Abstract
Structural transition can be induced in charged micelles by increasing the ionic strength of the medium. We have monitored the organization and dynamics of the functionally important tryptophan residues of gramicidin in spherical and rod-shaped sodium dodecyl sulfate micelles utilizing a combination of wavelength-selective fluorescence and related fluorescence approaches. Our results show that tryptophans in gramicidin, present in the single-stranded β6.3 conformation, experience slow solvent relaxation giving rise to red edge excitation shift in spherical and rod-shaped micelles. In addition, changes in fluorescence polarization with increasing excitation or emission wavelength reinforce that the gramicidin tryptophans are localized in motionally restricted regions of these micelles. Fluorescence quenching experiments using acrylamide as a quencher of tryptophan fluorescence show that there is reduced water penetration in rod-shaped micelles. Taken together, we show that gramicidin conformation and dynamics is sensitive to the salt-induced structural transition in charged micelles. In addition, these results demonstrate that deformation of the host assembly could modulate protein conformation and dynamics.
INTRODUCTION
Detergents are extremely important in studies of biological membranes due to their ability to solubilize membrane proteins (1,2). They are soluble amphiphiles and above a critical concentration (strictly speaking, a narrow concentration range), known as the critical micelle concentration (CMC), self-associate to form thermodynamically stable, noncovalent aggregates called micelles (3). Studies on micellar organization and dynamics assume special significance in light of the fact that the general principle underlying the formation of micelles (that is, the hydrophobic effect) is common to other related assemblies such as reverse micelles, bilayers, liposomes, and biological membranes (3,4). Micelles have been used as membrane mimetics to characterize membrane proteins and peptides (5). Micelles are highly cooperative, organized molecular assemblies of amphiphiles and are dynamic in nature (6). Further, they offer certain inherent advantages in fluorescence studies over membranes because micelles are smaller and optically transparent, have well-defined sizes, and are relatively scatter free.
Structural transition can be induced in charged micelles at a given temperature by increasing ionic strength of the medium or amphiphile concentration (7,8). Thus, spherical micelles of sodium dodecyl sulfate (SDS) that exist in water at concentrations higher than CMC assume an elongated rod-like structure in presence of high electrolyte (salt) concentrations when interactions among the charged headgroups are attenuated due to the added salt (see inset in Fig. 1). This is known as sphere-to-rod transition (9). This shape change induced by increased salt concentration is accompanied by a reduction in CMC (10,11). It has been suggested that large rod-shaped micelles are better models for biomembranes (12) and the hydrocarbon chains are more ordered in rod-shaped micelles compared to spherical micelles (7) giving rise to higher microviscosity in rod-shaped micelles (13). Micellar sphere-to-rod transitions can be explained in terms of the packing model described by Israelachvili (14).
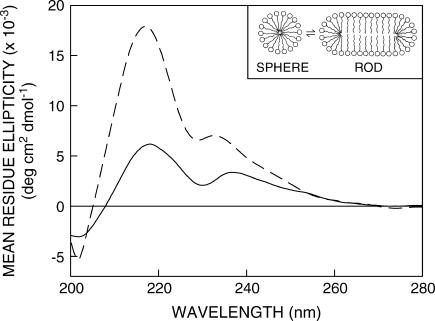
FIGURE 1.
Far-UV CD spectra of gramicidin in SDS micelles in the absence (solid line) and presence (dashed line) of 0.5 M NaCl. The ratio of gramicidin/SDS was 1:500 (mol/mol) and the concentration of gramicidin was 32 μM. See Materials and Methods for other details. The inset shows a schematic representation of the sphere-to-rod transition in charged SDS micelles induced by salt. Note that the headgroup spacing is reduced in rod-shaped micelles due to attenuation of interactions among the charged headgroups by the added salt.
Ion channels are crucial cellular components that are involved in the maintenance of appropriate ion balance across a biological membrane. The linear peptide gramicidin forms prototypical ion channels specific for monovalent cations and has been extensively used to study the organization, dynamics, and function of membrane-spanning channels (15–17). Gramicidin serves as an excellent model for transmembrane channels due to its small size, ready availability, and the relative ease with which chemical modifications can be performed. This makes gramicidin unique among small membrane-active peptides and provides the basis for its use to explore the principles that govern the folding and function of membrane-spanning channels in particular, and membrane proteins in general. The unique sequence of alternating l- and d-chirality renders gramicidin sensitive to the environment in which it is placed (18). Gramicidin therefore adopts a wide range of environment-dependent conformations (19). The most preferred (thermodynamically stable) conformation in membranes is the single-stranded β6.3 conformation (20). The head-to-head (amino terminal to amino terminal) single-stranded β6.3 helical dimer form is the cation conducting channel conformation of gramicidin in membranes (21). In this conformation, the carboxy terminus is exposed to the membrane-water interface and the amino terminus is buried in the hydrophobic core of the membrane. This places the tryptophan residues clustered at the membrane-water interface at the entrance to the channel (21–25).
In this article, we have explored the organization and dynamics of the functionally important tryptophan residues of gramicidin in SDS micelles utilizing a combination of wavelength-selective fluorescence and related fluorescence approaches. In addition, we have monitored the change in organization and dynamics of gramicidin tryptophans due to the salt-induced sphere-to-rod transition in SDS micelles. Wavelength-selective fluorescence comprises a set of approaches based on the red edge effect in fluorescence spectroscopy that can be used to directly monitor the environment and dynamics around a fluorophore in an organized molecular assembly (26,27). A shift in the wavelength of maximum fluorescence emission toward higher wavelengths, caused by a shift in the excitation wavelength toward the red edge of the absorption band, is termed red edge excitation shift (REES) (26–28). This effect is mostly observed with polar fluorophores in motionally restricted environments such as viscous solutions or condensed phases where the dipolar relaxation time for the solvent shell around a fluorophore is comparable to or longer than its fluorescence lifetime. REES arises due to slow rates of solvent relaxation (reorientation) around an excited state fluorophore, which is dependent on the motional restriction imposed on the solvent molecules in the immediate vicinity of the fluorophore. Utilizing this approach, it becomes possible to probe the mobility parameters of the environment itself (which is represented by the relaxing solvent molecules) using the fluorophore merely as a reporter group. This makes the use of REES in particular and the wavelength-selective fluorescence approach in general very useful because hydration plays a crucial modulatory role in a large number of important cellular events including protein folding, lipid-protein interactions, and ion transport (29). The unique feature about REES is that whereas all other fluorescence techniques such as fluorescence quenching, resonance energy transfer, and polarization measurements yield information about the fluorophore itself, REES provides information about the relative rates of solvent (water in biological systems) relaxation dynamics, which is not possible to obtain by other techniques.
