Abstract
A growing number of natively disordered proteins undergo a folding/binding process that is essential for their biological function. An interesting question is whether these proteins have incompletely solvated regions that drive the folding/binding process. Although the presence of predominantly hydrophobic buried regions can be easily ascertained by high-sensitivity differential scanning calorimetry analysis, the identification of those residues implicated in the burial requires NMR analysis. We have selected a partially solvated natively disordered fragment of Escherichia coli, thioredoxin, C37 (38–108), for full NMR spectral assignment. The secondary chemical shifts, temperature coefficients, and relaxation rates (R1 and R2) of this fragment indicate the presence of a flexible backbone without a stable hydrogen bond network near neutral pH. 1H-15N heteronuclear single quantum coherence analysis of the pH dependence of amide chemical shifts in fragment C37 within pH 2.0 and 7.0 suggests the presence of interactions between nonionizable residues and the carboxylate groups of four Asp and four Glu residues. The pH midpoints (pHm) of the amides in the ionizable residues (Asp or Glu) and, consequently, the shifts in the pHm (ΔpHm) of these residues with respect to model tetrapeptides, are sequence-dependent; and the nonionizable residues that show pH dependence cluster around the ionizable ones. The same pH dependence has been observed in two fragments: M37 (38–73) and C73 (74–108), ruling out the participation of long-range interactions. Our studies indicate the presence of a 15-residue pH-dependent segment with the highest density of ionizable sites in the disordered ensembles of fragments C37 and M37. The observed correlations between ionizable and nonionizable residues in this segment suggest the organization of the backbone and side chains through local and medium-range interactions up to nine residues apart, in contrast to only a few interactions in fragment C73. These results agree qualitatively with the predominantly hydrophobic buried surface detected only in fragments C37 and M37 by highly sensitive differential scanning calorimetry analysis. This work offers a sensitive and rapid new tool to obtain clues about local and nonlocal interactions between ionizable and nonionizable residues in the growing family of natively disordered small proteins with full NMR assignments.
INTRODUCTION
During the last decade a growing number of proteins variously labeled “natively unfolded” (1), “intrinsically disordered” (2,3), and “intrinsically unstructured” (4) have been found to play a key role in various biological functions that require folding/binding (5–7). For the sake of concreteness we will refer to them as “natively disordered proteins”. These proteins are easily recognized by a strong minimum around 200 nm in the far-ultraviolet circular dichroism spectrum and poorly dispersed amide proton resonances in a 1D-NMR spectrum. An intriguing question is whether these proteins have incompletely solvated regions which are essential for the folding/binding process and, most importantly, which residues are responsible for these regions. Although the presence of buried, predominantly hydrophobic regions can be easily ascertained by high-sensitivity DSC analysis, as has been done for natively disordered protein fragments (8,9), this technique lacks atomic detail, which requires the resolution provided by multidimensional NMR analysis (10–15). The latter readily provides information about the protein backbone (secondary chemical shifts; temperature coefficients; protection of amide protons to solvent exchange; NOE distance constraints; 15N relaxation rates of the backbone; 1H-15N heteronuclear, etc.). However, some “natively disordered proteins” do not exhibit rigid segments in the sequence and/or regions of secondary structure (1,4), but they may still have clusters of side chains mediated by local and nonlocal interactions whose detection requires NOESY experiments. Although these experimental approaches are standard for well folded proteins, in the case of denatured or natively disordered proteins, where an ensemble of conformations is likely to be present, the analysis requires caution (12–14,16) and more laborious approaches that combine NMR with other techniques, such as mutagenesis (17), paramagnetic spin labels (18), and selective labels (19). An excellent example is furnished by the recent NOESY experiments on the selectively labeled N-terminal SH3 domain of Drosophila (drkNSH3) (19), in which folded and unfolded states coexist in slow exchange under native conditions (20–22). These experiments demonstrate the presence of at least two conformers in the natively unfolded state of drkNSH3: a compact species with native-like residual secondary structure, and a less compact one with a nonnative hydrophobic cluster mediated by long-range interactions. A methodology based on simple NMR experiments that provide clues about local and nonlocal interactions in natively disordered proteins will be a useful tool for experimentalists.
Numerous studies on folded proteins that involve NMR titration, mutagenesis, and computational analysis (23,24) have provided evidence for local and nonlocal interactions between ionizable and nonionizable residues. The pKas of carboxylate side chains in folded proteins may be shifted upfield or downfield due to the presence of hydrophobic (25,26) or polar residues in the surroundings (27–29). The pH midpoints (pHm) of the backbone amides in folded proteins reflect the electrostatic environment produced by nearby carboxylates of Asp and Glu residues (27,28,30–33). In contrast, much less is known about the pH dependence of backbone amide chemical shifts and the pKa of ionizable side chains in natively disordered polypeptides (34–41) and denatured proteins (31,42). Studies on model host-guest tetrapeptides indicate that the pHm of the amides from Asp and Glu residues are very close to the pKa of their carboxylates (32,43). Studies on natively disordered peptides and natively unfolded proteins, on the other hand, show that the pKas of the carboxylate side chains are shifted relative to the model tetrapeptides in some cases (35–37), but they are very close to them in others, even in the case of sequential carboxylate side chains (39,40). Although more studies are needed to understand the pH dependence in natively disordered polypeptides, so far these results lead us to believe that the pH dependence of backbone amide chemical shifts and carboxylate side chains may be excellent probes to identify intramolecular interactions in these polypeptides.
During the last few years, we have been studying a family of natively disordered fragments of Escherichia coli Trx as a model for biologically active natively disordered proteins (8,9,44–49). These studies have produced interesting correlations between DSC (8) and NMR experiments (48). For example, the rather rigid hydrophobic helical region of fragment N73 (1–73) correlates with the estimate of its predominantly hydrophobic buried surface. Recently, high-sensitivity DSC studies on a family of natively disordered fragments of E. coli Trx again have provided evidence for the presence of incompletely solvated regions in some fragments (9), leading us to carry out structural and dynamic NMR experiments on a fragment with a significant hydrophobic buried surface. Here we report using the pH dependence of backbone amide chemical shifts as a probe to obtain clues about local and nonlocal interactions between ionizable and nonionizable residues in the natively disordered fragment C37 and two overlapping fragments.
METHODOLOGY
Standard structural and dynamic NMR parameters of fragments
Fragment C37 of E. coli Trx was generated according to previously reported procedures (9). Samples of the isolated 15N-13C-labeled fragment C37 were prepared in 25 mM potassium acetate buffer with 10% D2O and 0.01% sodium azide at three pH values: 3.75, 5.0, and 6.25 for 0.5 mM, 0.1 mM, and 0.1 mM fragments, respectively. NMR experiments were performed on an INOVA AS600 (Varian, Palo Alto, CA) or an Avance 800 spectrometer with triple resonance probes (Bruker, Billerica, MA). The 1H chemical shift was referenced to the temperature-dependent H2O frequency and corrected using sodium 2,2-dimethyl-,2-silapentane-5-sulfonate as an external reference. The 15N and 13C chemical shifts were indirectly referenced (50). The temperature was calibrated using methanol (51). NMR data were processed and analyzed using NMRPipe (52) and SPARKY (53), respectively. Sequential backbone resonance assignments were carried out using HNCA (54), HNCACB (55), HNCO (56), and 1H-15N HSQC experiments at pH 3.4, 5.0, and 6.25 and a range of temperatures from 3.5°C to 35°C.
