domain sequences are dispensable for viral growth in tissue culture (35), carboxy-terminal truncations of the NS1 protein of influenza A and B viruses result in attenuation in mice (8, 46). These observations indicate that the C-terminal sequences of the NS1 proteins contribute to the virulence of influenza viruses during infection.
Alterations in amino acid sequences, such as large truncations, could change the highly ordered structure of a protein. These changes may subsequently affect the functions of the remaining modules of a multifunctional protein. The dimerization or multimerization of the NS1 protein has been suggested to be a prerequisite for its ability to bind RNA (52). Although the RNA binding domain of the NS1 protein (contained within its first 73 amino acids) crystallized as a dimer (25), its effector domain was shown to significantly contribute to its dimerization in a yeast two-hybrid system (33). We have carried out experiments with rescued viruses to test the biological significance of this structural role of the NS1 effector domain for virus pathogenicity.
In this paper we demonstrate that while a deletion of the carboxy-terminal domains of the influenza A virus NS1 protein renders the mutant viruses attenuated, the replacement of these sequences with short heterologous dimerization domains from either yeast or Drosophila proteins restores the virulence of the viruses. These data suggest that one of the major roles of the carboxy-terminal sequences of the NS1 protein in vivo is to stabilize and/or facilitate formation of NS1 dimers and multimers and, therefore, to promote the RNA binding function of the NS1 amino-terminal domain.
MATERIALS AND METHODS
Viruses, cells, and animals.
Influenza A/PR/8/34 (PR8) (H1N1) virus was propagated in 10-day-old embryonated chicken eggs at 37°C. The delNS1 virus is a PR8-derived virus in which the NS1 gene is deleted (12). This virus was grown in 7-day-old embryonated chicken eggs, as described previously (46). Influenza A/WSN/33 (WSN) virus (H1N1) and the WSN isogenic mutant viruses WSN-NS1(1-126), WSN-NS1(1-126)DmNcd, WSN-NS1(1-126)PUT3, WSN-NS1(1-73)HA, and WSN-NS1(1-73)HA-DmNcd were propagated in Madin-Darby bovine kidney (MDBK) cells. Viral titers were obtained by plaque assay on MDBK cells. MDBK cells were maintained in enriched medium with 10% fetal calf serum and antibiotics. A549 cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal calf serum plus antibiotics. PKR, RNaseL, and Mx triple knockout mice were generated as previously described (55). Specific-pathogen-free C57BL/6 and BALB/c mice were purchased from Charles River Farms.
Animal infection.
Six-week-old BALB/c mice, C57BL/6 mice, and PKR, RNaseL, and Mx triple knockout mice were anesthetized and infected intranasally with 50 μl of phosphate-buffered saline (PBS) containing the indicated amounts of influenza A viruses. For viral lung titers, mice were sacrificed at either day 3 or 6 postinfection. Lungs were homogenized and resuspended in sterile PBS and titrated on MDBK cells. For other experiments, animals were monitored daily and were euthanized when observed in extremis. All procedures were in accordance with the National Institutes of Health guidelines on care and use of laboratory animals.
Plasmids.
The 12 constructs used in rescuing recombinant influenza A/WSN/33 virus were previously described (9). The construct pPolI-NS-RT was the only one replaced in transfections for rescue of NS1 recombinant viruses. For example, for rescue of WSN-NS1(1-126) virus, pPolI-NS1(1-126)-RT was used with the other 11 constructs instead of the pPolI-NS-RT construct. pPolI-NS1(1-126)-RT was made by introducing two stop codons and a PacI restriction site at nucleotides 405 to 418 of the NS1 open reading frame in the pPolI-NS-RT construct. pPolI-NS1(1-126)PUT3 and pPolI-NS1(1-126)DmNcd have sequences encoding a truncated NS1 protein of 126 amino acids fused to an AgeI site, a 6-glycine linker, and a dimerization-domain-encoding sequence. The sequence of the 28-amino-acid-long dimerization domain of the Saccharomyces cerevisiae PUT3 protein is IVVSTKYLQQLQKDLNDKTEENNRLKAL (50). The sequence of the 24-amino-acid-long dimerization domain of the Drosophila melanogaster protein DmNcd is KEQLFQSNMERKELHNTVMDLRGN (49). In the pPolI-NS1(1-73)HA-RT construct, a 9-amino-acid hemagglutinin (HA) tag, an AgeI site, a stop codon, and a PacI site were introduced by PCR after nucleotide 245 of the NS gene. pPolI-NS1(1-73)HA-DmNcd was identical to pPolI-NS1(1-73)HA except for the insertion of sequences encoding a 6-glycine linker and the DmNcd domain between AgeI and PacI restriction sites. The Gal4, VP16 fusion protein constructs pGal4-WSN-NS1, pVP16-WSN-NS1, pGal4-NS1(1-73)HA, pVP16-NS1(1-73)HA, pGal4-NS1(1-73)HA-DmNcd, and pVP16-NS1(1-73)HA-DmNcd used in the mammalian two-hybrid assay were made by in-frame fusion of the corresponding cDNAs to the SalI and EcoRI sites of Gal4/N and VP16/P plasmids as described previously (15). pG5-Luc (Stratagene) is a luciferase reporter plasmid under the control of five copies of the Gal4 binding site. pRL-TK (Promega) is a plasmid used as an internal control for monitoring transfection efficiency.
Generation of recombinant viruses.
Recombinant viruses were generated by a modified transfection-based reverse genetics protocol for influenza A virus (9). Briefly, 293T cells were transfected with 12 plasmids: 4 pCAGGS expression plasmids for viral polymerases PB1, PB2, and PA, and nucleoprotein NP; 7 pPolI-Rib plasmids for wild-type viral genomic segments PB1, PB2, PA, NP, HA, NA, and M; and 1 pPolI-Rib plasmid for the specific mutant NS segment. Each plasmid (0.5 μg) was mixed with Lipofectamine 2000 (2 μl/μg of plasmid) (Invitrogen). The transfection was done by following the manufacturer's recommendation. Forty-eight hours posttransfection the supernatant was harvested and inoculated on fresh MDBK cells. The viral NS segments from the recovered mutant viruses were analyzed by reverse transcription-PCR and sequencing (data not shown).
Western blot analysis.
Dishes (diameter, 35 mm) of confluent MDBK cells were mock-infected or infected with WSN, WSN-NS1(1-126), WSN-NS1(1-126)PUT3 WSN-NS1(1-126)DmNcd, WSN-NS1(1-73)HA, or WSN-NS1(1-73)HA-DmNcd virus at a multiplicity of infection (MOI) of 1. Sixteen hours postinfection, cells were lysed. Cell lysates were subjected to Western blot analysis by using anti-NS1, anti-HA-tag, or anti-NP antibodies as described previously (53).
Pulse-chase labeling experiments.
MDBK cells were infected with wild-type WSN, WSN-NS1(1-126), WSN-NS1(1-126)DmNcd, and WSN-NS1(1-126)PUT3 viruses at an MOI of 2. Four hours postinfection, cells were starved for cysteine and methionine for 20 min. These cells were subsequently labeled with medium containing 35S-Cys and 35S-Met for 30 min and were chased with cold medium containing cysteine and methionine. At 1, 2, 4, and 20 h postchase total cell extracts were made in radioimmunoprecipitation assay buffer and were immunoprecipitated with anti-NP and anti-NS1 antibodies. The immunoprecipitated proteins were separated by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis (SDS-12% PAGE) and were exposed to X-ray film.
Mammalian two-hybrid assays.