MATERIALS AND METHODS
Materials
Gramicidin A′ (from Bacillus brevis), SDS, 1,6-diphenyl-1,3,5-hexatriene (DPH), and trifluoroethanol (TFE) were purchased from Sigma Chemical (St. Louis, MO). Gramicidin A′, as obtained, is a mixture of gramicidins A, B, and C. The concentration of gramicidin was calculated from its molar extinction coefficient (ɛ) of 20,700 M−1cm−1 at 280 nm (20). Ultrapure-grade acrylamide was from Invitrogen Life Technologies (Carlsbad, CA). The purity of acrylamide was checked from its absorbance using its molar extinction coefficient (ɛ) of 0.23 M−1cm−1 at 295 nm and optical transparency beyond 310 nm (30). All other chemicals used were of the highest purity available. Water was purified through a Millipore (Bedford, MA) Milli-Q system and used throughout. The purity of SDS was checked by measuring its CMC and comparing with literature CMC. The CMC of SDS was determined fluorimetrically utilizing the enhancement of DPH fluorescence upon micellization (11). All experiments were done at room temperature (23°C).
Sample preparation
The concentration of SDS (16 mM) used was double its CMC to ensure that it is in the micellar state. The molar ratio of peptide/detergent was carefully chosen to give optimum signal/noise ratio with minimal perturbation to the micellar organization and negligible interprobe interactions. The maximum molar ratio of gramicidin/SDS used was 1:500 (mol/mol). To incorporate gramicidin into micelles, 48 nmol of gramicidin (6 nmol in experiments involving acrylamide quenching) from a TFE stock solution was dried under a stream of nitrogen while being warmed gently (∼35°C). After further drying under a high vacuum for at least 24 h, 1.5 mL of 16 mM SDS (in the presence or absence of 0.5 M NaCl) was added, and samples were vortexed for 3 min. The micellar suspension was then sonicated for 15 min in a Branson model 250 sonifier (Branson Ultrasonics, Danbury, CT) fitted with a microtip. Sonicated samples were then centrifuged at 15,000 rpm for 15 min at room temperature to remove any titanium particles shed from the microtip during sonication, and incubated at 65°C for 12 h to induce the β6.3 conformation that is typical of the channel conformation (20,31). Background samples were prepared the same way except that peptide was not added to them. All samples were equilibrated at room temperature in dark for 2 h after incubation at 65°C.
Steady-state fluorescence measurements
Steady-state fluorescence measurements were performed with a Hitachi F-4010 spectrofluorometer using 1-cm pathlength quartz cuvettes. Excitation and emission slits with a nominal bandpass of 5 nm were used for all measurements. Background intensities of samples in which gramicidin was omitted were negligible in most cases and were subtracted from each sample spectrum to cancel out any contribution due to the solvent Raman peak and other scattering artifacts. The spectral shifts obtained with different sets of samples were identical in most cases, or were within ±1 nm of the ones reported. Fluorescence polarization measurements were performed using a Hitachi polarization accessory. Polarization values were calculated from the equation (32):
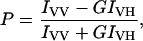 |
(1) |
where IVV and IVH are the measured fluorescence intensities (after appropriate background subtraction) with the excitation polarizer vertically oriented and emission polarizer vertically and horizontally oriented, respectively. G is the grating correction factor and is the ratio of the efficiencies of the detection system for vertically and horizontally polarized light, and is equal to IHV/IHH. All experiments were done with multiple sets of samples and average values of polarization are shown in Figs. 4 and 5.
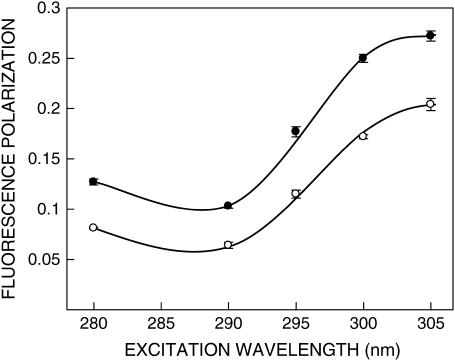
FIGURE 4.
Fluorescence polarization of gramicidin in SDS micelles in the absence (○) and presence (•) of 0.5 M NaCl as a function of excitation wavelength. The emission wavelength was 331 nm in all cases. The data points shown are the means ± SE of at least three independent measurements. All other conditions are as in Fig. 1. See Materials and Methods for other details.
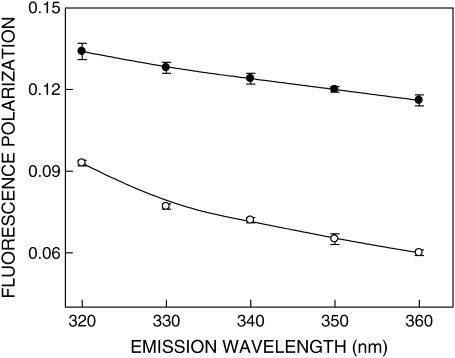
FIGURE 5.
Fluorescence polarization of gramicidin in SDS micelles in the absence (○) and presence (•) of 0.5 M NaCl as a function of emission wavelength. The excitation wavelength was 280 nm in all cases. The data points shown are the means ± SE of at least three independent measurements. All other conditions are as in Fig. 1. See Materials and Methods for other details.
Fluorescence quenching measurements
Acrylamide quenching experiments of gramicidin tryptophan fluorescence were carried out by measurement of fluorescence intensity after serial addition of small aliquots of a freshly prepared stock solution of 4 M acrylamide in water to a stirred sample followed by incubation for 3 min in the sample compartment in the dark (shutters closed). The excitation wavelength used was 295 nm and emission was monitored at 334 nm. Corrections for inner filter effect were made using the following equation (32):
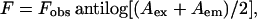 |
(2) |
where F is the corrected fluorescence intensity and Fobs is the background-subtracted fluorescence intensity of the sample (also corrected for dilution). Aex and Aem are the measured absorbances at the excitation and emission wavelengths. The absorbances of the samples were measured using a Hitachi U-2000 ultraviolet (UV)-visible absorption spectrophotometer. Quenching data were analyzed by fitting to the Stern-Volmer equation (32):
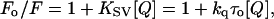 |
(3) |
where Fo and F are the fluorescence intensities in the absence and presence of the quencher, respectively, [Q] is the molar quencher concentration, and KSV is the Stern-Volmer quenching constant. The Stern-Volmer quenching constant KSV is equal to kqτo where kq is the bimolecular quenching constant and τo is the lifetime of the fluorophore in the absence of quencher.
Time-resolved fluorescence measurements
Fluorescence lifetimes were calculated from time-resolved fluorescence intensity decays using a Photon Technology International (London, Western Ontario, Canada) LS-100 luminescence spectrophotometer in the time-correlated single-photon counting mode. This machine uses a thyratron-gated nanosecond flash lamp filled with nitrogen as the plasma gas (17 ± 1 inches of mercury vacuum) and is run at 22–25 kHz. Lamp profiles were measured at the excitation wavelength using Ludox (colloidal silica) as the scatterer. To optimize the signal/noise ratio, 5000 photon counts were collected in the peak channel. The excitation wavelength used was 297 nm and emission was set at 340 nm. All experiments were performed using excitation and emission slits with a bandpass of 8 nm or less. The sample and the scatterer were alternated after every 10% acquisition to ensure compensation for shape and timing drifts occurring during the period of data collection. This arrangement also prevents any prolonged exposure of the sample to the excitation beam thereby avoiding any possible photodamage to the fluorophore. The data stored in a multichannel analyzer were routinely transferred to an IBM PC for analysis. Fluorescence intensity decay curves so obtained were deconvoluted with the instrument response function and analyzed as a sum of exponential terms:
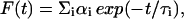 |
(4) |
where F(t) is the fluorescence intensity at time t and αi is a preexponential factor representing the fractional contribution to the time-resolved decay of the component with a lifetime τi such that Σi αi = 1. The decay parameters were recovered using a nonlinear least-squares iterative fitting procedure based on the Marquardt algorithm (33). The program also includes statistical and plotting subroutine packages (34). The goodness of the fit of a given set of observed data and the chosen function was evaluated by the reduced χ2 ratio, the weighted residuals (35), and the autocorrelation function of the weighted residuals (36). A fit was considered acceptable when plots of the weighted residuals and the autocorrelation function showed random deviation about zero with a minimum χ2 value generally not more than 1.2. Mean (average) lifetimes (i.e., intensity-averaged lifetimes) τ for biexponential decays of fluorescence were calculated from the decay times and preexponential factors using the following equation (32):
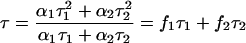 |
(5) |
where
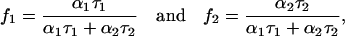 |
and represent fractional intensities corresponding to each lifetime component.