The transfer of backbone assignments from pH 3.4, 5.0, and 6.25 to other pH was based on 1H-15N HSQC experiments done every 0.25 pH units. Secondary chemical shifts of the amide proton (HN), α carbons (Cα), amide nitrogen (N), and carbonyl (C′) were calculated based on the full spectral assignments at pH 6.25 and 25°C, and the random coil database (57). These shifts were corrected for sequence (58) and temperature (59,60) dependence. The temperature coefficients of the HN and C′ were calculated based on the assigned 1H-15N HSQC and HNCO spectra at pH 6.25 and four temperature points (3.5, 5, 10, and 15°C). The 15N longitudinal (R1) and 15N transverse (R2) relaxation rates were acquired as previously described (61,62) with a Varian INOVA AS600 spectrometer. NMR relaxation data were collected at pH 6.25, 0°C. The spectra were processed using NMRPipe and analyzed using a nonlinear fitting to a single exponential function with SPARKY (53).
pH dependence of amide chemical shifts in fragments
The pH dependence of amide chemical shifts in 15N-labeled fragments C37, M37, and C73 was monitored using 1H-15N HSQC experiments every ∼0.25 pH units between pH 2 and 7 at 25°C. Measurements were performed with an Accumet (Ossining, NY) combination electrode of 3-mm diameter and 1.5-in flexible stem and used without correcting for deuterium isotope effect (63). Concentrations of fragments as low as 0.1 mM were used to circumvent aggregation. Initial samples of the isolated fragments were prepared in 25 mM potassium acetate at three different pH: 3.75 (0.5 mM of fragment); 4.5 (0.5 mM of C73, and 0.1 mM of C37 and M37); and 6.0 (same as for pH 4.5). The samples at pH 3.75 were titrated down to pH 2.0; those at pH 6.0 were titrated in both directions to fill the region between pH 5.0 and 7.0. The samples at pH 4.5 were titrated in both directions to cover the region between 3.75 and 5.0. The spectra at pH 3.75 and 5.0 were thus obtained in two different ways and served as control points for ionic strength changes. The full NMR assignments of fragment C37 at pH 3.4, 5.0, and 6.25 were used to resolve severe crosspeak overlaps that occurred among Glu residues, with the exception of Glu-85. Data were fitted to a modified Hill equation (64):
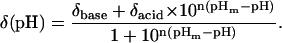 |
(1) |
The acidic (δacid) and basic (δbase) plateaus, Hill coefficient (n), and pHm were left as floating parameters during the nonlinear least-square analysis carried out using Origin v5.
RESULTS
Standard structural and dynamic NMR analysis of natively disordered fragments
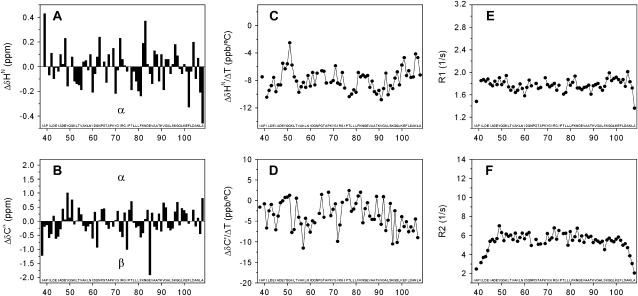
To find regions with secondary structure, a secondary chemical shift analysis of fragment C37 was conducted under native-like conditions (see Fig. 1, A and B). The absolute deviations of the chemical shifts from statistical random coil values are in general <0.3 ppm and 1.0 ppm for the amide proton (HN) and α carbons (Cα), respectively, and <2 ppm for both the amide nitrogen (HN) and the carbonyl (C′). Comparison of the secondary chemical shifts for HN, Cα, and other nuclei show the absence of sequential segments of five or more residues with the same tendency for a given secondary structure. These results indicate lack of secondary structure in the sequence of C37. Moreover, comparison of the secondary chemical shifts of fragment C37 against those of fragments M37 and C73 (see Fig. 2) shows that the secondary chemical shifts of the short fragments are essentially similar to those of the parent fragment, except near the cleavage site (65). The similarity among the secondary chemical shifts of the three fragments indicates that they may share local and nonlocal interactions, but none of them have regions of secondary structure. To obtain evidence for the presence of stable hydrogen bonds, the temperature coefficients of the amide protons and carbonyl groups were obtained for fragment C37 (see Fig. 1, C and D). Most of the coefficients for HN fall in the range −6 to −10 ppb per °C and are significantly different from the typical value (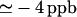 ) found in folded proteins (66). However, a sequential segment from 48 to 52, which includes Tyr-49, shows coefficients for HN close to −4 ppb per °C and even close to zero for Gly-51. The same segment exhibits similar coefficients for C′ between 0 and +2 ppb per °C, and relatively more positive values than the rest of the sequence. These results are in agreement with the effect of the aromatic ring current of Tyr on the temperature coefficients of HN from neighboring residues (66), which is expected to push those values toward zero. The temperature coefficients of segments 48–52 hint at the presence of local interactions between Tyr-49 and Gly-51. To determine whether any region of fragment C37 shows limited backbone mobility, relaxation measurements were carried out at low temperature (0°C) to stabilize transient hydrogen bonds and increase the solution viscosity, thereby enhancing the mobility differences. Despite the low temperature, the dispersion of the relaxation rates (R1 and R2) is small (see Fig. 1, E and F). These rates are relatively uniform along the sequence except at both N- and C-termini, and similar to the average values found in flexible peptides (48,62): 1.8/s (R1) and 6/s (R2). The NMR parameters of fragment C37 indicate the presence of a flexible backbone with local interactions between Tyr-49 and Gly-51, but without regular secondary structure. These results lead us to hypothesize that the predominantly hydrophobic buried surface estimated by DSC analysis arises from interactions involving rather hydrophobic side chains.
) found in folded proteins (66). However, a sequential segment from 48 to 52, which includes Tyr-49, shows coefficients for HN close to −4 ppb per °C and even close to zero for Gly-51. The same segment exhibits similar coefficients for C′ between 0 and +2 ppb per °C, and relatively more positive values than the rest of the sequence. These results are in agreement with the effect of the aromatic ring current of Tyr on the temperature coefficients of HN from neighboring residues (66), which is expected to push those values toward zero. The temperature coefficients of segments 48–52 hint at the presence of local interactions between Tyr-49 and Gly-51. To determine whether any region of fragment C37 shows limited backbone mobility, relaxation measurements were carried out at low temperature (0°C) to stabilize transient hydrogen bonds and increase the solution viscosity, thereby enhancing the mobility differences. Despite the low temperature, the dispersion of the relaxation rates (R1 and R2) is small (see Fig. 1, E and F). These rates are relatively uniform along the sequence except at both N- and C-termini, and similar to the average values found in flexible peptides (48,62): 1.8/s (R1) and 6/s (R2). The NMR parameters of fragment C37 indicate the presence of a flexible backbone with local interactions between Tyr-49 and Gly-51, but without regular secondary structure. These results lead us to hypothesize that the predominantly hydrophobic buried surface estimated by DSC analysis arises from interactions involving rather hydrophobic side chains.
FIGURE 1.
Standard structural and dynamic NMR analysis of fragment C37 of E. coli Trx. Δδ is defined as the difference between the observed chemical shifts and the corresponding random coil values. Secondary chemical shifts of the HN (A) and Cα (B) are given at pH 6.25 and 25°C. Temperature coefficients of the HN (C) and C′ (D) are given at pH 6.25. R1 (E) and R2 values (F) are given at pH 6.25 and 0°C.
