Abstract
Kinesins are dimeric motor proteins that move processively along microtubules. It has been proposed that the processivity of conventional kinesins is increased by electrostatic interactions between the positively charged neck of the motor and the negatively charged C-terminus of tubulin (E-hook). In this report we challenge this anchoring hypothesis by studying the motility of a fast fungal kinesin from Neurospora crassa (NcKin). NcKin is highly processive despite lacking the positive charges in the neck. We present a detailed analysis of how proteolytic removal of the E-hook affects truncated monomeric and dimeric constructs of NcKin. Upon digestion we observe a strong reduction of the processivity and speed of dimeric motor constructs. Monomeric motors with truncated or no neck display the same reduction of microtubule gliding speed as dimeric constructs, suggesting that the E-hook interacts with the head only. The E-hook has no effect on the strongly bound states of NcKin as microtubule digestion does not alter the stall forces produced by single dimeric motors, suggesting that the E-hook affects the interaction site of the kinesin·ADP-head and the microtubule. In fact, kinetic and binding experiments indicate that removal of the E-hook shifts the binding equilibrium of the weakly attached kinesin·ADP-head toward a more strongly bound state, which may explain reduced processivity and speed on digested microtubules.
INTRODUCTION
Kinesins are microtubule (MT)-based motor proteins that hydrolyze ATP to power various intracellular transport processes in eukaryotic organisms (1). Within the kinesin family of motor proteins, the best studied motors are the founding members (2,3), now referred to as conventional kinesins. In vivo, conventional animal kinesins are heterotetrameric motors, consisting of two heavy and two light chains (4). The globular, N-terminal domains of the heavy chains, called the heads, contain the MT-binding and ATP hydrolysis activities (5,6). Two heavy chains form a functional dimer through coiled-coil interactions in the neck and stalk domain, which are interrupted by flexible regions (7,8). These so-called hinges and kinks allow control of the motor activity through inhibitory back folding of the C-terminal regions, mainly the globular tail, which is also responsible for light chain and cargo binding (9–12). It could be shown in vitro that single conventional kinesin dimers comprising at least the head and neck domain have the remarkable ability to move processively for micrometer-long distances along MTs without detaching (13–17). On average such processive runs consist of ∼100 8 nm steps, each of which is tightly coupled to the hydrolysis of 1 ATP molecule (18–20). Recent direct evidence convincingly demonstrates that the two heads of kinesin interact with the MT in a hand-over-hand fashion such that the dimeric motor is bound to the MT during processive movement with at least one head at all times (21). To ensure this, the two heads need to communicate their respective nucleotide state, which is believed to be principally achieved by intramolecular strain (22,23). Events disrupting this cycle lead to complete dissociation of the motor from the MT. The distribution of run length of individual processive encounters follows Poisson statistics, resulting in a single exponential distribution with the average run length as a quantitative measure of the processivity (17,24,25). It was proposed that the extent of processivity is influenced through electrostatic interactions of the motor's neck with the flexible most C-terminal domains of the tubulin subunits of the MT (26,27).
The last 10 and 18 C-terminal residues of α- and β-tubulin are called E-hooks, as they are rich in negatively charged aspartic acid (Asp, D) and glutamic acid (Glu, E) residues (28). They are also believed to be mobile, because they are not resolved in crystal structures derived from two-dimensional electron microscopy (29). Specific proteolytic removal of the E-hooks by subtilisin (30,31) strongly reduced the processivity of conventional kinesin (and dynein), which suggested an important role of the E-hook in processive movement (26). Conversely, quadruplicating positively charged regions of the neck of human kinesin (HsKin) resulted in a motor with increased average processive run length, which in turn was sensitive to high salt concentrations and removal of the E-hook (27). These observations led to the hypothesis that the high processivity of wild-type (wt)-HsKin is maintained by strong electrostatic interactions between the E-hook and the neck domain (anchoring hypothesis). Although the charge distribution in the neck domain of animal conventional kinesins is largely conserved, it is markedly different in conventional kinesins from fungi like Neurospora crassa.
The homodimeric kinesin NcKin from the filamentous fungus N. crassa (32) is basically indistinguishable from its animal counterparts in its stepping behavior (33), but it possesses distinct structural and functional features which seem to be shared with other fungal kinesins: truncation studies revealed that homologous parts of the neck domain which lead to coiled-coil formation in animal kinesins are insufficient for dimerization of NcKin (34). Interestingly, this region of the neck contains a tyrosine residue (Y362), which is conserved in fugal kinesins and inhibits catalytic activity in monomeric constructs (35), possibly mimicking regulatory functions of animal kinesin light chains. Interestingly, so far light chains have not been identified for fungal kinesins. Furthermore, NcKin is unusually fast and twice as processive as conventional kinesin (24). Thus, natural variations in the structure and function of NcKin provide a sensitive tool to investigate and compare mechanisms underlying processivity and its regulation in kinesins in general. For example, the high processivity of NcKin as compared to conventional kinesins cannot be readily explained by an increased electrostatic interaction between its neck and the E-hook of tubulin because NcKin does not contain the cluster of positive charge found in the neck of conventional animal kinesins. As an alterative to the anchoring hypothesis, structural and functional effects of the E-hook could be mediated through interactions with the head domains.
Here we set out to experimentally challenge the anchoring hypothesis by testing if and how MT digestion affects a set of truncated monomeric and dimeric NcKin constructs in single molecule and bulk biochemical experiments. We observe a strong reduction of the processivity and speed of dimeric motor constructs on digested MTs (dMT). Monomeric motors with truncated or no neck display the same reduction of MT gliding speed as dimeric constructs, implying that the E-hook interacts with the head only. The E-hook has no effect on the strongly bound states of NcKin as MT digestion does not alter the stall forces produced by single dimeric motors, suggesting effects on the kinesin·ADP-head. In fact, kinetic and binding experiments of the minimal motor domain indicate that removal of the E-hook shifts the binding equilibrium of the weakly attached kinesin·ADP-head toward a more strongly bound state.
METHODS
Unless otherwise indicated, all chemicals were obtained from Sigma (St. Louis, MO) and all concentrations reported are final. All measurements were performed at room temperature.
Cloning and protein preparation
All expression clones use the pT7-7 vector backbone and are inserted using the NdeI and PstI restriction sites. Clones of pNK433, pNK383, pNK378, and pNK343 were kindly provided by A. Kallipolitou. Genes for pNK391 and pNK400 were amplified by polymerase chain reaction. The sequences of all plasmids were confirmed by sequencing.
Motors were expressed and purified as previously described (24) using Escherichia coli BL21(DE3) cells. Briefly, a freshly transformed colony was incubated overnight at 37°C in Luria Broth medium with ampicillin. With this preculture, a tryptone medium (24) was inoculated to an optical density of 0.1–0.2 and incubated at 22°C and 220 rpm. Expression was induced at growth densities between 0.5 and 1 optical density using 0.2–0.5 mM IPTG. Cells were harvested after 16–24 h and stored at −80°C. Cells were lysed in 20 mM Na-phosphate buffer, pH 7.4, using ultrasound. The motors were isolated from the clarified lysate by ion exchange chromatography (Sepharose fast-flow Hitrap columns, 5 ml, Amersham Biosciences, Piscataway, NJ), using a NaCl-step gradient. Fractions containing kinesin were pooled, supplemented with 5%–10% sucrose, and flash frozen in liquid, nitrogen for storage at −80°C. Motors were labeled with biotin or Cy3 using the cysteine-specific monofunctional maleimide esters. Labeling is expected to occur at the highly flexible and accessible cysteine tag (36,37).
Bovine tubulin was obtained by repeated polymerization and depolymerization cycles from clarified brain lysate. MAPs were removed by ion exchange chromatography (P11, Whatman, Florham Park, NJ) followed by another polymerization and depolymerization cycle. Cy5- and TMR-labeled tubulin was obtained by labeling polymerized MTs using a nonspecific monofunctional succinimidyl ester at mildly basic buffer conditions. Functional tubulin was again separated from dysfunctional monomers by polymerization and depolymerization cycles. Usually, MTs were polymerized using 1 mM GTP and 2 mM MgCl2 using 5% DMSO to promote polymerization. MTs were stabilized by 10 μM taxol (Paclitaxel, Calbiochem, San Diego, CA).