Circular dichroism measurements
Circular dichroism (CD) measurements were carried out at room temperature (23°C) on a JASCO J-715 spectropolarimeter that was calibrated with (+)-10-camphorsulfonic acid (37). The spectra were scanned in a quartz optical cell with a pathlength of 0.1 cm. All spectra were recorded in 0.5-nm wavelength increments with a 4-s response and a bandwidth of 1 nm. For monitoring changes in secondary structure, spectra were scanned from 200 to 280 nm at a scan rate of 100 nm/min. Each spectrum is the average of 12 scans with a full scale sensitivity of 10 mdeg. All spectra were corrected for background by subtraction of appropriate blanks and were smoothed making sure that the overall shape of the spectrum remains unaltered. Data are represented as mean residue ellipticities and were calculated using the formula:
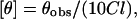 |
(6) |
where θobs is the observed ellipticity in mdeg, l is the pathlength in centimeters, and C is the concentration of peptide bonds in mol/L.
RESULTS
Gramicidin conformation in SDS micelles monitored by circular dichroism spectroscopy
Circular dichroism spectroscopy has been extensively used to characterize gramicidin conformations (20,25,31). The single-stranded β6.3 conformation has been earlier shown to be the preferred conformation in SDS micelles (38). To avoid any conformational heterogeneity in our samples, we chose to use gramicidin from a TFE stock solution that would allow direct incorporation of gramicidin in the single-stranded β6.3 conformation. Nonetheless, to further ensure that there is no conformational heterogeneity, we sonicated the samples followed by prolonged heat incubation at 65°C before measurements were made (see Material and Methods). The CD spectra of gramicidin incorporated in SDS micelles are shown in Fig. 1. The spectral characteristics in both spherical and rod-shaped micelles are typical of the single-stranded β6.3 conformation with two characteristic peaks of positive ellipticity at ∼218 and 235 nm and a valley at ∼230 nm. Gramicidin has previously been shown to exist as a head-to-head β6.3 dimer in SDS micelles (38,39). The aggregation state of gramicidin in the conditions described here is presumed to be dimeric although the surfactant/gramicidin molar ratio is higher. It should be noted that the peaks are red shifted in the spherical micelles compared to the corresponding peaks in the case of rod-shaped micelles. This could possibly be due to the increased water penetration in spherical micelles (see later) because far-UV CD spectra of gramicidin are known to be sensitive to solvent polarity (40). In addition, the ellipticity at 218 nm is nearly triple in rod-shaped micelles as compared to spherical micelles. This enhancement in ellipticity is probably due to tighter (bilayer-like) packing in the rod-shaped micelles that would reduce conformational heterogeneity and enhance secondary structural elements.
Fluorescence characteristics and red-edge excitation shift of gramicidin in SDS micelles
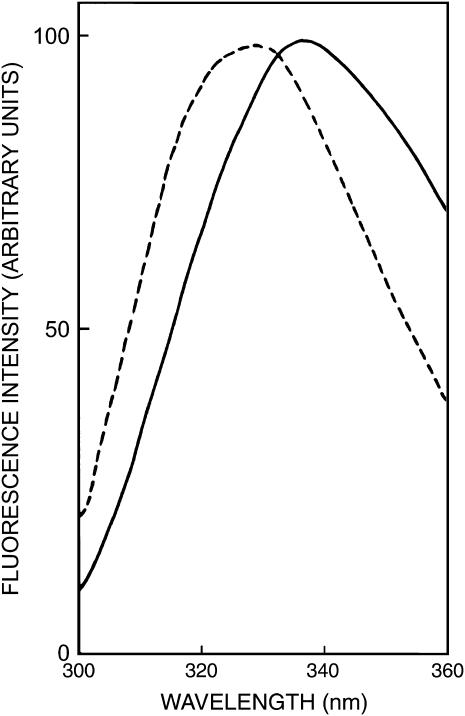
The fluorescence emission spectra of gramicidin in SDS micelles in the absence and presence of NaCl are shown in Fig. 2. Gramicidin tryptophans exhibit an emission maximum of 336 nm in SDS micelles in the absence of NaCl. The emission maximum of gramicidin in SDS micelles in the presence of NaCl, however, displays a blue shift and is at 327 nm. This indicates a reduction in polarity of the tryptophan environment in presence of NaCl, i.e, in rod-shaped micelles. This is possibly due to a decrease in water content as a result of tighter packing in rod-shaped micelles owing to neutralization of the charge on detergent headgroups by the counterions.
FIGURE 2.
Fluorescence emission spectra of gramicidin in SDS micelles in the absence (solid line) and presence (dashed line) of 0.5 M NaCl. The excitation wavelength used was 280 nm. All other conditions are as in Fig. 1. See Materials and Methods for other details.
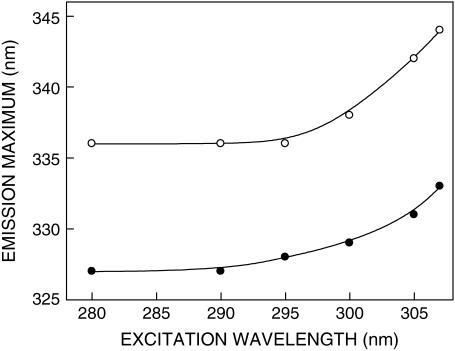
The shift in the maxima of fluorescence emission of the tryptophan residues of gramicidin in SDS micelles as a function of excitation wavelength is shown in Fig. 3. We have used the term maximum of fluorescence emission in a somewhat wider sense here. In every case, we have monitored the wavelength corresponding to maximum fluorescence intensity, as well as the center of mass of the fluorescence emission. In most cases, both these methods yielded the same wavelength. In cases where minor discrepancies were found, the center of mass of emission has been reported as the fluorescence maximum.
FIGURE 3.
Effect of changing excitation wavelength on the wavelength of maximum emission of gramicidin in SDS micelles in the absence (○) and presence (•) of 0.5 M NaCl. All other conditions are as in Fig. 1. See Materials and Methods for other details.