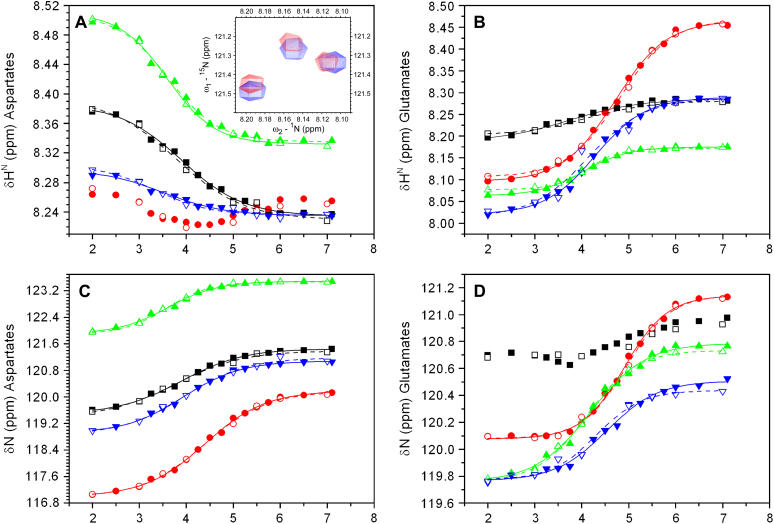
FIGURE 2.
Titration curves for Asp and Glu residues of fragments from E. coli Trx. Chemical shifts of both HN and N for Asp (A and C, respectively) and Glu (B and D, respectively) at 25°C are depicted as a function of pH for the short (open symbols) and long (solid symbols) fragments as follows: D43 and E44 (black squares); D47 and E48 (red circles); D61 and E85 (green triangles); and D104 and E101 (blue inverted triangles). The nonlinear fitting of the experimental chemical shift values to Eq. 1 (see text) is represented by solid (long fragment C37) and dashed (short fragments M37 and C73) lines. The inset in A shows the overlay of the HSQC spectrum of fragment C37 in 25 mM potassium acetate at pH 3.75 (blue) and the spectrum of a previously titrated solution of the same fragment from pH 4.5 to pH 3.75 (red) at 25°C.
pH Dependence of amide chemical shifts in natively disordered fragments
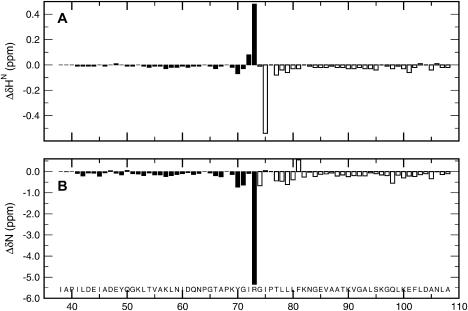
To obtain clues about which interactions account for the predominantly hydrophobic buried surface in fragment C37, we monitored the pH dependence of amide chemical shifts in the acidic regime by NMR analysis (see Fig. 3). The pH dependence was analyzed in terms of a modified Hill equation (64) (Eq. 1), in which the pHm and the so-called “Hill coefficient” (n) are the essential parameters. In the case of amides, the Hill coefficient measures the cooperativity of removing a proton from a carboxylic group in the vicinity of the amide being observed. A coefficient value of unity reflects the presence of one titrating carboxylate and a coefficient less than unity implies the interaction of more than one carboxylate (28). To evaluate the participation of long-range interactions between the N- and C-terminal regions of fragment C37, which was not revealed by the secondary chemical shifts, we monitored and analyzed the pH dependence of two complementary fragments, M37 (38–73) and C73 (74–108) (65) (see Fig. 3). Interestingly, the titration curves of the amide protons from fragments C37, M37, and C73 show the same trend observed in model tetrapeptides: the basic plateau for the amide of Asp is lower than the acidic one, and the reverse occurs for the amide of Glu (32).
FIGURE 3.
Chemical shift differences between the short and long fragments of E. coli Trx. The chemical shift differences (Δδ) between fragment C37 and fragments M37 and C73 are depicted by solid and open vertical bars, respectively. (A and B) Δδ values at pH 6.25 and 25°C for HN and N, respectively. The chemical shifts of M37 and C73 were obtained using previous spectral assignments under the same conditions (65).
The information obtained from the analysis of titration curves is given in Table 1. The majority of the curves display a simple sigmoidal pH dependence; however, several residues (e.g., Glu-44 and Asp-47) exhibit more complex shapes. Inspection of Table 1 indicates that
The amide proton and nitrogen of most residues have similar pHm and Hill coefficient within the experimental error;
The discrepancy between the pHm of a given residue is significantly lower than the shift in the pHm (ΔpHm) with respect to model tetrapeptides (32);
The ΔpHm and Hill coefficient of a given residue are essentially the same whether measured in the long (C37) or short (M37 or C73) fragments, with the exception of Glu-48 and Ser-95;
The pHm of the amide in the carboxy terminus of fragment M37 (Arg-73,
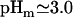 ) is significantly downshifted from that of C37 and C73 (Ala-108,
) is significantly downshifted from that of C37 and C73 (Ala-108, 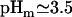 in both cases), suggesting that the interaction between the positively charged side chain of Arg-73 and its carboxylate produces a downshift of 0.5 pH units.
in both cases), suggesting that the interaction between the positively charged side chain of Arg-73 and its carboxylate produces a downshift of 0.5 pH units.
TABLE 1.
pH Dependence of amide reporters in fragments of E. coli Trx
| Amide proton (HN)
|
Amide nitrogen (N)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Residue | Fragment | pHm | ΔpHm | n | ΔδH | pHm | ΔpHm | n | ΔδN |
| Ionizable residues | |||||||||
| D43 | C37 | 3.90 ± 0.06 | +0.00 ± 0.16 | 0.65 ± 0.06 | −0.15 | 3.89 ± 0.04 | −0.01 ± 0.14 | 0.66 ± 0.04 | +1.9 |
| D43 | M37 | 4.03 ± 0.04 | +0.13 ± 0.14 | 0.65 ± 0.04 | −0.16 | 3.94 ± 0.02 | +0.04 ± 0.12 | 0.68 ± 0.02 | +1.9 |
| E44 | C37 | 3.73 ± 0.12 | −0.57 ± 0.22 | 0.56 ± 0.07 | +0.10 | * | |||
| E44 | M37 | 3.95 ± 0.06 | −0.35 ± 0.16 | 0.62 ± 0.05 | +0.08 | * | |||
| D47 | C37 | * | 4.40 ± 0.03 | +0.50 ± 0.13 | 0.68 ± 0.03 | +3.2 | |||
| D47 | M37 | * | 4.45 ± 0.03 | +0.55 ± 0.13 | 0.71 ± 0.03 | +3.1 | |||
| E48 | C37 | 4.70 ± 0.03 | +0.40 ± 0.13 | 0.79 ± 0.03 | +0.37 | 4.91 ± 0.03 | +0.61 ± 0.13 | 0.95 ± 0.05 | +1.1 |
| E48 | M37 | 4.79 ± 0.03 | +0.49 ± 0.13 | 0.86 ± 0.04 | +0.35 | 4.94 ± 0.03 | +0.64 ± 0.13 | 0.97 ± 0.06 | +1.1 |
| D61 | C37 | 3.62 ± 0.06 | −0.28 ± 0.16 | 0.81 ± 0.07 | −0.17 | 3.61 ± 0.05 | −0.29 ± 0.15 | 0.81 ± 0.06 | +1.6 |
| D61 | M37 | 3.70 ± 0.03 | −0.20 ± 0.13 | 0.80 ± 0.04 | −0.18 | 3.75 ± 0.02 | −0.15 ± 0.12 | 0.81 ± 0.03 | +1.6 |
| E85 | C37 | 4.03 ± 0.04 | −0.27 ± 0.14 | 0.90 ± 0.06 | +0.11 | 4.18 ± 0.05 | −0.12 ± 0.15 | 0.74 ± 0.06 | +1.0 |
