Abstract
Although the kinetics of hybridization between a soluble polynucleotide and an immobilized complementary sequence have been studied by others, it is almost universally assumed that the interaction between each probe/target pair can be treated as a separate event. This simplifies the mathematics considerably, but it can give a false picture of the extent of hybridization that one achieves at equilibrium as well as the relative quantities of each hybridized pair during the approach to equilibrium. Here we solve the relevant kinetics equations simultaneously using Mathematica as a simulation language. Among the interesting results of this study are that, for certain circumstances, the relative ratio of incorrect to correct hybrids can change dramatically with time; that the relative abundances of two pairs are not what one would expect based on their equilibrium dissociation constants; that the volume of a wash solution after hybridization can have a large effect on results; and the fact that a short wash is typically better than a long one. We show that an optimum wash time exists for a given set of conditions. In addition, the ratio of soluble to insoluble (spotted) molecules can influence results substantially. Finally, the true levels of rare transcripts can be masked by the presence of highly abundant ones. Code is supplied to enable others to study conditions beyond those presented in this article.
INTRODUCTION
As the field of microarray expression analysis has matured, there has been an increasing interest in the accuracy of measured concentrations and concentration ratios of species such as mRNA. Whereas previously experimental reproducibility was such that threefold ratios of concentration between two samples were considered the minimum that could be reliably distinguished, better equipment and more reliable techniques are now producing data of higher caliber. As better experiments are carried out, more anomalies are reported. Relogio et al. (1), for example, studied mixtures of two RNA samples, each labeled with a different dye, and varied the ratio between them from 1:1 to 100:1. The measured ratios ranged from 1.6:1 to 30:1 with a smooth and continuous change from overestimation to underestimation of the correct ratio. Yue et al. (2) did a similar experiment and found that ratios from 30:1 to 1:30 gave anomalous results unless a certain minimal amount of DNA was spotted on their arrays. Dorris et al. (3) also reported significant errors in a dilution series for commercial CodeLink (Context, Captiva Software, San Diego, CA) and even more so for GeneChip (Affymetrix, Santa Clara, CA) arrays, so the problems are not confined to homemade arrays. They also demonstrated that the errors in discriminating perfectly-matched from single-base mismatched hybrids diminished as the hybridization time increased. In fact, these examples cited appeared to be careful and thorough studies, so it is very likely that most if not all of these problems are real and not related to technique or some peculiarity due to the sample.
At the same time, there has been an increasing interest in the thermodynamics and kinetics of microarray hybridization. The former has been invoked in an attempt to understand why some sequences work well, i.e., hybridize strongly, and others function poorly, and the latter to understand whether different hybridization times, concentrations, etc. should provide better experimental data (and to explain anomalies such as those cited above). This article does not consider the molecular interaction factors affecting kinetic constants, but instead examines how different constants and species concentrations affect the observed results, particularly the time-dependent and competitive effects as one or two different solution-phase species bind complementary oligonucleotide or cDNA strands in two different microarray spots. Heterogeneous (solid/soluble as opposed to two soluble species) hybridization kinetics have been examined by a number of people. However, the competitive binding situation has not been examined as carefully, and certain limitations in published approaches have led to results that are not always accurate.
Theory for a simple system
Here, we examine several general cases involving one or two soluble species competing for one or two types of binding sites. We use S to represent a soluble species, I to represent immobilized surface-bound species, and SI to represent the hybridized pair in a microarray spot. Although the terms probe and target are often applied to these species, two opposite conventions defining which is which are currently in use, so we prefer this less ambiguous nomenclature. Subscripts A,B,C, etc. will be used for the different sequences of nucleic acid. As usual, the kinetics are represented by the familiar relationship
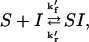 |
(1) |
where k′f represents the forward rate constant and k′r is the reverse constant. The equilibrium dissociation constant K′d is the ratio k′r/k′f. Since k′r ≪ k′f, K′d ≪ 1 and becomes smaller as the binding strength increases—the usual biochemical convention. For simplicity in these initial derivations, we will assume only a single set of species and omit the subscripts.
Unlike the case when both species are in solution, the situation is more complex when one of the two is immobilized on a surface. The relative amounts of the surface and solution phases must be taken into account. In addition, concentration units on the surface are generally measured in molecules/area (e.g., molecules/cm2), whereas those in solution are in moles/liter. This can be taken into account by dividing the surface phase concentration by v, the volume in liters of solution per square centimeter of surface area and by Avogadro's number, NAv, to convert from molecules to moles. Still more complications can arise if the layer of immobilized molecules does not cover the entire surface, or if the solution is unstirred, so that the effective volume that can equilibrate with DNA-covered surface differs from the total volume to total surface area ratio. This first of these complications can be handled by insuring that only the nucleotide-covered portion of the surface is used (spot area) rather the total surface. For purposes of this analysis, the last complication will be assumed not to exist, although in practice nonhomogeneous solution concentrations can be a significant problem. With these considerations, the differential equation describing adsorptive and desorptive events resulting in hybridization can be written as
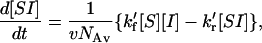 |
(2) |
where the square brackets denote concentrations in the units described above. When the soluble molecules are in substantial excess over the immobile ones, this equation can be written (using the subscript o to represent the initial concentration value at time = 0) as
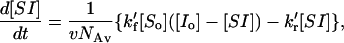 |
(3) |
leaving the hybrid concentration [SI] as the only variable. For simplicity, the primes on the rate constants (and on their corresponding dissociation constants) can be omitted and new constants used. These new kf1 and kr1 values should be understood to contain within their definition the values for the constants v and NAv as well; i.e., kf1 = k′f1 /v NAv. The value Kd, moles × ℓ−1, remains the same as K′d (see below), and the units of kf and kr have their usual units of liters × mol−1 × s−1 and s−1, respectively. These substitutions simplify the resulting equations. However, one must remember this substitution when interpreting their values. The recast Eq. 3 then becomes
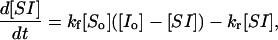 |
(4) |
which can be solved to give
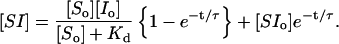 |
(5) |
In the usual case where initially there is no bound complex, the second term disappears, leaving
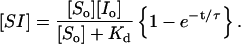 |
(6) |
In Eqs. 4 and 5, the term τ is defined below as
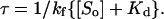 |
(7) |
Equation 4 is formally identical to that presented by Lauffenburger and Linderman (4) for receptor/ligand interaction except for some nomenclature changes and the fact that v Nav is embedded in the definition of the rate constants. Since both rate constants are divided by these constants, their ratio as the dissociation constant is the same as without them:
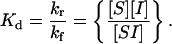 |
(8) |
It is worth roughly estimating the magnitude of 1/(v NAv). A typical microarray slide might have 10 μl of solution in contact with 1 cm2 of spot surface area. Therefore, 1/(v NAv) would be on the order of 10−18 cm2 × mole × liter−1 × molecule−1. A monolayer of DNA in a surface spot could contain up to ∼1012 molecules × cm−2 (although it is often considerably less; Graves, (5)). The equivalent of this concentration in liquid phase units would thus be ∼10−6 mole × liter−1 or less, not an unreasonable figure in comparison with what is likely to be present in the liquid phase.