As the excitation wavelength is changed from 280 to 307 nm, the emission maximum of gramicidin is shifted toward longer wavelengths in both cases. Fig. 3 shows that the emission maximum is shifted from 336 to 344 nm in case of gramicidin in spherical micelles, which corresponds to a REES of 8 nm. It is possible that there could be further red shift if excitation is carried out beyond 307 nm. We found it difficult to work in this wavelength range due to low signal/noise ratio and artifacts due to the solvent Raman peak that sometimes remained even after background subtraction. Such dependence of the emission maximum on excitation wavelength is characteristic of the red-edge excitation shift. Because gramicidin tryptophans are localized in the micellar interfacial region (39), such a result would imply that this region of the micellar assembly offers considerable restriction to reorientation of solvent dipoles around the excited state tryptophans. Earlier work from our group (24,25) and others (41) have shown that gramicidin tryptophans in the single-stranded β6.3 conformation are localized in a motionally restricted region of the membrane, consistent with the interfacial localization of these tryptophans in the membrane (21,23). Our results here show that the gramicidin tryptophans, on the average, are localized in a region of slow solvent relaxation (reorientation) in the micellar assembly, and reinforce their localization at the micellar interface. Importantly, the micellar interface is characterized by a motionally restricted environment (42,43) and is associated with ordered water that displays slow relaxation dynamics compared to bulk water (44,45). Because REES arises due to the dynamics of reorientational motion of solvent molecules, our results assume significance in the overall context of restricted micellar dynamics in the interface region. In the presence of 0.5 M NaCl (i.e., in the rod-shaped micelles), the emission maximum of gramicidin tryptophans was shifted from 327 to 333 nm when the excitation wavelength was shifted in the same wavelength range (from 280 to 307 nm) that corresponds to a REES of 6 nm (Fig. 3).
Fluorescence polarization of gramicidin in SDS micelles is dependent on excitation and emission wavelengths
The fluorescence polarization of gramicidin tryptophans in SDS micelles is shown in Table 1. The polarization values provide information on the local rotational motion of the tryptophan residues of gramicidin in these environments. The polarization value is low in spherical micelles in the absence of NaCl whereas polarization is relatively high in rod-shaped micelles formed in presence of NaCl. The gramicidin tryptophans that are interfacially localized are sensitive to the difference in packing caused by the sphere-to-rod transition induced by NaCl and this gives rise to the increase in polarization (also see below).
TABLE 1.
Fluorescence polarization of gramicidin in SDS micelles
| Host* | Fluorescence polarization † | τc‡ (ns) |
|---|---|---|
| SDS micelles | 0.072 ± 0.001 | 0.65 |
| SDS micelles (in the presence of 0.5 M NaCl) | 0.124 ± 0.002 | 1.06 |
The excitation wavelength was 280 nm; emission was monitored at 340 nm. All other conditions are as in Fig. 1. See Materials and Methods for other details.
Calculated using Eq. 1. The values shown are the means ± SE of three independent experiments.
Calculated using Eq. 7. See text for other details.
In addition to the shift in emission maximum on red-edge excitation, fluorescence polarization is also known to be dependent on excitation wavelength in motionally restricted media (46). Due to strong dipolar interactions with the surrounding solvent molecules, there is a decreased rotational rate of the fluorophore in the solvent relaxed state. Red-edge excitation results in selective excitation of this subclass of fluorophore. Because of strong interactions with the polar solvent molecules in the excited state, one may expect these “solvent relaxed” fluorophores to rotate more slowly, thereby increasing the polarization. The excitation polarization spectra (i.e., a plot of steady-state polarization versus excitation wavelength) of gramicidin tryptophans in SDS micelles are shown in Fig. 4. As observed earlier (Table 1), the polarization values for gramicidin tryptophans in spherical micelles are lower than the corresponding values in rod-shaped micelles at all excitation wavelengths. More importantly, the polarization values exhibit considerable change upon altering the excitation wavelength, with a sharp increase toward the red edge of the absorption band and a characteristic dip near 290 nm in both cases. Such an increase in polarization upon red-edge excitation for peptides and proteins containing tryptophans, especially in media of reduced mobility has been previously reported (24,47).
It is known that tryptophan has two overlapping So → S1 electronic transitions (1La and 1Lb), which are almost perpendicular to each other (48). Both So → 1La and So → 1Lb transitions occur in the 260–300-nm range. In nonpolar solvents, 1La has higher energy than 1Lb. However, in polar solvents, the energy level of 1La is lowered, making it the lowest energy state. This inversion is believed to occur because 1La transition has higher dipole moment (as it is directed through the ring −NH group), and can have dipole-dipole interactions with polar solvent molecules. Irrespective of whether 1La or 1Lb is the lowest S1 state, equilibration between these two states is believed to be very fast (of the order of 10−12 s), so that only emission from the lower S1 state is observed (49). In a motionally restricted polar environment, absorption at the red edge photoselects the lowest energy S1 (1La in this case), and thus the polarization is high because depolarization only due to small angular differences between the absorption and emission transition moments and solvent reorientation, if any, occurs. Excitation at the shorter wavelengths, however, populates both 1La and 1Lb states. Equilibration between these two states produces a depolarization due to the ∼90° angular difference between 1La and 1Lb moments. Thus, near 290 nm, there is a dip in polarization due to maximal absorption by the 1Lb state. Fig. 4 shows such a characteristic dip around 290 nm in the excitation polarization spectrum of gramicidin tryptophans. Thus, the sharp increase in polarization toward the red edge of the absorption band is probably because the extent of depolarization in gramicidin tryptophans is reduced at the red edge not only due to decreased rotational rate of the fluorophore in the solvent relaxed state, but also due to photoselection of the predominantly 1La transition, which in turn, reduces the contribution to depolarization because of 1Lb → 1La equilibration.
For fluorophores incorporated in motionally restricted media, fluorescence polarization is also known to be dependent on emission wavelength. Under such conditions, a steady decrease in polarization is observed with increasing emission wavelength (46). Fig. 5 shows variation in fluorescence polarization of gramicidin tryptophans in SDS micelles as a function of emission wavelength. As previously noted, the polarization values for gramicidin tryptophans in spherical micelles are in general lower than the corresponding values in rod-shaped micelles at all emission wavelengths. As seen from Fig. 5, there is a considerable reduction in polarization with increasing emission wavelength in both cases. The lowest polarization is observed toward the red edge where the solvent relaxed emission predominates. Taken together, the changes in fluorescence polarization of gramicidin in SDS micelles as a function of excitation and emission wavelengths reinforce the presence of a motionally restricted environment in the vicinity of the gramicidin tryptophans.