| E85 | C73 | 4.07 ± 0.04 | −0.23 ± 0.14 | 0.87 ± 0.06 | +0.12 | 4.06 ± 0.02 | −0.24 ± 0.12 | 0.75 ± 0.03 | +1.0 |
| E101 | C37 | 4.28 ± 0.04 | −0.02 ± 0.14 | 0.80 ± 0.06 | +0.27 | s | |||
| E101 | C73 | 4.19 ± 0.03 | −0.11 ± 0.13 | 0.72 ± 0.03 | +0.30 | 4.29 ± 0.03 | − 0.01 ± 0.13 | 0.67 ± 0.03 | +0.9 |
| D104 | C37 | s | 3.93 ± 0.03 | +0.03 ± 0.13 | 0.70 ± 0.04 | +2.2 | |||
| D104 | C73 | s | 3.89 ± 0.02 | − 0.01 ± 0.12 | 0.69 ± 0.02 | +2.3 | |||
| R73 | M37 | 2.97 ± 0.03 | 0.84 ± 0.03 | −0.64 | 2.98 ± 0.03 | 0.80 ± 0.03 | +4.4 | ||
| A108 | C37 | 3.48 ± 0.05 | 0.77 ± 0.05 | −0.48 | 3.54 ± 0.04 | 0.76 ± 0.04 | +4.9 | ||
| A108 | C73 | 3.47 ± 0.02 | 0.80 ± 0.02 | −0.47 | 3.53 ± 0.02 | 0.78 ± 0.02 | +4.8 | ||
| Nonionizable residues | |||||||||
| I45 | C37 | 4.76 ± 0.03 | 0.72 ± 0.04 | +0.13 | * | ||||
| I45 | M37 | 4.83 ± 0.04 | 0.74 ± 0.05 | +0.12 | * | ||||
| A46 | C37 | 4.89 ± 0.03 | 0.88 ± 0.05 | +0.09 | 4.94 ± 0.04 | 1.02 ± 0.08 | +1.6 | ||
| A46 | M37 | 4.92 ± 0.05 | 0.78 ± 0.06 | +0.09 | 4.92 ± 0.04 | 0.98 ± 0.07 | +1.6 | ||
| Q50 | C37 | * | 3.83 ± 0.05 | 0.69 ± 0.05 | −1.5 | ||||
| Q50 | M37 | * | 3.92 ± 0.04 | 0.60 ± 0.03 | −1.5 | ||||
| G51 | C37 | 3.95 ± 0.06 | 0.65 ± 0.06 | +0.19 | * | ||||
| G51 | M37 | 4.07 ± 0.05 | 0.54 ± 0.04 | +0.19 | * | ||||
| K52 | C37 | 4.11 ± 0.08 | 0.48 ± 0.06 | −0.15 | * | ||||
| K52 | M37 | 4.25 ± 0.05 | 0.57 ± 0.04 | −0.15 | * | ||||
| L53 | C37 | * | 3.87 ± 0.06 | 0.68 ± 0.07 | −0.9 | ||||
| L53 | M37 | * | 3.96 ± 0.04 | 0.55 ± 0.03 | −0.8 | ||||
| K82 | C37 | 4.00 ± 0.03 | 0.87 ± 0.06 | +0.12 | * | ||||
| K82 | C73 | 4.02 ± 0.02 | 0.89 ± 0.04 | +0.13 | * | ||||
| N83 | C37 | 3.97 ± 0.04 | 0.94 ± 0.07 | +0.08 | * | ||||
| N83 | C73 | 4.05 ± 0.02 | 0.93 ± 0.04 | +0.09 | * | ||||
| S95 | C37 | 4.00 ± 0.03 | 0.75 ± 0.04 | +0.14 | * | ||||
| S95 | C73 | 3.82 ± 0.03 | 0.71 ± 0.03 | +0.16 | * | ||||
The pH midpoint (pHm), Hill coefficient (n), and the chemical shift differences between the basic and acidic plateaus for HN (ΔδH) and N (ΔδN) are given for the ionizable and nonionizable residues of fragments C37, M37, and C73. The ΔpHm column indicates the difference between the pHm of an ionizable residue and the pKa of the same ionizable residue in model tetrapeptides (pKa of 3.9 ± 0.1 and 4.3 ± 0.1 for Asp and Glu, respectively) (32,43). The parameters were calculated using cutoffs of 0.08 and/or 0.8 ppm for the absolute ΔδH and/or ΔδN, respectively. Undetermined pHm due to problems with cutoff and complex pH dependence (not a simple sigmoidal) are marked with an “s” and an asterisk, respectively.
These results argue for the high sensitivity of amides to the electrostatic environment around neighboring carboxylates. They also argue against the relevance of long-range interactions between the N- and C-terminal regions (∼35 residues) of fragment C37, and lead us to interpret the pH dependence in terms of interactions between ionizable and nonionizable residues within the sequence of the short fragments.
DISCUSSION
Evidence for interactions with ionizable residues in natively disordered polypeptides
Three lines of evidence lead us to infer that intramolecular interactions between ionizable residues, which are negatively charged at neutral conditions, and nonionizable residues modulate the pHm of the amides (see Table 1). First, the pHm of the ionizable residues (Asp or Glu) depends on their position in the sequence. Consequently, the shifts in the pHm (ΔpHm) of these residues with respect to model tetrapeptides also depend on their position in the sequence. These position-dependent pHm, by themselves, are enough to strongly suggest the participation of interactions. Second, pH-dependent apolar and hydrogen acceptor/donor residues, with the exception of Ser-95, cluster around the ionizable residues. This is a strong indication that the electrostatic environment of the amide group in these nonionizable residues is affected by the charge state of the ionizable ones. Our third line of evidence is the presence (or, at least, intimation) of a correlation between the ΔpHm of an ionizable residue and the type (apolar or polar) of the associated nonionizable residues that share the same pHm. Interestingly, the same phenomena have been observed in folded proteins (67).
Correlation between ionizable and nonionizable residues in natively disordered polypeptides
Comparison of the pHm and Hill coefficients of ionizable residues (amide protons and nitrogens) with those of nonionizable ones (see Table 1) reveals correlations in which a given ionizable residue and a group of nonionizable residues have amides with similar pHm and Hill coefficient (see Table 2). Since the pHm and Hill coefficients of the sequence common to the long and short fragments are essentially unchanged, we can assume that correlations do not extend across the cleavage site (Arg-73). Based on that assumption, three robust sets of correlated residues are shown in Table 2: (Asp-43, Gln-50, Gly-51, and Leu-53); (Glu-48 and Ile-45); and (Glu-85, Lys-82, and Asn-83). If we also include residues whose pHm and Hill coefficients are within the experimental error, the correlation sets will be: (Asp-43, Gln-50, Gly-51, Lys-52, and Leu-53); (Glu-48, Ile-45, and Ala-46); (Glu-85, Lys-82, Asn-83, and Ser-95); and (Glu-44, Gly-51 and Lys-52).
TABLE 2.