Single molecule fluorescence assays
Single molecule fluorescence assays were performed as previously described for NcKin motors (24). Briefly, fluorescently labeled motor molecules were diluted to ∼5 nM in a P12 buffer (12 mM PIPES/KOH, 1 mM EGTA, 2m M MgCl2, pH 6.8) containing 30 mg/ml bovine serum albumin. Diluted motor protein was combined with equal parts (1.1 μL each) of the following three solutions: 1), glucose-oxidase (0.4 mg/ml) and catalase (0.32 mg/ml) in P12; 2), MgCl2 (4 mM), AMP-PNP (4–40μM), DTT (4 mM), and glucose (40 mM) in P12; and 3), Cy5-labeled MTs or dMTs (25-fold diluted in P12 with 40 μM taxol). The solutions were incubated for ∼3–15 min to allow the motor to bind to the MT before applying a 4 μL volume to a cleaned quartz slide. After identification of an MT with immobilized Cy3-labeled motors, an ATP-containing solution (4 mM ATP) was washed into the chamber and the ensuing movement of the motor molecules was recorded with a digital video camera. Electronically stored image sequences were analyzed using ImageJ.
Multiple molecule gliding assays
Multiple molecule gliding assays with biotinylated motors were performed following standard procedures (34). Briefly, flow chambers were precoated for 3 min with a BRB80 solution (80 mM PIPES/KOH, 1 mM EGTA, 2 mM MgCl2, pH 6.9) containing 1 mg/ml streptavidin and 0.8 mg/ml bovine serum albumin; then kinesin diluted in BRB80 with 0.05 mg/ml casein was introduced and allowed to incubate for 3 min. Subsequently, TMR-labeled MTs were diluted in an MgATP-containing oxygen-scavenger solution (0.1 mg/ml glucose-oxidase, 0.08 mg/ml catalase, 1 mM DTT, 10 mM glucose) with an additional 10 μM taxol and added to the flow chamber. MTs were observed using an epifluorescence microscope (Axiovert 135 TV, Zeiss, Göttingen, Germany).
Steady-state ATPases
Steady-state ATPase rates were determined spectroscopically by monitoring the oxidation of NADH, which is enzymatically coupled to the turnover of ATP by LDH, PK, and PEP (38,39). Assays were performed in ATPase buffer (12 mM ACES·KOH, 25 mM potassium acetate, 5 mM MgCl2, 0.5 mM EGTA, pH 6.8). ATP was used at a 2 mM final concentration, and reactions were started by the addition of a constant amount of motor protein. Individual rates were plotted against increasing MT concentrations and fit to a hyperbolic function. K0.5,MT is determined as the concentration of MT at which half-maximal ATP turnover occurs. Kcat is calculated from the extrapolated maximal turnover and the motor concentration in the assay.
MantADP release experiments
Nucleotide bound to kinesin was replaced with mantADP under conditions of basal ATP turnover. Motors were incubated with a two- to fourfold molar excess of mantATP for 15–30 min. Unbound nucleotide was removed from the kinesin·mantADP solution using passivated desalting columns (Sephadex G25 M, Amersham Biosciences). Motor concentrations and loading ratios were determined spectroscopically. Binding of mantADP·kinesin to varying substochiometric concentrations of MT and dMTs was observed as an exponential loss of fluorescence due to immediate unbinding of ADP (Aminco Bowman AB2 fluorimeter (Thermo Electron, Waltham, MA), excitation 365 nm, emission 445 nm, motor concentrations ∼100 nM). Rate constants of the exponential decays were plotted against the MT concentration. The bimolecular rate constant of productive encounter, kbi,ADP, was determined from a linear regression of the data (13).
Equilibrium-binding experiments
Equilibrium-binding experiments were essentially performed as described by Klumpp et al. (40). Motor-containing solutions were desalted as described for mantADP-release experiments to remove ATP present after purification. A total of 2 μM motor was then preincubated with varying concentrations of MgADP (0–20 mM) for 30 min, 4 μM MT or dMTs were added to yield a final volume of 110 μl, and the mixture was allowed to react for 30 min at 22°C before pelleting MT and bound motor by centrifugation (Airfuge, rotor A-100, Beckman, Fullerton, CA; 30 psi, 5 min). To determine the fraction of unbound motor, a 55 μl aliquot of the supernatant was removed and mixed with 50 μl 2× SDS-PAGE sample buffer. After carefully removing the residual supernatant, the pellet was briefly washed with 50 μl buffer and then resuspended in 55 μl 2× sample buffer and 50 μl buffer was added. Equivalent amounts of supernatant and pellet were submitted to SDS-PAGE. Using ImageJ for quantification, the fraction of motor bound to the MT was plotted against the ADP concentration.
Laser-trapping experiments
Biotinylated NK433 was specifically attached to streptavidin-coated latex spheres, which were obtained by covalent modification of carboxylated beads (Molecular Probes, Eugene, OR; 1 μm, dark red) with EDC and Biotin-XX-cadaverine and subsequent incubation with streptavidin. Streptavidin beads, sparsely coated with biotinylated motor, were trapped in a single beam laser trap and presented to an unspecifically immobilized MT. Position data of the trapped bead was acquired by back-focal plane interferometry using a quadrant photo detector. Dilution series with decreasing amounts of motor/bead were assayed, and data were acquired from dilutions in which only one out of two–five beads generated movement. As a control, we also recorded events from beads coated with kinesin at a 10-fold lower motor/bead ratio; apart from a much lower fraction of active beads, there was no discernible difference in the interactions, ensuring that data were acquired from single motor molecules.
Velocity information at increasing force was determined (custom-developed C-code) by fitting a series of short linear segments of variable duration (20 ms or longer, displacement 20 nm) to individual kinesin runs. Data from many such events were binned in 0.5 pN intervals, and the average motor speed and force were calculated. The resulting data were combined in a force-velocity diagram, and stall forces were determined by extrapolation to zero velocity.
RESULTS
Removal of the E-hook reduces the speed and processivity of NcKin dimers
To determine how sensitive NcKin is to the removal of the E-hook, we analyzed the processive movement of single, Cy3-labeled NcKin dimers (NK433) on dMT (Fig. 1). We observe smooth and unidirectional processive movement along dMT as previously observed by us for HsKin and NcKin on undigested MTs (Fig. 2) (24). Surprisingly, single NcKin dimers move only at ∼½ the speed on dMTs as compared to MTs (0.97 μm/s and 1.70 μm/s, respectively, Table 1, Supplementary Material Sup_1 and Sup_2). This reduction is more pronounced than observed in previous studies on the movement of single animal kinesins on dMT (26). We cannot rule out with complete certainty that this reduction in speed is caused by local defects in the MT lattice, but the smooth, continuous motility in our single molecule experiments and structural studies by others (41) do not support the existence of such defects. Individual processive runs were combined in a histogram, and the average run length was determined by fitting a single exponential function to the data (Fig. 3). We observe a reduction of the average processive run length of NcKin on dMTs by ∼50% as compared to MTs (0.89 μm and 1.75 μm, respectively, Table 1). To confirm these unexpected findings with independent methods and to further test for a possible influence of regions of the hinge domain, we performed steady-state ATPase and mantADP-release experiments of stable dimeric NcKin constructs of decreasing length (NK433, NK400, and NK391). For all three dimers we find a reduction of ATP turnover of ∼30% from ∼70 ATP/head*s on MT to ∼50 ATP/head*s on dMT (Table 2, Fig. 4). The apparent overall affinity of the dimers to dMTs is significantly reduced, as indicated by increased k0.5,dMT-values for all three dimeric constructs. Furthermore, the determination of the rate of mantADP release on MT and dMT allows us to calculate the number of ATP molecules turned over per productive diffusional encounter, i.e., the biochemical processivity. Removal of the E-hook results in a strong reduction of biochemical processivity of 30-50% for the three dimeric motors (Table 2, Fig. 4), confirming our results from single molecule fluorescence assays. Although NcKin does not contain positively charged clusters in the neck domain, the removal of the E-hook nonetheless has a strong effect on the processive movement of NcKin dimers. This suggests that the E-hook does not exert its effects via an interaction with the neck, but rather via an interaction with the head domain of kinesin.