Expected values for the forward, reverse, and dissociation constants can be estimated from literature values (6–9). These are typically between 104 and 5 × 106 ℓ × mole−1 s−1 for kf, 0.1–10−5 s−1 for kr, and 10−7–10−11 mole × ℓ−1 for Kd. If for convenience we convert these to a micromolar basis, they become 0.01–5 ℓ × μmole−1 s−1 for kf and 10−1–10−5 μmole × ℓ−1 for Kd (kr unchanged). It has been estimated that when the two strands are mismatched, the relative affinities decrease ∼10- to 100-fold (i.e., kr and Kd both increase by these amounts) (7). Liquid phase concentrations also can be given in μmol × ℓ−1 and immobilized phase concentrations converted to the same units using the 1/(v NAv) factor. These definitions and substitutions for constants and concentrations can be used in the following sections of this article to give values that generally range from 10−3 to 103, but we make no attempt to cover the entire range of reasonable values. Furthermore, one should recall that the effects of diffusion have been totally ignored in the simulations that follow, so the timescale in the simulated results, which should be in seconds to be consistent with these other units, shows results that change much too quickly in comparison to real-world data. Livshits and Mirzabekov show that diffusion can dramatically slow the attainment of hybridization equilibrium (10). It is safest to view all the results that follow as qualitative indications about how simultaneous hybridizations will behave and interfere with one another. For this reason, units have been omitted from the results. Furthermore, many of the important results are related to the state that exists at equilibrium, where absolute rates are not particularly relevant.
Extension of theory for more than one equilibrium state
The exponential approach to an equilibrium value predicted by Eqs. 5 and 6 is not surprising, and a curve representing the time course of this behavior will not be presented here. However, more interesting and surprising results are seen when a soluble species is distributed between two immobilized sequences. For these and more complex situations, simultaneous differential equations must be solved, and the complexity of this situation dictates that a more efficient computerized solution be used. This method is less prone to human error, even if analytical solutions could be found in some cases. We have used Mathematica (Wolfram Research, Champaign, IL) to aid in this process, and now can remove the previous limitation of an excess of soluble over insoluble material. For soluble species Sc interacting with two different immobilized species IA and IB, respectively to create the immobilized complexes ICA and ICB, the pertinent differential equations are
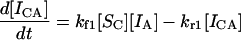 |
(9) |
and
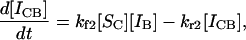 |
(10) |
where the subscripts 1 and 2 are used in the rate constants to denote the first and second competing reactions. The first reaction, with subscripts 1, refers to the A/C reaction and 2 to the B/C reaction with interactions analogous to those in Eq. 1. This set of equations was solved for the simple case of equimolar amounts of SC, IA, and IB and for rate constants kf1 = kf2 = 1, kr1 = 0.01, and kr2 = 0.1. These rate constants mean that the dissociation constants Kd1 and Kd2 are also respectively 0.01 and 0.1; in other words, the soluble species binds 10 times more strongly to immobilized A than it does to B. Therefore, CA will be considered the correct perfect-match hybrid, and CB represents the incorrect hybrid.
RESULTS
The initial binding event for a single soluble species on two spots
One might assume that for this case (with equilibrium constants that differ by a factor of 10), when the system comes to equilibrium, the concentration of the spot ICA would be 10 times as high as that of ICB. However, this is not the case. Fig. 1 a shows that after 50 arbitrary time units, the concentration of ICA is ∼0.74 and ICB is 0.23. Mathematica can carry out the calculation to an arbitrary number of decimal places, and typically we used ∼20. After 10,000 time units, ICA is 0.7448477… and ICB is 0.22595997… giving a ratio CA/CB closer to 3 than to 10, and showing that the apparent equilibrium at a time as short as 50 is indeed close to the correct value. Furthermore, ICB initially overshoots its final equilibrium value before dropping back to this value. This behavior is due primarily to two factors: the equal forward rate constants, and the depletion of material from the pool of soluble C species. That these results are correct is confirmed by the corresponding result that SC = 0.0291923…. Using the definition of Kd1 = SC · IA/ICA or the corresponding ratio for Kd2, these three values of concentration, it can be shown that they give very precisely the required values for Kd1 and Kd2,
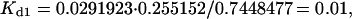 |
(11) |
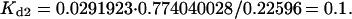 |
(12) |
In other words, these results show that one cannot assume a soluble species will partition between its perfect-match partner and a mismatch partner in a ratio proportional to the dissociation constant ratio. Once a molecule has hybridized to an incorrect spot, it is virtually impossible to get most of it to find its correct partner, even in this idealized model where there are no diffusional barriers to slow down the reequilibration. The results are even more striking if one sets the initial quantity in region B to 10 times that in region A, as we will see shortly. Physically, this latter situation is quite reasonable, since the amount of immobilized DNA, perhaps in a number of almost-complementary spots that could bind C, would be much greater than that in the single perfect-match spot. Now, the amount of incorrectly hybridized C exceeds that of the correct hybrid by a ratio of ∼3:2.