Correlations between ionizable and nonionizable residues in fragments of E. coli Trx
| Residue | pHm (HN/N) | ΔpHm | I45 | A46 | Q50 | G51 | K52 | L53 | K82 | N83 | S95 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| D43 | 3.97/3.92 | 0 | —/3.88 | 4.01/— | 4.18*/— | —/3.92 | |||||
| E44 | 3.84/— | −0.46 | 4.01*/— | 4.18*/— | |||||||
| D47 | —/4.43 | +0.53 | |||||||||
| E48 | 4.75/4.93 | +0.54 | 4.80/— | 4.91*/4.93 | |||||||
| D61 | 3.66/3.68 | −0.23 | |||||||||
| R73 | 2.97/2.98 | −0.78† | |||||||||
| E85 | 4.05/4.12 | −0.22 | 4.01/— | 4.03/— | 4.00/— | ||||||
| E101 | 4.24/— | 0 | |||||||||
| D104 | —/3.91 | 0 | |||||||||
| A108 | 3.48/3.54 | — |
The parameters of the ionizable and nonionizable residues and the correlations between them are displayed along the vertical and horizontal axes, respectively. The ΔpHm column indicates whether the pHm (averaged over measurements for fragments C37, M37, and C73) of a given ionizable residue is upshifted (>0), downshifted (<0) or unshifted (0, within the experimental error), with respect to the pKa of the same ionizable residue in model tetrapeptides (pKa of 3.9 ± 0.1 and 4.3 ± 0.1 for Asp and Glu, respectively) (43,32). The filled boxes represent correlated residues in which the pHm and n value of an ionizable and a nonionizable residue are similar (see Table 1). A dash indicates an undetermined pHm due to problems with cutoff and/or complex pH dependence (not a simple sigmoidal). Correlations between ionizable and nonionizable residues from different short fragments are not given.
The difference between the pHm and Hill coefficients is just within the experimental error (see Table 1).
The ΔpHm was calculated using a value of 3.75 for the carboxy terminus pKa (73).
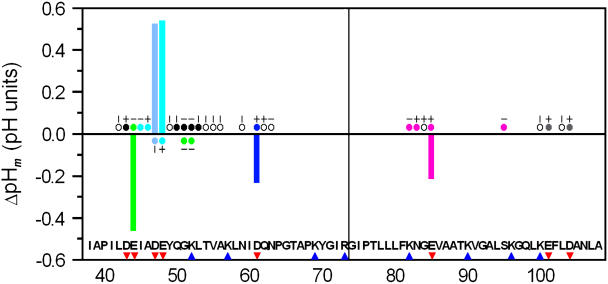
Analysis of the ΔpHm for the ionizable residues in terms of the type of associated nonionizable residues in the long and short fragments (see Fig. 4) reveals the following:
When the pHm of a given ionizable residue is upshifted with respect to the model tetrapeptides (i.e., Glu-48), the associated nonionizable residues, if any, are apolar (Ile-45 and Ala-46).
When the pHm is downshifted (i.e., Glu-85), the associated nonionizable residues, if any, are predominantly hydrogen bond acceptors/donors (Lys-82 and Asn-83).
When the pHm is essentially unshifted, such as for Asp-43, the associated nonionizable residues, if any, include both hydrogen bond acceptors/donors (Gln-50 and Lys-52) and apolar side chains (Leu-53), suggesting the presence of compensatory effects, as previously reported for folded proteins (67).
FIGURE 4.
Differences in pH dependence between the fragments of E. coli Trx and model tetrapeptides. The shifts in the averaged pHm (ΔpHm) of Asp or Glu with respect to model tetrapeptides (see Table 2) are represented by vertical bars. The basic and acidic residues are labeled underneath the sequence with blue triangles and red inverted triangles, respectively. The vertical line between R73 and G74 indicates the C- and N-termini of M37 and C73, respectively. The pH-dependent residues are labeled with circles. Residues whose titration curves for HNand N behave like simple sigmoidals are labeled with short vertical and horizontal lines above the circles. Residues whose ΔδH and ΔδN are lower than the cutoff are labeled with open circles. Sets of correlated residues are distinguished by the color of the circles. The uncorrelated ionizable residues are labeled with gray circles.
The Hill coefficients provide an indication of cooperativity. Coefficients <1 are associated with “negative cooperativity” suggesting that the observed pHms are influenced by multiple ionizable residues that broaden the observed transition curves. Isolated ionizable residues (i.e., Asp-61 and Glu-85, see Fig. 4) would be expected to have amides with coefficients close to unity unless there are significant through-space interactions with other carboxylates. In contrast, clusters of ionizable residues (Asp-43, Glu-44, Asp-47, and Glu-48; Glu-101 and Asp-104; see Fig. 4) would be expected to have amides with significantly lower coefficients. Inspection of the Hill coefficients in Table 1 shows values lower than unity, although some of them are much closer to unity than the average, which indicates that in general these expectations are confirmed.
In summary, most ionizable residues (amide protons and nitrogens) with significant ΔpHm in the three fragments, with the exception of the upshifted pHm of Asp-47, show a correlation between the sign of the ΔpHm and the type of associated nonionizable residues. Moreover, ionizable residues with Hill coefficients significantly lower than unity seem to correlate with the presence of neighboring interacting carboxylates in the sequence.
Medium-range interactions between ionizable and nonionizable residues and buried surfaces in natively disordered polypeptides
Both fragments M37 and C37 contain a pH-dependent 15-residue segment from Leu-42 to Ala-56 that exhibits the highest density of ionizable sites and the most complex behavior, as shown by the presence of correlations between residues that are up to nine residues apart. Although the observed correlations await confirmation by mutagenesis and NMR analysis, we can attempt to interpret them. Since the amide of an ionizable residue should be affected by the environment of its own carboxylate side chain, the pHm of the amide should be close to the pKa of its own carboxylate unless the amide is affected by more than one carboxylate (directly or indirectly) (32,43). The Hill coefficient should distinguish between amides near an isolated carboxylate and amides interacting with multiple carboxylates. However, the expected conformational exchange within the ensemble seems to complicate the interpretation. Since the amides of nonionizable residues will not show pH dependence unless they are affected by the environment of one or more carboxylates (directly or indirectly), the simplest interpretation for the presence of pairs containing one ionizable and one nonionizable residue with similar pH dependence, is that the pair of residues interact with the same electrostatic environment in at least one sizable subensemble. For each set of correlated residues, there are several such pairs, each one associated with a possibly different subensemble, which raises the question of the overlap between these subensembles. Although the answer is unknown, the similarity in the pH dependence of the nonionizable residues suggests that the overlap is large and thus there is a sizable subensemble in which all the correlated residues from a set have the same electrostatic environment.
The pH-dependent segment from Leu-42 to Ala-56 contains three robust sets of correlated residues. Two of them are associated with medium-range interactions (which we arbitrarily define as interactions between residues that are six to nine residues apart) among residues. The first set includes Asp-43, Gln-50, Gly-51, Lys-52, and Leu-53; the second includes Glu-44, Gly-51, and Lys-52. In contrast, the third set is associated only with local interactions among Glu-48, Ile-45, and Ala-46. The fact that two sets include Gly-51 and Lys-52 is tantalizing and the simplest interpretation is that the 15-residue pH-dependent segment adopts a hairpin-like conformation in a sizable subensemble, which accommodates a hydrophobic cluster (Leu-42, Ile-45, Ala-46, hydrophobic portion of Lys-52, Leu-53, Val-55, and Ala-56) and a polar cluster of hydrogen acceptor/donor side chains (Tyr-49, Gln-50, Lys-52, and Thr-54). The hydrophobic clusters would shield the carboxylate groups of Asp-43, Asp-47, and Glu-48, and the polar cluster would form transient hydrogen bonds with Asp-43 and Glu-44. This hairpin may even be poised to form salt bridges among Asp-43, Glu-44, and Lys-52. Due to the conformational exchange in an ensemble, to establish the relative population and conformational bias for a hairpin of a sizable subensemble is not a simple task and would require NMR experiments on frozen conformational ensembles (68,69). The presence of a sizable subensemble with a hairpin conformation is consistent with the unexpected temperature coefficient of Gly-51 (see Fig. 1), which suggests the presence of local interactions between the aromatic ring of Tyr-49 and Gly-51 in the middle of the putative hairpin. The presence of that subensemble is also consistent with previous studies on Tyr-X-Gly triads in disordered polypeptides which indicate aromatic-backbone interactions between Tyr and Gly (70). It is also known that local and nonlocal interactions between ionizable and nonionizable residues can be responsible for the pH dependence of β-hairpin peptides (71), and local aromatic-backbone interactions are widely observed in folded proteins (72).