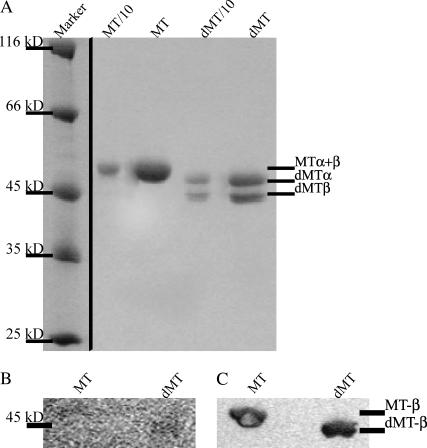
FIGURE 1.
MT digestion. Panel A shows an SDS-PAGE analysis of native MTs and subtilisin dMTs. Undigested α- and β-subunits of tubulin do not separate under standard conditions. Digestion, however, leads to clear separation of the subunits. We confirmed complete digestion of β-tubulin using a Western blot with monoclonal anti-β-tubulin antibodies. Panel B shows the Ponceau S-stained blot, and panel C the Western blot after detection of the primary antibody with a secondary, alkaline phosphatase-conjugated antibody and incubation with NBT/BCIP as substrate.
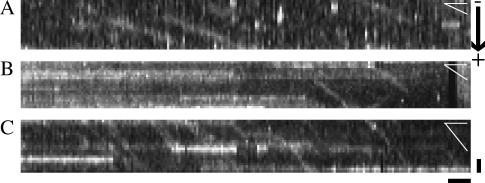
FIGURE 2.
Kymographs of single molecule fluorescence events. Kymographs were generated from unprocessed image sequences using Metamorph software. From each frame a single row of pixels along the length of the MT was extracted. The resulting slices from sequential frames were aligned side-by-side such that the vertical axis of the kymographs represents the direction along the MT (scale bar = 1 μm) and the horizontal axis represents the time (scale bar corresponds to 1 s). Events of single fluorescently labeled motor molecules moving along an MT emerge as inclined bright lines, and immotile motors or fluorescent particles lead to horizontal streaks. In this panel, all MTs are oriented with the plus end of the MT pointing downward. Panels A and C show examples of HsKin560 and NK433 motors, respectively, moving along an MT. Panel B shows examples of single NK433 motors moving along a dMT. White angles in the far upper corner of each panel illustrate the different slopes of the events, reflecting the speed of the motors.
TABLE 1.
Single molecule fluorescence data
| MT† | dMT | |
|---|---|---|
| Run length [μm]* | 1.75 ± 0.09 (182) | 0.89 ± 0.08 (67) |
| Velocity [μm/s] | 1.70 ± 0.05 (182) | 0.97 ± 0.45 (67) |
All errors given as ± se, number of events given in parenthesis.
Run length determined as the decay constant of a single exponential fit to histogram representation of individual runs without correction for bleaching.
Data taken from Lakamper et al. (24).
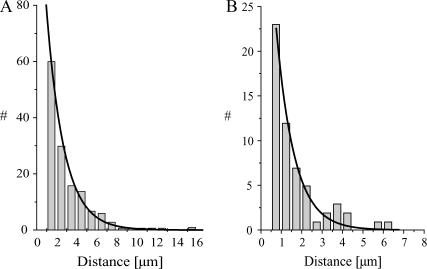
FIGURE 3.
Histogram analysis of single molecule fluorescence assays. The distances traveled in individual events of processive movement along the MTs were combined in a histogram. The data were fit by a single exponential, the time constant of which represents the average run length. Panel A shows data for NcKin on MTs (data from Lakamper et al. (24); N = 182; run length 1.75 ± 0.09 μm). Panel B shows the histogram for NcKin motors moving along subtilisin-treated MTs (N = 67; run length 0.89 ± 0.08 μm, Table 1).
TABLE 2.
Summary of kinetic data for native and digested microtubules
 |
 |
 |
 |
 |
 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NK343
|
NK378
|
NK383
|
NK391
|
NK400
|
NK433
|
|||||||
| Motor | MT | dMT | MT | dMT | MT | dMT | MT | dMT | MT | dMT | MT | dMT |
| kcat [ATP/head*s] | 247.6 ± 22.2 (7) | 249.4 ± 17.0 (6) | 18.81 ± 1.69 (12) | 33.64 ± 2.89 (10) | 20.53 ± 2.71 (10) | 32.92 ± 5.56 (8) | 71.72 ± 5.16 (8) | 50.01 ± 4.10 (8) | 68.59 ± 7.35 (7) | 49.84 ± 6.00 (7) | 64.18 ± 4.45 (13) | 46.10 ± 4.16 (11) |
| k0.5,MT [μM] | 0.94 ± 0.35 (6) | 0.76 ± 0.21 (5) | 7.44 ± 1.21 (12) | 13.13 ± 1.54 (10) | 6.76 ± 1.50 (10) | 11.72 ± 3.14 (8) | 0.21 ± 0.05 (5) | 0.31 ± 0.08 (5) | 0.11 ± 0.01 (6) | 0.16 ± 0.05 (6) | 0.23 ± 0.06 (13) | 0.46 ± 0.13 (10) |
| kbi,ADP [μM−1s−1] | 19.6 ± 0.94 (13) | 24.75 ± 2.78 (12) | 2.58 ± 0.30 (4) | 2.05 ± 0.08 (4) | 3.44 ± 0.13 (4) | 3.18 ± 0.13 (4) | 14.16 ± 1.02 (4) | 14.99 ± 0.72 (4) | 20.44 ± 1.55 (4) | 14.12 ± 0.65 (4) | 16.57 ± 1.47 (7) | 9.77 ± 0.99 (7) |
| kbi, ratio [#]* | 13.45 | 13.26 | 0.98 | 1.24 | 0.88 | 0.88 | 24.11 | 10.76 | 30.51 | 22.06 | 16.84 | 10.26 |
All errors given as ± se, number of independent assays given in parenthesis.
kbi,ratio,MT is defined as (kcat,MT/ K0.5, MT) / (kbi,ADP, MT), representing the number of ATP hydrolyzed per productive diffusional encounter of motor with MT.
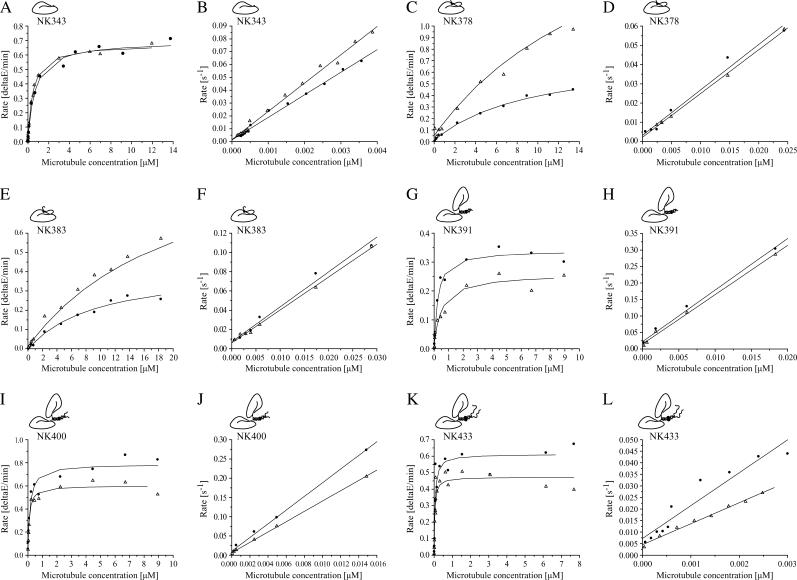
FIGURE 4.