Transfections for the mammalian two-hybrid assays were done by using Lipofectamine 2000 (Invitrogen). One microgram of pG5-Luc together with 0.5 μg of Gal4-WSN-NS1 and 0.5 μg of VP16-WSN-NS1, 0.5 μg of Gal4-NS1(1-73)HA and 0.5 μg of VP16-NS1(1-73)HA, or 0.5 μg of Gal4-NS1(1-73)HA-DmNcd and 0.5 μg of VP16-NS1(1-73)HA-DmNcd were cotransfected into 293T cells. pRL-TK (0.2 μg) was included in each transfection as an internal control to normalize transfection efficiency. After 48 h cells were lysed and luciferase activities were determined with a Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's recommendation.
Northern blot analysis.
Total RNA was extracted with TRIzol reagent (Molecular Research Center) according to the manufacturer's recommendation from A549 cells mock infected or infected with PR8, delNS1, WSN, WSN-NS1(1-126), WSN-NS1(1-126)DmNcd, WSN-NS1(1-126)PUT3, WSN-NS1(1-73)HA, or WSN-NS1(1-73)HA-DmNcd virus at an MOI of 1. Ten micrograms of total RNA was used for human IFN-β-specific Northern blot analysis. The cDNAs used for labeling specific probes were described previously (53).
RESULTS
Attenuation of influenza A/WSN/33 virus after deletion of its NS1 protein effector domain.
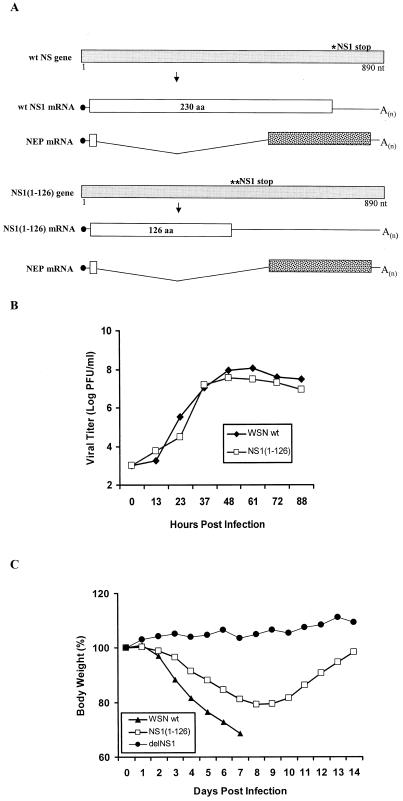
We have shown that a recombinant influenza A virus expressing only the first 126 amino acids of the NS1 protein in an influenza A/PR8/34 virus background prevented the induction of IFN-α/β mRNA in 293 cells as measured by Northern blot analysis (53). This suggested that the N-terminal sequence of influenza A virus NS1 protein is sufficient for its IFN-α/β antagonist function, at least in 293 cells. However, mutant influenza A/PR8/34 viruses (8, 46) with deletions of the NS1 C terminus have previously been shown to be attenuated in vivo. To further evaluate the role of the carboxy-terminal sequences of NS1 in the pathogenicity of influenza A virus, a recombinant WSN-NS1(1-126) virus was generated from cDNA (Fig. 1A). Similar to NS1(1-126)/PR8 virus, the WSN-NS1(1-126) virus lacking the C-terminal 127- to 230-amino-acid sequence of its NS1 protein did not show apparent growth defects in tissue culture (MDBK cells) (Fig. 1B). Also, the truncated NS1 protein, NS1(1-126), was expressed in virus-infected cells to levels similar to those of wild-type virus (Fig. 2C). However, consistent with previous studies, we found a clear difference in the pathogenicity between this mutant virus and the wild-type WSN virus in mice. Six-week-old BALB/c mice were intranasally infected with 2 × 104 PFU of either the wild-type WSN virus or the mutant WSN-NS1(1-126) virus per mouse. Several disease symptoms, such as body weight drop, decreased movement, and appearance of ruffled fur, developed rapidly in the five mice infected with wild-type WSN virus. All of these mice died in 9 days. The five mice infected with WSN-NS1(1-126) virus showed disease symptoms as evidenced by loss of body weight; however, body weight losses were less dramatic than those in wild-type WSN virus-infected mice, and all WSN-NS1(1-126) virus-infected mice recovered within 2 weeks (Fig. 1C). No signs of disease were detected in mice infected with the same dosage of delNS1 virus, a recombinant influenza A/PR8/34 virus with a complete deletion of the NS1 gene (Fig. 1C). These data demonstrate that although the carboxy-terminal half of the NS1 protein appeared to be dispensable for influenza A virus growth in MDBK cells, both the amino-terminal and carboxy-terminal sequences of this protein play a role in the pathogenicity of influenza A/WSN/33 virus in vivo.
FIG. 1.
Wild-type influenza A/WSN/33 virus and recombinant WSN-NS1(1-126) virus. (A) Schematic representation of the NS genes and gene transcripts for wild-type (wt) influenza WSN virus and recombinant WSN-NS1(1-126) virus. Viral NS genes are indicated by light gray boxes, with nucleotide (nt) length indicated in numbers below the gene segments. Stop codons for wild-type NS1 and NS1(1-126) open reading frames are indicated by an asterisk above the viral genes. Viral nuclear export protein (NEP) mRNAs are also shown, with spotted boxes representing the specific open reading frames of the viral NEP mRNA transcripts. (B) Growth curves of WSN-NS1(1-126) and WSN viruses on MDBK cells. Confluent cell monolayers in 35-mm-diameter dishes were infected with wild-type influenza A/WSN/33 virus and with the mutant WSN-NS1(1-126) virus at an MOI of 0.001. At the indicated time points, infectious particles present in the media were titrated by plaque assay in MDBK cells. (C) Pathogenicity of WSN-NS1(1-126) virus in mice. Groups of five mice were infected intranasally with either wild-type WSN virus or WSN-NS1(1-126) virus at a dose of 2 × 104 PFU/mouse. In addition, four mice were infected with 2 × 104 PFU of delNS1 virus/mouse. Animals were weighed dailyfollowing infection. The body weights on each day postinfection are given as the percentage of the original body weight (on day 0). The average body weight percentages of animals per group are represented. aa, amino acid.
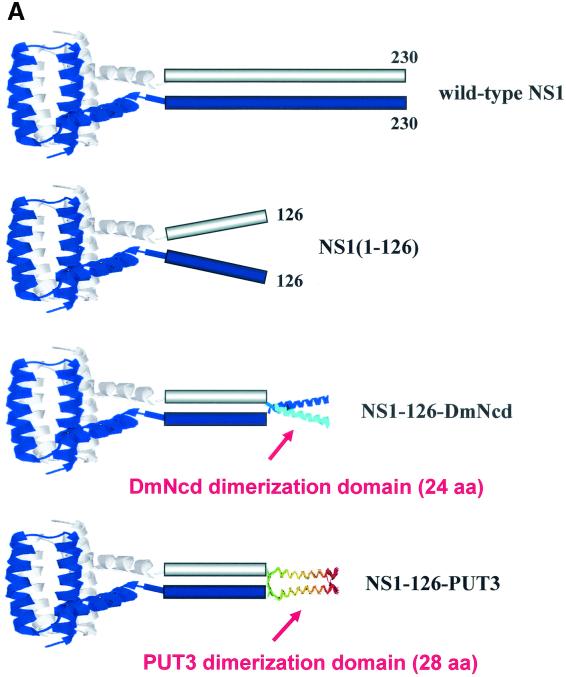
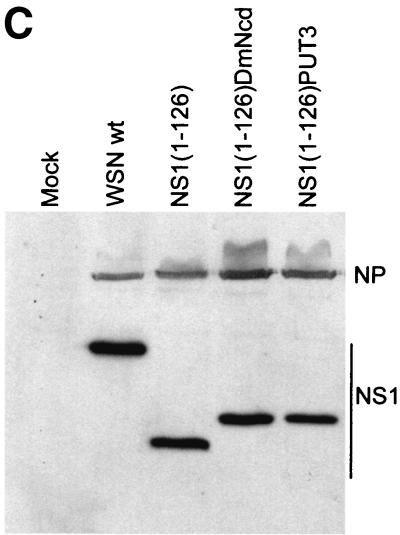
FIG. 2.