Regardless of the presence or absence of the hairpin and the degree of overlap of the subensembles, the observed set of correlated residues (see Table 2 and Fig. 4) in fragments C37 and M37 suggests the presence of medium-range interactions in a sizable subensemble, in contrast to only a few interactions in fragment C73. Our NMR results also agree qualitatively with the high-sensitivity DSC analysis of the natively disordered fragments C37 and M37 under conditions close to neutral pH (9). Although DSC analysis could not be conducted for these fragments at strongly acidic conditions due to problems with aggregation, our results indicate that 1H-15N HSQC experiments permit a broad pH range to be characterized and constitute a sensitive and rapid tool to obtain clues about local and nonlocal interactions between ionizable and nonionizable residues in disordered ensembles, which could explain the presence of predominantly hydrophobic buried surfaces.
CONCLUSION
Although mutagenesis and NMR experiments in folded proteins have demonstrated that amide protons are reporters of long-range interactions with carboxylate groups (67), amide protons of natively disordered model tetrapeptides have so far reported only through-bond and through-space interactions between ionizable residues and their first nonionizable neighbors (32). Here we have shown that monitoring the pH dependence of amide chemical shifts in the disordered ensembles of E. coli Trx fragments via standard 1H-15N HSQC experiments provides clues on (possibly transient) local and medium-range interactions among carboxylate groups and neighboring hydrophobic and/or hydrogen bond acceptor/donor residues. The accumulated data on the pH dependence of these fragments should provide leads to design mutagenesis and NMR experiments to confirm the presence of nonlocal interactions in these disordered ensembles. This work offers a sensitive and rapid new tool to obtain clues on nonlocal interactions between ionizable and nonionizable residues in the growing family of natively disordered small proteins with full NMR assignments.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
The authors acknowledge instrument time on the 800-MHz and 600-MHz spectrometers of the New York Structural Biology Center and Weill Medical College of Cornell University, respectively. We thank Marilyn Gunner and David Eliezer for insightful comments.
This work was supported by a National Science Foundation award (MCB-0118252) to M.L.T., a National Institutes of Health grant (5G12RR03060 from the National Center for Research Resources) for the 600-MHz spectrometer of The City College of New York, and an American Heart Association award to C.B. C.B. and M.L.T. are members of the New York Structural Biology Center supported by National Institutes of Health grant GM66354.
Abbreviations used: DSC, differential scanning calorimetry; ppm, parts per million; ppb, parts per billion; HSQC, heteronuclear single quantum coherence; NOE, nuclear Overhauser enhancement; NOESY, NOE spectroscopy; Trx, thioredoxin; C37, fragment 38–108; M37, fragment 38–73; C73, fragment 74–108.
References
- 1.Zeev-Ben-Mordehai, T., E. H. Rydberg, A. Solomon, L. Toker, V. J. Auld, I. Silman, S. Botti, and J. L. Sussman. 2003. The intracellular domain of the Drosophila cholinesterase-like neural adhesion protein, gliotactin, is natively unfolded. Proteins. 15:758–767. [DOI] [PubMed] [Google Scholar]
- 2.Longhi, S., V. Receveur-Bréchot, D. Karlin, K. Johansson, H. Darbon, D. Bhella, R. Yeo, S. Finet, and B. Canard. 2003. The C-terminal domain of the measles virus nucleoprotein is intrinsically disordered and folds upon binding to the C-terminal moiety of the phosphoprotein. J. Biol. Chem. 278:18638–18648. [DOI] [PubMed] [Google Scholar]
- 3.Thapar, R., G. A. Mueller, and W. F. Marzluff. 2004. The N-terminal domain of the Drosophila histone mRNA binding protein, SLBP, is intrinsically disordered with nascent helical structure. Biochemistry. 43:9390–9400. [DOI] [PubMed] [Google Scholar]
- 4.Green, T. B., O. Ganesh, K. Perry, L. Smith, L. H. Phylip, T. M. Logan, S. J. Hagen, B. M. Dunn, and A. S. Edison. 2004. IA3, an aspartic proteinase inhibitor from Saccharomyces cerevisiae, is intrinsically unstructured in solution. Biochemistry. 43:4071–4081. [DOI] [PubMed] [Google Scholar]
- 5.Wright, P. E., and H. J. Dyson. 1999. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 293:321–331. [DOI] [PubMed] [Google Scholar]
- 6.Dyson, H. J., and P. E. Wright. 2002. Coupling of folding and binding for unstructured proteins. Curr. Opin. Struct. Biol. 12:54–60. [DOI] [PubMed] [Google Scholar]
- 7.Gunasekaran, K., C.-J. Tsai, S. Kumar, D. Zanuy, and R. Nussinov. 2003. Extended disordered proteins: targeting function with less scaffold. Trends Biochem. Sci. 28:81–85. [DOI] [PubMed] [Google Scholar]
- 8.Georgescu, R. E., M. D. M. García-Mira, M. L. Tasayco, and J. M. Sánchez-Ruiz. 2001. Heat capacity analysis of oxidized Escherichia coli thioredoxin fragments (1–73, 74–108) and their noncovalent complex. Evidence for the burial of apolar surface in protein unfolded states. Eur. J. Biochem. 268:1–10. [DOI] [PubMed] [Google Scholar]
- 9.Mendoza, C., F. Figueirido, and M. L. Tasayco. 2003. DSC studies of a family of natively disordered fragments from Escherichia coli thioredoxin; surface burial in intrinsic coils. Biochemistry. 42:3349–3358. [DOI] [PubMed] [Google Scholar]
- 10.Shortle, D. R. 1996. Structural analysis of non-native states of proteins by NMR methods. Curr. Opin. Struct. Biol. 6:24–30. [DOI] [PubMed] [Google Scholar]
- 11.Zhang, O., J. D. Forman-Kay, D. Shortle, and L. E. Kay. 1997. Triple-resonance NOESY-based experiments with improved spectral resolution: applications to structural characterization of unfolded, partially folded and folded proteins. J. Biomol. NMR. 9:181–200. [DOI] [PubMed] [Google Scholar]
- 12.Dyson, H. J., and P. E. Wright. 2001. Nuclear magnetic resonance methods for elucidation of structure and dynamics in disordered states. Methods Enzymol. 339:258–270. [DOI] [PubMed] [Google Scholar]
- 13.Dyson, H. J., and P. E. Wright. 2002. Insights into structure and dynamics of unfolded proteins from nuclear magnetic resonance. Adv. Protein Chem. 62:311–340. [DOI] [PubMed] [Google Scholar]
- 14.Dyson, H. J., and P. E. Wright. 2004. Unfolded proteins and protein folding studied by NMR. Chem. Rev. 8:3607–3622. [DOI] [PubMed] [Google Scholar]
- 15.Bracken, C. 2001. NMR spin relaxation methods for characterization of disorder and folding in proteins. J. Mol. Graph. Model. 19:3–12. [DOI] [PubMed] [Google Scholar]