Kinetic analysis of NcKin motors interacting with MTs and dMTs. Panels A–L show typical ATPase and ADP-release diagrams of MTs (circles) and dMTs (triangles) for the motor constructs used in this study ordered in increasing length. MT digestion leaves the kcat of the minimal motor domain, NK343, unaffected (A; MT 247.6 ± 22.2 ATP/head*s; dMT 249.4 ± 17.0 ATP/head*s). The slight reduction of the k0.5,MT-value upon digestion of MTs from 0.94 ± 0.35 μM to 0.76 ± 0.21 μM is statistically not significant. (B) The rates of mantADP release for NK343 increase from 19.6 ± 0.94 μM−1s−1 on MTs to 24.75 ± 2.78 μM−1s−1. ATP turnover of NK378 on dMTs (C) is increased from 18.81 ± 1.69 ATP/head*s to 33.64 ± 2.89 ATP/head*s, whereas k0.5,MT is doubled from 7.44 ± 1.21 μM to 13.13 ± 1.54 μM on dMTs. Kbi,ADP-values drop from 2.58 ± 0.30 μM−1s−1 on MTs to 2.05 ± 0.08 μM−1s−1 on dMTs (D). Similarly, ATP turnover of NK383 on dMTs (E) is increased from 20.53 ± 2.71 ATP/head*s to 32.92 ± 5.56 ATP/head*s, whereas k0.5,MT is doubled from 6.76 ± 1.50 μM to 11.72 ± 3.14 μM on dMTs. kbi,ADP-values drop from 3.44 ± 0.13μM−1s−1 on MTs to 3.18 ± 0.13 μM−1s−1 on dMTs (F). ATP turnover of dimeric NK391 on dMTs (G) is reduced from 71.72 ± 5.16 ATP/head*s to 50.01 ± 4.10 ATP/head*s, whereas k0.5,MT is increased from 0.21 ± 0.05 μM to 0.31 ± 0.08 μM on dMT. kbi,ADP-values remain unchanged at 14.16 ± 1.02 μM−1s−1 and 14.99 ± 0.72 μM−1s−1 (H). ATP turnover of NK400 on dMTs similarly drops from 68.59 ± 7.35 ATP/head*s to 49.84 ± 6.00 ATP/head*s, k0.5,MT increases from 0.11 ± 0.01 μM to 0.16 ± 0.05 μM on dMTs (I), whereas kbi,ADP-values drop from 20.44 ± 1.55 μM−1s−1 on MTs to 14.12 ± 0.65 μM−1s−1 on dMTs (J). ATP turnover of NK433 on dMTs (K) is decreased from 64.18 ± 4.45 ATP/head*s to 46.10 ± 4.16 ATP/head*s, whereas k0.5,MT is doubled from 0.23 ± 0.06 μM to 0.46 ± 0.13 μM on dMTs. kbi,ADP values for NK433 drop from 16.57 ± 1.47 μM−1s−1 to 9.77 ± 0.99 μM−1s−1 (L) upon digestion.
Multiple molecule gliding assays exclude the neck as the E-hook interaction site
To further test if the E-hook interacts with the head rather than with the neck, we performed multiple molecule gliding assays with the previously described dimers and also with monomeric constructs containing parts (NK383 and NK378) or no neck (NK343). To ensure the best comparable conditions for each motor, we performed multiple molecule gliding assays using a mixture of clearly distinguishable, brightly labeled MTs and dimly labeled dMTs in the same assay. We observe MT speeds very similar to previously reported values (Table 3, Supplementary Material Sup_3, (34)). Removal of the E-hook results in a striking reduction of gliding speeds by on average 20% for all motor constructs, independent on whether the full, part, or no neck was present (Table 3). This neck-independent reduction of gliding speed strongly supports the hypothesis that the E-hook interacts with the head, not the neck, domain of kinesin. Such an E-hook-head interaction hypothesis is structurally plausible from docking experiments of the crystal structure of kinesins into cryoelectron microscopy maps of MTs decorated with motors and foot-printing experiments (42,43). These experiments revealed that a major site of interaction between kinesin's head and the MT is helix 12 on the β-tubulin subunit, which forms the most C-terminal region that is ordered in the tubulin structure and is immediately followed by the highly mobile E-hook. The proposed E-hook-head interaction would explain the reduced affinity of the head to the dMT, as suggested by the previously mentioned increase of k0.5,MT and the decrease of kbi,ADP for dimeric motors on dMT (Table 2, Fig. 4).
TABLE 3.
Multiple molecule gliding assays with digested microtubules show similar reductions in speed for all constructs
 |
 |
 |
 |
 |
 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NK343
|
NK378
|
NK383
|
NK391
|
NK400
|
NK433
|
|||||||
| Motor | MT | dMT | MT | dMT | MT | dMT | MT | dMT | MT | dMT | MT | dMT |
| Speed [μm/s] | 0.31 ± 0.01 (132) | 0.26 ± 0.01 (138) | 0.68 ± 0.03 (72) | 0.52 ± 0.02 (83) | 0.64 ± 0.03 (44) | 0.54 ± 0.03 (38) | 0.71 ± 0.02 (141) | 0.53 ± 0.01 (133) | 1.53 ± 0.05 (147) | 1.26 ± 0.04 (152) | 2.00 ± 0.09 (92) | 1.53 ± 0.03 (90) |
| Reduction upon digestion* | - | −16 % | - | −23 % | - | −16% | - | −25 % | - | −18 % | - | −24 % |
Average speeds ± SE of the mean were calculated for the number of individual microtubules given in parenthesis. Three independent motor preparations were used for all constructs, except NK383cys, for which only two independent preparations were used.
Taking speed of native microtubules (MT) as 100%.
The removal of the E-hook does not affect the strongly bound states of NcKin
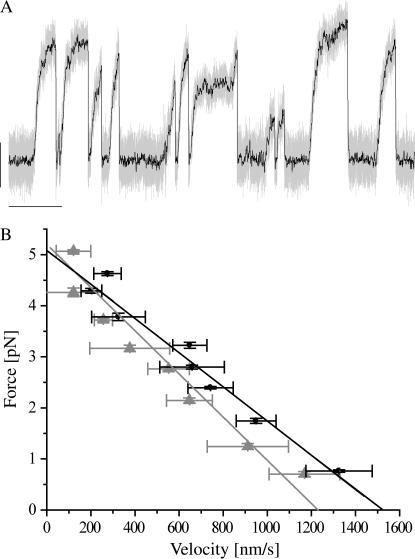
To test if the removal of the E-hook reduces the affinity of the head to the MT, we performed single molecule trapping experiments with dimeric NcKin motors and established a force-velocity relationship for single dimeric NcKin motors on MTs and dMTs. The relationship between force and speed of conventional kinesins is approximately inversely linear (but see detailed work by Visscher et al. (44)), which is generally believed to be based on a reduced rate of binding of the tethered head to the next binding site when substantial force is pulling the entire motor backwards. Therefore, stall forces are predominantly determined by 1), the binding affinity of the strongly bound states (ATP bound and nucleotide free) of the head to the MT, and 2), the ATP-binding kinetics to the nucleotide-free head as ATP-binding is expected to be followed by fast turnover and dissociation in the ADP-state.
Streptavidin-modified latex beads which are very sparsely coated with biotinylated dimeric NcKin motors produce displacements on both MTs and dMTs, which are consistent in appearance and frequency with previously described single molecule trapping experiments. In agreement with the gliding assays, speeds at—or near—zero-load conditions are significantly reduced by ∼20% on dMTs as compared to MT (Table 4, Fig. 5 B). Linear regressions of our force-velocity relationships show that the speed is proportionally reduced over the complete force range, and the average stall force extrapolates to the same value of ∼5 pN force for both MTs and dMTs (Table 4, Fig. 5). These observations argue that the affinity of the strong binding states of NcKin are unaffected by MT digestion. They furthermore rule out a previously proposed model (27) in which an interaction between the neck and MTs was proposed to occur only at zero or near-zero forces whereas the increase of the external load opposing kinesin movement disrupts the interactions. This model requires a nonlinear force-velocity relationship, which is in disagreement with our findings for both MTs and dMTs. Our results point toward an effect of the E-hook on the kinetics of the ADP state of kinesin: in the simplest scenario the proportional reduction in speed on dMTs is entirely due to changes in the binding and release of the kinesin·ADP. Indications that ADP kinetics is affected by MT digestion are further substantiated by steady-state measurements of monomeric NcKin constructs.