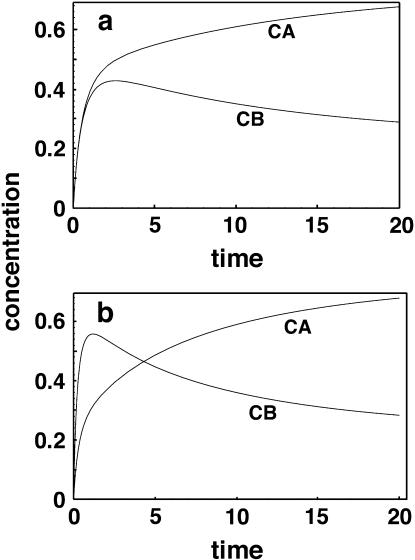
FIGURE 1.
(a) Time course for competitive hybridization for correct hybrids (CA) and incorrect hybrids (CB). Forward rate constants are equal to 1 and reverse constants are 0.01 and 0.1 respectively. (b) Similar plot but with forward and reverse constants for CB increased threefold. Note that in this latter case the incorrect hybrid can temporarily exceed the correct hybrid in concentration, although the eventual result is the same as in a.
In response to a reviewer's suggestion, we developed an entirely different Mathematica solution to find equilibrium values for the species concentrations. It involved optimizing concentrations to minimize the differences between forward and reverse rates using a built-in function of the mathematical language. This method culminated in results for this case and others that follow, indistinguishable from those found by the kinetic analysis. Code for both methods is provided.
This exercise was repeated with other values of the reverse rate constants and relative quantities of material A, B, and C, and the results are shown in Table 1. Note that this analysis considers only the initial hybridization step and not the washing step, which will be covered later. These calculations were carried out with dissociation constants of 0.01 and 0.001 for species A (the preferred immobilized material), while the constant for species B (the incorrect hybrid) remained fixed at 0.1. Some of the conclusions that can be drawn looking at the results in this table are as follows:
When the relative amounts of A and B are similar and there is not an excess of the soluble species (rows 2 and 7), the relative amount of C hybridized to the two spots does not vary as much as one might expect. Even when the ratio of equilibrium dissociation constants is 100 (row 7), the expected fluorescence signals differ only by a factor of ∼10.
The time required to reach equilibrium is generally long unless there is a large excess of soluble species over that which is immobilized.
When there is a large excess of soluble material (rows 3, 5, 8, and 10), the value of the dissociation constant plays very little role in determining the relative amounts of correct and incorrect hybrid formed. Both types of spots become saturated regardless of their dissociation constants.
When the incorrect immobilized material and soluble species are both in excess (rows 5 and 10), hybridization to the incorrect spot can greatly exceed that to the correct partner.
An illustration of why this type of analysis is necessary and how different the results can be from those obtained by considering the equilibrium of two spots as separate events (as has been done by others) can be seen by looking again at Fig. 1 a. With kf1 = kf2 = 1, kr1 = 0.01, and kr2 = 0.1, both species initially hybridize at similar rates, so that their concentrations are virtually identical. However, as both species start to reach equilibrium, the reverse rates make themselves felt and the less-strongly bound species B begins to come off the surface again, reversing the rate of adsorption to a net desorption. Clearly, any attempt to measure relative affinities or concentrations too early in the process would result in very inappropriate conclusions. Furthermore, simple exponential adsorption curves would not represent the actual kinetics well at all.
TABLE 1.
Relative hybridization to two immobilized spots
| Rev. rate const. kr1
|
Initial molar amounts of
|
% Hybridized
|
Expected signal on
|
Time to equilibrium
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Row | Immob. A (correct) | Immob. B (incorrect) | C (sol.) | A | B | C | A (correct) | B (incorrect) | ||
| 1 | 0.01 | 10 | 1 | 1 | 9.8 | 1.1 | 99.9 | 0.98 | 0.01 | long |
| 2 | 0.01 | 1 | 1 | 1 | 74.5 | 22.6 | 97.1 | 0.745 | 0.226 | long |
| 3 | 0.01 | 1 | 1 | 10 | 99.9 | 98.8 | 19.9 | 1 | 0.99 | short |
| 4 | 0.01 | 1 | 10 | 1 | 39.1 | 6 | 99.4 | 0.39 | 0.6 | long |
| 5 | 0.01 | 1 | 10 | 10 | 98.2 | 84.7 | 94.5 | 0.98 | 8.47 | short |
| 6 | 0.001 | 10 | 1 | 1 | 10 | 0.1 | 100 | 1 | 0.001 | long |
| 7 | 0.001 | 1 | 1 | 1 | 90.4 | 8.6 | 99.1 | 0.9 | 0.086 | long |
| 8 | 0.001 | 1 | 1 | 10 | 100 | 98.8 | 19.9 | 1 | 0.988 | short |
| 9 | 0.001 | 1 | 10 | 1 | 73.2 | 2.7 | 99.7 | 0.73 | 0.27 | long |
| 10 | 0.001 | 1 | 10 | 10 | 99.8 | 84.5 | 94.5 | 0.99 | 8.45 | short |
The rate constants for these simulations were kf1 = kf2 = 1; kr2 = 0.1. Short equilibrium times denote values of ≤5, while long times are >50.
If one were to make kf2 = 3 and kr2 = 0.3, so that the dissociation constant for the incorrect species pair remains the same but the on- and off-rates are three times higher, it is even possible to have the less strongly bound species temporarily at higher concentration than the more strongly bound species (Fig. 1 b). In fact, there is some experimental evidence that the initial formation of incorrect hybrids can occur faster than that of correct hybrids (11), and such a reversal makes sense intuitively. It is easier for a DNA or RNA chain to find a partial match than a perfect one. Those who claim that it is not necessary to be at or near equilibrium to get accurate estimates of the relative expression of mRNAs would be hard-pressed to justify their position in light of these results. If the binding constants are much stronger but in the same ratio (for example, kr1 = 10−6 and kr2 = 10−5), virtually all of the soluble C will be taken up rapidly and in a ratio proportional to the amounts of immobilized A and B present. Eventually, however, over long times the ratio will adjust to an equilibrium value with more material in CA (data not shown). These results suggest that the two-dye method (where a standard sample is modified by one dye, the test sample by a second dye and the two samples mixed before hybridization) is likely to provide better information on relative abundances when one cannot afford to wait for a slow equilibration. This question will be considered again in more detail later.