- 16.Gardner, K. H., and L. E. Kay. 1998. The use of 2H,13C,15N multidimensional NMR to study the structure and dynamics of proteins. Annu. Rev. Biophys. Biomol. Struct. 27:357–408. [DOI] [PubMed] [Google Scholar]
- 17.Klein-Seetharaman, J., M. Oikawa, S. B. Grimshaw, J. Wirmer, E. Duchardt, T. Ueda, T. Imoto, L. J. Smith, C. M. Dobson, and H. Schwalbe. 2002. Long-range interactions within a nonnative protein. Science. 295:1719–1722. [DOI] [PubMed] [Google Scholar]
- 18.Lietzow, M. A., M. Jamin, H. J. Dyson, and P. E. Wright. 2002. Mapping long-range contacts in a highly unfolded protein. J. Mol. Biol. 322:655–662. [DOI] [PubMed] [Google Scholar]
- 19.Crowhurst, K. A., and J. D. Forman-Kay. 2003. Aromatic and methyl NOEs highlight hydrophobic clustering in the unfolded state of an SH3 domain. Biochemistry. 42:8687–8695. [DOI] [PubMed] [Google Scholar]
- 20.Zhang, O., and J. D. Forman-Kay. 1995. Structural characterization of folded and unfolded states of an SH3 domain in equilibrium in aqueous buffer. Biochemistry. 34:6784–6794. [DOI] [PubMed] [Google Scholar]
- 21.Crowhurst, K. A., M. Tollinger, and J. D. Forman-Kay. 2002. Cooperative interactions and a non-native buried Trp in the unfolded state of an SH3 domain. J. Mol. Biol. 322:163–178. [DOI] [PubMed] [Google Scholar]
- 22.Choy, W.-Y., D. R. Shortle, and L. E. Kay. 2003. Side chain dynamics in unfolded protein states: an NMR based 2H spin relaxation study of Δ131Δ. J. Am. Chem. Soc. 125:1748–1758. [DOI] [PubMed] [Google Scholar]
- 23.Forsyth, W. R., J. Antosiewicz, and A. D. Robertson. 2002. Empirical relationships between protein structure and carboxyl pKa values in proteins. Proteins. 48:388–403. [DOI] [PubMed] [Google Scholar]
- 24.Li, H., A. D. Robertson, and J. H. Jensen. 2004. The determinants of carboxyl pKa values in turkey ovomucoid third domain. Proteins. 55:689–704. [DOI] [PubMed] [Google Scholar]
- 25.Qin, J., G. M. Clore, and A. M. Gronenborn. 1996. Ionization equilibria for side-chain carboxyl groups in oxidized and reduced human thioredoxin and in the complex with its target peptide from the transcription factor NFB. Biochemistry. 35:7–13. [DOI] [PubMed] [Google Scholar]
- 26.Chivers, P. T., K. E. Prehoda, B. F. Volkman, B.-M. Kim, J. L. Markley, and R. T. Raines. 1997. Microscopic pKa values of Escherichia coli thioredoxin. Biochemistry. 36:14985–14991. [DOI] [PubMed] [Google Scholar]
- 27.Sundd, M., N. Iverson, B. Ibarra-Molero, J. M. Sánchez-Ruiz, and A. D. Robertson. 2002. Electrostatic interactions in ubiquitin: stabilization of carboxylates by lysine amino groups. Biochemistry. 41:7586–7596. [DOI] [PubMed] [Google Scholar]
- 28.Song, J., M. Leskowski, M. A. Qasim, and J. L. Markley. 2003. NMR determination of pKa values for Asp, Glu, His, and Lys mutants at each variable contiguous enzyme-inhibitor contact position of the turkey ovomucoid third domain. Biochemistry. 42:2847–2856. [DOI] [PubMed] [Google Scholar]
- 29.Laurents, D. V., B. M. P. Huyghues-Despointes, M. Bruix, R. L. Thurlkill, D. Schell, S. Newson, G. R. Grimsley, K. L. Shaw, S. Treviño, M. Rico, J. M. Briggs, J. M. Antosiewicz, M. Scholtz, and C. N. Pace. 2003. Charge-charge interactions are key determinants of the pK values of ionizable groups in ribonuclease Sa (pI = 3.5) and a basic variant (pI = 10.2). J. Mol. Biol. 325:1077–1092. [DOI] [PubMed] [Google Scholar]
- 30.Schaller, W., and A. D. Robertson. 1995. pH, ionic strength, and temperature dependences of ionization equilibria for the carboxyl groups in turkey ovomucoid third domain. Biochemistry. 34:4714–4723. [DOI] [PubMed] [Google Scholar]
- 31.Oliveberg, M., V. L. Arcus, and A. R. Fersht. 1995. pKa values of carboxyl groups in the native and denatured states of barnase: The pKa values of the denatured state are on average 0.4 units lower than those of model compounds. Biochemistry. 34:9424–9433. [DOI] [PubMed] [Google Scholar]
- 32.Bundi, A., and K. Wüthrich. 1979. Use of amide 1H-NMR titration shifts for studies of polypeptide conformation. Biopolymers. 18:299–311. [Google Scholar]
- 33.Forsyth, W. R., and A. D. Robertson. 2000. Insensitivity of perturbed carboxyl pKa values in the ovomucoid third domain to charge replacement at a neighboring residue. Biochemistry. 39:8067–8072. [DOI] [PubMed] [Google Scholar]
- 34.Fairman, R., K. R. Shoemaker, E. J. York, J. M. Stewart, and R. L. Baldwin. 1990. The Glu 2- …Arg 10+ side-chain interaction in the C-peptide helix of ribonuclease A. Biophys. Chem. 37:107–119. [DOI] [PubMed] [Google Scholar]
- 35.Horng, J. C., V. Moroz, D. J. Rigotti, R. Fairman, and D. P. Raleigh. 2002. Characterization of large peptide fragments derived from the N-terminal domain of the ribosomal protein L9: Definition of the minimum folding motif and characterization of local electrostatic interactions. Biochemistry. 41:13360–13369. [DOI] [PubMed] [Google Scholar]
- 36.Kuhlman, B., D. L. Luisi, P. Young, and D. P. Raleigh. 1999. pKa values and the pH dependent stability of the N-terminal domain of L9 as probes of electrostatic interactions in the denatured state. Differentiation between local and nonlocal interactions. Biochemistry. 38:4896–4903. [DOI] [PubMed] [Google Scholar]
- 37.Marti, D. N., I. Jelesarov, and H. R. Bosshard. 2000. Interhelical ion pairing in coiled coils: solution structure of a heterodimeric leucine zipper and determination of pKa values of Glu side chains. Biochemistry. 39:12804–12818. [DOI] [PubMed] [Google Scholar]
- 38.Kao, Y.-H., C. A. Fitch, S. Bhattacharya, C. J. Sarkisian, J. T. J. Lecomte, and B. E. García-Moreno. 2000. Salt effects on ionization equilibria of histidines in myoglobin. Biophys. J. 79:1637–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tollinger, M., J. D. Forman-Kay, and L. E. Kay. 2002. Measurement of side-chain carboxyl pKa values of glutamate and aspartate residues in an unfolded protein by multinuclear NMR spectroscopy. J. Am. Chem. Soc. 124:5714–5717. [DOI] [PubMed] [Google Scholar]
- 40.Tollinger, M., K. A. Crowhurst, L. E. Kay, and J. D. Forman-Kay. 2003. Site-specific contributions to the pH dependence of protein stability. Proc. Natl. Acad. Sci. USA. 100:4545–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dlugosz, M., J. Antosiewicz, and A. D. Robertson. 2004. Constant-pH molecular dynamics study of protonation-structure relationship in a heptapeptide derived from ovomucoid third domain. Phys. Rev. E. 69:21915–21925. [DOI] [PubMed] [Google Scholar]
- 42.Tan, Y.-J., M. Oliveberg, B. Davis, and A. R. Fersht. 1995. Perturbed pKa values in the denatured sates of proteins. J. Mol. Biol. 254:980–992. [DOI] [PubMed] [Google Scholar]