TABLE 4.
Single molecule trapping data
| MT | dMT | |
|---|---|---|
| Stall force [pN] | 5.07 ± 0.21 (5) | 4.97 ± 0.18 (5) |
| Initial velocity [μm/s] | 1.70 ± 0.08 (5) | 1.36 ± 0.04 (5) |
All errors given as ± se, number of measurements given in parenthesis.
FIGURE 5.
Single molecule trapping experiments. Panel A shows a representative tracing of a trapped, kinesin-coated bead powered by a single NK433 dimer along a dMT (unfiltered position signal, light shaded; filtered at 50 Hz, solid; vertical scale 50 nm, horizontal scale 2 s, trap stiffness 0.049 pN/nm). Using custom-written C-code, we fit a series of short linear regressions (20 nm displacement, duration >20 ms) to individual runs of kinesin and computed the average speed and force for each segment. Speeds were averaged over 0.5 pN force intervals and combined to a force-velocity diagram. Panel B shows examples of force-velocity diagrams for single NK433 motors interacting with MTs (solid) and dMTs (shaded) at 2 mM ATP. These two force-velocity relations were averaged from traces containing a large number of individual events all obtained from a single kinesin-coated bead. The extrapolated initial velocity at zero-load conditions for dMTs is ∼20% lower (1.52 ± 0.10 μm/s) than for MTs (1.22 ± 0.12 μm/s). At higher forces, the two curves converge to indistinguishable stall forces of 5.08 ± 0.20 pN and 5.20 ± 0.23 pN for MTs and dMTs, respectively (the data from five such measurements are summarized in Table 4).
The E-hook affects the ADP kinetics of monomeric kinesins containing a conserved tyrosine
Compared to dimeric constructs, the monomeric constructs NK383 and NK378 display strongly reduced ATP turnover (kcat) on, and very low affinities (k0.5,MT and kbi,ADP) to, native MTs. The calculated biochemical processivity of these constructs is in complete agreement with the model that mechanical processivity of conventional kinesins requires two heads (45). Recent mutational studies revealed that the strongly reduced ATP turnover is due to the inhibitory effect of the benzene ring of the fungal-specific, conserved tyrosine residue in the neck domain, Y362 (35). It strictly controls the rate of ADP release, kmax,ADP, from the motor head. Surprisingly, removal of the E-hook increases steady-state ATP turnover of both constructs (Table 2, Fig. 4). However, in these constructs k0.5,MT is increased to very high levels, whereas kbi,ADP is almost unaffected, together leading to a biochemical processivity very close to 1. Consequently, the increase in ATP turnover can only be brought about by an increase in kmax,ADP on dMTs. Interestingly, this drastic increase of in-solution ATP turnover is not reflected in increased gliding speeds. Although it is difficult and speculative to fully explain the kinetic behavior of these constructs in the context of the proposed processive stepping cycle (see Discussion), these data argue that the E-hook has important effects on the ADP kinetics of kinesin. Consequently, we investigated the kinetic behavior of the construct NK343, which comprises only the head and neck-linker domain and constitutes the minimal motor unit of NcKin.
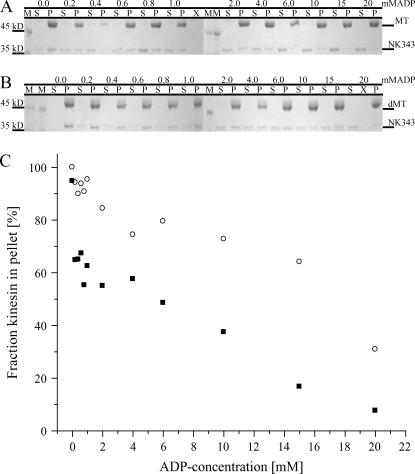
MT digestion shifts the binding equilibrium of the minimal motor domain of NcKin toward the bound state
Minimal motor constructs of both conventional and NcKin have been previously reported to display extremely high ATP turnovers while producing comparatively low MT speeds in surface gliding assays. This behavior has been interpreted as multiple futile turnovers (34,46,47). Interestingly, and in contrast to both dimeric and monomeric constructs containing parts of the neck, the removal of the E-hook does not lead to a discernable change in steady-state ATP turnover of the minimal motor domain of NcKin. The k0.5,MT is slightly reduced but the kbi,ADP is significantly increased, which is in stark contrast to the longer constructs (Table 2, Fig. 4). This increased kbi,ADP suggests altered ADP kinetics of the motor head. Following similar work of others (41) on the behavior of the minimal motor domain of rat kinesin on dMTs, we performed equilibrium-binding experiments of NK343 on MT and dMT at increasing ADP concentrations. Upon removal of the E-hook, we observe a strong shift of the binding equilibrium of the minimal motor toward the bound state (Fig. 6). This effect is most pronounced within the range 0.1–4 mM ADP. At higher ADP concentrations, additional dissociation from both the native and dMTs is observed, which we ascribed to a particularly high salt sensitivity of NcKin. At any concentration, however, larger fractions of motor remain bound to the dMTs as compared to MTs. These observations pinpoint to interactions of the E-hook on the neckless motor head and allow us to further hypothesize by which mechanisms the described altered kinetics of the kinesin·ADP-head domain reduce processivity and speed on dMTs.
FIGURE 6.
Equilibrium-binding experiments with NK343. Panels A and B show the results from binding experiments of NK343 to MTs and dMTs, respectively (M = marker, S = supernatant, P = pellet). Panel C illustrates the fraction of motor bound to the MTs (▪) and dMTs (○) as a function of the ADP concentration. Notice that much larger fractions of motor partition to the dMTs at any ADP concentration.
DISCUSSION
Previous work on the influence of tubulin's E-hook on the processivity of animal kinesin has led to a working hypothesis that postulates an electrostatic interaction between the E-hook and kinesins neck domain (26,27). In contrast to animal conventional kinesins, the neck of the fast and highly processive fungal conventional kinesin NcKin carries no net positive charge (48). As the electrostatic anchoring hypothesis predicts that such uncharged necks will not interact with the E-hook, processivity of NcKin-dimers should not be affected by proteolytic removal of the E-hook. Surprisingly, our single molecule fluorescence processivity assays show that removal of the E-hook has a strong effect on the speed and processivity of NcKin dimers (NK433, Table 1). This effect could be confirmed by independent, biochemical measurements of speed (ATP turnover, kcat) and processivity (kbi,ratio) and is independent of the length or presence of the hinge domain (NK433, NK400, and NK391, Table 2). Furthermore, the gliding speed of dMTs of the three dimeric constructs is consistently reduced, suggesting that the E-hook does not influence kinesin motility following a simple electrostatic interaction model (Table 3). However, these observations on dimeric motors in themselves do not completely rule out an interaction between neck and E-hook, as a sequence comparison of fungal and animal kinesins reveals two conserved lysine residues (K346 and K351, NcKin numbering) in the positions e and c of the first and second heptad of the predicted coiled-coil. In the crystal structure of the dimeric rat kinesin (49), these positively charged residues protrude away from the coiled-coil, consistent with a possible electrostatic interaction with the E-hook. Interestingly, an animal kinesin construct with a neck containing a quadruplicated first coiled-coil heptad (1HQ, (27)) was shown to be highly processive, which was interpreted as support for the anchoring hypothesis. However the spatially specific interaction of the two lysines in this construct is actually disrupted, as the second lysine is part of the second heptad. This suggests that the increased processivity of the 1HQ-mutant must be based on a different mechanism and is not caused by an interaction of the E-hook with the lysine residues in the neck (see also the discussion below on the force-velocity relationship and processivity). More importantly, in multiple molecule gliding assays using monomeric NcKin constructs with shortened or no neck at all, we also observed reductions of gliding speeds of dMTs which are strikingly similar to those of dimeric motors. The latter experiments not only rule out interactions of the E-hook with the neck but provide additional, strong evidence that the E-hook influences motility of NcKin through interactions with the head.