Fluorescence lifetime and acrylamide quenching of gramicidin tryptophans in SDS micelles
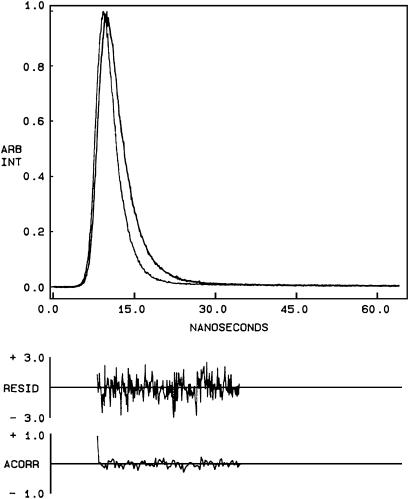
Fluorescence lifetime serves as a sensitive indicator for the local environment and polarity in which a given fluorophore is placed (50). A typical decay profile of gramicidin tryptophans in spherical micelles of SDS with its biexponential fitting and the various statistical parameters used to check the goodness of the fit is shown in Fig. 6. The fluorescence lifetimes of gramicidin tryptophans in SDS micelles are shown in Table 2. We chose to use the mean fluorescence (intensity-averaged) lifetime as an important parameter for describing the behavior of gramicidin tryptophans in SDS micelles because it is independent of the number of exponentials used to fit the time-resolved fluorescence decay. The mean fluorescence lifetimes of gramicidin tryptophans in SDS micelles calculated using Eq. 5 are shown in Table 2. In general, tryptophan lifetimes are known to be reduced when exposed to polar environments (51). Because the hydrocarbon chains in rod-shaped micelles are more ordered, water penetration is relatively reduced as compared to spherical micelles. In other words, the spherical micelles would allow more water penetration into the micellar interior (see acrylamide quenching results below) and this, in principle, could lead to a reduction in tryptophan lifetime. Surprisingly, the mean fluorescence lifetime of gramicidin tryptophans is reduced in rod-shaped micelles (0.91 ns as compared to 1.46 ns in spherical micelles). This would not be expected on the basis of the dependence of tryptophan lifetime on polarity alone. However, there are other factors that need to be considered while interpreting changes in fluorescence lifetime. It has been previously suggested that aromatic-aromatic (stacking) interactions between Trp-9 and Trp-15 could reduce the mean fluorescence lifetime of gramicidin incorporated in membranes (24). Such an interaction will be more predominant in membrane or membrane-mimetic environments (such as in rod-shaped micelles) than in spherical micelles (38,52). This could result in reduced tryptophan lifetime in rod-shaped micelles.
FIGURE 6.
Time-resolved fluorescence intensity decay of gramicidin in spherical SDS micelles. Excitation wavelength was at 297 nm, which corresponds to a peak in the spectral output of the nitrogen lamp. Emission was monitored at 340 nm. The sharp peak on the left is the lamp profile. The relatively broad peak on the right is the decay profile, fitted to a biexponential function. The two bottom plots show the weighted residuals and the autocorrelation function of the weighted residuals. All other conditions are as in Fig. 1. See Materials and Methods for other details.
TABLE 2.
Fluorescence lifetimes of gramicidin in SDS micelles
The difference in fluorescence lifetimes in spherical and rod-shaped micelles could influence the observed fluorescence polarization values (Table 1). To ensure that the observed change in steady-state polarization is not due to any change in lifetime, we calculated the apparent (average) rotational correlation times for gramicidin tryptophans in spherical and rod-shaped micelles using Perrin's equation (32):
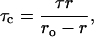 |
(7) |
where ro is the limiting anisotropy of tryptophan, r is the steady-state anisotropy (derived from the polarization values using r = 2P/(3 − P)), and τ is the mean fluorescence (intensity-averaged) lifetime taken from Table 2. The values of the apparent rotational correlation times, calculated this way using a value of ro of 0.16 (53), are shown in Table 1. As can be seen from the table, there is a considerable increase in the apparent rotational correlation time of gramicidin tryptophans upon sphere-to-rod transition. This clearly shows that the observed change in polarization values were not due to any lifetime-induced artifacts.
It should be mentioned here that the apparent rotational correlation times reported by us are not exact because Perrin's equation is strictly applicable only in case of isotropic rotors (32). Nonetheless, it is assumed here that this equation will apply to a first approximation. The presence of multiple tryptophans could be an additional complication. However, this would be minimized because we have used mean fluorescence lifetimes for calculating apparent rotational correlation times.
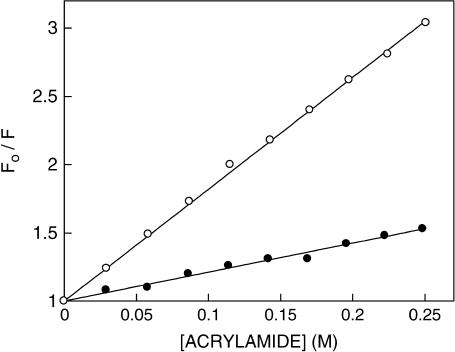
Acrylamide quenching of tryptophan fluorescence is widely used to monitor tryptophan environments in proteins (54). Fig. 7 shows representative Stern-Volmer plots of acrylamide quenching of gramicidin tryptophans in spherical and rod-shaped micelles. The slope (KSV) of such a plot is related to the accessibility (degree of exposure) of the tryptophans to the quencher. The quenching parameters obtained by analyzing the Stern-Volmer plot are shown in Table 3. The Stern-Volmer constant (KSV) for acrylamide quenching of gramicidin tryptophans in spherical micelles was found to be 8.46 M−1 whereas the value in rod-shaped micelles was found to be 2.20 M−1. However, interpretation of the Stern-Volmer constant is complicated this way due to its intrinsic dependence on fluorescence lifetime (see Eq. 3). The bimolecular quenching constant (kq) for acrylamide quenching is therefore a more accurate measure of the degree of exposure because kq takes into account differences in fluorescence lifetime. The bimolecular quenching constants, calculated using Eq. 3, are shown in Table 3. The kq values show that the tryptophans in spherical micelles are considerably more accessible to acrylamide. This could be a result of reduced water penetration (and therefore reduced accessibility to the aqueous quencher acrylamide) in the more tightly packed rod-shaped micelles formed in the presence of NaCl.
FIGURE 7.
Representative data for Stern-Volmer analysis of acrylamide quenching of gramicidin fluorescence in SDS micelles in the absence (○) and presence (•) of 0.5 M NaCl. Fo is the fluorescence in the absence of quencher, and F is the corrected fluorescence in the presence of quencher. The excitation wavelength was fixed at 295 nm and emission was monitored at 334 nm. Concentration of SDS was 16 mM and the ratio of gramicidin/SDS was 1:4000 (mol/mol). See Materials and Methods for other details.
TABLE 3.
Acrylamide quenching of gramicidin in SDS micelles
| Host* | KSV† (M−1) | kq (× 10−9)‡ (M−1s−1) |
|---|---|---|
| SDS micelles | 8.46 ± 0.51 | 5.79 |
| SDS micelles (in the presence of 0.5 M NaCl) | 2.20 ± 0.07 | 2.42 |
The ratio of gramicidin/SDS was 1:4000 (mol/mol). The excitation wavelength was 295 nm; emission was monitored at 334 nm. See Materials and Methods for other details.
DISCUSSION
Structural transitions involving shape changes play an important role in cellular physiology. For example, the shape of erythrocytes (red blood cells) has been shown to change with the pH and ionic strength of the medium (55). The shape of the erythrocyte is thought to be maintained by the membrane skeleton in close interaction with the plasma membrane (56). Investigations into the role of the membrane in such shape changes have revealed that modification of either the membrane composition or the structure of its individual constituents can lead to shape changes (57). Thus, alteration of the cholesterol composition, selective removal of phospholipids from the outer membrane leaflet, pH and membrane potential alterations, metabolic depletion, and introduction of lysophospholipids, fatty acids, and charged amphipathic agents in membranes leads to shape changes in erythrocytes (57–59). Shape changes can be induced even in liposomes by mechanical stress, temperature or pH variation, osmotic shock, and by asymmetric transmembrane distribution of phospholipids (60). Interestingly, SDS micellar shape change induced by chlorpromazine, an amphiphilic cationic phenothiazine drug, has recently been reported (61).