Washing of hybridized spots
These results have not yet considered the effect of a washing step after hybridization. This was done by using Mathematica to solve Eqs. 9 and 10 again following transformation in the manner shown in Eq. 4 to consider the initial amounts of immobilized complex. The initial concentration values for complexes CA and CB were taken from the results in Table 1, and the initial amount of C in the washing solution was set at zero. One additional complication for this analysis is that the wash solution volume is generally much larger than that used during hybridization. A dilution term symbolized as “dil” was used in the calculations to dilute the effective concentration of C as it returns to the solution phase. For illustration purposes, this factor was set arbitrarily at 100 to generate the results shown in Table 2. Now, in several cases where the concentration of the incorrect hybrid was comparable to or exceeded the properly matching one (rows 2, 3, 5, and 8–10), the situation has been at least partially corrected by washing. Only in rows 5 and 10 is the situation still rather poor. In those cases where the relative ratio during hybridization was favorable (rows 1, 6, and 7) the situation is approximately the same or slightly improved relative to the prewashing results.
TABLE 2.
Hybridization washing results
| Initial molar amounts of
|
Expected signal before washing
|
Expected signal after long washing
|
Expect. signal ratio after washing
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Row | Rev. rate const. kr1 | Immob. A (correct) | Immob. B (incorrect) | C (sol.) | A (correct) | B (incorrect) | A (correct) | B (incorrect) | for opt. time | at t = 1000 | Opt. wash time |
| 1 | 0.01 | 10 | 1 | 1 | 0.98 | 0.01 | 0.883 | 0.0096 | 144.4 | 92 | 9 |
| 2 | 0.01 | 1 | 1 | 1 | 0.745 | 0.226 | 0.359 | 0.053 | 15.03 | 6.77 | 32 |
| 3 | 0.01 | 1 | 1 | 10 | 1 | 0.99 | 0.567 | 0.116 | 6.66 | 4.9 | 40 |
| 4 | 0.01 | 1 | 10 | 1 | 0.39 | 0.6 | 0.269 | 0.354 | 1.1 | 0.758 | 17 |
| 5 | 0.01 | 1 | 10 | 10 | 0.98 | 8.47 | 0.839 | 3.42 | 0.251 | 0.245 | 24 |
| 6 | 0.001 | 10 | 1 | 1 | 1 | 0.001 | 0.99 | 0.001 | 1526 | 902 | 9 |
| 7 | 0.001 | 1 | 1 | 1 | 0.9 | 0.086 | 0.714 | 0.024 | 79.7 | 29.5 | 43 |
| 8 | 0.001 | 1 | 1 | 10 | 1 | 0.988 | 0.908 | 0.09 | 10.99 | 10.09 | 60 |
| 9 | 0.001 | 1 | 10 | 1 | 0.73 | 0.27 | 0.644 | 0.176 | 5.17 | 3.65 | 24 |
| 10 | 0.001 | 1 | 10 | 10 | 0.99 | 8.45 | 0.981 | 3.372 | 0.291 | 0.291 | 45 |
The rate constants for these simulations were kf1 = kf2 = 1; kr2 = 0.1. The washing dilution factor was set at 100. All washings were done for a time of 1000. By this time, CB had come to a virtual equilibrium, but CA generally was still decreasing.
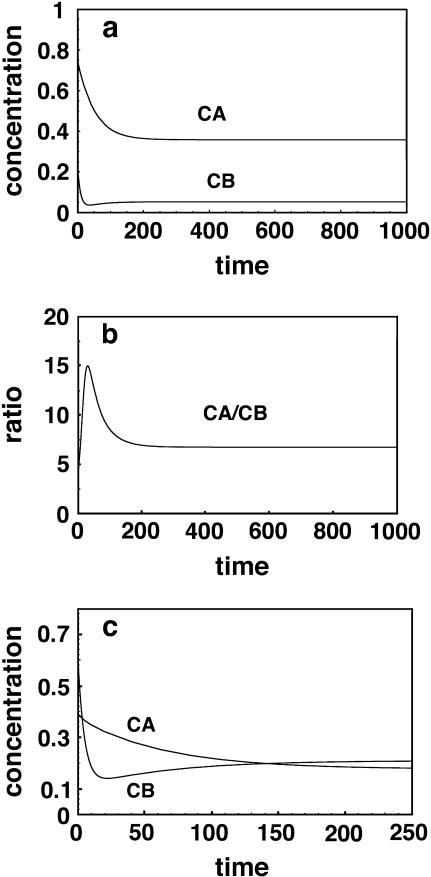
An extreme example is shown in row 4, where the correct to incorrect ratio increases from an unfavorable (CA/CB < 1) to a favorable value (CA/CB > 1) but then deteriorates again to an unfavorable value with continued washing, presumably because the poor binding of the CB complex is more than offset by the much larger amount of B relative to A. It is interesting to note that, in general, one is better off conducting the wash step for only a limited time. Since the incorrect CB complex initially dissociates faster than the correctly hybridized CA complex, it will reach a low value relatively quickly. If the wash is terminated at this point in time, the CA/CB ratio will be higher than it is when final equilibrium is reached and additional CA has had time to dissociate. An optimum wash time exists, as shown in the last column. The previous two columns in Table 2 show the ratio at the optimum time and a time of 1000, which effectively represents infinite time. Fig. 2, a and b, shows the rate of approach of the two complexes to the equilibrium values and at what value of the washing time the ratio between the two is at an optimum value. The unusual case represented by the row-4 data, where the relative abundance of the two products reverses twice, is shown in Fig. 2 c. Again, one should recognize that none of this behavior could be predicted by considering kinetics of the two products separately and independently.
FIGURE 2.
(a) Time course for washing hybrids CA and CB with 100 times the initial hybridization volume using clean solution. (b) Note that an optimum time (maximum CA/CB ratio) exists for washing. Extended washing removes C from CA as well as from the incorrect CB. (c) For the case given by row 4 in Table 2, the CA/CB ratio changes from unfavorable to favorable before returning to an unfavorable value again at long times.