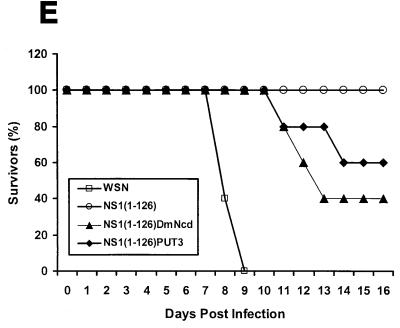
WSN-NS1(1-126)PUT3 and WSN-NS1(1-126)DmNcd viruses. (A) Schematic representation of the dimeric structures of the wild-type and mutant NS1(1-126), NS1(1-126)DmNcd, and NS1(1-126)PUT3 proteins. The crystal structure of the first 73 amino acids (aa) of the NS1 protein is adapted from reference 25. The crystal structure of the dimerization domain of DmNcd is adapted from reference 49. The structure of the dimerization domain of PUT3 is adapted from reference 50. The unknown structures are indicated by boxes. (B) Schematic representation of the NS genes and gene transcripts for influenza A WSN-NS1(1-126) dimerization domain-containing viruses, WSN-NS1(1-126)DmNcd and WSN-NS1(1-126)PUT3. NS genes and mRNA transcripts of the top two viruses are schematically represented as in Fig. 1. The 6-glycine linker region in WSN-NS1(1-126)DmNcd and WSN-NS1(1-126)PUT3 viruses is indicated by the striped box. The 24-amino-acid-long and 28-amino-acid-long dimerization domains from the D. melanogaster DmNcd protein or the S. cerevisiae PUT3 protein are represented by the checkered boxes. NEP, nuclear export protein; aa, amino acid; nt, nucleotide. (C) Western blot analysis of wild-type and mutant NS1 proteins in infected cells. MDBK cells were mock infected (mock) or were infected with wild-type (wt) WSN virus, recombinant WSN-NS1(1-126), WSN-NS1(1-126)DmNcd, or WSN-NS1(1-126)PUT3 virus at an MOI of 1. Sixteen hours postinfection, cell extracts were made and subjected to Western blot analysis with antibodies against the viral NP and NS1 proteins. (D) Pulse-chase analysis of wild-type NS1 and mutant NS1 proteins. MDBK cells were either mock infected or infected with wild-type WSN virus, WSN-NS1(1-126), WSN-NS1(1-126)DmNcd, or WSN-NS1(1-126)PUT3 virus at an MOI of 2. Four hours postinfection, cells were starved for methionine and cysteine and then pulse-labeled with 35S-Met and 35S-Cys for 30 min. At 1, 2, 4, and 20 h postchase total cell extracts were immunoprecipitated with antibodies against influenza NP and NS1 proteins. Samples were then boiled in SDS loading buffer and were separated by SDS-12% PAGE. The gel was then dried and subjected to autoradiography. h.p.c., hours postchase. (E) Pathogenicity of WSN-NS1(1-126)DmNcd and WSN-NS1(1-126)PUT3 viruses in mice. Groups of five mice were intranasally infected with 2 × 104 PFU of either wild-type WSN, WSN-NS1(1-126), WSN-NS1(1-126)DmNcd, or WSN-NS1(1-126)PUT3 virus/mouse. Animals were inspected daily following infection. The percentage of surviving animals as a function of time is represented.
Heterologous dimerization domains compensate for the loss of the C-terminal 104 amino acids of the NS1 protein.
Although the C-terminal region of the NS1 protein has been reported to play a role in blocking host mRNA processing and transport, we decided to study the biological significance of the structural role of the C-terminal sequences of NS1. We generated two mutant viruses, each of which has a short dimerization domain substituting for the C-terminal 104 amino acids of their NS1 proteins (Fig. 2A). The first dimerization domain comprises 28 amino acids from the S. cerevisiae transcription factor PUT3 (50). For the second dimerization domain we used 24 amino acids from the α-7 coiled coil of the D. melanogaster kinesin motor protein, DmNcd (49). A 6-glycine peptide linker was added in between the first 126 amino acids of the NS1 protein and the dimerization domains to reduce the chances of perturbing the folding of the NS1 N-terminal region (Fig. 2B). The dimerization domain-containing viruses, WSN-NS1(1-126)PUT3 and WSN-NS1(1-126)DmNcd, were generated by using a plasmid-based influenza A virus rescue system (9, 34). The viruses grew to titers similar to those of wild-type parental WSN virus in MDBK cells (data not shown). Expression of the mutant NS1 proteins in infected cells was confirmed by Western blot analysis (Fig. 2C). These data suggested that the introduction of short foreign peptide sequences into the NS1 protein did not interfere with the replication of influenza A virus in tissue culture.
The deletion of amino acids 127 to 230 from the NS1 protein and/or the addition of foreign sequences after the first 126 amino acids of the NS1 protein might affect the stability of this protein. To test this possibility, we performed pulse-chase assays. MDBK cells were infected with either wild-type WSN, WSN-NS1(1-126), WSN-NS1(1-126)DmNcd, or WSN-NS1(1-126)PUT3 virus at an MOI of 2. Four hours postinfection cells were pulse-labeled with medium containing 35S-Met and 35S-Cys for 30 min and subsequently were chased with cold medium. At different times postchase, total cell extracts were prepared and immunoprecipitated with anti-NP and anti-NS1 antibodies. The immunoprecipitated proteins were analyzed by SDS-PAGE. No apparent stability differences were found among the wild-type and mutant NS1 proteins (Fig. 2D).
To determine whether the introduction of heterologous dimerization domains compensated for the function of the C-terminal sequences of NS1 protein in vivo, 6-week-old BALB/c mice were intranasally infected with 2 × 104 PFU of mutant WSN-NS1(1-126)PUT3 and WSN-NS1(1-126)DmNcd viruses per mouse, which was the same dosage used with the WSN-NS1(1-126) virus. Interestingly, these two mutant viruses were virulent in mice. Two out of five mice infected with WSN-NS1(1-126)PUT3 virus and three out of five mice infected with WSN-NS1(1-126)DmNcd died (Fig. 2E). With the same infection dose all five mice infected with WSN-NS1(1-126) virus survived (Fig. 2E). These observations indicated that the addition of short heterologous dimerization domains could compensate, at least partially, for the function in vivo of the carboxy-terminal sequences of the wild-type NS1 protein.
Generation and attenuation of a mutant influenza A/WSN/33 virus expressing only the first 73 amino acids of the NS1 protein.