Biochemical measurements, namely, a marked increase of k0.5,MT and a decrease in kbi,ADP, indicated that the observed influence of the E-hook on speed and processivity might be due to a reduced affinity of the heads of dimeric NcKin to the dMT (Table 2, Fig. 4), which might provide an explanation of reduced processivity. Similarly, strong increases of the k0.5,MT of the monomeric constructs containing parts of the neck underline this interpretation. A reduction of the heads' affinity is readily conceivable, as the E-hook is immediately adjacent to important structures of the motor binding site (H12) and might form essential parts of the motor binding site, possibly even in a helical conformation (50). However, the kinetic behavior of the minimal motor itself, comprising only head and neck-linker (NK343), suggests an increase in affinity to the dMT, as removal of the E-hook slightly decreases k0.5,MT and increases kbi,ADP drastically (Table 2, Fig. 4). To decide if and in which state the affinity of the motor is changed, we determined the stall forces of the dimeric NcKin constructs on MTs and dMTs. We expect that changes in the strong binding states (nucleotide free and ATP) of the heads due to the removal of the E-hook lead to discernable differences in the stall forces. Surprisingly and in contrast to our initial expectations, we find 1), no significant change in the average stall force of NcKin on MTs or dMTs, and 2), a proportional reduction in gliding speed over the entire force range (Table 4, Fig. 5). In the simplest scenario these two effects are due to changes in the slowest, rate-limiting step of the cycle, the dissociation of the ADP-bound rear head from a weakly bound state. Although it is possible that more processes are affected, we believe this to be highly unlikely as the data converge to the same stall force without distinct deviation from a linear fit. In all, our force-velocity data are inconsistent with the working model presented by Thorn et al. (27), which suggests that the E-hook-neck interaction strongly affects processivity, but mainly at zero or low loads, whereas the interaction is relieved when the kinesin motor is strained by external loads. Despite the fact that their model does not clearly define the mechanism(s) through which the E-hook influences kinesin's velocity, it inherently predicts a nonlinear force-velocity relationship for NcKin motors interacting with dMTs. At low loads the motor's speed would be significantly reduced, but at moderate or high forces (1–1.5 pN and beyond, based on Thorn et al. (27)) the E-hook's influence on speed would be reduced and the force-velocity diagrams for MTs and dMTs would superimpose. Our data, however, do not show such nonlinear changes with increasing force but reveal a constant proportional reduction of speed for dMTs at all force levels (Fig. 5). This observation provides further evidence that the E-hook influences the head domain directly, most likely by altering the kinetics of the weakly bound kinesin·ADP complex.
In agreement with this interpretation, ADP equilibrium-binding experiments with the minimal motor domain (NK343) reveal that in the presence of excess ADP substantially larger fractions of motor partition to dMTs as compared to MTs (Fig. 6). These findings—but also the result from ATPase and ADP-release experiments—are consistent with a recent report by Skiniotis et al. (41), which demonstrates altered binding behavior of the minimal motor domain of rat kinesin (rK354) to dMTs: they showed that the shift in the ADP-binding equilibrium toward a more strongly bound state is caused by the trapping of an otherwise transient kinesin·ADP state on the dMT. Accordingly, our findings of a reduced rate of ADP release for dimeric NcKin constructs on dMTs are due to a slower dissociation of ADP from the motor upon binding to the MT. Together, the E-hook is believed to facilitate dissociation of the ADP·kinesin-head form the MT through a so far unknown structural interaction. On the basis of differential cryo-electron microscopy maps of decorated dMTs and MTs, Skiniotis et al. put forth an intriguing structural hypothesis postulating interactions of the E-hook with the switch-II domain of kinesin: the presence of both the E-hook and ADP in kinesin triggers structural rearrangements of the switch II cluster which facilitate dissociation of the head. These structural projections and our results allow us to hypothesize at which step in the processive stepping cycle the removal of the E-hook might interfere to cause the observed effects.
Before discussing the effects of the E-hook on the stepping cycle, it is necessary to briefly discuss differences of our kinetic data of the NcKin constructs on native MTs to previous reports; overall, our observations with undigested MTs agree well with published data (34): all dimeric motors display a similar kcat (∼70 ATP/head* s), whereas the long monomers NK378 and NK383 display strongly reduced ATP turnover and the minimal motor domain, NK343, shows very high turnover rates. The k0.5,MT-values are higher than reported earlier, but the differences between the constructs follow previously reported trends. Interestingly, the bimolecular rate for ADP-release for the dimeric NcKin constructs is very similar to that of the minimal motor domain, but ∼3–5 times higher than previously reported (Table 2, Fig. 4, (34)). Our results indicate that the reduced kbi,ADP for the long monomers is mainly due to the drastically reduced dissociation rate of ADP (kmax,ADP) caused by the inhibition brought about by the residue Y362 (35): the overall collision rate of the motors is dominated by their diffusivity, which for the dimeric and monomeric motor constructs used here should differ by a factor of 2 or less (the Stokes-Einstein relation and Chen-Wilke correlation predict differences of 1.3–1.5). Thus, the actual collision rates should only differ by the same (small) factor. As fast ADP release upon collision is prerequisite for strong binding of the motor in the nucleotide-free state, the effective rate of inhibition in the monomers by the exposed Y362 is drastically reduced. Conversely, the dimeric motors show high rates of productive encounters, which implies that the inhibitory Y362 is buried in the coiled-coil and is therefore inactive. This suggests two alternative interpretations:
If a Y362-inhibited head (as NK378 or NK383) is a structural intermediate in the stepping cycle of the dimeric motor, it can only be part of the cycle after the initial binding from solution, as kbi,ADP of the dimeric motors is high. This hypothesis suggests that the neck coiled-coil unwinds after motor binding and during stepping. Although previous studies of animal kinesins have shown this to be unlikely (51), the unusual neck domain of NcKin might possibly allow unwinding.
Alternatively, it is conceivable that the inhibition of the ADP-release—and therefore the binding of the motor to the MT—in the long monomers represent a regulatory mechanism, which mimics the function of the kinesin light chain of conventional kinesins.
Kirchner et al. (52) could show for NcKin that the inhibitory tail domain binds in the hinge/neck transition region rather than the head directly. This region is likely to form a helix-capping motif in both conventional (53) and fungal kinesins (F. Bathe, K. Hahlen, R. Dombi, L. Driller, M. Schliwa, and G. Woehlke, University of Muenchen, personal communication, 2005). It is therefore possible that structural interference after binding of the tail to the neck/hinge domain leads to disruption of the coiled-coil, which is the structural prerequisite for processive stepping. Interestingly, new data suggest that the tail forces the two heads of conventional kinesin apart (D. Cai and K. Verhey, University of Michigan, personal communication, 2005). In the case of conventional kinesins, the binding of the motor to the MT is furthermore inhibited through interactions with the kinesin light chains (D. Cai). As kinesin light chains have not been identified yet in fungi, it is likely that the Y362-inhibition represents the mechanistic equivalent of the inhibitory functions of kinesin light chains in conventional kinesins. Following this latter hypothesis, we suggest that the behavior of NK378 and NK383 does not reflect an actual stepping intermediate. However, detailed analysis of these constructs might still allow valuable insight in the molecular mechanisms of how the motor controls its activity. Interestingly, these long monomeric constructs display a dramatic increase in ATP turnover upon MT digestion. This can only be brought about by an increase in kmax,ADP, which is controlled by the inhibitory Y362 (35). Thus, the behavior of these monomers on dMTs further indicates that removal of the E-hook affects the head's ADP-kinetics. Together with the previously discussed effects of MT digestion, this allows us to formulate a hypothesis for the role of the E-hook in the processive stepping of kinesin.