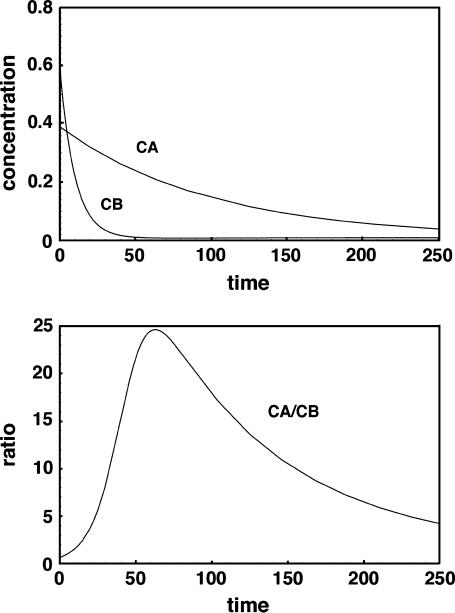
By increasing the dilution factor to a large value, for example 10,000, a good ratio of correct to incorrect hybrid can be obtained even in these extreme cases, but only if the wash is conducted for a restricted period of time. The results corresponding to row 4 but with dil = 10,000 (not shown) give a CA/CB ratio of 24.6 at the optimum time of 63, compared to the ratio shown of 1.1 at an optimum wash time of 17 when the dilution factor is 100. At long times such as 1000 with a wash dilution of 10,000, the ratio returns to a poor value of approximately unity and both complexes are washed off the surface to very low concentrations. Fig. 3 shows that for large-volume short time washes there is a tradeoff: Although one would not want to exceed a wash time of 63 for this particular case, even shorter times than this might be preferable since the signal intensities drop off significantly with time. For example, at a time of 25, the CA/CB ratio is still a fairly respectable 5.63, whereas the CA signal is 0.304 rather than the value of 0.209 that it assumes at the optimum time of 63. This result emphasizes how important a large-volume, but relatively short washing step can be, in eliminating incorrect hybrids.
FIGURE 3.
Time course for washing of hybridized species at 10,000 times the initial hybridization volume. Times shorter than the optimal value of 63 may be useful to prevent too much of the correct hybrid CA from dissociating.
These calculations could easily be carried out with the derived equations for more complex cases such as those in Southern blotting, where several different stringencies (simulated by different values of kr and kf) are used in a washing sequence. They also suggest that the real value of multiple washes may be that they are needed to dilute the removed soluble molecule more thoroughly than would be possible in a single wash step. As an example, dil was set at 3000 and two consecutive washes were simulated, each for their optimum times. After two washes, the CA/CB ratio was 52.5, more than twice as good as a single wash with dil at 10,000. However, the final CA concentration in this case was only 0.174, slightly less than the 0.209 value for the single wash; and the total time, 83, for wash 1 and wash 2 optimal times of 50 and 33, respectively, was larger than the time of 63 required for a single wash. The total wash volume of 6000 is obviously less than the single wash volume of 10,000 assumed in the previous case.
Competitive binding between two immobilized molecules and two soluble molecules
Although the case of two immobilized and one soluble species is quite revealing, a simulation with two of each species is closer to the actual set of complex competitive processes taking place within real microarray systems. In this situation the relevant set is equations is as follows:
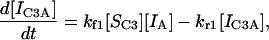 |
(13) |
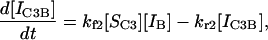 |
(14) |
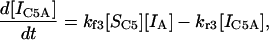 |
(15) |
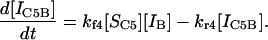 |
(16) |
We first consider a special case of this reaction set where the two soluble species have identical binding properties for the two immobilized species (i.e., kf1 = kf3, kr1 = kr3, kf2 = kf4, kr2 = kr4). This simulates the familiar two-dye experiment where presumably the dyes represent a standard and test sample that have been separately labeled and then mixed together. The premise of the two-color method is that even if hybridization is not carried out to equilibrium, the relative amounts of standard and test molecules that hybridize (as shown by the two dyes) will be proportional to their initial concentrations. This enables one to determine whether a particular gene has been up- or downregulated in a test sample relative to the standard. We use C3 and C5 to represent Cy 3- and Cy 5-labeled samples of the soluble species. Several cases were examined with binding constants and initial concentrations covering ranges similar to those studied in the previous section with one soluble species. In all cases and for all times, the relative amounts of surface complexes formed were indeed directly proportional to the initial amounts of C3 and C5 in solution (results not shown). Each adsorption curve was proportionally mirrored by its twin. This is exactly as expected and is as much a validation of the mathematical model itself as it is a validation of the two-color experiment. Of course, in the real world, the different relative sizes of the dye molecules or their labeling efficiencies, etc., will probably affect the kinetic constants and/or results, so dye-swap experiments, where the dyes used to identify standard and test samples are interchanged, are still necessary. Washing experiments were not simulated for this case since they were not expected to provide any further useful information.
A more interesting case is one in which all eight binding constants are allowed to assume independent values. Of course with eight constants and four initial species concentrations it is much more difficult to study a reasonable subset of possible conditions. One interesting case that was studied assumed that C1 (the first soluble species) was supposed to bind to A and C2 (the second) to B, each pair with equal strength. However, each could also bind the incorrect immobilized partner (C1 to B and C2 to A) more weakly but again with equal strength. This should be a fair representation of real experiments, since immobilized species are generally designed to have approximately equal binding affinities for their complements. Note that the soluble species nomenclature has been modified from C3 and C5 to C1 and C2 to avoid confusion. These species no longer represent two different dyes but simply two different gene products. We have already shown that the dyes are expected to sort according to values of the kinetic constants.
Just as was the case for the earlier simulation, the second type of solution method (concentration optimization to minimize forward and reverse rates) was carried out to verify the apparent equilibrium values obtained at long times. This optimization was very demanding on the algorithm with so many variables, and three of the cases (given by lines 5, 7, and 9 in Table 3) did not converge. Two others started to converge (lines 2 and 11 in Table 3) but did not completely regenerate all the correct equilibrium constants. All other cases converged and gave results indistinguishable from the differential equation solution method. For those that did not converge properly, a third method was used. Equilibrium concentrations predicted by the differential equation solver (our first method) were substituted into all four equilibrium relations and the resulting constants were compared with the values originally supplied as data. Again, in all cases results were indistinguishable from the originals. Since all results have been verified by at least one independent method (several were tested by both methods two and three), we are quite confident of their correctness and accuracy.