- 43.Bundi, A., and K. Wüthrich. 1979. 1H-NMR parameters for the common amino acid residues measured in aqueous solutions of the linear tetrapeptides H-Gly-Gly-X-l-Ala-OH. Biopolymers. 18:285–297. [Google Scholar]
- 44.Tasayco, M. L., and K. Chao. 1995. NMR study of the reconstitution of the β-sheet of thioredoxin by fragment complementation. Proteins. 22:41–44. [DOI] [PubMed] [Google Scholar]
- 45.Yang, X.-M., R. Georgescu, J.-H. Li, W. F. Yu, Haierhan, and M. L. Tasayco. 1999. Recognition between disordered polypeptide chains from cleavage of an α/β domain: Self- versus non-self association. Proc. Pac. Symp. Biocomput. 4:590–600. [DOI] [PubMed] [Google Scholar]
- 46.Yang, X.-M., W.-F. Yu, J.-H. Li, J. A. Fuchs, J. Rizo, and M. L. Tasayco. 1998. NMR evidence for the reassembly of an α/β domain after cleavage of an α-helix: Implications for protein design. J. Am. Chem. Soc. 120:7985–7986. [Google Scholar]
- 47.Yu, W.-F., C.-S. Tung, H. Wang, and M. L. Tasayco. 2000. NMR analysis of cleaved E. coli thioredoxin (1–73/74–108) and its P76A variant: Cis/trans peptide isomerization. Protein Sci. 9:20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daughdrill, G. W., P. D. Vise, H. Zhou, X.-M. Yang, W.-F. Yu, M. L. Tasayco, and D. F. Lowry. 2004. Reduced spectral density mapping of a partially folded fragment of E. coli thioredoxin. J. Biomol. Struct. Dyn. 21:663–670. [DOI] [PubMed] [Google Scholar]
- 49.Marulanda, D., M. L. Tasayco, A. McDermott, M. Cataldi, V. Arriarán, and T. Polenova. 2004. Magic angle spinning solid-state NMR spectroscopy for structural studies of protein interfaces. Resonance assignments of differentially enriched Escherichia coli thioredoxin reassembled by fragment complementation. J. Am. Chem. Soc. 22:16608–16620. [DOI] [PubMed] [Google Scholar]
- 50.Wishart, D. S., C. G. Bigam, J. Yao, F. Abildgaard, H. J. Dyson, E. Oldfield, J. L. Markley, and B. D. Sykes. 1995. 1H, 13C and 15N chemical shifts referencing in biomolecular NMR. J. Biomol. NMR. 6:135–140. [DOI] [PubMed] [Google Scholar]
- 51.Cavanagh, J., W. J. Fairbrother, A. G. Palmer, and N. J. Skelton. 1996. Protein NMR Spectroscopy: Principles and Practice. Academic Press, New York.
- 52.Delaglio, F., S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer, and A. D. Bax. 1995. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 6:277–293. [DOI] [PubMed] [Google Scholar]
- 53.Goddard, T. D. and Kneller, D. G. SPARKY 3.0. University of California, San Francisco.
- 54.Farmer, B. T., R. A. Venters, L. D. Spicer, M. Wittekind, and L. A. Müller. 1992. A refocused and optimized HNCA: increased sensitivity and resolution in large macromolecules. J. Biomol. NMR. 2:195–202. [DOI] [PubMed] [Google Scholar]
- 55.Wittekind, M., and L. A. Müller. 1993. HNCACB, a high sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with α- and proton and nitrogen resonances with α- and β-carbon resonances in proteins. J. Magn. Reson. Ser. B. 101:201–205. [Google Scholar]
- 56.Kay, L. E., G.-Y. Xu, and T. Yamazaki. 1994. Enhanced sensitivity triple-resonances spectroscopy with minimal H2O saturation. J. Magn. Reson. Ser. A. 109:129–133. [Google Scholar]
- 57.BMRB (BioMagResBank). http://www.bmrb.wisc.edu.
- 58.Schwarzinger, S., G. J. A. Kroon, T. R. Foss, J. Chung, P. E. Wright, and H. J. Dyson. 2001. Sequence-dependent correction of random coil NMR chemical shifts. J. Am. Chem. Soc. 123:2970–2978. [DOI] [PubMed] [Google Scholar]
- 59.Merutka, G., H. J. Dyson, and P. E. Wright. 1995. Random coil 1H chemical shifts obtained as a function of temperature and trifluoroethanol concentration for the peptide series GGXGG. J. Biomol. NMR. 5:14–24. [DOI] [PubMed] [Google Scholar]
- 60.Shalongo, W. M., L. Dugad, and E. Stellwagen. 1994. Analysis of the thermal transition of a model helical peptide using 13C NMR. J. Am. Chem. Soc. 116:2500–2507. [Google Scholar]
- 61.Skelton, N. J., A. G. Palmer, M. Akke, J. Kordel, M. Rance, and W. J. Chazin. 1993. Practical aspects of two-dimensional proton-detected 15N spin relaxation measurements. J. Magn. Reson. B. 102:253–264. [Google Scholar]
- 62.Cao, W., C. Bracken, N. R. Kallenbach, and M. Lu. 2004. Helix formation and the unfolded state of a 52-residue helical protein. Protein Sci. 13:177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bunton, C. A., and V. J. Shiner. 1961. Isotope effects in deuterium oxide solution. I. Acid-base equilibria. J. Am. Chem. Soc. 83:42–47. [Google Scholar]
- 64.Markley, J. L. 1975. Observation of histidine residues in proteins by means of nuclear magnetic resonance spectroscopy. Acc. Chem. Res. 8:70–80. [Google Scholar]
- 65.Tasayco, M. L., J. A. Fuchs, X.-M. Yang, D. Dyalram, and R. E. Georgescu. 2000. Interaction between two discontiguous chain segments from the β-sheet of E. coli thioredoxin suggests initiation site for folding. Biochemistry. 39:10613–10618. [DOI] [PubMed] [Google Scholar]
- 66.Otlewski, J., and T. Cierpicki. 2001. Amide proton temperature coefficients as hydrogen bond indicators in proteins. J. Biomol. NMR. 21:249–261. [DOI] [PubMed] [Google Scholar]
- 67.Sundd, M., and A. D. Robertson. 2003. Rearrangement of charge-charge interactions in variant ubiquitins as detected by double-mutant cycles and NMR. J. Mol. Biol. 332:927–936. [DOI] [PubMed] [Google Scholar]
- 68.De Dios, A. C., D. N. Sears, and R. Tycko. 2004. NMR studies of peptide T, an inhibitor of HIV infectivity, in an aqueous environment. J. Pept. Sci. 10:622–630. [DOI] [PubMed] [Google Scholar]
- 69.Havlin, R. H., and R. Tycko. 2005. Probing site-specific conformational distributions in protein folding with solid-state NMR. Proc. Natl. Acad. Sci. USA. 102:3284–3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kemmink, J., and T. E. Creighton. 1995. The physical properties of local interactions of tyrosine residues in peptides and unfolded proteins. J. Mol. Biol. 245:251–260. [DOI] [PubMed] [Google Scholar]
- 71.de Alba, E., F. J. Blanco, M. A. Jiménez, M. Rico, and J. L. Nieto. 1995. Interactions responsible for the pH dependence of the β-hairpin conformational population formed by a designed linear peptide. Eur. J. Biochem. 233:283–292. [DOI] [PubMed] [Google Scholar]
- 72.Toth, G., C. R. Watts, R. F. Murphy, and S. Lovas. 2001. Significance of aromatic-backbone amide interactions in protein structure. Proteins. 43:373–381. [DOI] [PubMed] [Google Scholar]
- 73.Edsall, J. T., and J. Wyman. 1958. Polybasic acids, bases and ampholytes, including proteins. Biophys. Chem. 1:536. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.