Shape changes in cellular membranes that occur due to modifications of membrane composition (57–59) can directly affect the function of membrane proteins such as mechanosensitive channels that respond to changes in membrane curvature (62). Interestingly, the function of the gramicidin channel has been shown to be sensitive to curvature changes of the membrane bilayer (63). In light of the fact that membrane packing properties are being increasingly recognized as important targets for ion channel toxins (64), the effects of such structural changes on the gramicidin channel, an important model for ion channels, become relevant.
Gramicidin represents a useful model for the realistic determination of conformational preference in a membrane or membrane-mimetic environment despite the alternating sequence of l-d chirality generally not encountered in naturally occurring peptides and proteins. This is due to the fact that the dihedral angle combinations generated in the conformation space by various gramicidin conformations are “allowed” according to the Ramachandran plot (65). Importantly, gramicidin channels share important structural features with other naturally occurring channel proteins like the bacterial KcsA K+ channel. These features include membrane interfacial localization of tryptophan residues, the channel interior being made of the peptide backbone, and ion selectivity arising out of backbone interactions (17).
In this article, we have explored the organization and dynamics of the functionally important tryptophan residues of gramicidin in spherical and rod-shaped SDS micelles utilizing a combination of wavelength-selective fluorescence and related fluorescence approaches. We show that gramicidin tryptophans show increased REES in spherical micelles (8 nm) as compared to rod-shaped micelles (6 nm). This difference in REES could reflect difference in microenvironment and packing experienced by the gramicidin tryptophans in these cases. Interestingly, we have previously shown that gramicidin in the channel conformation exhibits a REES of 4 nm in membrane bilayers under similar conditions (24,25). Because gramicidin is a multitryptophan peptide, any REES information obtained would be indicative of the average environment experienced by the tryptophans. The contributions of individual tryptophan residues of gramicidin to the observed fluorescence therefore becomes important. Earlier work using fluorescence (24,66) and molecular dynamics simulations (52) have clearly indicated the heterogeneity of the contributing tryptophan residues. Although fluorescence measurements provide evidence for stacking interactions among Trp-9 and Trp-15 when incorporated in membranes at least in the fluorescence timescale (∼ nanoseconds) (24), molecular dynamics simulations point out motional flexibility giving rise to conformational heterogeneity of Trp-9 (52). Moreover, recent results from other groups have indicated that conformational difference could exist for Trp-9 in micellar and membrane environments (38,52). This could influence REES results obtained for gramicidin tryptophans in these environments. In addition, fluorescence lifetime experiments indicate increased aromatic-aromatic stacking interactions in rod-shaped micelles, which would possibly influence the conformational heterogeneity of Trp-9 as discussed above.
As mentioned earlier, REES arises due to slow relaxation (reorientation) of solvent dipoles around the excited state dipole moment of the fluorophore, and provides useful information about environmental dynamics. It is important to note here that we have previously shown that the dynamics of the environment reported by REES is well correlated to the organization and dynamics of the fluorophore incorporated in such an environment (26,27). For example, we earlier reported that whereas tryptophan residues in a protein (such as tubulin) exhibit REES in the native (folded) state, no REES is observed upon denaturation (67). This is due to the loss of native structure in the denatured protein, because the folded protein matrix around the tryptophan residues increases motional restriction leading to slowing down of solvent dipole relaxation in the excited state. This clearly illustrates that dynamics of fluorophores in a macromolecular assembly is related to the dynamics of the environment (solvent dipolar relaxation). Interestingly, this relationship assumes special relevance in case of anisotropic molecular assemblies such as membranes and micelles. We have previously shown, using anthroyloxy probes located at a graded series of depths in the membrane, that the motional anisotropy in the z axis of the membrane bilayer along the phospholipid acyl chains (68) is correlated with the rate of solvent relaxation along this axis (69). This result reinforces the fact that there is significant coupling between the environment (solvent dipoles) and the fluorophore itself. More importantly, we have recently shown that REES serves as an indicator of the conformational status of the membrane-bound gramicidin (25). The dipolar relaxation around the tryptophan residues is therefore related to the dynamics of the peptide.
Our results show that gramicidin conformation, particularly the dynamics of the tryptophan residues, is sensitive to the salt-induced structural transition in charged micelles and in particular, the tighter packing of rod-shaped micelles as compared to more dynamic spherical micelles. In conclusion, our results using the well-characterized ion channel gramicidin, demonstrate that deformation of the host assembly could modulate protein conformation and dynamics.
Acknowledgments
We thank Y. S. S. V. Prasad and G. G. Kingi for technical help and members of our laboratory for critically reading the manuscript.
This work was supported by the Council of Scientific and Industrial Research, government of India. A.C. is an honorary faculty member of the Jawaharlal Nehru Centre for Advanced Scientific Research, Bangalore, India. S.S.R. and D.A.K. thank the Council of Scientific and Industrial Research for the award of Senior Research Fellowships.
Satinder S. Rawat's present address is Rm. 211, Bldg. 469, Laboratory of Experimental and Computational Biology, NCI-FCRDC, National Institutes of Health, Frederick, MD 21702-1201.
References
- 1.Chattopadhyay, A., K. G. Harikumar, and S. Kalipatnapu. 2002. Solubilization of high affinity G-protein-coupled serotonin1A receptors from bovine hippocampus using pre-micellar CHAPS at low concentration. Mol. Membr. Biol. 19:211–220. [DOI] [PubMed] [Google Scholar]
- 2.Seddon, A. M., P. Curnow, and P. J. Booth. 2004. Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta. 1666:105–117. [DOI] [PubMed] [Google Scholar]
- 3.Tanford, C. 1978. The hydrophobic effect and the organization of living matter. Science. 200:1012–1018. [DOI] [PubMed] [Google Scholar]
- 4.Israelachvili, J. N., S. Marcelja, and R. G. Horn. 1980. Physical principles of membrane organization. Q. Rev. Biophys. 13:121–200. [DOI] [PubMed] [Google Scholar]
- 5.Sham, S. S., S. Shobana, L. E. Townsley, J. B. Jordan, J. Q. Fernandez, O. S. Andersen, D. V. Greathouse, and J. F. Hinton. 2003. The structure, cation binding, transport and conductance of Gly15-gramicidin A incorporated into SDS micelles and PC/PG vesicles. Biochemistry. 42:1401–1409. [DOI] [PubMed] [Google Scholar]
- 6.Menger, F. M. 1979. The structure of micelles. Acc. Chem. Res. 12:111–117. [Google Scholar]
- 7.Heerklotz, H., A. Tsamaloukas, K. Kita-Tokarczyk, P. Strunz, and T. Gutberlet. 2004. Structural, volumetric, and thermodynamic characterization of a micellar sphere-to-rod transition. J. Am. Chem. Soc. 126:16544–16552. [DOI] [PubMed] [Google Scholar]
- 8.Geng, Y., L. S. Romsted, S. Froehner, D. Zanette, L. J. Magid, I. M. Cuccovia, and H. Chaimovich. 2005. Origin of the sphere-to-rod transition in cationic micelles with aromatic counterions: specific ion hydration in the interfacial region matters. Langmuir. 21:562–568. [DOI] [PubMed] [Google Scholar]
- 9.Missel, P. J., N. A. Mazer, M. C. Carey, and G. B. Benedek. 1982. Thermodynamics of the sphere-to-rod transition in alkyl sulfate micelles. In Solution Behavior of Surfactants: Theoretical and Applied Aspects, Vol. 1. K. L. Mittal and E. J. Fendler, editors. Plenum Press, New York, NY. 373–388.