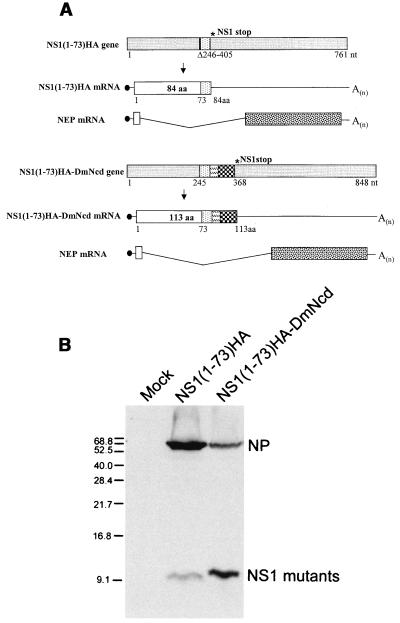
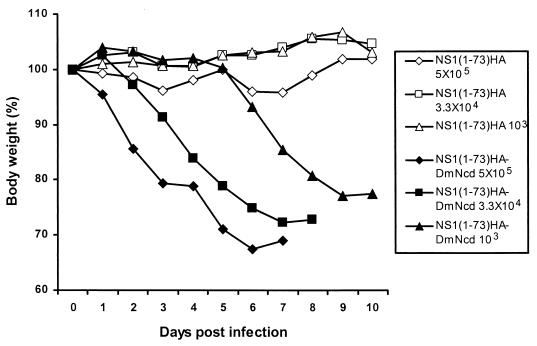
We next generated mutant WSN-NS1(1-73)HA virus, which expresses only the first 73 amino acids of the NS1 protein plus a 9-amino-acid HA tag (Fig. 3A, upper panel). WSN-NS1(1-73)HA virus replicated well and grew to titers comparable to that of the wild-type parental WSN virus in MDBK cells, with both viruses reaching between 107 and 108 PFU/ml at 60 h postinfection with an MOI of 0.001 (data not shown). The expression of the mutant NS1 protein was detected by Western blot analysis by using anti-HA-tag antibody (Fig. 3B, middle lane). In contrast to WSN-NS1(1-126), the truncated NS1 protein of WSN-NS1(1-73)HA virus lacks not only the NS1 effector domain but also amino acids involved in NS1-eIF4GI interactions, and it retains only its amino-terminal RNA binding domain. To determine the pathogenicity of the WSN-NS1(1-73)HA virus, groups of five C57BL/6 mice were intranasally infected with different doses of either WSN-NS1(1-73)HA or wild-type WSN viruses. Deletion of amino acids 74 to 230 of the NS1 protein rendered WSN-NS1(1-73)HA virus highly attenuated in mice. Intranasal infection with a dose as high as 5 × 105 PFU/mouse did not cause any signs of disease in mice, whereas infection with only 103 PFU of wild-type WSN virus/mouse killed all five mice (Table 1 and Fig. 4). Lung titers for mice infected with 3.3 × 104 PFU were measured at 3 and 6 days postinfection by plaque assay on MDBK cells. Mice infected with wild-type WSN virus had average lung titers of 107 PFU/ml and 4 × 106 PFU/ml at 3 and 6 days postinfection, respectively. However, neither at 3 nor 6 days postinfection were viral titers detectable in mice infected with WSN-NS1(1-73)HA virus (Table 2).
FIG. 3.
WSN-NS1(1-73)HA and WSN-NS1(1-73)HA-DmNcd viruses. (A) Schematic representation of the NS genes and gene transcripts for influenza A WSN-NS1(1-73)HA and WSN-NS1(1-73)HA-DmNcd viruses. Viral NS genes and mRNA transcripts are schematically represented as in Fig. 1. A deletion of 159 nucleotides (nt) in the NS1(1-73)HA gene is indicated by a vertical bar. A 9-amino-acid (aa) HA tag in WSN-NS1(1-73)HA and WSN-NS1(1-73)HA-DmNcd viral NS genes is presented by the dotted bar. The 6-glycine linker region in WSN-NS1(1-73)DmNcd virus is indicated by the striped box. The dimerization domain is represented by the checkered boxes as in Fig. 2. NEP, nuclear export protein. (B) Western blot analysis of mutant NS1 proteins in infected cells. MDBK cells were either mock infected (mock) or were infected with WSN-NS1(1-73)HA or WSN-NS1(1-73)HA-DmNcd virus at an MOI of 1. Sixteen hours postinfection, cell extracts were made and were subjected to Western blot analysis with antibodies against the viral NP protein and the HA tag. It should be noted that at the electrophoretic conditions used (SDS-12% PAGE), both NS1(1-73)HA and NS1(1-73)HA-DmNcd run with the front and do not show significant differences in electrophoretic mobility. Numbers at the left are molecular sizes.
TABLE 1.
Survival of 6-week-old C57BL/6 mice infected with wild-type WSN and recombinant WSN-NS1(1-73)HA viruses
| Virus or medium | Dosage (PFU) | Survivorsa (no. survived/total inoculated) |
|---|---|---|
| Wild-type WSN | 5 × 105 | 0/5 |
| 3.3 × 104 | 0/5 | |
| 1 × 103 | 0/5 | |
| NS1(1-73)HA | 5 × 105 | 5/5 |
| 3.3 × 104 | 5/5 | |
| 1 × 103 | 5/5 | |
| NS1(1-73)HA-DmNcd | 5 × 105 | 0/5 |
| 3.3 × 104 | 0/5 | |
| 1 × 103 | 3/5 | |
| PBS | 0 | 2/2 |
Mice were inspected and weighed daily for four weeks.
FIG. 4.
Pathogenicity of WSN-NS1(1-73)HA and WSN-NS1(1-73)HA-DmNcd viruses in mice. Groups of five C57BL/6 mice were intranasally infected with 5 × 105, 3.3 × 104, or 1 × 103 PFU of either WSN-NS1(1-73)HA or WSN-NS1(1-73)HA-DmNcd virus. Animals were weighed daily following infection, and body weights were compared with those from the day of infection. The average body weight percentages of animals per group are represented.
TABLE 2.
Lung titers of C57BL/6 micea and PKR, RNaseL, and Mx triple knockout mice immunized with 3.3 × 104 PFU of wild-type WSN, WSN-NS1(1-73)HA, or WSN-NS1(1-73)HA-DmNcd virus/mouse
| Virus and mouse type | Titers (PFU/ml) and index values at:
|
|||
|---|---|---|---|---|
| 3 Days postinfection
|
6 Days postinfection
|
|||
| Lung titerb | PKR, RNase L indexc | Lung titerb | PKR, RNase L indexc | |
| Wild-type WSN, C57BL/6 | 1.3 × 107, 1.0 × 107 | 5.2 | 3 × 106, 2 × 106, 7 × 106 | 1.8 |
| Wild-type WSN, triple null | 5 × 107, 7 × 107 | 5.2 | 6 × 106, 8 × 106, 8 × 106 | 1.8 |
| WSN-NS1(1-73)HA, C57BL/6 | <10, <10, <10 | >1,700 | <10, <10, <10 | >270 |
| WSN-NS1(1-73)HA, triple null | 2 × 104, 2 × 104, 1 × 104 | >1,700 | 4 × 103, 1 × 103, 3 × 103 | >270 |
| WSN-NS1(1-73)HA-DmNcd, C57BL/6 | 5 × 106, 2 × 106, 4 × 106 | 13 | 4 × 104, 1 × 104, 1 × 104 | 21 |
| WSN-NS1(1-73)HA-DmNcd, triple null | 5 × 107, 3 × 107, 6 × 107 | 13 | 4 × 105, 2.5 × 105, 6 × 105 | 21 |
These mice have a naturally occurring deletion in their Mx genes.
Numbers represent titers (PFU/ml) from individual infected mice.
Ratio of average lung titers between triple knockout and C57BL/6 mice.
WSN-NS1(1-73)HA virus replicates in lungs of PKR, RNaseL, and Mx triple knockout mice.