TABLE 3.
Double hybridization results
| Rev. rate const. kr1,kr4
|
Initial molar amounts of
|
Expected (correct/incorrect) signal ratio before washing
|
Actual concentrations of correct hybrids
|
Error from C1B on C2B
|
|||||
|---|---|---|---|---|---|---|---|---|---|
| Row | C1 (solution) | C2 (solution) | A, B (surface) | C1A/C2A | C2B/C1B | C1A | C2B | ||
| 1 | 0.01 | 1 | 1 | 1 | 10 | 10 | 0.826 | 0.826 | 10% |
| 2 | 0.01 | 1 | 0.1 | 1 | 225 | 0.444 | 0.754 | 0.095 | 310% |
| 3 | 0.01 | 0.1 | 0.1 | 1 | 10 | 10 | 0.09 | 0.09 | 10% |
| 4 | 0.01 | 0.1 | 0.01 | 1 | 102 | 0.949 | 0.089 | 0.009 | 108% |
| 5 | 0.01 | 0.01 | 0.001 | 1 | 100.7 | 0.993 | 0.009 | 0.0009 | 101% |
| 6 | 0.001 | 1 | 1 | 1 | 100 | 100 | 0.959 | 0.959 | 1% |
| 7 | 0.001 | 1 | 0.1 | 1 | 8171 | 1.22 | 0.909 | 0.1 | 100% |
| 8 | 0.001 | 0.1 | 0.1 | 1 | 100 | 100 | 0.099 | 0.099 | 1% |
| 9 | 0.001 | 0.1 | 0.01 | 1 | 1095 | 9.13 | 0.099 | 0.0099 | 11% |
| 10 | 0.001 | 0.01 | 0.001 | 1 | 1098 | 9.91 | 0.0099 | 0.00099 | 10% |
| 11 | 0.001 | 1 | 0.001 | 1 | 8.70E+05 | 0.0116 | 0.9043 | 0.000997 | 9515% |
The rate constants for these simulations were kf1 = kf2 = 1 = kf3 = kf4 = 1. Reverse constant kr2 = kr3 = 0.1.
The first question to be asked for the four-species cross-hybridization case was whether a large amount of the incorrect species could distort the apparent concentration of the correct species. Table 3 shows results where all four forward rate constants kf1, kf2, kf3, and kf4 were set equal to 1. Two of the reverse constants, kr2 and kr3, were set equal to 0.1. The other two constants were set to the values shown in the table. The quantities of immobilized material A and B in the two types of spots were set equal to one another, and the soluble species C1 and C2 were varied as shown. When the concentrations of the soluble species are comparable (rows 1, 3, 6, and 8), the correct to incorrect ratios at long times (C1A/C2A and C2B/C1B) are excellent (≫1). However, when the ratio of correct to incorrect binding constants was only 10 (kr1 and kr4 = 0.01; rows 1–5) and the C1 and C2 concentrations differed, the rarer nucleotide complex suffered (rows 2, 4, and 5). Note that the situation becomes somewhat better as the total amount of soluble material decreases relative to the amount of material in the spots, but the ratio never reaches even a value of 1.
The situation improves in rows 6–10, where the binding constant ratio becomes 100 rather than 10. However, the situation is still not good in row 7, where both soluble species are relatively abundant in comparison to the immobilized spot concentration. Row 11 was added to show what would happen in the case of a very rare nucleotide in the presence of a large amount of a common one. Now, even though the binding constant ratio of correct to incorrect is very favorable (100), the large concentration difference completely overcomes this advantage. The C2B/C1B ratio indicates that the proper amounts of C2 and C1 in solution would not be registered by hybridization to their respective immobilized complements.
The last three columns in the table represent the actual concentrations of the correct hybrids C1A and C2B and the error in perceived value due to the additional binding of the incorrect species. In other words, both C1A and C2A would fluoresce, so the perceived signal on spot A would be incorrectly high (likewise on spot B). Note that when the ratios in columns 6 and 7 are well above unity, correct results are seen. However, when they become small, errors can be large. This is especially apparent in the row-11 data, where the C2 product is present at only 0.001 of the amount of C1. Here, the incorrect hybrid completely swamps the signal, giving a value >95 times that of the correct one even though it binds 100 times less strongly than the correct one. This result suggests that the measured concentrations of rare gene products may be much higher than their true values in typical microarray experiments. It would seem that even two-dye labeling would not help resolve the issue, since all mRNAs in a given sample (test or standard) would have the same label. One would expect that the relative abundances of such rare products (test versus standard) would follow the ratio of an incorrect but plentiful product that also hybridizes slightly to the complementary immobilized spot rather than assuming their true values. However, it must be remembered that we have not yet considered the washing step.
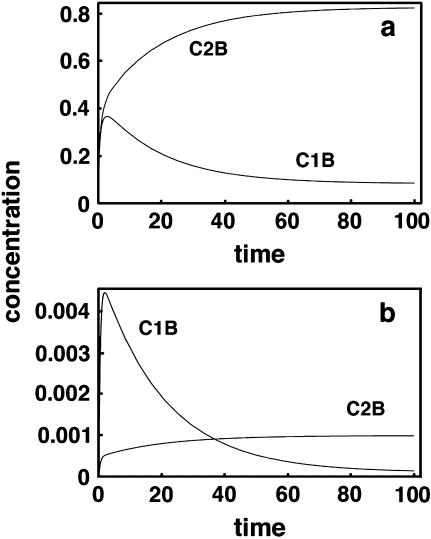
Fig. 4 shows two representative sets of results for some of these simulations. Fig. 4 a represents results for the data in row 1 and Fig. 4 b for that in row 10. Although the curves are labeled C1B and C2B, they really represent all four species. Since the kinetic constants chosen were symmetrical, curve C1B also represents C2A (both being the incorrect hybrids) and C2B represents C1A (the correct hybrids). Note that in the first panel, where the soluble species C1 and C2 are initially present in equal amounts, the correct hybrid always exceeds the incorrect one in concentration. However, in the second panel, one can see that because C1 is present in solution at 10 times the concentration of C2, initially the incorrect hybrid C1B forms faster than C2B. Any attempt to measure their relative amounts before ∼40 time units would give completely erroneous information. As stated above, this has implications for rare transcripts relative to the abundant ones in a mixture.