- 10.Reynolds, J. A., and C. Tanford. 1970. Binding of dodecyl sulfate to proteins at high binding ratios. Possible implications for the state of proteins in biological membranes. Proc. Natl. Acad. Sci. USA. 66:1002–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chattopadhyay, A., and E. London. 1984. Fluorimetric determination of critical micelle concentration avoiding interference from detergent charge. Anal. Biochem. 139:408–412. [DOI] [PubMed] [Google Scholar]
- 12.Rawat, S. S., and A. Chattopadhyay. 1999. Structural transition in the micellar assembly: a fluorescence study. J. Fluoresc. 9:233–244. [Google Scholar]
- 13.Jönsson, B., B. Lindman, K. Holmberg, and B. Kronberg. 1998. Surfactants and Polymers in Aqueous Solution. John Wiley, New York, NY.
- 14.Israelachvili, J. N. 1991. Intermolecular and Surface Forces, 2nd Ed. Academic Press, London, UK.
- 15.Killian, J. A. 1992. Gramicidin and gramicidin-lipid interactions. Biochim. Biophys. Acta. 1113:391–425. [DOI] [PubMed] [Google Scholar]
- 16.Andersen, O. S., and R. E. Koeppe. 1992. Molecular determinants of channel function. Physiol. Rev. 72:89–158. [DOI] [PubMed] [Google Scholar]
- 17.Wallace, B. A. 2000. Common structural features in gramicidin and other ion channels. Bioessays. 22:227–234. [DOI] [PubMed] [Google Scholar]
- 18.Chattopadhyay, A., and D. A. Kelkar. 2005. Ion channels and D-amino acids. J. Biosci. 30:147–149. [DOI] [PubMed] [Google Scholar]
- 19.Veatch, W. R., E. T. Fossel, and E. R. Blout. 1974. The conformation of gramicidin A. Biochemistry. 13:5249–5256. [DOI] [PubMed] [Google Scholar]
- 20.Killian, J. A., K. U. Prasad, D. Hains, and D. W. Urry. 1988. The membrane as an environment of minimal interconversion. A circular dichroism study on the solvent dependence of the conformational behavior of gramicidin in diacylphosphatidylcholine model membranes. Biochemistry. 27:4848–4855. [DOI] [PubMed] [Google Scholar]
- 21.O'Connell, A. M., R. E. Koeppe, and O. S. Andersen. 1990. Kinetics of gramicidin channel formation in lipid bilayers: transmembrane monomer association. Science. 250:1256–1259. [DOI] [PubMed] [Google Scholar]
- 22.Hu, W., K.-C. Lee, and T. A. Cross. 1993. Tryptophans in membrane proteins: indole ring orientations and functional implications in the gramicidin channel. Biochemistry. 32:7035–7047. [DOI] [PubMed] [Google Scholar]
- 23.Ketchem, R. R., W. Hu, and T. A. Cross. 1993. High-resolution conformation of gramicidin A in a lipid bilayer by solid-state NMR. Science. 261:1457–1460. [DOI] [PubMed] [Google Scholar]
- 24.Mukherjee, S., and A. Chattopadhyay. 1994. Motionally restricted tryptophan environments at the peptide-lipid interface of gramicidin channels. Biochemistry. 33:5089–5097. [DOI] [PubMed] [Google Scholar]
- 25.Rawat, S. S., D. A. Kelkar, and A. Chattopadhyay. 2004. Monitoring gramicidin conformations in membranes: a fluorescence approach. Biophys. J. 87:831–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chattopadhyay, A. 2003. Exploring membrane organization and dynamics by the wavelength-selective fluorescence approach. Chem. Phys. Lipids. 122:3–17. [DOI] [PubMed] [Google Scholar]
- 27.Raghuraman, H., D. A. Kelkar, and A. Chattopadhyay. 2005. Novel insights into protein structure and dynamics utilizing the red edge excitation shift approach. In Reviews in Fluorescence 2005, Vol. 2. C. D. Geddes and J. R. Lakowicz, editors. Springer, New York, NY. 199–214.
- 28.Demchenko, A. P. 2002. The red-edge effects: 30 years of exploration. Luminescence. 17:19–42. [DOI] [PubMed] [Google Scholar]
- 29.Mentré, P. 2001. Water in the cell. Cell. Mol. Biol. 47:709–970. [PubMed] [Google Scholar]
- 30.Eftink, M. R. 1991. Fluorescence quenching reactions: probing biological macromolecular structure. In Biophysical and Biochemical Aspects of Fluorescence Spectroscopy. T. G. Dewey, editor. Plenum Press, New York, NY. 1–41.
- 31.LoGrasso, P. V., F. Moll, and T. A. Cross. 1988. Solvent history dependence of gramicidin A conformations in hydrated lipid bilayers. Biophys. J. 54:259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lakowicz, J. R. 1999. Principles of Fluorescence Spectroscopy. Kluwer-Plenum Press, New York, NY.
- 33.Bevington, P. R. 1969. Data Reduction and Error Analysis for the Physical Sciences. McGraw-Hill, New York, NY.
- 34.O'Connor, D. V., and D. Phillips. 1984. Time-Correlated Single Photon Counting. Academic Press, London, UK.