In order to understand whether attenuation of WSN-NS1(1-73)HA virus in mice is dependent on the IFN-α/β system, we investigated its replication in PKR, RNaseL, and Mx triple knockout mice. These mice lack three of the most important antiviral enzymes induced by IFN-α/β (55). Five to six 6-week-old PKR, RNaseL, and Mx triple knockout mice were infected with either WSN or WSN-NS1(1-73)HA virus. Two or three mice were sacrificed at either 3 or 6 days postinfection, and viral lung titers were determined. The average lung titer of wild-type WSN virus-infected mice was 6 × 107 PFU/mouse and 7.3 × 106 PFU/ml at 3 and 6 days postinfection, respectively (Table 2). These numbers were similar to those detected in infected wild-type C57BL/6 mice (Table 2). Remarkably, while no virus was detected in WSN-NS1(1-73)HA virus-infected C57BL/6 mouse lungs (Table 2), the average lung titers of triple knockout mice infected with this mutant virus were 1.7 × 104 PFU/ml at 3 days postinfection and 2.7 × 103 PFU/ml at 6 days postinfection (Table 2). These observations suggest that the main reason for the lack of replication of WSN-NS1(1-73)HA virus in mice is its inability to inhibit the IFN-α/β system in these animals.
A 24-amino-acid-long dimerization domain compensates for the loss of the C-terminal 157 amino acids of the NS1 protein.
Short heterologous dimerization domains compensate for the loss of the last 104 amino acids of the NS1 protein in mice. To determine whether a short dimerization domain can also compensate for longer deletions of C-terminal NS1 sequences, we generated a mutant virus with the 24-amino-acid-long dimerization domain of DmNcd substituting for the C-terminal 74 to 230 amino acids of the NS1 protein (Fig. 3A). Similar to the previous design of dimerization-domain-containing viruses, a 6-glycine linker was inserted between the HA tag and the dimerization domain. WSN-NS1(1-73)HA-DmNcd virus expressed a truncated NS1 protein of the expected length, as determined by Western blot analysis of infected cells by using anti-HA antibody (Fig. 3B). Of interest, steady-state levels of the NS1(1-73)HA-DmNcd protein were significantly higher than those detected in cells infected with mutant WSN-NS1(1-73)HA virus lacking the DmNcd dimerization domain. To test the pathogenicity of WSN-NS1(1-73)HA-DmNcd virus, C57BL/6 mice were intranasally infected with 2.5 × 105, 3.3 × 104, or 1 × 103 PFU/mouse. Interestingly, WSN-NS1(1-73)HA-DmNcd virus was found to be virulent. Mice infected with this virus had dramatic body weight loss, while no apparent disease was detected in mice infected with WSN-NS1(1-73)HA virus (Fig. 4). All five mice infected with 2.5 × 105 and 3.3 × 104 PFU of WSN-NS1(1-73)HA-DmNcd virus died within 9 days (Table 1). At a dose of 103 PFU/mouse, two out of five infected mice died (Table 1). In addition, all of the three mice that survived infection showed significant body weight losses for at least 12 days postinfection (data not shown). Viral lung titers were also determined after intranasal infection with 3.3 × 104 PFU of WSN-NS1(1-73)HA-DmNcd virus/mouse. As summarized in Table 2, the average lung titers of C57BL/6 mice infected with this virus was 2.8 × 106 PFU/ml and 2 × 104 PFU/ml at 3 and 6 days postinfection, respectively. Although these titers are three times and one hundred times lower than those of wild-type WSN virus-infected mice, no virus was detected in WSN-NS1(1-73)HA-infected mice (Table 2). The contribution of PKR and RNaseL pathways on inhibiting viral replication in lungs was significantly higher for WSN-NS1(1-73)HA virus than for WSN-NS1(1-73)HA-DmNcd virus (Table 2). These observations suggest that the C-terminal region (amino acids 74 to 230) of the influenza A virus NS1 protein contributes to the inhibition of the IFN-α/β system in vivo mainly by promoting and/or stabilizing the dimeric structure formed by the amino-terminal RNA binding domain of this protein.
WSN-NS1(1-73)HA-DmNcd mutant protein retains its ability to dimerize.
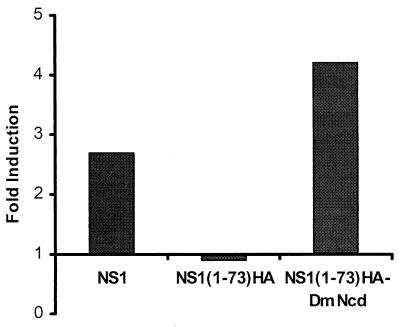
To investigate whether the fusion of heterologous dimerization domains to the RNA binding domain of NS1 protein promotes its dimerization, a mammalian two-hybrid system was utilized (15). In this assay, wild-type WSN-NS1 or the mutants WSN-NS1(1-73)HA and WSN-NS1(1-73)HA-DmNcd were fused in frame to the DNA binding domain of the transcription factor Gal4 or the transcription activation domain of herpesvirus VP16, respectively (15). 293T cells were cotransfected with plasmids expressing the Gal4 and VP16 fusion proteins and reporter gene pG5-luc, in which the expression of luciferase is under the control of five copies of Gal4 binding sites. pRL-TK was also included in each transfection as an internal control to normalize transfection efficiency. Forty-eight hours posttransfection a luciferase assay was performed to determine the interaction of wild-type and mutant NS1 proteins as an indication of their ability to form dimers and multimers. Expression of wild-type NS1 fusion proteins resulted in an approximately 2.6-fold induction of the reporter gene (Fig. 5). In contrast, expression of the fusion proteins of NS1(1-73)HA could not stimulate the reporter gene (Fig. 5). Introduction of the DmNcd heterologous dimerization domain restored the reporter gene activity to levels slightly higher than those of wild-type NS1 fusion proteins (Fig. 5). These results suggested that the DmNcd domain can functionally replace the C-terminal domain of NS1 (74 to 230 amino acids) to promote NS1 dimerization and multimerization.
FIG. 5.
Dimerization and/or multimerization of wild-type NS1 and mutant NS1(1-73)HA and NS1(1-73)HA-DmNcd proteins, as measured by using a mammalian two-hybrid assay. 293T cells were cotransfected with pG5-Luc, pRL-TK, and Gal4-WSN-NS1 and VP16-WSN-NS1 (NS1, first column), Gal4-NS1(1-73)HA and VP16-NS1(1-73)HA [NS1(1-73)HA, second column], or Gal4-NS1(1-73)HA-DmNcd and VP16-NS1(1-73)HA-DmNcd plasmids [NS1(1-73)HA-DmNcd, third column]. Forty-eight hours posttransfection, cell extracts were made and luciferase activity was determined.
Pathogenicity of NS1 mutant influenza viruses correlates with their ability to prevent IFN-α/β mRNA induction.
In order to investigate whether there is a correlation between the virulence and the IFN-β inhibitory activity of NS1 mutant influenza A viruses, we determined levels of IFN-β mRNA in infected A549 cells. The A549 cell line is a human epithelial alveolar lung cell line derived from lung carcinomatous tissue. A549 cells were mock infected or were infected with wild-type PR8, delNS1, wild-type WSN, WSN-NS1(1-126), WSN-NS1(1-126)DmNcd, WSN-NS1(1-126)PUT3, WSN-NS1(1-73)HA, or WSN(1-73)HA-DmNcd virus at an MOI of 1. At 10 or 20 h postinfection total RNA was extracted and subjected to IFN-β mRNA-specific Northern blot analysis. Consistent with findings of previous studies, delNS1 virus infection but not wild-type PR8 virus infection induced high levels of IFN-β mRNA (53). No IFN-β mRNA was detected by Northern blot analysis in WSN virus-infected cells. Interestingly, levels of IFN-β mRNA in cells infected with WSN-NS1(1-126) and WSN-NS1(1-73)HA viruses correlated with their attenuated properties in mice, and WSN-NS1(1-73)HA virus induced higher levels of IFN-β mRNA than did the WSN-NS1(1-126) virus. Also, the introduction of short, heterologous dimerization domains after either the N-terminal 126 amino acids or the N-terminal 73 amino acids of the NS1 protein decreased the induction of IFN-β mRNA to levels close to those of wild-type-virus-infected cells (Fig. 6).