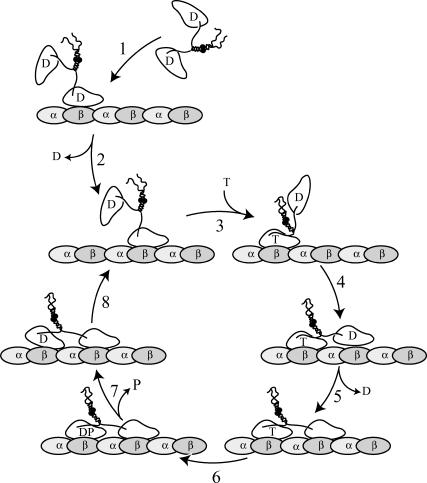
Our working model is summarized in Fig. 7. Upon collision with the MT (1), the dimeric motor quickly looses the ADP molecule from the bound head (2). Structural constraints require binding of ATP and reorientation of the neck-linker (3), before the second head can bind to the next available binding site (4) and release ADP (5) to generate an internally strained motor that is bound with both head domains to the MT. It has been shown that the rearward strain on the leading head prevents ATP binding (23), and we hypothesize that the forward strain on the rear head accelerates ATP hydrolysis (6). This strain leads to rapid phosphate release (7) and dissociation of the ADP·kinesin-head (8), which allows the cycle to start again. Our results indicate two possible points of interaction with the E-hook, namely, step 8 and step 5. Slowing of the rear head detachment (step 8) by a trapped ADP state would readily explain the reduced speed in gliding assays and lower ATP turnover. In this case the decreased processivity is brought about by the increased lifetime of this ADP state and a concurrent increased likelihood of front head dissociation due to either nucleotide binding or spontaneous dissociation in the strongly bound nucleotide-free state. Alternatively, step 5 could be slowed in the absence of the E-hook, such that the front head binds weakly to the MT without losing its ADP. We hypothesize that the intramolecular strain resulting from binding of the kinesin·ADP-head in the absence of the E-hook leads to a modest acceleration of ATP hydrolysis in the rear head as compared to the strongly bound nucleotide-free head. Consequently, ATP turnover and gliding speed are expected to be slower without the E-hook interaction; this scenario also provides an attractive explanation for the reduced processivity on dMTs: although less effective than the nucleotide-free state, the trapped ADP state triggers ATP hydrolysis and phosphate release from the rear head. This leads to a motor-MT complex with both heads in comparatively weakly bound ADP states, which is prone to dissociation. Therefore, our working model predicts a relative decrease in the frequency of single molecule trapping interactions that reach maximum stalling forces of 5 pN if the E-hook affects indeed the weakly to strongly bound transition of step 5. Although this quantification is experimentally challenging, it provides an attractive, testable hypothesis.
FIGURE 7.
Schematic working hypothesis for the processive movement of NcKin. Proposed influence of the E-hook on the chemomechanical cycle of kinesin. See Discussion for a detailed description of the model. In the native MT-motor system, the E-hook modulates (reduces) the affinity of the kinesin·ADP complex such that weak-binding intermediates facilitate fast, processive movement of kinesin. Digestion of the MT (removal of the E-hook) creates a trapped ADP-state(s) of kinesin: we propose that the forward rates of steps 5 and/or 8 in our model are severely slowed in the absence of the E-hook, leading to the observed changes in gliding speed and processivity.
Future work, including detailed cross-linking studies and single molecule experiments, should now be directed toward identifying the exact interaction site for the E-hook in the head of kinesin and the molecular mechanisms by which the E-hook functions. Interestingly, the E-hook is the predominant site of various posttranslational modifications of both α- and β-tubulin (54) and based on recent studies it had been suggested that these modifications affect the binding of motors (55). Therefore, in depth single molecule studies of the interaction of the E-hook with kinesin should provide valuable insights, not only into the molecular mechanism of kinesin-based motility, but also into illuminating possible roles of the E-hook in the regulation of MT-based motor molecules.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
We thank M. Schliwa, G. Woehlke, and F. Bathe for helpful discussions.
This work was in part supported by the Defense Advanced Research Projects Agency, the Deutsche Forschungsgemeinschaft, and the National Science Foundation.
Stefan Lakämper's present address is Dept. of Physics of Complex Systems, Vrije Universiteit, Amsterdam, The Netherlands.
References
- 1.Vale, R. D. 2003. The molecular motor toolbox for intracellular transport. Cell. 112:467–480. [DOI] [PubMed] [Google Scholar]
- 2.Vale, R. D., T. S. Reese, and M. P. Sheetz. 1985. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 42:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brady, S. T. 1985. A novel brain ATPase with properties expected for the fast axonal transport motor. Nature. 317:73–75. [DOI] [PubMed] [Google Scholar]
- 4.Kuznetsov, S. A., E. A. Vaisberg, N. A. Shanina, N. N. Magretova, V. Y. Chernyak, and V. I. Gelfand. 1988. The quaternary structure of bovine brain kinesin. EMBO J. 7:353–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scholey, J. M., J. Heuser, J. T. Yang, and L. S. Goldstein. 1989. Identification of globular mechanochemical heads of kinesin. Nature. 338:355–357. [DOI] [PubMed] [Google Scholar]
- 6.Bloom, G. S., M. C. Wagner, K. K. Pfister, and S. T. Brady. 1988. Native structure and physical properties of bovine brain kinesin and identification of the ATP-binding subunit polypeptide. Biochemistry. 27:3409–3416. [DOI] [PubMed] [Google Scholar]
- 7.de Cuevas, M., T. Tao, and L. S. Goldstein. 1992. Evidence that the stalk of Drosophila kinesin heavy chain is an alpha-helical coiled coil. J. Cell Biol. 116:957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kosik, K. S., L. D. Orecchio, B. Schnapp, H. Inouye, and R. L. Neve. 1990. The primary structure and analysis of the squid kinesin heavy chain. J. Biol. Chem. 265:3278–3283. [PubMed] [Google Scholar]
- 9.Grummt, M., G. Woehlke, U. Henningsen, S. Fuchs, M. Schleicher, and M. Schliwa. 1998. Importance of a flexible hinge near the motor domain in kinesin-driven motility. EMBO J. 17:5536–5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hackney, D. D., J. D. Levitt, and J. Suhan. 1992. Kinesin undergoes a 9 S to 6 S conformational transition. J. Biol. Chem. 267:8696–8701. [PubMed] [Google Scholar]
- 11.Hackney, D. D., and M. F. Stock. 2000. Kinesin's IAK tail domain inhibits initial microtubule-stimulated ADP release. Nat. Cell Biol. 2:257–260. [DOI] [PubMed] [Google Scholar]
- 12.Seiler, S., J. Kirchner, C. Horn, A. Kallipolitou, G. Woehlke, and M. Schliwa. 2000. Cargo binding and regulatory sites in the tail of fungal conventional kinesin. Nat. Cell Biol. 2:333–338. [DOI] [PubMed] [Google Scholar]
- 13.Hackney, D. D. 1995. Highly processive microtubule-stimulated ATP hydrolysis by dimeric kinesin head domains. Nature. 377:448–450. [DOI] [PubMed] [Google Scholar]
- 14.Huang, T. G., J. Suhan, and D. D. Hackney. 1994. Drosophila kinesin motor domain extending to amino acid position 392 is dimeric when expressed in Escherichia coli. J. Biol. Chem. 269:16502–16507. [PubMed] [Google Scholar]
- 15.Gilbert, S. P., M. L. Moyer, and K. A. Johnson. 1998. Alternating site mechanism of the kinesin ATPase. Biochemistry. 37:792–799. [DOI] [PubMed] [Google Scholar]
- 16.Howard, J., A. J. Hudspeth, and R. D. Vale. 1989. Movement of microtubules by single kinesin molecules. Nature. 342:154–158. [DOI] [PubMed] [Google Scholar]
- 17.Vale, R. D., T. Funatsu, D. W. Pierce, I. Romberg, Y. Harada, and T. Yanagida. 1996. Direct observation of single kinesin molecules moving along microtubules. Nature. 380:451–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Svoboda, K., C. F. Schmidt, B. J. Schnapp, and S. M. Block. 1993. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 365:721–727. [DOI] [PubMed] [Google Scholar]