FIGURE 4.
Time course for a system with two soluble and two insoluble polynucleotides. C1 is the complement of A and C2 of B. However, C1 and cross-hybridize with B and C2 with A. Although only B hybrids are shown, the constants used are symmetrical so that C2B represents C1A and C1B represents C2A as well. (a) Results from the constants given in Table 3, row 1. (b) Results for Table 3, row 10.
Washing the hybrid products of two immobilized molecules and two soluble molecules
In a manner analogous to that used for a single soluble species, simulated washing of the hybrids between two soluble and two immobilized species was carried out. Table 4 presents results for washing the long-time products predicted by the binding simulation for conditions in Table 3. Thus, results for a given row in Table 3 representing long hybridization times were used directly as starting conditions for the simulations presented in the corresponding row in Table 4. As in the case of washing a single soluble species from the hybrids, an optimum washing time exists. Before this time, the incorrect hybrid is being removed faster than the correct one, and afterwards the correct hybrid dissociates faster (because of the combined effects of concentration and dissociation rate for each species). It is interesting to note that the optimum times fall into a narrow range for all the conditions in this table. Although this is not entirely unexpected since the dissociation rate constant for the incorrect species was held constant for the entire set, it is a bit surprising since the reverse rate constant for the correct species was changed by a factor of 10 between rows 5 and 6. Note that when this array has been washed for the optimum time, all ratios of correct to incorrect hybrids (columns 5 and 6) have been corrected to a value of 10 or better except for the last row. Since this row represents a rare transcript in the presence of a much larger amount of another one, the potential problem this represents in actual microarray studies is worrisome.
TABLE 4.
Double hybridization washing results
| Rev. rate const. kr1,kr4
|
Initial molar amounts of
|
Opt. (correct/incorrect) signal ratio after wash
|
Conc. of correct hybrids at opt. time
|
Opt. wash time
|
C2B error (%)
|
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Row | C1 (solution) | C2 (solution) | A, B (surface) | C1A/C2A | C2B/C1B | C1A | C2B | |||
| 1 | 0.01 | 1 | 1 | 1 | 240 | 240 | 0.5098 | 0.5098 | 49 | 0.42 |
| 2 | 0.01 | 1 | 0.1 | 1 | 2502 | 10 | 0.4678 | 0.059 | 40 | 38.22 |
| 3 | 0.01 | 0.1 | 0.1 | 1 | 139 | 139 | 0.0582 | 0.0582 | 45 | 0.72 |
| 4 | 0.01 | 0.1 | 0.01 | 1 | 1398 | 13 | 0.0576 | 0.00583 | 45 | 8.92 |
| 5 | 0.01 | 0.01 | 0.001 | 1 | 1339 | 13 | 0.00589 | 0.00059 | 44 | 7.56 |
| 6 | 0.001 | 1 | 1 | 1 | 12662 | 12662 | 0.899 | 0.899 | 65 | 0.01 |
| 7 | 0.001 | 1 | 0.1 | 1 | 1.58E+05 | 47 | 0.871 | 0.0959 | 43 | 12.46 |
| 8 | 0.001 | 0.1 | 0.1 | 1 | 1931 | 1931 | 0.0946 | 0.0946 | 47 | 0.05 |
| 9 | 0.001 | 0.1 | 0.01 | 1 | 1.99E+04 | 174 | 0.0947 | 0.0095 | 46 | 0.89 |
| 10 | 0.001 | 0.01 | 0.001 | 1 | 1.86E+04 | 178 | 0.00948 | 0.00095 | 45 | 0.56 |
| 11 | 0.001 | 1 | 0.001 | 1 | 1.55E+07 | 0.407 | 0.8675 | 0.00096 | 42 | 281.21 |
The rate constants for these simulations were kf1 = kf2 = 1 = kf3 = kf4 = 1. Reverse constant kr2 = kr3 = 0.1.
It is interesting that Sakai et al. fractionated a population of cDNA fragments to produce subsets richer in rare transcripts and found that they were able to identify 10 times as many differentially expressed genes as they could with the unfractionated product (12). This may represent experimental confirmation that the potential problem we have identified is also a real one. Another very recent article shows similar results, and the authors comment that differentially expressed genes in a fractionated sample were more readily detected even when their absolute intensities had not been enhanced by the specific PCR primers relative to the initial unfractionated sample (13). Another less-specific but perhaps relevant article was published by Miklos and Maleszka (14). These authors compared up- and downregulated genes in schizophrenia obtained with synthesized Affymetrix arrays, with spotted oligonucleotide arrays, and those genes identified in clinical studies. Only one identified gene was common to the two types of arrays out of 89 and 49 found by the two methods individually. For the combined arrays (138 total identified genes), only eight were found to be in common with the 97 up- or downregulated genes identified clinically. The authors also stated that the genes identified in some cases depended on the particular bioinformatic tools used in array analysis. Thus, there is a real question whether cross-hybridization may be confounding hybridization results in a typical experiment.
DISCUSSION OF RESULTS AND CONCLUSIONS
It is clear that when there is the possibility that soluble DNA species can interact with one or more immobilized species, the simple exponential approaches to equilibrium expected for a single hybrid pair are no longer seen (and a number of analyses presented previously in the literature are thus shown to be incorrect). We have developed fairly simple code based on the Mathematica language that can be used to investigate multiple simultaneous hybridization and washing phenomena. The investigation has revealed a number of interesting and important results that have not been identified by others in previous work.