FIG. 6.
IFN-β gene induction by viruses expressing wild-type NS1 protein and mutant NS1 proteins. A549 cells were either mock infected (mock) or were infected with wild-type PR8, delNS1, wild-type WSN, WSN-NS1(1-126), WSN-NS1(1-126)DmNcd, WSN-NS1(1-126)PUT3, WSN-NS1(1-73)HA, or WSN-NS1(1-73)HA-DmNcd virus at an MOI of 1. At 10 and 20 h postinfection RNAs were extracted, and 10 μg of total RNAs from each sample was used for Northern blot analysis by using probes specific for the human IFN-β gene. As a control, 28S and 18S rRNAs were visualized by ethidium bromide staining.
DISCUSSION
The NS1 protein of influenza A virus appears to be a multifunctional protein with at least three described functional domains. Its amino-terminal 73 amino acids contain an RNA binding domain that is critical for the IFN-α/β antagonist function of NS1 (53). On the other hand, its carboxy-terminal region (effector domain) has been reported to inhibit processing (24, 32) and nuclear-cytoplasmic transport of host mRNA (26), and these functions have also been suggested to play a major role in inhibiting the host innate antiviral response (22). A third domain, located in the middle of the protein, appears to enhance translation of viral mRNA through interaction with the cellular translation initiation factor eIF4GI (2). Although deletions of the effector and eIF4GI binding domains did not significantly affect the ability of the corresponding mutant viruses to replicate in tissue culture, these viruses became attenuated in mice (Fig. 2C and Table 2). These data demonstrate an important biological role for the two domains during replication of influenza virus in an animal host. However, these data do not exclude the possibility that attenuation of NS1-truncated viruses is due to a structural alteration of the truncated NS1 proteins rather than due to a loss of their abilities to inhibit host mRNA processing and to enhance viral mRNA translation. Importantly, although the N-terminal RNA binding domain is able to dimerize in vitro, the C-terminal region of NS1 also appears to contribute to efficient NS1 dimerization in a yeast two-hybrid system, possibly through interactions with a cellular protein (33).
In order to understand the structural role of the carboxy-terminal domains of the NS1 protein, we generated recombinant influenza A viruses expressing chimeric NS1 proteins. Our experiments demonstrated for the first time that substitution of the C-terminal region of the NS1 protein with short heterologous sequences from either yeast PUT3 or Drosophila DmNcd proteins restored virus lethality in mice. These heterologous sequences were previously known to mediate homodimerization of the PUT3 and DmNcd proteins. In addition, a PUT3 dimerization domain has previously been used to force specific dimeric orientations of the E5 protein from bovine papillomavirus (30). The recombinant dimerization domain-containing viruses, WSN-NS1(1-126)DmNcd, WSN-NS1(1-126)PUT3, and WSN-NS1(1-73)HA-DmNcd, regained virulence and lethality in mice (Fig. 2E and Table 1). These observations not only indicate that the carboxy-terminal sequences of the NS1 protein are actively involved in the formation of a dimeric and/or multimeric NS1 structure during infection but also suggest that the main biological role of the carboxy-terminal two-thirds of the NS1 protein is the enhancement of its dimerization abilities.
We have previously shown that the main biological role of the NS1 protein of influenza A virus is the inhibition of the IFN-α/β system of the host (12). Previous results also demonstrated that during influenza A virus infection, the NS1 protein prevents the induction of high levels of IFN-α/β mRNA (53) as well as the activation of the IFN-inducible PKR kinase (3), a translational inhibitor with potent antiviral activity. No apparent growth defects were observed for WSN-NS1(1-126) and WSN-NS1(1-73)HA viruses in MDBK cells. However, these viruses are attenuated in mice (Fig. 2E and Table 2). No viruses were detected in lungs of C57BL/6 mice infected with WSN-NS1(1-73)HA virus at days 3 and 6 postinfection (Table 2). A possible explanation for this finding is that the IFN-α/β system restricts viral replication more effectively in the mouse than it does in tissue culture. The carboxy-terminal-truncated NS1 proteins might then be less efficient than wild-type NS1 protein in their ability to inhibit the IFN-α/β system in mice. Consistent with this hypothesis, WSN-NS1(1-73)HA virus regained the ability to replicate in lungs of PKR, RNaseL, and Mx triple knockout mice, which lack three important IFN-inducible enzymes with essential roles in the IFN-mediated antiviral responses (55). Our data suggest that deletion of the carboxy-terminal sequences of the NS1 protein interferes with its anti-IFN activity. Consistently, infections with both WSN-NS1(1-73)HA and WSN-NS1(1-126) viruses, but not with wild-type WSN and PR8 viruses, induced the upregulation of IFN-β mRNA levels in A549 cells (Fig. 6). Strikingly, there was a clear correlation between the ability of NS1 mutant viruses to prevent the induction of IFN-β mRNA and their virulence in mice. We also noticed that the steady-state levels of the NS1(1-73)HA protein in infected cells were lower than those of the NS1(1-126) or wild-type NS1 proteins (Fig. 3B and 2C), suggesting that sequences between amino acids 74 to 126 might contribute to the stability of the NS1 protein.
At least two domains have been deleted in the NS1 protein of the attenuated WSN-NS1(1-73)HA virus, the eIF4GI binding domain and the effector domain. These domains have been proposed to be important for the translational and posttranscriptional regulation of viral and cellular gene expression. Intriguingly, the substitution of the last 157 amino acids containing these two domains with a 24-amino-acid-long heterologous dimerization sequence from Drosophila DmNcd protein restored the virulence of this virus to almost wild-type levels (Table 2). Our results also demonstrate that the amino-terminal 73 amino acids of the NS1 protein are able to inhibit IFN-α/β synthesis during influenza A virus infection when in the context of a strong NS1 dimer or multimer (Fig. 6). Since it is highly unlikely that the NS1(1-73)HA-DmNcd protein gains the ability to bind to eIF4GI and to inhibit host mRNA processing, our results suggest that the main function of the last 157 amino acids of the NS1 protein is the stabilization and/or facilitation of NS1 dimerization. Consistent with this suggestion, the fusion of the DmNcd dimerization domain to the RNA binding domain of the NS1 protein resulted in a chimeric NS1 that was able to interact with itself in mammalian cells. The strength of these NS1-NS1 interactions was comparable to (or even higher than) that of wild-type NS1 protein, as measured by a mammalian two-hybrid assay (Fig. 5). Interestingly, while carboxy-terminal-truncated NS1 proteins dimerize in vitro with similar efficiency to wild-type NS1 protein, we and others (33) have found that the strength of dimerization is affected by deletions in the carboxy-terminal region in vivo.
There are several possible explanations for the fact that the WSN-NS1(1-73)HA-DmNcd virus, lacking the carboxy-terminal domains of NS1, is virulent in mice. First, the posttranscriptional and translational regulatory functions attributed to the carboxy-terminal eIF4GI binding and effector domains of the NS1 protein may play only minor roles in the pathogenicity of influenza A virus in mice, the main function of these sequences being the stabilization and/or facilitation of the dimeric NS1. Second, the posttranscriptional and translational regulatory functions associated with the carboxy-terminal domains of the NS1 protein may be critical for the biological fitness of influenza A virus required for its evolutionary success but not for its virulence or growth in mice. The NS1 amino-terminal RNA binding domain, on the other hand, is essential for all these functions. Finally, there might be a functional redundancy of the N-terminal domain and the posttranscriptional and translational regulatory functions associated with C-terminal domains of the NS1 protein as it relates to pathogenicity of influenza A virus. This possibility is presently under investigation.