- 19.Hua, W., E. C. Young, M. L. Fleming, and J. Gelles. 1997. Coupling of kinesin steps to ATP hydrolysis. Nature. 388:390–393. [DOI] [PubMed] [Google Scholar]
- 20.Schnitzer, M. J., and S. M. Block. 1997. Kinesin hydrolyses one ATP per 8-nm step. Nature. 388:386–390. [DOI] [PubMed] [Google Scholar]
- 21.Yildiz, A., M. Tomishige, R. D. Vale, and P. R. Selvin. 2004. Kinesin walks hand-over-hand. Science. 303:676–678. [DOI] [PubMed] [Google Scholar]
- 22.Hancock, W. O., and J. Howard. 1999. Kinesin's processivity results from mechanical and chemical coordination between the ATP hydrolysis cycles of the two motor domains. Proc. Natl. Acad. Sci. USA. 96:13147–13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenfeld, S. S., P. M. Fordyce, G. M. Jefferson, P. H. King, and S. M. Block. 2003. Stepping and stretching. How kinesin uses internal strain to walk processively. J. Biol. Chem. 278:18550–18556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lakämper, S., A. Kallipolitou, G. Woehlke, M. Schliwa, and E. Meyhöfer. 2003. Single fungal kinesin motor molecules move processively along microtubules. Biophys. J. 84:1833–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Howard, J. 1993. Molecular motors. One giant step for kinesin. Nature. 365:696–697. [DOI] [PubMed] [Google Scholar]
- 26.Wang, Z., and M. P. Sheetz. 2000. The C-terminus of tubulin increases cytoplasmic dynein and kinesin processivity. Biophys. J. 78:1955–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorn, K. S., J. A. Ubersax, and R. D. Vale. 2000. Engineering the processive run length of the kinesin motor. J. Cell Biol. 151:1093–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Redeker, V., R. Melki, D. Prome, J. P. Le Caer, and J. Rossier. 1992. Structure of tubulin C-terminal domain obtained by subtilisin treatment. The major alpha and beta tubulin isotypes from pig brain are glutamylated. FEBS Lett. 313:185–192. [DOI] [PubMed] [Google Scholar]
- 29.Nogales, E., S. G. Wolf, and K. H. Downing. 1998. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 391:199–203. [DOI] [PubMed] [Google Scholar]
- 30.Mandelkow, E. M., M. Herrmann, and U. Ruhl. 1985. Tubulin domains probed by limited proteolysis and subunit-specific antibodies. J. Mol. Biol. 185:311–327. [DOI] [PubMed] [Google Scholar]
- 31.Bhattacharyya, B., D. L. Sackett, and J. Wolff. 1985. Tubulin, hybrid dimers, and tubulin S. Stepwise charge reduction and polymerization. J. Biol. Chem. 260:10208–10216. [PubMed] [Google Scholar]
- 32.Steinberg, G., and M. Schliwa. 1996. Characterization of the biophysical and motility properties of kinesin from the fungus Neurospora crassa. J. Biol. Chem. 271:7516–7521. [DOI] [PubMed] [Google Scholar]
- 33.Crevel, I., N. Carter, M. Schliwa, and R. Cross. 1999. Coupled chemical and mechanical reaction steps in a processive Neurospora kinesin. EMBO J. 18:5863–5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kallipolitou, A., D. Deluca, U. Majdic, S. Lakamper, R. Cross, E. Meyhofer, L. Moroder, M. Schliwa, and G. Woehlke. 2001. Unusual properties of the fungal conventional kinesin neck domain from Neurospora crassa. EMBO J. 20:6226–6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schafer, F., D. Deluca, U. Majdic, J. Kirchner, M. Schliwa, L. Moroder, and G. Woehlke. 2003. A conserved tyrosine in the neck of a fungal kinesin regulates the catalytic motor core. EMBO J. 22:450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Inoue, Y., A. Hikikoshi Iwane, T. Miyai, E. Muto, and T. Yanagida 2001. Motility of single one-headed kinesin molecules along microtubules. Biophys. J. 81:2838–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inoue, Y., Y. Y. Toyoshima, A. H. Iwane, S. Morimoto, H. Higuchi, and T. Yanagida. 1997. Movements of truncated kinesin fragments with a short or an artificial flexible neck. Proc. Natl. Acad. Sci. USA. 94:7275–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang, T. G., and D. D. Hackney. 1994. Drosophila kinesin minimal motor domain expressed in Escherichia coli. Purification and kinetic characterization. J. Biol. Chem. 269:16493–16501. [PubMed] [Google Scholar]
- 39.Grummt, M., S. Pistor, F. Lottspeich, and M. Schliwa. 1998. Cloning and functional expression of a ‘fast’ fungal kinesin. FEBS Lett. 427:79–84. [DOI] [PubMed] [Google Scholar]
- 40.Klumpp, L. M., K. M. Brendza, J. M. Rosenberg, A. Hoenger, and S. P. Gilbert. 2003. Motor domain mutation traps kinesin as a microtubule rigor complex. Biochemistry. 42:2595–2606. [DOI] [PubMed] [Google Scholar]
- 41.Skiniotis, G., J. C. Cochran, J. Muller, E. Mandelkow, S. P. Gilbert, and A. Hoenger. 2004. Modulation of kinesin binding by the C-termini of tubulin. EMBO J. 23:989–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mandelkow, E., and A. Hoenger. 1999. Structures of kinesin and kinesin-microtubule interactions. Curr. Opin. Cell Biol. 11:34–44. [DOI] [PubMed] [Google Scholar]
- 43.Song, Y. H., and E. Mandelkow. 1993. Recombinant kinesin motor domain binds to beta-tubulin and decorates microtubules with a B surface lattice. Proc. Natl. Acad. Sci. USA. 90:1671–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Visscher, K., M. J. Schnitzer, and S. M. Block. 1999. Single kinesin molecules studied with a molecular force clamp. Nature. 400:184–189. [DOI] [PubMed] [Google Scholar]
- 45.Hancock, W. O., and J. Howard. 1998. Processivity of the motor protein kinesin requires two heads. J. Cell Biol. 140:1395–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moyer, M. L., S. P. Gilbert, and K. A. Johnson. 1996. Purification and characterization of two monomeric kinesin constructs. Biochemistry. 35:6321–6329. [DOI] [PubMed] [Google Scholar]
- 47.Ma, Y. Z., and E. W. Taylor. 1995. Kinetic mechanism of kinesin motor domain. Biochemistry. 34:13233–13241. [DOI] [PubMed] [Google Scholar]
- 48.Steinberg, G., and M. Schliwa. 1995. The Neurospora organelle motor: a distant relative of conventional kinesin with unconventional properties. Mol. Biol. Cell. 6:1605–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kozielski, F., S. Sack, A. Marx, M. Thormahlen, E. Schonbrunn, V. Biou, A. Thompson, E. M. Mandelkow, and E. Mandelkow. 1997. The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell. 91:985–994. [DOI] [PubMed] [Google Scholar]
- 50.Jimenez, M. A., J. A. Evangelio, C. Aranda, A. Lopez-Brauet, D. Andreu, M. Rico, R. Lagos, J. M. Andreu, and O. Monasterio. 1999. Helicity of alpha(404–451) and beta(394–445) tubulin C-terminal recombinant peptides. Protein Sci. 8:788–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomishige, M., and R. D. Vale. 2000. Controlling kinesin by reversible disulfide cross-linking. Identifying the motility-producing conformational change. J. Cell Biol. 151:1081–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kirchner, J., S. Seiler, S. Fuchs, and M. Schliwa. 1999. Functional anatomy of the kinesin molecule in vivo. EMBO J. 18:4404–4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tripet, B., and R. S. Hodges. 2002. Helix capping interactions stabilize the N-terminus of the kinesin neck coiled-coil. J. Struct. Biol. 137:220–235. [DOI] [PubMed] [Google Scholar]
- 54.Westermann, S., and K. Weber. 2003. Post-translational modifications regulate microtubule function. Nat. Rev. Mol. Cell Biol. 4:938–947. [DOI] [PubMed] [Google Scholar]
- 55.Rosenbaum, J. 2000. Cytoskeleton: functions for tubulin modifications at last. Curr. Biol. 10:R801–R803. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.