First, the relative abundances of two hybrid pairs that form simultaneously can change dramatically with time, and an initial incorrect hybrid even can be present temporarily at a higher level than the correct one. Therefore, microarray data taken too early in the equilibration process are likely to be in error. Even when equilibrium has been reached, the relative abundances of hybrid complexes are not in the same ratios as one might expect from the relative equilibrium dissociation constants. If both equilibria are favored, the hybrids tend to be more similar in concentration than one might expect. Therefore, the probability that cross-hybridization is significant is also higher than one might expect. Second, the results obtained from a microarray experiment will depend strongly on the conditions used during the washing cycle: how many times the cycle is repeated with fresh solution, how effective mixing is during the washing process, and what volume of wash solution is employed. We have shown that an optimum washing time exists, which, to our knowledge, has not been demonstrated theoretically before. Although good experimentalists undoubtedly have an intuitive feeling that too little washing will fail to remove the incorrect hybrids and too long a wash cycle will remove both incorrect and correct hybrids, leading to weak signals, our technique provides quantitative values for optimum wash times given approximate values for the binding constants and volume of wash solution. Although considerable effort on equilibration and discussion of the effects of different equilibration times is seen in the literature, washing has not received the attention it deserves, nor has its importance been generally recognized.
Third, we show that two-dye experiments are more likely to provide correct answers in microarray experiments than single-dye experiments, particularly where the solution and microspot phases have not come to equilibrium. Here the experimentalists' intuitive feeling about how to improve a microarray analysis has been accurate.
Finally, and perhaps most importantly, cross-hybridization can be an especially significant problem when the incorrect soluble species is much higher in concentration than the correct soluble species. This result, in combination with the earlier stated tendency for hybrids to be similar in concentration even when their equilibrium constants differ significantly, suggests that high expression level mRNAs (or their cDNA representatives) can overpower the low expression level molecules by what amounts to a law-of-mass-action effect. The practical significance of this result is that the way most microarray experiments is currently being carried out may lead to completely erroneous results for some of the rare transcripts. Two articles are cited in which this effect may already have been observed. In these works, the authors fractionated mRNA populations to eliminate some of the high expression level molecules and they saw more differentially expressed genes in the microarray analysis. This result should be of concern to all those who use microarrays to understand cellular function. Unfortunately, since a given sample will have all mRNAs labeled with a particular dye, two-dye experiments are just as likely to suffer from this problem as the simpler single dye experiment.
The Mathematica programs used to obtain these results are available to permit others to study situations not addressed by the cases we have presented here. They are straightforward to use even by those with little familiarity with the language. Other questions and situations in which this type of analysis is useful undoubtedly will arise, and the simulation method and code provided here should prove useful in addressing them. The equilibrium results we present have been verified by use of at least two, and in some cases three, entirely different solution methods. Thus, we have considerable confidence in their accuracy.
SUPPLEMENTAL MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
This work was supported by National Science Foundation grants No. BES-9986834 and No. BES-0314265.
References
- 1.Relogio, A., C. Schwager, A. Richter, W. Ansorge, and J. Valcarcel. 2002. Optimization of oligonucleotide-based DNA microarrays. Nucleic Acids Res. 30:e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yue, H., P. S. Eastman, B. B. Wang, J. Minor, M. H. Doctolero, R. L. Nuttall, R. Stack, J. W. Becker, J. R. Montgomery, M. Vainer, and R. Johnston. 2001. An evaluation of the performance of cDNA microarrays for detecting changes in global mRNA expression. Nucleic Acids Res. 29:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorris, D. R., A. Nguyen, L. Gieser, R. Lockner, A. Lublinsky, M. Patterson, E. Touma, T. J. Sendera, R. Elghanian, and A. Mazumder. 2003. Oligodeoxyribonucleotide probe accessibility on a three-dimensional DNA microarray surface and the effect of hybridization time on the accuracy of expression ratios. BMC Biotechnol. 3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lauffenburger, D. A., and J. J. Linderman. 1993. Receptors. Oxford University Press, Oxford, UK.
- 5.Graves, D. J. 1999. Microarrays: powerful tools for genetic analysis come of age. Trends Biotechnol. 17:127–134. [DOI] [PubMed] [Google Scholar]
- 6.Persson, B., K. Stenhag, P. Nilsson, A. Larsson, M. Uhlen, and P.-A. Hygren. 1997. Analysis of oligonucleotide probe affinities using surface plasmon resonance: a means for mutational scanning. Anal. Biochem. 246:34–44. [DOI] [PubMed] [Google Scholar]
- 7.Wang, S. S., A. E. Friedman, and E. T. Kool. 1995. Origins of high sequence selectivity: a stopped-flow kinetics study of DNA/RNA hybridization by duplex- and triplex-forming oligonucleotides. Biochemistry. 34:9774–9784. [DOI] [PubMed] [Google Scholar]
- 8.Stillman, B. A., and J. L. Tonkinson. 2001. Expression microarray hybridization kinetics depend on length of the immobilized DNA but are independent of immobilization substrate. Anal. Biochem. 295:149–157. [DOI] [PubMed] [Google Scholar]
- 9.Tawa, K., and W. Knoll. 2004. Mismatching base-pair dependence of the kinetics of DNA-DNA hybridization studied by surface plasmon fluorescence spectroscopy. Nucleic Acids Res. 32:2372–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Livshits, M. A., and A. D. Mirzabekov. 1996. Theoretical analysis of the kinetics of DNA hybridization with gel immobilized oligonucleotides. Biophys. J. 71:2795–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai, H., M. Meyer, S. Stepaniants, M. Ziman, and R. Stoughton. 2002. Use of hybridization kinetics for differentiating specific from non-specific binding to oligonucleotide microarrays. Nucleic Acids Res. 30:e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakai, K., H. Higuchi, K. Matsubara, and K. Kato. 2000. Microarray hybridization with fractionated cDNA: enhanced identification of differentially expressed genes. Anal. Biochem. 287:32–37. [DOI] [PubMed] [Google Scholar]
- 13.Rondeau, G., M. McClelland, T. Nguyen, R. Risques, Y. Wang, M. Judex, A. H. Cho, and J. Welsh. 2005. Enhanced microarray performance using low complexity representations of the transcriptome. Nucleic Acids Res. 33:e100–e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miklos, G. L., and R. Maleszka. 2004. Microarray reality checks in the context of a complex disease. Nat. Biotechnol. 22:615–621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.