IFN was discovered as a factor released by cells treated with heat-inactivated influenza A viruses (21). By contrast, live influenza A viruses are poor inducers of IFN (20). Consistent with earlier observations, our results show low levels of IFN-β mRNA in A549 cells infected with wild-type influenza A viruses. It was previously found that inhibition of IFN synthesis by influenza A viruses is due to the viral NS1 protein, which inhibits the activation of transcription factors involved in IFN-β mRNA expression (27, 45, 53). In addition, the NS1 protein also inhibits the activation of the PKR kinase (3, 17, 26, 47), which is an important component of the host antiviral response induced by IFN. Nevertheless, most influenza A virus strains are not able to completely inhibit the IFN-α/β system (13, 45) and, therefore, induce some levels of IFN, especially in vivo (11, 31, 41). Thus, highly pathogenic influenza virus strains may express NS1 proteins with more efficient IFN antagonistic properties that those from typical influenza virus strains. Interestingly, this seems to be the case for the highly pathogenic 1918 (13) and Hong Kong (H5N1) influenza virus strains (19).
dsRNA generated during viral replication is believed to be a strong inducer of IFN-α/β. Sequestering of dsRNA by the RNA binding domain of the NS1 protein could be one of the mechanisms for its IFN-α/β antagonist function (53). Although in vitro assays showed that the NS1 amino-terminal domain was sufficient for RNA binding activity (38), additional sequences might be required for optimal RNA binding properties in the context of viral infection. Our results strongly suggest that the carboxy-terminal two-thirds of the NS1 protein are required for optimal dimerization of the NS1 in vivo. Since NS1 dimerization is critical for NS1 RNA binding activity, it is likely that the carboxy-terminal two-thirds of the NS1 protein are also required for optimal RNA binding activity in vivo. However, mere sequestration of dsRNA may not completely explain the NS1 inhibitory properties on the IFN-α/β system. First, it has been reported that the RNA binding domain of NS1 has low affinity for its RNA targets (24). Second, other viral effector molecules, in addition to dsRNA, appear to trigger the IFN-α/β system during viral infections (43). A more likely scenario that may explain the IFN-α/β antagonistic function of NS1 could involve NS1 interacting with and inhibiting the cellular kinase(s) that triggers IFN-α/β production. Since this unknown kinase(s) is also likely to recognize dsRNA, dsRNA might cooperatively enhance interactions between this cellular protein(s) and the NS1 protein. Dimerization and multimerization might then also be critical for the interaction between the NS1 protein and the uncharacterized essential component(s) of these pathways.
Similar to influenza A virus NS1 protein, the human immunodeficiency virus Rev protein has an N-terminal RNA binding domain and a C-terminal effector domain and functions as a dimer or multimer. In addition to the essential role of the Rev effector domain in mediating Rev export through a nuclear export signal, both the N-terminal and C-terminal regions of Rev are involved in its multimerization in vivo (28). A close functional relative of the influenza virus NS1 protein is the vaccinia virus E3L protein (6). Both E3L and NS1 proteins are dsRNA binding proteins involved in the inhibition of PKR as well as in the prevention of activation of IRF transcription factors involved in IFN-α/β synthesis (43). A conserved carboxy-terminal domain in the E3L protein is responsible for its dsRNA-binding and IFN-α/β antagonistic properties. This domain is required for the broad host range of vaccinia virus, whereas its N-terminal domain is dispensable in cell culture systems. However, a recombinant vaccinia virus containing a deletion at the N terminus of its E3L protein was found to be attenuated in mice (4). Although the attenuation of this mutant might be mediated by the loss of a Z-DNA binding domain located in the E3L N-terminal region, it is also possible that the N-terminal sequences of E3L, similar to the C-terminal sequences of NS1, have a structural role in stabilizing the RNA binding domain.
The development of live attenuated influenza virus vaccine strains could provide more effective and cost-efficient immunizations to protect against influenza virus infection. Previous studies with delNS1 and NS1-99 viruses have shown that vaccination with these viruses protect mice from lethal challenge with wild-type virus (46). Consistent with this finding, we found that in spite of its highly attenuated phenotype, infection of WSN-NS1(1-73)HA virus with a dose as low as 103 PFU/mouse was able to protect mice from lethal challenge with wild-type WSN virus (data not shown). Findings in this paper provide insights for the development of novel live attenuated influenza vaccine strains. In addition, since dimerization of NS1 appears to be essential for its role in antagonizing the IFN system, this structural requirement of NS1 may provide an attractive target for antiviral therapy against influenza viruses.
Acknowledgments
We thank members of the A.G.-S. and P.P. laboratories for critical discussions. We gratefully acknowledge Estanislao Nistal-Villán for excellent technical assistance.
This work was supported by NIH research grants to B.R.G.W. (AI34039), R.H.S. (CA44059), P.P., and A.G.-S.
REFERENCES
- 1.Alcami, A., and U. H. Koszinowski. 2000. Viral mechanisms of immune evasion. Mol. Med. Today 6:365-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aragón, T., S. de La Luna, I. Novoa, L. Carrasco, J. Ortín, and A. Nieto. 2000. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cell. Biol. 20:6259-6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann, M., A. García-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandt, T. A., and B. L. Jacobs. 2001. Both carboxy- and amino-terminal domains of the vaccinia virus interferon resistance gene, E3L, are required for pathogenesis in a mouse model. J. Virol. 75:850-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buonagurio, D. A., M. Krystal, P. Palese, D. C. DeBorde, and H. F. Maassab. 1984. Analysis of an influenza A virus mutant with a deletion in the NS segment. J. Virol. 49:418-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang, H. W., J. C. Watson, and B. L. Jacobs. 1992. The E3L gene of vaccinia virus encodes an inhibitor of the interferon-induced, double-stranded RNA-dependent protein kinase. Proc. Natl. Acad. Sci. USA 89:4825-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, Z., Y. Li, and R. M. Krug. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 18:2273-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egorov, A., S. Brandt, S. Sereinig, J. Romanova, B. Ferko, D. Katinger, A. Grassauer, G. Alexandrova, H. Katinger, and T. Muster. 1998. Transfectant influenza A viruses with long deletions in the NS1 protein grow efficiently in Vero cells. J. Virol. 72:6437-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fodor, E., L. Devenish, O. G. Engelhardt, P. Palese, G. G. Brownlee, and A. García-Sastre. 1999. Rescue of influenza A virus from recombinant DNA. J. Virol. 73:9679-9682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fortes, P., A. Beloso, and J. Ortín. 1994. Influenza virus NS1 protein inhibits pre-mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 13:704-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García-Sastre, A., R. K. Durbin, H. Zheng, P. Palese, R. Gertner, D. E. Levy, and J. E. Durbin. 1998. The role of interferon in the tropism of influenza virus. J. Virol. 72:8550-8558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 13.Geiss, G. K., M. Salvatore, T. M. Tumpey, V. S. Carter, X. Wang, C. F. Basler, J. K. Taubenberger, R. E. Bumgarner, P. Palese, M. G. Katze, and A. García-Sastre. 2002. Cellular transcriptional profiling in influenza A virus infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. USA 99:10736-10741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 15.Harty, R. N., and P. Palese. 1995. Measles virus phosphoprotein (P) requires the NH2- and COOH-terminal domains for interactions with the nucleoprotein (N) but only the COOH terminus for interactions with itself. J. Gen. Virol. 76:2863-2867. [DOI] [PubMed] [Google Scholar]