- 35.Lampert, R. A., L. A. Chewter, D. Phillips, D. V. O'Connor, A. J. Roberts, and S. R. Meech. 1983. Standards for nanosecond fluorescence decay time measurements. Anal. Chem. 55:68–73. [Google Scholar]
- 36.Grinvald, A., and I. Z. Steinberg. 1974. On the analysis of fluorescence decay kinetics by the method of least-squares. Anal. Biochem. 59:583–598. [DOI] [PubMed] [Google Scholar]
- 37.Chen, G. C., and J. T. Yang. 1977. Two-point calibration of circular dichrometer with d-10-camphorsulphonic acid. Anal. Lett. 10:1195–1207. [Google Scholar]
- 38.Townsley, L. E., W. A. Tucker, S. Sham, and J. F. Hinton. 2001. Structures of gramicidin A, B, and C incorporated into sodium dodecyl sulfate micelles. Biochemistry. 40:11676–11686. [DOI] [PubMed] [Google Scholar]
- 39.Arseniev, A. S., I. L. Barsukov, V. F. Bystrov, A. L. Lomize, and Y. A. Ovchinnikov. 1985. 1H-NMR study of gramicidin A transmembrane ion channel. Head-to-head right-handed, single stranded helices. FEBS Lett. 186:168–174. [DOI] [PubMed] [Google Scholar]
- 40.Chen, Y., and B. A. Wallace. 1997. Solvent effects on the conformation and far UV CD spectra of gramicidin. Biopolymers. 42:771–781. [DOI] [PubMed] [Google Scholar]
- 41.Koeppe, R. E., H. Sun, P. C. A. van der Wel, E. M. Scherer, P. Pulay, and D. V. Greathouse. 2003. Combined experimental/theoretical refinement of indole ring geometry using deuterium magnetic resonance and ab initio calculations. J. Am. Chem. Soc. 125:12268–12276. [DOI] [PubMed] [Google Scholar]
- 42.Gruen, D. W. R. 1985. A model for the chains in amphiphilic aggregates. 2. Thermodynamic and essential comparisons for aggregates of different shape and size. J. Phys. Chem. 89:153–163. [Google Scholar]
- 43.Bogusz, S., R. M. Venable, and R. W. Pastor. 2001. Molecular dynamics simulations of octyl glucoside micelles: dynamic properties. J. Phys. Chem. B. 105:8312–8321. [Google Scholar]
- 44.Bruce, C. D., S. Senapati, M. L. Berkowitz, L. Perera, and M. D. E. Forbes. 2002. Molecular dynamics simulations of sodium dodecyl sulfate micelle in water: the behavior of water. J. Phys. Chem. B. 106:10902–10907. [Google Scholar]
- 45.Pal, S., S. Balasubramanian, and B. Bagchi. 2003. Identity, energy, and environment of interfacial water molecules in a micellar solution. J. Phys. Chem. B. 107:5194–5202. [Google Scholar]
- 46.Mukherjee, S., and A. Chattopadhyay. 1995. Wavelength-selective fluorescence as a novel tool to study organization and dynamics in complex biological systems. J. Fluoresc. 5:237–246. [DOI] [PubMed] [Google Scholar]
- 47.Valeur, B., and G. Weber. 1978. A new red-edge effect in aromatic molecules: anomaly of apparent rotation revealed by fluorescence polarization. J. Chem. Phys. 69:2393–2400. [Google Scholar]
- 48.Callis, P. R. 1997. 1La and 1Lb transitions of tryptophan: applications of theory and experimental observations to fluorescence of proteins. Methods Enzymol. 278:113–150. [DOI] [PubMed] [Google Scholar]
- 49.Ruggiero, A. J., D. C. Todd, and G. R. Fleming. 1990. Subpicosecond fluorescence anisotropy studies of tryptophan in water. J. Am. Chem. Soc. 112:1003–1014. [Google Scholar]
- 50.Prendergast, F. G. 1991. Time-resolved fluorescence techniques: methods and applications in biology. Curr. Opin. Struct. Biol. 1:1054–1059. [Google Scholar]
- 51.Kirby, E. P., and R. F. Steiner. 1970. The influence of solvent and temperature upon the fluorescence of indole derivatives. J. Phys. Chem. 74:4480–4490. [Google Scholar]
- 52.Allen, T. W., O. S. Andersen, and B. Roux. 2003. Structure of gramicidin A in a lipid bilayer environment determined using molecular dynamics simulations and solid-state NMR data. J. Am. Chem. Soc. 125:9868–9877. [DOI] [PubMed] [Google Scholar]
- 53.Eftink, M. R., L. A. Selvidge, P. R. Callis, and A. A. Rehms. 1990. Photophysics of indole derivatives: experimental resolution of La and Lb transitions and comparison with theory. J. Phys. Chem. 94:3469–3479. [Google Scholar]
- 54.Eftink, M. R. 1991. Fluorescence quenching: theory and applications. In Topics in Fluorescence Spectroscopy, Vol. 2: Principles. J. R. Lakowicz, editor. Plenum Press, New York, NY. 53–126.
- 55.Rasia, M., and A. Bollini. 1998. Red blood cell shape as a function of medium's ionic strength and pH. Biochim. Biophys. Acta. 1372:198–204. [DOI] [PubMed] [Google Scholar]
- 56.Luna, E. J., and A. L. Hitt. 1992. Cytoskeleton-plasma membrane interactions. Science. 258:955–963. [DOI] [PubMed] [Google Scholar]
- 57.Kuypers, F. A., B. Roelofsen, W. Berendsen, J. A. F. Op den Kamp, and L. L. M. van Deenen. 1984. Shape changes in human erythrocytes induced by replacement of the native phosphatidylcholine with species containing various fatty acids. J. Cell Biol. 99:2260–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Backman, L., J. B. Jonasson, and P. Hörstedt. 1998. Phosphoinositide metabolism and shape control in sheep red blood cells. Mol. Membr. Biol. 15:27–32. [DOI] [PubMed] [Google Scholar]
- 59.Gedde, M. M., and W. H. Huestis. 1997. Membrane potential and human erythrocyte shape. Biophys. J. 72:1220–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Farge, E., and P. F. Devaux. 1992. Shape changes of giant liposomes induced by an asymmetric transmembrane distribution of phospholipids. Biophys. J. 61:347–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Caetano, W., E. L. Gelamo, M. Tabak, and R. Itri. 2002. Chlorpromazine and sodium dodecyl sulfate mixed micelles investigated by small angle X-ray scattering. J. Colloid Interface Sci. 248:149–157. [DOI] [PubMed] [Google Scholar]
- 62.Perozo, E., A. Kloda, D. M. Cortes, and B. Martinac. 2002. Physical principles underlying the transduction of bilayer deformation forces during mechanosensitive channel gating. Nat. Struct. Biol. 9:696–703. [DOI] [PubMed] [Google Scholar]
- 63.Lundbaek, J. A., A. M. Maer, and O. S. Anderesen. 1997. Lipid bilayer electrostatic energy, curvature stress, and assembly of gramicidin channels. Biochemistry. 36:5695–5701. [DOI] [PubMed] [Google Scholar]
- 64.Suchyna, T. M., S. E. Tape, R. E. Koeppe, O. S. Andersen, F. Sachs, and P. A. Gottlieb. 2004. Bilayer-dependent inhibition of mechanosensitive channels by neuroactive peptide enantiomers. Nature. 430:235–240. [DOI] [PubMed] [Google Scholar]
- 65.Andersen, O. S., G. Saberwal, D. V. Greathouse, and R. E. Koeppe. 1996. Gramicidin channels: a solvable membrane “protein” folding problem. Indian J. Biochem. Biophys. 33:331–342. [PubMed] [Google Scholar]
- 66.Kelkar, D. A., and A. Chattopadhyay. 2005. Effect of graded hydration on the dynamics of an ion channel peptide: a fluorescence approach. Biophys. J. 88:1070–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guha, S., S. S. Rawat, A. Chattopadhyay, and B. Bhattacharyya. 1996. Tubulin conformation and dynamics: a red edge excitation shift study. Biochemistry. 35:13426–13433. [DOI] [PubMed] [Google Scholar]
- 68.Seelig, J. 1977. Deutrium magnetic resonance: theory and application to lipid membranes. Q. Rev. Biophys. 10:353–418. [DOI] [PubMed] [Google Scholar]
- 69.Chattopadhyay, A., and S. Mukherjee. 1999. Depth-dependent solvent relaxation in membranes: wavelength-selective fluorescence as a membrane dipstick. Langmuir. 15:2142–2148. [Google Scholar]