- 16.Hatada, E., and R. Fukuda. 1992. Binding of influenza A virus NS1 protein to dsRNA in vitro. J. Gen. Virol. 73:3325-3329. [DOI] [PubMed] [Google Scholar]
- 17.Hatada, E., S. Saito, and R. Fukuda. 1999. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J. Virol. 73:2425-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatada, E., S. Saito, N. Okishio, and R. Fukuda. 1997. Binding of the influenza virus NS1 protein to model genome RNAs. Virology 78:1059-1063. [DOI] [PubMed] [Google Scholar]
- 19.Heui Seo, S., E. Hoffmann, and R. G. Webster. 2002. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat. Med. 8:950-954. [DOI] [PubMed]
- 20.Isaacs, A., and D. C. Burke. 1958. Mode of action of interferon. Nature 4642:1073-1076. [DOI] [PubMed] [Google Scholar]
- 21.Isaacs, A., and J. Lindenmann. 1957. Virus interference. 1. The interferon. Proc. R. Soc. Lond. B. 147:258-267.13465720 [Google Scholar]
- 22.Kim, M. J., A. G. Latham, and R. M. Krug. 2002. Human influenza viruses activate an interferon-independent transcription of cellular antiviral genes: outcome with influenza A virus is unique. Proc. Natl. Acad. Sci. USA 99:10096-10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levy, D. E., and A. García-Sastre. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143-156. [DOI] [PubMed] [Google Scholar]
- 24.Li, Y., Z. Y. Chen, W. Wang, C. C. Baker, and R. M. Krug. 2001. The 3′-end-processing factor CPSF is required for the splicing of single-intron pre-mRNAs in vivo. RNA 7:920-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, J., P. A. Lynch, C. Y. Chien, G. T. Montelione, R. M. Krug, and H. M. Berman. 1997. Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat. Struct. Biol. 4:896-899. [DOI] [PubMed] [Google Scholar]
- 26.Lu, Y., M. Wambach, M. G. Katze, and R. M. Krug. 1995. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 214:222-228. [DOI] [PubMed] [Google Scholar]
- 27.Ludwig, S., X. Wang, C. Ehrhardt, H. Zheng, D. Nicola, O. Planz, S. Pleschka, A. García-Sastre, G. Heins, and T. Wolff. 2002. The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J. Virol. 76:11166-11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madore, S. J., L. S. Tiley, M. H. Malim, and B. R. Cullen. 1994. Sequence requirements for Rev multimerization in vivo. Virology 202:186-194. [DOI] [PubMed] [Google Scholar]
- 29.Marión, R. M., T. Aragón, A. Beloso, A. Nieto, and J. Ortín. 1997. The N-terminal half of the influenza virus NS1 protein is sufficient for nuclear retention of mRNA and enhancement of viral mRNA translation. Nucleic Acids Res. 25:4271-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mattoon, D., K. Gupta, J. Doyon, P. J. Loll, and D. DiMaio. 2001. Identification of the transmembrane dimer interface of the bovine papillomavirus E5 protein. Oncogene 20:3824-3834. [DOI] [PubMed] [Google Scholar]
- 31.Murphy, B. R., S. Baron, E. G. Chalhub, C. P. Uhlendorf, and R. M. Chanock. 1973. Temperature-sensitive mutants of influenza virus. IV. Induction of interferon in the nasopharynx by wild-type and a temperature-sensitive recombinant virus. J. Infect. Dis. 128:488-493. [DOI] [PubMed] [Google Scholar]
- 32.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1:991-1000. [DOI] [PubMed] [Google Scholar]
- 33.Nemeroff, M. E., X. Y. Qian, and R. M. Krug. 1995. The influenza virus NS1 protein forms multimers in vitro and in vivo. Virology 212:422-428. [DOI] [PubMed] [Google Scholar]
- 34.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norton, G. P., T. Tanaka, K. Tobita, S. Nakada, D. A. Buonaugurio, D. Greenspan, M. Krystal, and P. Palese. 1987. Infectious influenza A and B virus variants with long carboxyl terminal deletions in the NS1 polypeptides. Virology 156:204-213. [DOI] [PubMed] [Google Scholar]
- 36.Park, Y. W., and M. G. Katze. 1995. Translational control by influenza virus. Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J. Biol. Chem. 270:28433-28439. [DOI] [PubMed] [Google Scholar]
- 37.Qian, X. Y., F. Alonso-Caplen, and R. M. Krug. 1994. Two functional domains of the influenza virus NS1 protein are required for regulation of nuclear export of mRNA. J. Virol. 68:2433-2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian, X. Y., C. Y. Chien, Y. Lu, G. T. Montelione, and R. M. Krug. 1995. An amino-terminal polypeptide fragment of the influenza virus NS1 protein possesses specific RNA-binding activity and largely helical backbone structure. RNA 1:948-956. [PMC free article] [PubMed] [Google Scholar]
- 39.Qiu, Y., and R. M. Krug. 1994. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A). J. Virol. 68:2425-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qiu, Y., M. Nemeroff, and R. M. Krug. 1995. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA 1:304-316. [PMC free article] [PubMed] [Google Scholar]
- 41.Richman, D. D., B. R. Murphy, S. Baron, and C. Uhlendorf. 1976. Three strains of influenza A virus (H3N2): interferon sensitivity in vitro and interferon production in volunteers. J. Clin. Microbiol. 3:223-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimizu, K., A. Iguchi, R. Gomyou, and Y. Ono. 1999. Influenza virus inhibits cleavage of the HSP70 pre-mRNAs at the polyadenylation site. Virology 254:213-219. [DOI] [PubMed] [Google Scholar]
- 43.Smith, E. J., I. Marié, A. Prakash, A. García-Sastre, and D. E. Levy. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or IκB kinase but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 276:8951-8957. [DOI] [PubMed] [Google Scholar]
- 44.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 45.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. García-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Talon, J., M. Salvatore, R. E. O'Neill, Y. Nakaya, H. Zheng, T. Muster, A. García-Sastre, and P. Palese. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. USA 97:4309-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan, S. L., and M. G. Katze. 1998. Biochemical and genetic evidence for complex formation between the influenza A virus NS1 protein and the interferon-induced PKR protein kinase. J. Interferon Cytokine Res. 18:757-766. [DOI] [PubMed] [Google Scholar]
- 48.Treanor, J. J., R. Buja, and B. R. Murphy. 1991. Intragenic suppression of a deletion mutation of the nonstructural gene of an influenza A virus. J. Virol. 65:4204-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wade, R. H., and F. Kozielski. 2000. Structural links to kinesin directionality and movement. Nat. Struct. Biol. 7:456-460. [DOI] [PubMed] [Google Scholar]
- 50.Walters, K. J., K. T. Dayie, R. J. Reece, M. Ptashne, and G. Wagner. 1997. Structure and mobility of the PUT3 dimer. Nat. Struct. Biol. 4:744-750. [DOI] [PubMed] [Google Scholar]
- 51.Wang, W., and R. M. Krug. 1998. U6atac snRNA, the highly divergent counterpart of U6 snRNA, is the specific target that mediates inhibition of AT-AC splicing by the influenza virus NS1 protein. RNA 4:55-64. [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, W., K. Riedel, P. Lynch, C. Y. Chien, G. T. Montelione, and R. M. Krug. 1999. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA 5:195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. García-Sastre. 2000. Influenza A virus NS1 protein prevents the activation of NF-κB and induction of type I IFN. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 55.Zhou, A., J. M. Paranjape, S. D. Der, B. R. Williams, and R. H. Silverman. 1999. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology 258:435-440. [DOI] [PubMed] [Google Scholar]